Abstract
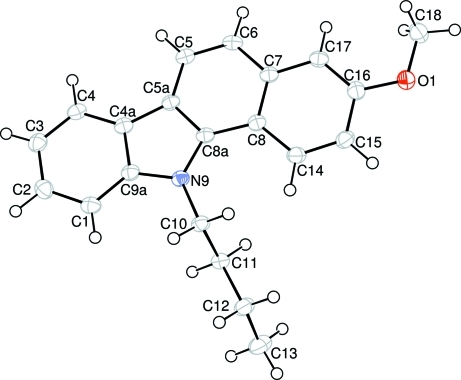
The title compound, C21H21NO, consists of a carbazole skeleton with a methoxybenzene ring fused to the carbazole, and a butyl group attached to the carbazole N atom. The carbazole skeleton is nearly planar [maximum deviation = 0.078 (2) Å], and it is oriented at a dihedral angle of 4.22 (4)° with respect to the adjacent methoxybenzene ring.
Related literature
For the biological activity of carbazole derivatives, see: Knölker & Reddy (2002 ▶). For the use of carbazole derivatives in the syntheses of indole alkaloids, see: Routier et al. (2001 ▶). For the use of benzo[a]carbazoles in cancer treatment, see: Carini et al. (2001 ▶). For the antitumor activity of a series of simple benzo[a]carbazoles against mammary tumors of rats, leukemia, renal tumors, colon cancer and malignant melanoma tumor cell lines, see: von Angerer & Prekajac (1986 ▶); Pindur & Lemster (1997 ▶). For the extensive application of benzo[a]carbazole derivatives as photographic materials, see: Oliveira et al. (2005 ▶. For tetrahydrocarbazole systems present in the frameworks of a number of indole-type alkaloids of biological interest, see: Phillipson & Zenk (1980 ▶); Saxton (1983 ▶); Abraham (1975 ▶). For related structures, see: Hökelek et al. (1994 ▶, 1998 ▶, 1999 ▶, 2004 ▶, 2006 ▶); Patır et al. (1997 ▶); Hökelek & Patır (1999 ▶, 2002 ▶); Çaylak et al. (2007 ▶). For bond-length data, see: Allen et al. (1987 ▶).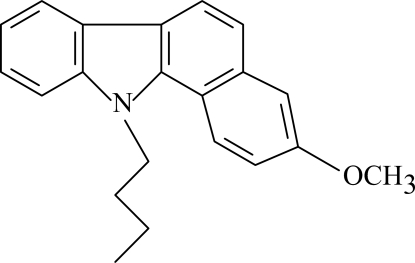
Experimental
Crystal data
C21H21NO
M r = 303.39
Monoclinic,
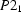
a = 10.7263 (6) Å
b = 5.5562 (3) Å
c = 13.8967 (7) Å
β = 97.841 (2)°
V = 820.46 (8) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 100 K
0.48 × 0.39 × 0.35 mm
Data collection
Bruker Kappa APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.965, T max = 0.974
7947 measured reflections
2248 independent reflections
2001 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.091
S = 1.04
2248 reflections
270 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.25 e Å−3
Δρmin = −0.16 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810021963/xu2773sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810021963/xu2773Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are indebted to Anadolu University and the Medicinal Plants and Medicine Research Centre of Anadolu University, Eskişehir, Turkey, for the use of the diffractometer.
supplementary crystallographic information
Comment
Carbazole derivatives display a range of biological activities, making them attractive compounds to synthetic and medicinal chemists (Knölker & Reddy, 2002). They also have an important role in the syntheses of indole alkaloids, in which their synthetic derivatives, possessing useful pharmacological properties, are currently under investigation (Routier et al., 2001). Benzo -annulated carbazole ring systems are found only rarely in natural products. The benzo[a]carbazoles, containing an aromatic ring fused to the a-face of the carbazole nucleus, are potential candidates for cancer treatment as a result of DNA intercelative binding properties (Carini et al., 2001). A series of simple benzo[a]carbazoles have been shown to bind to estrogen receptors and inhibit the growth of mammary tumors of rats (Angerer & Prekajac, 1986). Some benzo[a]carbazoles exhibit a pronounced antitumor activity against leukemia, renal tumor, colon cancer, and malignant melanoma tumor cell lines (Pindur & Lemster, 1997). Benzo[a]carbazole derivatives have also found extensive application as photographic materials (Oliveira et al., 2005).
