Abstract
Blue-light-receptor cryptochrome (CRY), which mediates cotyledon expansion, increased accumulation of anthocyanin, and inhibition of hypocotyl elongation, was first identified in Arabidopsis. Two Arabidopsis cryptochromes (AtCRY1 and AtCRY2) have been reported to be localized to the nucleus. However, there is no information on the cryptochromes in monocotyledons. In this study, we isolated two cryptochrome cDNAs, OsCRY1 and OsCRY2, from rice (Oryza sativa) plants. The deduced amino acid sequences of OsCRY1 and OsCRY2 have a photolyase-like domain in their N termini and are homologous to AtCRY1. To investigate the function of OsCRY1, we overexpressed a green fluorescence protein-OsCRY1 fusion gene in Arabidopsis and assessed the phenotypes of the resulting transgenic plants. When the seedlings were germinated in the dark, no discernible effect was observed. However, light-germinated seedlings showed pronounced inhibition of hypocotyl elongation and increased accumulation of anthocyanin. These phenotypes were induced in a blue-light-dependent manner, indicating that OsCRY1 functions as a blue-light-receptor cryptochrome. We also examined the intracellular localization of green fluorescence protein-OsCRY1 in the transgenic plants. It was localized to both the nucleus and the cytoplasm. We identified two nuclear localization domains in the primary structure of OsCRY1. We discuss the relationship between the function and intracellular localization of rice cryptochromes by using additional data obtained with OsCRY2.
Blue-light-receptor cryptochrome was first identified in a T-DNA insertion mutant of Arabidopsis allelic to hy4 (Ahmad and Cashmore, 1993). The mutant seedlings were deficient in their response to blue/UV-A light and showed long hypocotyls. The isolated gene was designated AtCRY1. Subsequently, a second cryptochrome gene, AtCRY2, was isolated from Arabidopsis (Lin et al., 1996, 1998), and orthologs were isolated from various organisms, including animals, the fern Adiantum capillus-veneris (Kanegae and Wada, 1998), tomato (Lycopersicon esculentum; Ninu et al., 1999), and fruitfly (Drosophila melanogaster; Stanewsky et al., 1998). A photolyase-like domain, which binds flavin chromophores, has been found in the N terminus of cryptochrome molecules. Photolyase is a DNA-repairing enzyme that mediates a redox reaction in response to the absorption of blue/UV-A light, but photolyase activity has not been found in cryptochromes (Lin, 2002).
Cryptochromes perceive blue-light signals and cause various photomorphogenic responses, such as cotyledon expansion and inhibition of hypocotyl elongation, as well as anthocyanin accumulation and chalcon synthetase gene expression (Briggs and Huala, 1999; Cashmore et al., 1999; Lin, 2002; Lin and Shalitin, 2003). Recently, two functional domains have been demonstrated in the photolyase-like region and the C terminus of crypotochromes (Yang et al., 2000). The C terminus of AtCRY1 or AtCRY2, fused with β-glucuronidase (GUS), mediated photomorphogenesis-like phenotypes even in complete darkness. This constitutive photomorphogenic phenotype suggests that the C-terminal domain of Arabidopsis cryptochromes is maintained in an active state. Blue light has been interpreted to relieve the repression through a redox change of the N-terminal photolyase-like domain.
The molecular mechanism of cryptochrome-mediated signal transduction has not been elucidated yet, but recent studies suggest the interaction of cryptochrome with other proteins. It has been reported that cryptochrome interacts with a negative regulator of photomorphogenesis, COP1 (Wang et al., 2001; Yang et al., 2001), which is a downstream factor of cryptochrome. AtCRY1 and its C terminus were both shown to interact with a WD40 repeat domain of COP1 under either dark or light conditions. Arabidopsis cryptochromes also were demonstrated to interact with red/far-red light receptors (phytochromes; Ahmad et al., 1998; Mas et al., 2000).
The protein-protein interactions between cryptochromes and other proteins appear to play an important role in blue-light signal transduction. The regulation of the interaction should be explored. The determination of the intracellular localization of cryptochromes will provide insight into the exact mechanism. To this end, fusion molecules of cryptochromes and marker proteins such as GUS and green fluorescence protein (GFP) have been expressed in transgenic plants, and the intracellular localization has been examined. In transgenic Arabidopsis, fusion constructs of GUS with full-length AtCRY2 or its C terminus were localized to the nucleus under both light and dark conditions (Guo et al., 1999; Kleiner et al., 1999; Yang et al., 2000). However, a construct in which the C terminus of AtCRY1 was fused to GUS was localized to the cytoplasm in light but to the nucleus in the dark (Yang et al., 2000). The intracellular localization of cryptochromes has been examined also in a fern, Adiantum capillus-veneris (Imaizumi et al., 2000). Five cryptochromes have been identified in A. capillus-veneris, and three of them (AcCRY1, AcCRY2, and AcCRY5) have been shown to be localized to both the nucleus and the cytoplasm. AcCRY4 was localized to the nucleus under both light and dark conditions, whereas the localization of AcCRY3 shifted depending on light versus dark. Thus, we have not obtained the definitive knowledge of the mechanism that determines the intracellular localization of the cryptochromes. Here, we isolated two cryptochrome homologs from monocot rice (Oryza sativa) and designated them OsCRY1 and OsCRY2. We evaluated the function of monocot cryptochromes in transgenic Arabidopsis, and we examined the intracellular localization of OsCRY1 and OsCRY2 by using a GFP tag in transgenic plants as well as in a transient expression assay system.
