Abstract
Experimental and clinical data support a growth inhibitory role for HER4 in breast cancer. Clinically HER4 expression is extinguished during breast tumorigenesis supporting a tumor suppressor function for HER4, however, a molecular mechanism to explain the selective loss of HER4 expression has remained elusive. Epigenetic mechanisms, for example, aberrant gene promoter hypermethylation, have been shown to ablate tumor suppressor gene expression in breast carcinomas. We identified a CpG island within the HER4 promoter and show by pyrosequencing of bisulfite-treated DNA an inverse correlation between HER4 expression and the extent of promoter methylation. Treatment of the HER4-negative BT20 cell line with the DNA demethylating agent 5-aza-2′-deoxycytidine (DAC)-enhanced HER4 expression, confirming a role for DNA methylation in suppressed HER4 expression. DAC treatment to reactive HER4 expression in combination with the HER4 ligand heregulin-β1 (HRG) resulted in apoptosis of BT20 cells providing a novel therapeutic strategy for triple-negative tumors. The BT20 cells were rescued from apoptosis when preincubated with HER4 small interfering RNA, thereby confirming a role for HER4 in DAC/HRG-induced apoptosis. We verified HER4 promoter methylation in primary breast carcinomas and detected a significant increase in HER4 promoter methylation in HER4-negative breast tumors (P<0.001). Furthermore, increased levels of HER4 promoter methylation were significantly associated with worse patient prognosis (P=0.0234). Taken together, our data support a tumor suppressor function for HER4, which is epigenetically suppressed in breast tumors through promoter hypermethylation.
Keywords: breast cancer, EGFR-family, epigenetics, apoptosis, heregulin
Introduction
The HER4/ERBB4 receptor tyrosine kinase belongs to the epidermal growth factor receptor-family of potent breast oncogenes. However, HER4 expression in breast carcinomas independently predicts improved patient survival (Thor et al., 2009) while inversely correlating with tumor grade, metastasis and disease recurrence (Sundvall et al., 2008; Kreike et al., 2009). Furthermore, when examined in the context of the recently defined molecular subtypes of breast tumors, HER4 expression is most prevalent in the luminal subtypes associated with the best overall patient survival while HER4 is rarely expressed in the HER2-positive or triple-negative tumor subtypes with the worst patient prognosis (Perou et al., 2000; Sorlie et al., 2001; Hoadley et al., 2007). When expressed in tumor subtypes associated with poor prognosis, HER4 predicts improved patient response to multiple therapeutic interventions (Naresh et al., 2008; Sassen et al., 2009).
Experimental models provide mechanistic support for these clinical observations and further suggest that HER4 has potent anti-tumor activity in the breast. For example, HER4 activation inhibits proliferation of multiple breast tumor cell lines by promoting differentiation and/or apoptosis (Naresh et al., 2006; Muraoka-Cook et al., 2006b; Jones, 2008). In preclinical xenograft models, HER4 apoptotic activity is essential for tamoxifen-mediated tumor cell killing and patients with tumor expression of HER4 respond to tamoxifen therapy with no failures after 14 years (Naresh et al., 2008). Ligand-activated HER4 has also been shown to cooperate with BRCA1 to induce a G2/M delay during breast tumor cell cycle progression (Muraoka-Cook et al., 2006a). Although under some experimental conditions HER4 has also been shown to promote breast tumor cell proliferation (Zhu et al., 2006; Jones, 2008; Muraoka-Cook et al., 2009) these results require clinical verification.
Although loss of HER4 expression during breast tumor progression to higher grade and metastatic carcinomas is supported clinically, (Sundvall et al., 2008) a mechanism for the selective loss of HER4 activity during breast tumorigenesis remains elusive. Independent laboratories have identified somatic mutations, which potentially attenuate HER4 activity (Soung et al., 2006; Rokavec et al., 2007; Tvorogov et al., 2008). These mutations, however, were only observed in a small percentage (1–5%) of breast tumors and therefore cannot account for the absence of HER4 expression observed in 32% of all breast tumors (Thor et al., 2009) and 90% of the most aggressive breast carcinomas (Hoadley et al., 2007). Epigenetic mechanisms of gene suppression are commonly observed in breast tumors and one epigenetic event involving hypermethylation of gene promoter CpG sites has been shown to abolish expression of multiple tumor suppressors in the breast including BRCA1 (Esteller, 2007). In this study, we show that hypermethylation of a CpG island in the HER4 promoter is associated with suppressed HER4 expression in breast tumors. Reactivation of HER4 expression results in tumor cell apoptosis supporting a tumor suppressor function for HER4 in the breast.