Tetrahydrocarbazole systems are present in the framework of a number of indole-type alkaloids of biological interest (Phillipson & Zenk, 1980; Saxton, 1983; Abraham, 1975). The structures of tricyclic, tetracyclic and pentacyclic ring systems with dithiolane and other substituents of the tetrahydrocarbazole core, have been the subject of much interest in our laboratory. These include 1,2,3,4-tetrahydrocarbazole-1-spiro-2'-[1,3]dithiolane, (II) (Hökelek et al., 1994), N-(2-methoxyethyl)-N-{2,3,4,9-tetrahydrospiro[1H-carbazole-1, 2-(1,3)dithiolane]-4-yl}benzene-sulfonamide, (III) (Patır et al., 1997), spiro[carbazole-1(2H),2'-[1,3]-dithiolan]-4(3H)-one, (IV) (Hökelek et al., 1998), 9-acetonyl-3-ethylidene-1,2,3,4-tetrahydrospiro[carbazole-1,2'-[1,3] dithiolan]-4-one, (V) (Hökelek et al., 1999), N-(2,2-dimethoxyethyl)-N -{9-methoxymethyl-1,2,3,4-tetrahydrospiro[carbazole-1,2'-[1,3]dithiolan] -4-yl}benzamide, (VI) (Hökelek & Patır, 1999), 3a,4,10,10b-tetrahydro-2H -furo[2,3-a]carbazol-5(3H)-one, (VII) (Çaylak et al., 2007); also the pentacyclic compounds 6-ethyl-4-(2-methoxyethyl)-2,6-methano-5-oxo-hexahydro- pyrrolo(2,3 - d)carbazole-1-spiro-2'-(1,3)dithiolane, (VIII) (Hökelek & Patır, 2002), N-(2-benzyloxyethyl)-4,7-dimethyl-6-(1,3-dithiolan-2-yl)-1,2, 3,4,5,6-hexahydro-1,5-methano-2-azocino[4,3-b]indol-2-one, (IX) (Hökelek et al., 2004) and 4-ethyl-6,6-ethylenedithio-2-(2-methoxyethyl)-7-methoxy- methylene-2,3,4,5,6,7-hexahydro-1,5-methano-1H-azocino[4,3-b]indol-3-one, (X) (Hökelek et al., 2006). The title compound, (I), may be considered as a synthetic precursor of tetracyclic indole alkaloids of biological interests. The present study was undertaken to ascertain its crystal structure.
The title compound consists of a carbazole skeleton with a methoxy benzoato group fused to the a-face of the carbazole nucleus, and a butyl group attached to atom N9 (Fig. 1), where the bond lengths (Allen et al., 1987) and angles are within normal ranges, and generally agree with those in compounds (II)-(X). In all structures atom N9 is substituted.
An examination of the deviations from the least-squares planes through individual rings shows that rings A (C1—C4/C4a/C9a), B (C4a/C5a/C8a/N9/C9a), C (C5a/C5—C8/C8a) and D (C7/C8/C14—C17) are planar. The carbazole skeleton, containing the rings A, B and C are also nearly planar [with a maximum deviation of 0.078 (2) Å for atom C2] with dihedral angles of A/B = 2.37 (6), A/C = 5.01 (5) and B/C = 2.81 (5) ° Ring D is oriented with respect to the planar carbazole skeleton at a dihedral angle of 4.22 (4) °. So, it is nearly coplanar with the carbazole skeleton. Atoms O1 and C18 displaced by 0.010 (1) and -0.045 (2) Å from the plane of ring D, respectively, while atom C10 is 0.092 (2) Å away from the plane of the carbazole skeleton.