RESULTS
Primary Structure of Rice Cryptochromes
We found a rice expressed sequence tag (EST) clone (S4586) with similarity to Arabidopsis cryptochromes. Using the EST clone as the probe, we screened a rice (O. sativa cv Nipponbare) cDNA library, and isolated two cryptochrome cDNA clones, OsCRY1 and OsCRY2 (DNA data bank of Japan accession nos. AB024337 for OsCRY1 and AB098568 for OsCRY2). OsCRY1 was 2,797 bp in length and contained an open reading frame encoding a predicted protein of 681 amino acids with a calculated mass of 75.2 kD; OsCRY2 was 2,650 bp long with an open reading frame that encoded a 568-amino acid predicted protein of 64.7 kD. We aligned the deduced amino acid sequences of both rice cryptochromes with those from Arabidopsis (Fig. 1). OsCRY1 showed 71.0% similarity with AtCRY1 and 56.1% with AtCRY2, and OsCRY2 had 64.9% similarity with AtCRY1 and 59.6% with AtCRY2. The similarity between the two rice cryptochromes was 78.8% overall, higher than any similarity with Arabidopsis cryptochromes. This similarity was even greater between residues 214 to 504 of OsCRY1 and 81 to 370 of OsCRY2. Like other cryptochromes from various organisms, the N-terminal regions of the deduced amino acid sequences of OsCRY1 and OsCRY2 each contained a photolyase-like domain, and the C-terminal regions contained three conserved motifs, referred as the DAS domain (Lin, 2002; Lin and Shalitin, 2003).
Figure 1.
Amino acid sequences of rice cryptochromes OsCRY1 and OsCRY2. We compared the deduced amino acid sequences of the rice cryptochromes (DNA data bank of Japan accession nos. AB024337 for OsCRY1 cDNA and AB098568 for OsCRY2 cDNA) with those of the Arabidopsis cryptochromes AtCRY1 (Ahmad and Cashmore, 1993) and AtCRY2 (Lin et al., 1996). Black boxes with white characters are identical amino acid residues in all sequences, and gray boxes with black characters are identical in three. NLS-like sequences (asterisks) in OsCRY1 are indicated above the alignment, and those in AtCRY2 are indicated below it. DAS domains in the C termini of cryptochromes are enclosed with boxes. Arrows indicate the regions of the fragments OsCRY1/N, OsCRY1/M, OsCRY1/C that we used for the analysis of intracellular localization (Fig. 6).
Inhibition of Hypocotyl Elongation in GFP-OsCRY1 Transgenic Arabidopsis Plants
To elucidate the function of rice cryptochromes, we constructed a chimeric gene encoding a GFP-OsCRY1 fusion protein and inserted it into the transformation vector pIG121-Hm (Ohta et al., 1990). The resulting construct was introduced into Agrobacterium tumefaciens, and wild-type Arabidopsis plants were transformed. We confirmed the transfer of the chimeric gene, GFP-OsCRY1, through PCR amplifications using a primer set recognizing the OsCRY1-encoded sequence. We obtained three lines of transgenic plants (L1–L3), which had very similar phenotypes.
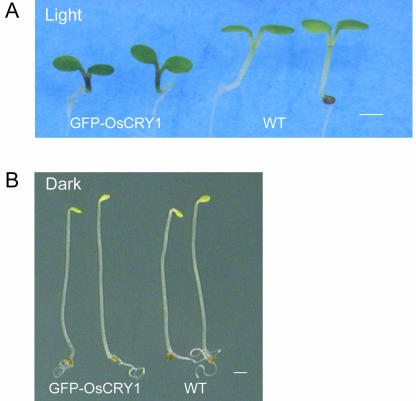
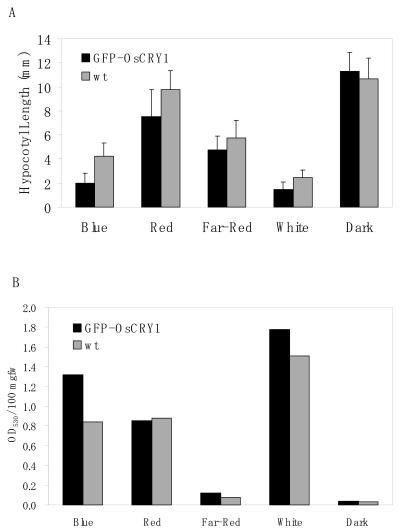
We compared the morphology of transgenic GFPOsCRY1 Arabidopsis seedlings grown in white light with that of wild-type plants. The transgenic seedlings had shorter hypocotyls than the wild type had (Fig. 2). However, when the seedlings were grown in complete darkness, the hypocotyl length did not differ significantly between GFP-OsCRY1 and wild-type plants. These results indicate that the GFP-OsCRY1 fusion protein inhibits hypocotyl elongation in light. To examine whether the inhibitory effect is specific to blue light, GFP-OsCRY1 seedlings were grown under continuous blue, red, or far-red light (Fig. 3A). Hypocotyl elongation in both GFP-OsCRY1 and wild-type plants was remarkably inhibited under blue-light conditions, and this effect was more pronounced in the transgenic plants than in the wild-type plants. The inhibitory effects of red and far-red light were far less dramatic, and no distinct differences were found between GFP-OsCRY1 and wild-type plants. These results indicate that OsCRY1 may function as a blue-light receptor to regulate responses such as inhibition of coleoptile elongation in rice.
Figure 2.
Phenotypes of transgenic Arabidopsis plants overexpressing the rice cryptochrome OsCRY1. A, Arabidopsis seedlings germinated for 5 d in 16-h light/8-h dark. Left, GFP-OsCRY1 transformants; right, wild-type (Columbia) seedlings. Bar = 1 mm. B, Arabidopsis seedlings germinated for 5 d in complete darkness. Left, GFP-OsCRY1 transformants; right, wild-type (Columbia) seedlings. Bar = 1 mm.
Figure 3.
Inhibition of hypocotyl elongation and enhanced accumulation of anthocyanin contents by blue light in transgenic Arabidopsis plants overexpressing the rice cryptochrome OsCRY1. We assessed the inhibition of hypocotyl elongation (A) and enhanced anthocyanin accumulation (B) of transgenic GFP-OsCRY1 (black bars) and wild-type (gray bars) seedlings grown under continuous blue, red, far-red, or white light or darkness. Plants were germinated for 5 d at 25°C. Error bars in A indicate sd. These experiments were repeated at least three times, and typical results are shown here.
Anthocyanin Accumulation in GFP-OsCRY1 Transgenic Arabidopsis Plants
The hypocotyls of light-grown GFP-OsCRY1 seedlings showed purple color darker than those of wild-type plants, suggesting enhanced accumulation of anthocyanin. Therefore, we determined the anthocyanin content of OsCRY1 transgenic and wild-type plants grown under conditions of continuous blue, red, or far-red light (Fig. 3B). GFP-OsCRY1 seedlings showed enhanced accumulation of anthocyanin when grown under blue light; this effect did not occur under red or far-red light conditions. Therefore, we have shown that OsCRY1 has a role in anthocyanin accumulation in a blue-light-dependent manner as well as in inhibition of hypocotyl elongation in Arabidopsis plants.
Localization of GFP-OsCRY1 in Stable Transformants of Arabidopsis Plants
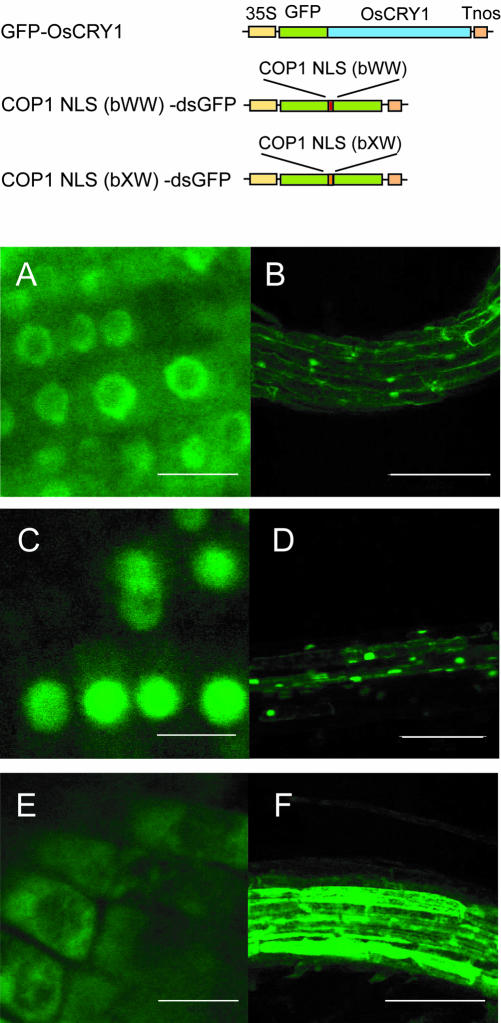
To study the relationship between the function of OsCRY1 and its intracellular localization, we examined the localization of GFP-OsCRY1 in transgenic Arabidopsis plants. To determine the intracellular localization of the GFP-OsCRY1 fusion protein, we used the GFP fusion proteins, COP1 NLS(bWW)dsGFP as a control for nuclear localization and COP1 NLS(bXW)-dsGFP for cytoplasmic localization of GFP (Jiang et al., 2001). We germinated COP1 NLS(bWW)-dsGFP, COP1 NLS(bXW)-dsGFP, and GFP-OsCRY1 transgenic Arabidopsis seeds and evaluated the intracellular localization of the fusion proteins. COP1 NLS(bWW)-dsGFP was accumulated in the nucleus but was scarcely detected in the cytoplasm, and COP1 NLS(bXW)-dsGFP was localized to the cytoplasm (Fig. 4, C–F). GFP-OsCRY1 seemed to be localized to both the nucleus and the cytoplasm (Fig. 4, A and B). Recently, Yang et al. (2000) indicated a light-mediated change in the intracellular localization (nucleus versus cytoplasm) of the C terminus of Arabidopsis cryptochrome fused with GUS. However, when we assessed the possible effect of light on the intracellular localization of GFPOsCRY1, we discerned no appreciable difference between light- and dark-grown seedlings (data not shown).