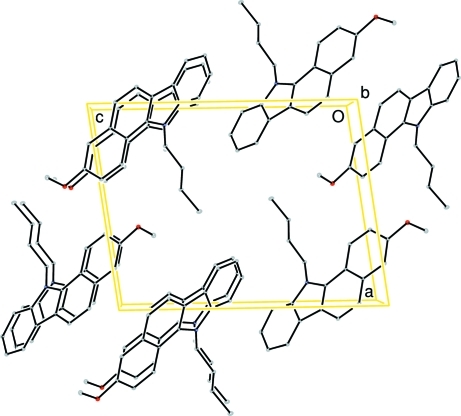
In the crystal structure, molecules are alongated along the c axis and stacked along the b axis (Fig. 2).
Experimental
For the preparation of the title compound, (I), a solution of 3-methoxy-11H -benzo[a]carbazole (1.00 g, 4.0 mmol) in dichloromethane (20 ml) was cooled to 273 K, and then sodium hydroxide (2 ml, 50%), tetrabutylammonium hydrogen sulfate (0.10 g) and butyl bromide (0.62 g, 4.5 mmol) were added. The mixture was stirred for 1 h at 273 K, and then 2 h at 298 K. It was washed with hydrochloric acid (50 ml, 10%) and the organic layer was dried with anhydrous magnesium sulfate. The solvent was evaporated under reduced pressure and the residue was crystallized from methanol (yield; 1.15 g, 93%, m.p. 369 K).
Refinement
H13A, H13B, H13C, H18A, H18B and H18C atoms (for methyl groups) were positioned geometrically with C—H = 0.98 Å and constrained to ride on their parent atoms, Uiso(H) = 1.5Ueq(C). The remaining H atoms were located in difference Fourier maps and refined isotropically. Friedel pairs were merged because of the weak anomalous scatterer of the compound, and the absolute structure was not determined.
Figures
Fig. 1.
The molecular structure of the title molecule with the atom-numbering scheme. The displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
A partial packing diagram.
Crystal data
| C21H21NO | F(000) = 324 |
| Mr = 303.39 | Dx = 1.228 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2yb | Cell parameters from 3840 reflections |
| a = 10.7263 (6) Å | θ = 2.3–28.4° |
| b = 5.5562 (3) Å | µ = 0.08 mm−1 |
| c = 13.8967 (7) Å | T = 100 K |
| β = 97.841 (2)° | Block, colorless |
| V = 820.46 (8) Å3 | 0.48 × 0.39 × 0.35 mm |
| Z = 2 |
Data collection
| Bruker Kappa APEXII CCD area-detector diffractometer | 2248 independent reflections |
| Radiation source: fine-focus sealed tube | 2001 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| φ and ω scans | θmax = 28.4°, θmin = 1.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −13→14 |
| Tmin = 0.965, Tmax = 0.974 | k = −7→6 |
| 7947 measured reflections | l = −17→18 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.035 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.091 | w = 1/[σ2(Fo2) + (0.0528P)2 + 0.1088P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 2248 reflections | Δρmax = 0.25 e Å−3 |