Figure 4.
Intracellular localization of GFP-OsCRY1 fusion protein in Arabidopsis transgenic plants. A and B, GFP-OsCRY1; C and D, COP1 NLS(bWW)-dsGFP as control for nuclear localization; E and F, COP1 NLS(bXW)-dsGFPs as control for cytoplasmic localization. A, C, and E, Root-tip cells; B, D, and F, the root cells near the hypocotyls. The seeds of transgenic plants were germinated in 16-h light/8-h dark. The seedlings were fixed in 3.7% (w/v) formaldehyde for 5 min and washed twice with phosphate-buffered saline before the analysis. Bar = 10 μm (A, C, and E) or 100 μm (B, D, and F). Schematic diagrams of the constructs used are shown at the top.
Transient Expression of GFP-OsCRY Chimeric Genes in Rice Cells
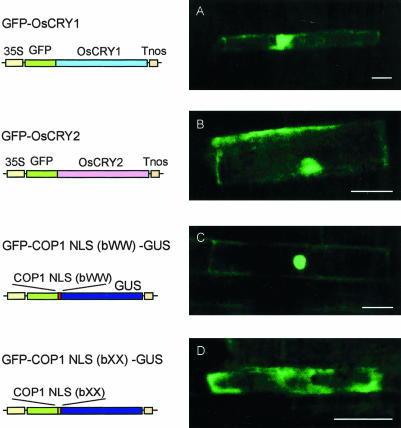
To define the intracellular localization of rice cryptochromes in rice cells, we introduced GFP-OsCRY1 into rice root cells by using particle bombardment. GFP-OsCRY2 chimeric gene was also constructed in the same way as GFP-OsCRY1 and introduced into rice cells. For control experiments of nuclear and cytoplasmic localization, GFP-COP1 NLS(bWW)GUS and GFP-COP1 NLS (bXX)-GUS chimeric genes were also expressed transiently in rice cells. By using the intracellular localization of these controls for comparison, we concluded that OsCRY1 fusion protein and OsCRY2 fusion protein were localized to both the nucleus and the cytoplasm in rice root cells (Fig. 5, A–D). These results are consistent with the intracellular localization of the GFP-OsCRY1 protein in Arabidopsis plants.
Figure 5.
Intracellular localization of GFP-OsCRY1 and GFPOsCRY2 transiently expressed in rice cells. GFP-OsCRY1 (A), GFPOsCRY2 (B), GFP-COP1 NLS(bWW)-GUS (control for nuclear localization; C), and GFP-COP1 NLS(bXX)-GUS (control for cytoplasmic localization; D). Bar = 20 μm. Schematic diagrams of the chimeric genes are shown at the left of the each figure.
Functional Domain That Determines the Intracellular Localization of OsCRY1 in Plant Cells
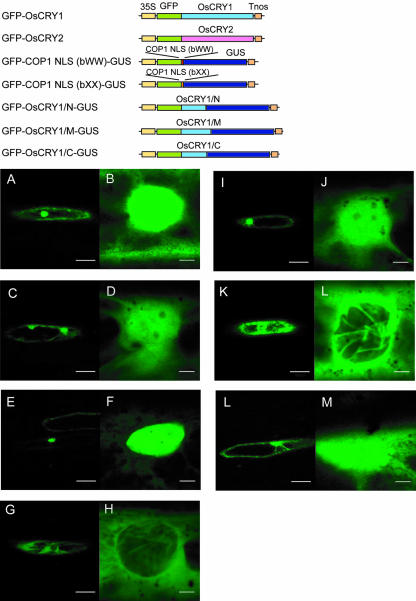
To identify the domain that determines the intracellular localization of cryptochromes, we divided OsCRY1 cDNA into three fragments encoding 1 through 213 amino acids (OsCRY1/N), 214 through 504 amino acids (OsCRY1/M), and 446 through 681 amino acids (OsCRY1/C) and then inserted each fragment between copies of the GFP and GUS genes. We transiently expressed these fusion genes in-frame under the control of the cauliflower mosaic virus (CaMV) 35S promoter in onion epidermal cells. For control experiments, we also expressed GFPOsCRY1, GFP-OsCRY2, GFP-COP1 NLS(bWW)GUS, and GFP-COP1 NLS (bXX)-GUS. Like GFPOsCRY1, GFP-OsCRY1/N-GUS and GFP-OsCRY1/C-GUS were localized to both the nucleus and the cytoplasm, but GFP-OsCRY1/M-GUS was accumulated only in the cytoplasm (Fig. 6, I–M). The estimated size of GFP-OsCRY1/N-GUS is 119 kD, GFPOsCRY1/M-GUS is 130 kD, and GFP-OsCRY1/CGUS is 123 kD. Consequently the nuclear localization of GFP-OsCRY1/N-GUS and GFP-OsCRY1/C-GUS might not be due to diffusion but to active transport.
Figure 6.
Intracellular localization of various GFP-OsCRY constructs transiently expressed in onion epidermal cells. GFP-OsCRY1 (A and B), GFP-OsCRY2 (C and D), GFP-COP1 NLS(bWW)GUS (E and F), GFP-COP1 NLS(bXX)-GUS (G and H), GFP-OsCRY1/N-GUS (I and J), GFP-OsCRY1/M-GUS (K and L), and GFP-OsCRY1/C-GUS (L and M). Left, Complete view of the cell; right, close-up. Bar = 100 μm (A, C, E, G, I, K, and L) or 10 μm (B, D, F, H, J, L, and M). Schematic diagrams of the chimeric genes are shown at the top of the figure.
DISCUSSION
We isolated two cryptochrome cDNA clones, OsCRY1 and OsCRY2, from rice. The N-terminal region of the deduced amino acid sequences of OsCRY1 and OsCRY2 each contained a photolyase-like domain, which is well conserved in other cryptochromes from various organisms. From the sequence data, it is indicated that both OsCRY1 and OsCRY2 are more closely related to AtCRY1 than AtCRY2. Gene duplication may have occurred after the divergence of monocotyledons and dicotyledons.
GFP-OsCRY1 transgenic Arabidopsis plants exhibited the phenotypes of short hypocotyls and increased anthocyanin accumulation, which are typical phenotypes induced by blue-light-receptor cryptochromes (Figs. 2 and 3). These phenotypes were observed when seedlings were grown in blue or white light but not when they were grown in red or far-red light or in the dark. The phenotypes of the transgenic plants indicate that OsCRY1 is a blue-light-receptor cryptochrome. Ours is the first functional characterization of a cryptochrome from monocotyledonous plants.
The blue-light signal transduction likely is mediated by various protein-protein interactions during the process from perception of a blue-light signal to gene expression in plant cells. When a protein interacts with another protein, they both must be in the same intracellular location. Therefore, investigation of the cellular localization of signaling molecules is a prerequisite for the elucidation of the signal transduction pathway. In this study, we examined the intracellular localization of rice cryptochromes by using GFP tags. We used COP1 bipartite nuclear localization signal (NLS) as a control (Jiang et al., 2001). COP1 NLS(bWW)-dsGFP or GFP-COP1 NLS(bWW)-GUS was used as a control for nuclear localization, and COP1 NLS(bXW)-dsGFP or GFPCOP1 NLS(bXX)-GUS, containing a mutated COP1 NLS, was used as a control for cytoplasmic localization of GFP. By using these controls for comparison, we concluded that GFP-OsCRY1 was localized to both the nucleus and the cytoplasm of cells in Arabidopsis plants (Fig. 4). Previously, Arabidopsis cryptochromes were reported to be localized to the nucleus (Cashmore et al., 1999; Guo et al., 1999; Kleiner et al., 1999; Yang et al., 2000), but our data on the localization of the rice cryptochrome seemingly contradict those findings. To confirm the intracellular localization of OsCRY1 in rice plants, GFP-OsCRY1 was expressed transiently in rice cells. GFP-OsCRY1 was localized to both the nucleus and the cytoplasm in rice cells (Fig. 5), consistent with the intracellular localization of GFP-OsCRY1 in transgenic Arabidopsis plants.
Transient expression system using onion epidermis has been used frequently for the investigation of intracellular localization of proteins, such as the Arabidopsis cryptochrome, AtCRY1 (Cashmore et al., 1999), and Arabidopsis COP1 (von Arnim and Deng, 1994). In this study, GFP-OsCRY1 was also transiently expressed in onion epidermal cells and shown to be localized to both the nucleus and the cytoplasm (Fig. 6). The intracellular localization of GFP-OsCRY1 to the nucleus and the cytoplasm has been demonstrated in three species of plants; Arabidopsis, rice, and onion. On the other hand, when the NLS sequence identified from COP1 protein was fused with GFP or GUS, the fusion proteins accumulated in the nucleus. This nuclear accumulation was not observed when the COP1 NLS was mutated. These NLS-dependent nuclear localization also has been demonstrated in the cells of the three plant species. Therefore, the mechanisms determining the intracellular localization of proteins seem to be conserved among a variety of plant species, and it was suggested that rice cryptochrome, OsCRY1, is localized to both nucleus and cytoplasm in rice plants.
Additionally, we examined the intracellular localization of OsCRY2, the second member of rice cryptochromes we identified. GFP-OsCRY2 was localized to the nucleus and the cytoplasm in rice cells (Fig. 5B) and in onion epidermal cells (Fig. 6, C and D). The localization of GFP-OsCRY2 is also examined in transgenic Arabidopsis plants, and a similar result was revealed (data not shown). Both of rice cryptochromes, OsCRY1 and OsCRY2, were localized to both the nucleus and the cytoplasm.
Using a transient expression assay system (Fig. 6, I–M), we noted pronounced nuclear accumulation of the GFP-fusion proteins GFP-OsCRY1/N-GUS and GFP-OsCRY1/C-GUS, but negligible nuclear accumulation of GFP-OsCRY1/M-GUS, which was apparent only as “threads” of GFP that suggested cytoplasmic stream on the nuclear surface. These results suggest that OsCRY1 has at least two domains in its N and C termini directing nuclear localization. This feature may be a characteristic of rice cryptochromes distinct from Arabidopsis cryptochromes, which harbor a single nuclear targeting domain in the C terminus (Guo et al., 1999).