| 270 parameters | Δρmin = −0.16 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.59965 (11) | 0.9420 (3) | −0.11895 (9) | 0.0299 (3) | |
| C1 | 1.02958 (17) | 0.2221 (4) | 0.41676 (13) | 0.0246 (4) | |
| H1 | 0.9764 (19) | 0.094 (5) | 0.4258 (15) | 0.030 (6)* | |
| C2 | 1.14448 (17) | 0.2475 (4) | 0.47538 (13) | 0.0276 (4) | |
| H2 | 1.166 (2) | 0.136 (5) | 0.5266 (17) | 0.042 (7)* | |
| C3 | 1.22609 (17) | 0.4381 (4) | 0.46185 (13) | 0.0271 (4) | |
| H3 | 1.3035 (19) | 0.447 (4) | 0.5040 (14) | 0.022 (5)* | |
| C4 | 1.19492 (16) | 0.6061 (4) | 0.38921 (13) | 0.0245 (4) | |
| H4 | 1.2520 (17) | 0.742 (4) | 0.3796 (13) | 0.021 (5)* | |
| C4A | 1.08080 (15) | 0.5806 (4) | 0.32759 (12) | 0.0217 (4) | |
| C5 | 1.06152 (16) | 0.9086 (4) | 0.19246 (13) | 0.0232 (4) | |
| H5 | 1.1411 (18) | 0.992 (4) | 0.2181 (13) | 0.020 (5)* | |
| C5A | 1.02208 (15) | 0.7091 (3) | 0.24338 (12) | 0.0210 (4) | |
| C6 | 0.98899 (16) | 0.9848 (4) | 0.10954 (13) | 0.0238 (4) | |
| H6 | 1.0136 (18) | 1.120 (5) | 0.0755 (14) | 0.025 (5)* | |
| C7 | 0.87121 (15) | 0.8724 (4) | 0.07581 (12) | 0.0215 (4) | |
| C8 | 0.82600 (15) | 0.6754 (4) | 0.12711 (12) | 0.0205 (4) | |
| C8A | 0.90768 (15) | 0.5921 (4) | 0.21092 (12) | 0.0197 (3) | |
| C9A | 0.99973 (15) | 0.3887 (4) | 0.34259 (12) | 0.0214 (4) | |
| N9 | 0.89451 (13) | 0.3987 (3) | 0.27259 (10) | 0.0209 (3) | |
| C10 | 0.79576 (15) | 0.2165 (4) | 0.26464 (13) | 0.0221 (4) | |
| H101 | 0.8331 (18) | 0.076 (5) | 0.2927 (14) | 0.021 (5)* | |
| H102 | 0.7675 (17) | 0.185 (4) | 0.1949 (14) | 0.021 (5)* | |
| C11 | 0.68345 (16) | 0.2861 (4) | 0.31519 (13) | 0.0228 (4) | |
| H111 | 0.7121 (18) | 0.304 (4) | 0.3868 (14) | 0.023 (5)* | |
| H112 | 0.6483 (19) | 0.442 (5) | 0.2886 (15) | 0.027 (6)* | |
| C12 | 0.58043 (17) | 0.0964 (4) | 0.29886 (15) | 0.0290 (4) | |
| H121 | 0.6196 (19) | −0.064 (5) | 0.3171 (14) | 0.025 (5)* | |
| H122 | 0.5517 (19) | 0.077 (5) | 0.2304 (17) | 0.036 (6)* | |
| C13 | 0.47038 (18) | 0.1518 (5) | 0.35479 (16) | 0.0397 (5) | |
| H13A | 0.4050 | 0.0290 | 0.3400 | 0.060* | |
| H13B | 0.4355 | 0.3104 | 0.3355 | 0.060* | |
| H13C | 0.5002 | 0.1516 | 0.4247 | 0.060* | |
| C14 | 0.70388 (16) | 0.5850 (4) | 0.09343 (13) | 0.0248 (4) | |
| H14 | 0.668 (2) | 0.458 (5) | 0.1282 (15) | 0.032 (6)* | |
| C15 | 0.63327 (16) | 0.6779 (4) | 0.01272 (13) | 0.0264 (4) | |
| H15 | 0.549 (2) | 0.613 (5) | −0.0092 (17) | 0.044 (7)* | |
| C16 | 0.68014 (16) | 0.8672 (4) | −0.03944 (12) | 0.0242 (4) | |
| C17 | 0.79658 (16) | 0.9641 (4) | −0.00876 (12) | 0.0235 (4) | |
| H17 | 0.8282 (16) | 1.097 (4) | −0.0415 (13) | 0.015 (5)* | |
| C18 | 0.63988 (18) | 1.1414 (4) | −0.17108 (14) | 0.0325 (5) | |
| H18A | 0.5741 | 1.1833 | −0.2245 | 0.049* | |
| H18B | 0.7172 | 1.0988 | −0.1974 | 0.049* | |
| H18C | 0.6561 | 1.2794 | −0.1272 | 0.049* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0241 (6) | 0.0399 (9) | 0.0257 (6) | 0.0017 (6) | 0.0027 (5) | 0.0051 (7) |
| C1 | 0.0267 (9) | 0.0213 (10) | 0.0275 (8) | 0.0008 (8) | 0.0096 (7) | −0.0004 (8) |
| C2 | 0.0292 (9) | 0.0284 (11) | 0.0259 (9) | 0.0058 (8) | 0.0057 (7) | 0.0023 (8) |
| C3 | 0.0228 (8) | 0.0342 (12) | 0.0250 (8) | 0.0021 (8) | 0.0062 (7) | −0.0029 (9) |
| C4 | 0.0212 (8) | 0.0280 (11) | 0.0253 (8) | −0.0024 (8) | 0.0071 (6) | −0.0025 (8) |
| C4A | 0.0222 (8) | 0.0219 (9) | 0.0223 (8) | 0.0003 (7) | 0.0077 (6) | −0.0039 (8) |
| C5 | 0.0215 (8) | 0.0229 (9) | 0.0263 (8) | −0.0032 (7) | 0.0075 (7) | −0.0025 (8) |
| C5A | 0.0197 (7) | 0.0218 (9) | 0.0229 (8) | 0.0002 (7) | 0.0072 (6) | −0.0024 (7) |
| C6 | 0.0262 (8) | 0.0209 (9) | 0.0261 (9) | −0.0032 (8) | 0.0100 (7) | 0.0006 (8) |
| C7 | 0.0215 (8) | 0.0226 (10) | 0.0220 (8) | 0.0009 (7) | 0.0088 (7) | −0.0025 (7) |
| C8 | 0.0207 (7) | 0.0203 (9) | 0.0215 (8) | 0.0008 (7) | 0.0065 (6) | −0.0024 (7) |
| C8A | 0.0203 (7) | 0.0178 (9) | 0.0225 (8) | 0.0001 (7) | 0.0081 (6) | −0.0021 (7) |
| C9A | 0.0199 (7) | 0.0217 (9) | 0.0239 (8) | 0.0016 (7) | 0.0076 (6) | −0.0015 (7) |
| N9 | 0.0204 (6) | 0.0186 (8) | 0.0240 (7) | −0.0017 (6) | 0.0045 (5) | 0.0000 (6) |
| C10 | 0.0210 (8) | 0.0189 (9) | 0.0269 (9) | −0.0034 (7) | 0.0051 (7) | −0.0001 (8) |
| C11 | 0.0212 (8) | 0.0226 (10) | 0.0252 (9) | −0.0026 (7) | 0.0054 (6) | 0.0003 (7) |
| C12 | 0.0230 (8) | 0.0338 (12) | 0.0307 (10) | −0.0073 (8) | 0.0051 (7) | −0.0015 (9) |
| C13 | 0.0246 (9) | 0.0471 (15) | 0.0491 (12) | −0.0051 (10) | 0.0112 (8) | 0.0041 (11) |
| C14 | 0.0247 (8) | 0.0265 (10) | 0.0240 (8) | −0.0033 (8) | 0.0062 (6) | 0.0006 (8) |
| C15 | 0.0201 (8) | 0.0323 (11) | 0.0272 (9) | −0.0018 (8) | 0.0049 (7) | −0.0012 (8) |
| C16 | 0.0235 (8) | 0.0286 (10) | 0.0211 (8) | 0.0045 (7) | 0.0051 (7) | −0.0009 (8) |
| C17 | 0.0246 (8) | 0.0240 (10) | 0.0234 (8) | 0.0005 (7) | 0.0087 (6) | −0.0008 (8) |
| C18 | 0.0324 (9) | 0.0366 (12) | 0.0283 (9) | 0.0040 (9) | 0.0035 (8) | 0.0064 (9) |
Geometric parameters (Å, °)
| O1—C16 | 1.371 (2) | C10—C11 | 1.525 (2) |