Nuclear localization of proteins is interpreted as the consequence of import to the nucleus after their translation on ribosomes. Nuclear proteins containing NLS are recognized and imported into the nucleus through nuclear pores by virtue of the NLS-receptor complex, which includes importin-α and -β. The NLSs of plant nuclear proteins have been identified in their primary structures and are classified into three types: monopartite NLS, consisting of a short stretch of basic amino acids; bipartite NLS, consisting of two clusters of basic amino acids; and yeast Mat α2-like NLS, which has hydrophobic as well as basic amino acids (Yamamoto and Deng, 1999). To address the mechanisms determining the intracellular localization of cryptochrome, we searched for NLS sequences in the primary structure of cryptochrome molecules of rice and Arabidopsis. The results of a PSORT (http://psort.nibb.ac.jp) homology search (Table I) predicted four NLS-like sequences in the primary structure of OsCRY1. Three putative NLSs, two bipartite NLSs, and a monopartite NLS were found in the N-terminal region (OsCRY1/N), and a putative monopartite NLS was found in the C-terminal region (OsCRY1/C). Whereas pronounced amount of both GFP-OsCRY1/N-GUS and GFP-OsCRY1/C-GUS accumulated in the nucleus, GFP-OsCRY1/M-GUS did not (Fig. 6). From these data, we surmise that these four NLS-like sequences may play an important role in the accumulation of OsCRY1 in the nucleus. However, the primary structure of OsCRY2 contains no NLS-like sequences, although GFP-OsCRY2 was localized to both nucleus and cytoplasm. Therefore, the possible involvement of NLS might be excluded from the mechanism determining the intracellular localization of these proteins. This interpretation is supported by the following three reasons. (a) Although Arabidopsis cryptochromes, AtCRY1 and AtCRY2, were reported to be localized to the nucleus, a putative NLS was found in AtCRY2 but not in AtCRY1. (b) Although putative NLSs occur in the primary structure of OsCRY1, these are atypical. (c) Monopartite NLSs (e.g. SV40 T-antigen NLS) work well in plant cells, but few monopartite NLSs have been identified among plant nuclear proteins (e.g. maize bZIP protein Opaque-2 and Arabidopsis COP1 protein), in which bipartite NLSs are more common (Varagona et al., 1992; Jiang et al., 2001).
Table I.
Putative NLSs and intracellular localization of plant cryptochromes NLSs were predicted by using the PSORT program (http://psort.nib.ac.jp). The underlining indicates basic amino acids. N, Nuclear localization; C, cytoplasmic localization; n.f., not found; n.d., not determined.
| Putative NLSs
|
Intracellular Localization
|
|||||
|---|---|---|---|---|---|---|
| Monopartite | Bipartite | Transient Assay | Stable Transformants | Reference | ||
| Rice | OsCRY1 | |||||
| OsCRY1 full length | N/C | N/C | This report | |||
| OsCRY1/N | KRRRRRR | RRQQARHPPLRRRRRRAHRAR QRRPAVRAMGGGRRRRLP | N/C | n.d. | This report | |
| OsCRY1/M | n.f. | n.f. | C | n.d. | This report | |
| OsCRY1/C | RRRR | n.f. | N/C | n.d. | This report | |
| OsCRY2 | n.f. | n.f. | N/C | n.d. | This report | |
| Arabidopsis | AtCRY1 | n.f. | n.f. | N | N | Cashmore et al. (1999) |
| AtCRY2 | n.f. | KRVKPEEEEERDMKKSR | n.d. | N | Guo et al. (1999); Kleiner et al. (1999) | |
During this decade, protein transport across nuclear pore has been well characterized in plant species (Yamamoto and Deng, 1999), and the Leu-rich nuclear export signals of various proteins and the receptor protein exportin were also shown to be important in protein export from the nucleus to the cytoplasm of plant cells (Haasen et al., 1999). Although the possible export of plant cryptochrome from nucleus to cytoplasm should be incorporated into hypotheses of the mechanism for intracellular localization, no nuclear export signals occur in OsCRY1 and OsCRY2. To explain the intracellular localization of OsCRY1, a possible mechanism other than direct import by importin-α and -β can be argued. The intracellular localization of the Arabidopsis homeotic proteins APETALA3 (AP3) and PISTALLA (PI) was examined using the GUS reporter (McGonigle et al., 1996). AP3-GUS and PI-GUS were localized to the cytoplasm when the fusion construct alone was expressed in the onion epidermal cells of a transient expression assay system. However, the coexpression of AP3-GUS with PI or of PI-GUS with AP3 resulted in nuclear localization of GUS activity. Those results suggest that the interaction of two or more proteins might cause a conformational change via the formation of a complex and result in the unmasking of NLSs. Recently, various cryptochrome molecules have been reported to interact with a specific partner molecule. For example, the plant cryptochrome AtCRY1 was shown to interact with a negative regulator of photomorphogenesis, COP1 (Yang et al., 2001), and mouse cryptochromes interact with mPER proteins in the negative limb of the circadian clock feedback loop (Kume et al., 1999). Therefore, rice cryptochromes also may interact with specific partner molecules, and their interactions may regulate intracellular localization to accomplish the physiological roles of perception and transduction of blue-light signals.