| O1—C18 | 1.422 (3) | C10—H101 | 0.94 (2) |
| C1—C2 | 1.389 (3) | C10—H102 | 0.991 (19) |
| C1—H1 | 0.93 (2) | C11—C12 | 1.521 (3) |
| C2—C3 | 1.403 (3) | C11—H111 | 1.006 (19) |
| C2—H2 | 0.95 (3) | C11—H112 | 0.99 (3) |
| C3—C4 | 1.382 (3) | C12—C13 | 1.530 (3) |
| C3—H3 | 0.95 (2) | C12—H121 | 1.00 (2) |
| C4—H4 | 0.99 (2) | C12—H122 | 0.97 (2) |
| C4A—C4 | 1.402 (2) | C13—H13A | 0.9800 |
| C4A—C9A | 1.410 (3) | C13—H13B | 0.9800 |
| C5—C5A | 1.411 (3) | C13—H13C | 0.9800 |
| C5—H5 | 1.00 (2) | C14—C8 | 1.421 (2) |
| C5A—C4A | 1.440 (2) | C14—H14 | 0.97 (2) |
| C6—C5 | 1.367 (3) | C15—C14 | 1.366 (3) |
| C6—H6 | 0.95 (2) | C15—H15 | 0.98 (2) |
| C7—C6 | 1.430 (2) | C16—C15 | 1.408 (3) |
| C8—C7 | 1.427 (2) | C16—C17 | 1.373 (3) |
| C8—C8A | 1.435 (2) | C17—C7 | 1.423 (3) |
| C8A—C5A | 1.408 (2) | C17—H17 | 0.96 (2) |
| C9A—C1 | 1.390 (3) | C18—H18A | 0.9800 |
| N9—C8A | 1.393 (2) | C18—H18B | 0.9800 |
| N9—C9A | 1.387 (2) | C18—H18C | 0.9800 |
| N9—C10 | 1.458 (2) | ||
| C16—O1—C18 | 116.45 (15) | N9—C10—H101 | 106.4 (12) |
| C2—C1—C9A | 117.62 (18) | N9—C10—H102 | 108.7 (12) |
| C2—C1—H1 | 120.6 (13) | C11—C10—H101 | 109.8 (12) |
| C9A—C1—H1 | 121.7 (13) | C11—C10—H102 | 109.9 (11) |
| C1—C2—C3 | 121.20 (18) | H101—C10—H102 | 108.3 (19) |
| C1—C2—H2 | 119.0 (15) | C10—C11—H111 | 109.1 (11) |
| C3—C2—H2 | 119.8 (15) | C10—C11—H112 | 109.4 (12) |
| C2—C3—H3 | 117.8 (14) | C12—C11—C10 | 111.04 (16) |
| C4—C3—C2 | 120.97 (17) | C12—C11—H111 | 109.5 (13) |
| C4—C3—H3 | 121.2 (14) | C12—C11—H112 | 108.4 (13) |
| C3—C4—C4A | 118.82 (18) | H112—C11—H111 | 109.4 (18) |
| C3—C4—H4 | 121.4 (11) | C11—C12—C13 | 112.32 (18) |
| C4A—C4—H4 | 119.8 (11) | C11—C12—H121 | 107.9 (12) |
| C4—C4A—C5A | 134.01 (18) | C11—C12—H122 | 110.6 (15) |
| C4—C4A—C9A | 119.40 (17) | C13—C12—H121 | 112.1 (12) |
| C9A—C4A—C5A | 106.56 (14) | C13—C12—H122 | 111.0 (13) |
| C5A—C5—H5 | 119.3 (12) | H121—C12—H122 | 103 (2) |
| C6—C5—C5A | 119.41 (17) | C12—C13—H13A | 109.5 |
| C6—C5—H5 | 121.3 (12) | C12—C13—H13B | 109.5 |
| C5—C5A—C4A | 132.01 (16) | C12—C13—H13C | 109.5 |
| C8A—C5A—C4A | 107.26 (16) | H13A—C13—H13B | 109.5 |
| C8A—C5A—C5 | 120.68 (16) | H13A—C13—H13C | 109.5 |
| C5—C6—C7 | 121.13 (17) | H13B—C13—H13C | 109.5 |
| C5—C6—H6 | 120.2 (12) | C8—C14—H14 | 120.6 (13) |
| C7—C6—H6 | 118.5 (12) | C15—C14—C8 | 121.32 (18) |
| C8—C7—C6 | 121.11 (16) | C15—C14—H14 | 118.1 (13) |
| C17—C7—C6 | 119.12 (17) | C14—C15—C16 | 120.52 (17) |
| C17—C7—C8 | 119.75 (16) | C14—C15—H15 | 120.0 (16) |
| C7—C8—C8A | 116.20 (15) | C16—C15—H15 | 119.5 (15) |
| C14—C8—C7 | 117.85 (17) | O1—C16—C15 | 114.31 (16) |
| C14—C8—C8A | 125.91 (17) | O1—C16—C17 | 125.26 (18) |
| C5A—C8A—C8 | 121.33 (16) | C17—C16—C15 | 120.42 (17) |
| N9—C8A—C5A | 108.44 (15) | C7—C17—H17 | 118.4 (11) |
| N9—C8A—C8 | 130.23 (16) | C16—C17—C7 | 120.08 (18) |
| C1—C9A—C4A | 121.96 (16) | C16—C17—H17 | 121.5 (11) |
| N9—C9A—C1 | 129.06 (17) | O1—C18—H18A | 109.5 |
| N9—C9A—C4A | 108.98 (15) | O1—C18—H18B | 109.5 |
| C8A—N9—C10 | 128.47 (14) | O1—C18—H18C | 109.5 |
| C9A—N9—C8A | 108.74 (14) | H18A—C18—H18B | 109.5 |
| C9A—N9—C10 | 122.57 (15) | H18A—C18—H18C | 109.5 |
| N9—C10—C11 | 113.60 (15) | H18B—C18—H18C | 109.5 |
| C18—O1—C16—C15 | 176.75 (16) | N9—C8A—C5A—C4A | 0.40 (19) |
| C18—O1—C16—C17 | −2.2 (3) | N9—C8A—C5A—C5 | 178.18 (16) |
| C9A—C1—C2—C3 | 1.6 (3) | C8—C8A—C5A—C4A | −179.64 (15) |
| C1—C2—C3—C4 | −0.4 (3) | C8—C8A—C5A—C5 | −1.9 (3) |
| C2—C3—C4—C4A | −1.2 (3) | N9—C9A—C1—C2 | 177.47 (17) |
| C5A—C4A—C4—C3 | −176.09 (19) | C4A—C9A—C1—C2 | −1.3 (3) |
| C9A—C4A—C4—C3 | 1.5 (2) | C9A—N9—C8A—C5A | −1.06 (18) |
| C4—C4A—C9A—N9 | −179.24 (16) | C9A—N9—C8A—C8 | 178.97 (17) |
| C4—C4A—C9A—C1 | −0.2 (2) | C10—N9—C8A—C5A | −175.70 (16) |
| C5A—C4A—C9A—N9 | −1.06 (18) | C10—N9—C8A—C8 | 4.3 (3) |
| C5A—C4A—C9A—C1 | 177.96 (16) | C8A—N9—C9A—C1 | −177.60 (17) |
| C6—C5—C5A—C4A | 175.63 (18) | C8A—N9—C9A—C4A | 1.33 (18) |
| C6—C5—C5A—C8A | −1.5 (3) | C10—N9—C9A—C1 | −2.6 (3) |
| C5—C5A—C4A—C4 | 0.8 (3) | C10—N9—C9A—C4A | 176.34 (15) |
| C5—C5A—C4A—C9A | −177.03 (18) | C9A—N9—C10—C11 | 96.69 (19) |
| C8A—C5A—C4A—C4 | 178.20 (19) | C8A—N9—C10—C11 | −89.3 (2) |
| C8A—C5A—C4A—C9A | 0.40 (19) | N9—C10—C11—C12 | 176.39 (15) |
| C7—C6—C5—C5A | 2.7 (3) | C10—C11—C12—C13 | 175.83 (17) |
| C8—C7—C6—C5 | −0.6 (3) | C15—C14—C8—C7 | 2.1 (3) |
| C17—C7—C6—C5 | 177.50 (17) | C15—C14—C8—C8A | 179.73 (17) |
| C8A—C8—C7—C6 | −2.6 (2) | C16—C15—C14—C8 | −0.1 (3) |
| C8A—C8—C7—C17 | 179.29 (16) | O1—C16—C15—C14 | 179.77 (17) |
| C14—C8—C7—C6 | 175.24 (16) | C17—C16—C15—C14 | −1.3 (3) |
| C14—C8—C7—C17 | −2.8 (2) | O1—C16—C17—C7 | 179.31 (16) |
| C7—C8—C8A—N9 | −176.22 (16) | C15—C16—C17—C7 | 0.5 (3) |
| C7—C8—C8A—C5A | 3.8 (2) | C16—C17—C7—C6 | −176.50 (16) |
| C14—C8—C8A—N9 | 6.1 (3) | C16—C17—C7—C8 | 1.6 (3) |
| C14—C8—C8A—C5A | −173.86 (17) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU2773).