We have shown that OsCRY1 functions as a blue-light receptor (Figs. 2 and 3) and that rice cryptochromes are localized to both the nucleus and the cytoplasm (Figs. 4, 5, 6). Nuclear-localized GFPOsCRY1 might perform the same physiological roles in Arabidopsis cells as do the intrinsic Arabidopsis cryptochromes, which are localized solely to the nucleus. The GFP-OsCRY1 in the cytoplasm might be interpreted as overflow from the nucleus after the high expression of the construct due to the strong CaMV 35S promoter and therefore might have no biological relevance. However, we surmise that rice cryptochromes have multiple functions and that the multiple intracellular localization patterns correspond to these functions. This intracellular localization scheme requires multiple partner molecules and will enable cryptochromes to play a role in numerous plant responses to blue light. Analysis of intracellular localization will give us some insight of the mechanism of blue-light signaling, which might be conserved or differentiated between Arabidopsis and rice. We now plan to identify the various proteins interacting with cryptochromes in rice.
MATERIALS AND METHODS
Cloning of OsCRY1 and OsCRY2 cDNAs
We found a rice (Oryza sativa) EST cDNA clone S4586 (accession no. D41779), which showed sequence homology with AtCRY1, and determined its complete nucleotide sequence. However, because it did not contained the entire open reading frame, we screened a rice etiolated seedling leaf cDNA library constructed in λZAPII vector with cDNA probe of the S4586 clone. Several clones were isolated, and the longest two clones were subjected to the determination of complete sequences. We designated them as OsCRY1 and OsCRY2, respectively.
Plasmid Constructions and Transformation
GFP-OsCRY1
To generate the GFP-OsCRY1 fusion gene, the OsCRY1 cDNA sequence was modified by the addition of an ApaI site at the 5′ end of the open reading frame, and the resultant fragment was cloned into the ApaI and SmaI sites of the previously constructed psGFPcs plasmid (Jiang et al., 2001), which produces GFP fusion proteins under the control of the CaMV 35S promoter. To add an ApaI site, the 0.7-kb 5′ end fragment was PCR-amplified by using the primers 5′-atgggcccatgtcagcgtcgccgtcgtc-3′ (the created ApaI site is underlined) and 5′-aactagtcccccgctcggactcgtcctc-3′. The fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced to confirm correct amplification. The fragment was ligated into the SphI site of OsCRY1 cDNA. The full-length OsCRY1 cDNA was excised at the ApaI and EcoRI sites, the EcoRI site was filled in to produce a blunt end, and the fragment was cloned into the ApaI and SmaI sites of psGFPcs. The resultant gene was designated GFP-OsCRY1 and was used in the transient expression assay.
GFP-OsCRY2
The GFP-OsCRY2 fusion gene was constructed in a similar way as GFPOsCRY1. To add an ApaI site at the 5′ end of the OsCRY2 cDNA sequence, the 0.3-kb 5′ end fragment was PCR-amplified by using the primers 5′-ttgggcccatgatggcagcggagggcatc-3′ and 5′-atcccggggtgccttgtctcgattctgc-3′ (ApaI and SmaI sites underlined). To generate a SmaI site at the 3′ end, we PCR-amplified the 0.5-kb 3′ end fragment using the primers 5′-atgtcgacgctgacaacagctcgaagac-3′ and 5′-ggcccgggtcacaatgactgggataattg-3′ (SalI and SmaI sites underlined). The amplified fragments were ligated into the native SalI and KpnI sites of the OsCRY2 cDNA, and the full-length OsCRY2 cDNA sequence was introduced into psGFPcs. The resultant gene was designated GFP-OsCRY2 and used in the transient expression assay.
Subcloning of GFP-OsCRY Chimeric Genes into a Ti Plasmid
For the transformation of plants, GFP-OsCRY1 and GFP-OsCRY2 were each inserted into the pIG121-Hm vector (Ohta et al., 1990) downstream of the CaMV 35S promoter.
Subcloning of OsCRY1 Fragments between GFP and GUS Genes
To produce GFP-GUS fusion proteins, the XmaI-EcoRI GUS fragment from pBI221.3 (BD Biosciences Clontech, Palo Alto, CA) was inserted into the XmaI and EcoRI sites of psGFPcs, resulting in psGFP-GUS/pUC18. The OsCRY1 fragments OsCRY1/N, OsCRY1/M, and OsCRY1/C encoding the N-terminal, middle, and C-terminal regions of OsCRY1 were PCR-amplified by using 5′-atgggcccatgtcagcgtcgccgtcgtc-3′ and 5′-aactagtcccccgctcggactcgtcctc-3′ for OsCRY1/N,5′-ttgggcccagcaacgcgctgctggcgag-3′ and 5′-gactagtctcgagctcccacatctctg-3′ for OsCRY1/M, and 5′-atgggcccctgccggagctggcaaggctg-3′ and 5′-cactagttcggcagcgtagtgatctgag-3′ for OsCRY1/C (created ApaI site is underlined). The PCR-amplified fragments were each cloned into pGEM-T Easy and sequenced to verify correct amplification. Each fragment was then inserted in-frame into the ApaI and SmaI sites of psGFP-GUS/pUC18 to produce the GFP-GUS fusion proteins. The resultant plasmids were designated GFP-OsCRY1/N-GUS, GFP-OsCRY1/M-GUS, and GFP-OsCRY1/C-GUS and used in the transient expression assay.