References
- Abraham, D. J. (1975). The Catharanthus Alkaloids, edited by W. I. Taylor & N. R. Fransworth, chs. 7 and 8. New York: Marcel Decker.
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Angerer, E. von & Prekajac, J. (1986). J. Med. Chem.29, 380–386. [DOI] [PubMed]
- Bruker (2005). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Carini, D. J., Kaltenbach, R. F. III, Liu, J., Benfield, P. A., Boylan, J., Boisclair, M., Brizuela, L., Burton, C. R., Cox, S., Grafstorm, R., Harrison, B. A., Harrison, K., Akamike, E., Markwalder, J. A., Nakano, Y., Seitz, S. P., Sharp, D. M., Trainor, G. L. & Sielecki, T. M. (2001). Bioorg. Med. Chem. Lett.11, 2209–2211. [DOI] [PubMed]
- Çaylak, N., Hökelek, T., Uludağ, N. & Patır, S. (2007). Acta Cryst. E63, o3913–o3914.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Hökelek, T., Gündüz, H., Patır, S. & Uludağ, N. (1998). Acta Cryst. C54, 1297–1299.
- Hökelek, T. & Patır, S. (1999). Acta Cryst. C55, 675–677.
- Hökelek, T. & Patır, S. (2002). Acta Cryst. E58, o374–o376.
- Hökelek, T., Patır, S., Gülce, A. & Okay, G. (1994). Acta Cryst. C50, 450–453.
- Hökelek, T., Patır, S. & Uludağ, N. (1999). Acta Cryst. C55, 114–116.
- Hökelek, T., Uludağ, N. & Patır, S. (2004). Acta Cryst. E60, o25–o27.
- Hökelek, T., Uludağ, N. & Patır, S. (2006). Acta Cryst. E62, o791–o793.
- Knölker, H.-J. & Reddy, K. R. (2002). Chem. Rev.102, 4303–4327. [DOI] [PubMed]
- Oliveira, M. M., Salvador, M. A., Coelho, P. J. & Carvalho, L. M. (2005). Tetrahedron, 61, 1681–1691.
- Patır, S., Okay, G., Gülce, A., Salih, B. & Hökelek, T. (1997). J. Heterocycl. Chem.34, 1239–1242.
- Phillipson, J. D. & Zenk, M. H. (1980). Indole and Biogenetically Related Alkaloids, ch 3. New York: Academic Press.
- Pindur, U. & Lemster, T. (1997). Recent Res. Devel. Org. Bioorg. Chem. pp. 33–54.
- Routier, S., Coudert, G. & Merour, J.-Y. (2001). Tetrahedron Lett.42, 7025–7028.
- Saxton, J. E. (1983). Editor. Heterocyclic Compounds, Vol. 25, The Monoterpenoid Indole Alkaloids, chs. 8 and 11. New York: Wiley.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810021963/xu2773sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810021963/xu2773Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report