Transformation and Analysis of Transgenic Arabidopsis Plants
GFP-OsCRY1 cloned into the pIG-Hm transformation vector was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and the transformed bacteria were vacuum-infiltrated into Arabidopsis Columbia ecotype (Bechtold et al., 1993). The transgenic plants were selected on germination media plates (Valvekens et al., 1988) containing hygromycin (50 mg L-1).
To elucidate the characteristics of transgenic plants, seeds were germinated at 22°C for 5 d under various light conditions (described later) and in constant dark, and their hypocotyl length and amounts of anthocyanin were measured. Anthocyanin was extracted from seedlings in 0.1% (w/v) HClmethanol at 4°C overnight, and the amount was determined by measuring OD530.
Light Conditions
Blue light (1.1 W m-2 s-1) was obtained from FL20S.B fluorescent tubes (Toshiba, Tokyo). Red light (4.4 W m-2 s-1) was obtained from FL20SRF fluorescent tubes (National, Osaka) filtered through a red plastic filter (Shinkolite A no. 102, Mitsubishi Rayon, Tokyo), and far-red light (1.9 W m-2 s-1) was obtained from FL20S-FR74 fluorescent tubes (Toshiba) wrapped with one layer of Polycolor no. 22 and one layer of Polycolor no. 72 film (Tokyo Butai Shomei, Tokyo). The intensities of monochromatic lights were measured by using a light meter (model LI-189, LI-COR, Lincoln, NE) and a ryranometer sensor (LI-COR).
Transient Expression Assay
Intracellular localization of GFP fusion proteins was also examined in the transient expression assay using rice roots or onion epidermis. Rice roots were prepared from 8-d-old seedlings, which were water-cultured as previously reported (Shoji et al., 1998). Plasmids were introduced by particle bombardment (Jiang et al., 2001). The roots and epidermis were incubated at 22°C on Murashige and Skoog agar medium overnight.
Fluorescence Microscopy
Intracellular localization of GFP in transgenic Arabidopsis was examined under a confocal laser-scanning microscope (TCS NT, Leica, Wetzlar, Germany).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material.
Acknowledgments
We are grateful to the Rice Genome Research Program, National Institute of Agrobiological Sciences for providing the rice EST cDNA clone, S4586. We thank T. Itou and K. Haga for having carried out the initial experiments of this study. We also thank A. Baba for providing the plasmids, GFP-COP1 NLS (bWW)-GUS and GFP-COP1 NLS (bXX)-GUS.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025759.
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162-166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR (1998) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell 1: 939-948 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci 316: 1194-1199 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Huala E (1999) Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15: 33-62 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760-765 [DOI] [PubMed] [Google Scholar]
- Guo H, Duong H, Ma N, Lin C (1999) The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J 19: 279-287 [DOI] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695-705 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kanegae T, Wada M (2000) Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell 12: 81-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Shoji K, Matsuki R, Baba A, Inagaki N, Ban H, Iwasaki T, Imamoto N, Yoneda Y, Deng X et al. (2001) Molecular cloning of a novel importin α homologue from rice, by which constitutive photomorphogenic 1 (COP1) nuclear localization signal (NLS)-protein is preferentially nuclear imported. J Biol Chem 276: 9322-9329 [DOI] [PubMed] [Google Scholar]
- Kanegae T, Wada M (1998) Isolation and characterization of homologues of plant blue-light photoreceptor (cryptochrome) genes from the fern Adiantum capillus-veneris. Mol Gen Genet 259: 345-353 [DOI] [PubMed] [Google Scholar]
- Kleiner O, Kircher S, Harter K, Batschauer A (1999) Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J 19: 289-296 [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 23: 193-205 [DOI] [PubMed] [Google Scholar]
- Lin C (2002) Blue light receptors and signal transduction. Plant Cell Suppl 14: S207-S225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Chan J, Cashmore AR (1996) CRY2: a second member of the Arabidopsis cryptochrome gene family. Plant Physiol 110: 10478819875 [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469-496 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686-2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, Kay SA (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207-211 [DOI] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF (1996) Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev 10: 1812-1821 [DOI] [PubMed] [Google Scholar]
- Ninu L, Ahmad M, Miarelli C, Cashmore AR, Giuliano G (1999) Cryptochrome 1 controls tomato development in response to blue light. Plant J 18: 551-556 [DOI] [PubMed] [Google Scholar]
- Ohta S, Mita S, Hatttori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31: 805-813 [Google Scholar]
- Shoji K, Iwasaki T, Matsuki R, Miyao M, Yamamoto N (1998) Cloning of a cDNA encoding an importin-α and down-regulation of the gene by light in rice leaves. Gene 212: 279-286 [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC (1998) The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681-692 [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536-5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4: 1213-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035-1045 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154-158 [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Deng XW (1999) Protein nucleocytoplasmic transport and its light regulation in plants. Genes Cells 4: 489-500 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573-2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815-827 [DOI] [PubMed] [Google Scholar]