Results and discussion
The HER4 promoter is hypermethylated in HER4-negative breast tumor cell lines
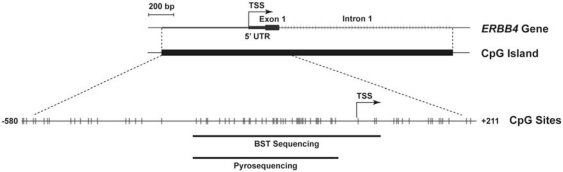
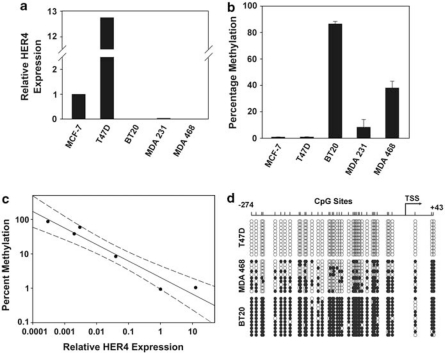
In breast cancer, aberrant hypermethylation of CpG islands within gene promoters is a common mechanism of tumor suppressor inactivation (Esteller, 2007). To determine if promoter hypermethylation regulates HER4 expression in breast tumor cells, we first analyzed the HER4 gene for the presence of CpG islands using UCSC Genome Bioinformatics. One CpG island was identified positioned at −580 to +1107 relative to the HER4 transcriptional start site (Figure 1). Extension of the CpG island into the first exon–intron sequence is potentially significant because these CpG sites can affect chromatin structure and gene regulatory transcription factor binding (Strathdee et al., 2004). In addition, exon–intron CpG sites may serve as ‘seed' sequences for upstream CpG methylation and gene silencing (Nguyen et al., 2001). An analysis of HER4 promoter methylation by pyrosequencing of bisulfite-treated DNA isolated from multiple breast tumor cell lines revealed a striking inverse correlation between HER4 expression and the extent of promoter methylation (Figures 2a–c).
Figure 1.
Identification of an ERBB4 CpG island. A CpG island was identified within the ERBB4 locus between −580 and +1107 relative to the transcriptional start site (TSS) using UCSC Genome Bioinformatics software. The lower portion shows a schematic of the CpG island between −580 and +211 relative to the TSS with CpG sites indicated by vertical black lines. Regions amplified by PCR for BST sequencing and pyrosequencing are indicated.
Figure 2.
The HER4 promoter is hypermethylated in breast tumor cells. (a) Relative HER4 expression determined by quantitative reverse transcriptase (qRT)–PCR with each cell line normalized to MCF-7 cells. Total RNA was isolated from cell lines using Trizol Reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturers' instructions. Superscript III (Invitrogen Life Technologies) was used to generate complementary DNA and HER4 expression was determined using ERBB4 TaqMan PCR primers and probe (assay ID Hs00171783_m1) with TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in a ABI Prism 7000 (Applied Biosystems). HER4 expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (assay ID Hs99999905_m1) and the 2−ΔΔCt method was used to calculate relative HER4 expression levels. (b) The CpG sites between −274 and −26 were analyzed by pyrosequencing and the mean methylation was determined. Data represent the mean and standard error of three independent DNA purifications. For pyrosequencing, the region of the ERBB4 CpG island between −130 and −62 (CpG sites 24–38) was amplified from bisulfite-treated DNA using primers 5′-GTTYGTTTTGGGAGTYGTTATAT-3′ and biotinylated 5′-ATCCAAATAACATATCCCCCTTT-3′. The sequencing primer 5′-GTTTTGGGAGTYGTTATA-3′ was used for pyrosequencing at EpigenDx (Worcester, MA, USA). (c) Simple regression plot with 95% confidence interval in which each point represents one breast tumor cell line. (d) Bisulfite sequencing analysis of HER4 CpG sites between −274 and +43 relative to the transcriptional start site (TSS) from 10 independent DNA clones for each cell line. Open boxes indicate unmethylated CpG sites and closed boxes indicate methylated CpG sites. DNA was isolated from cultured cells and bisulfite treated using EZ DNA Methylation Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. A region of the ERBB4 CpG island between −274 and +43 relative to the transcriptional start site (TSS) was PCR amplified using primers designed for bisulfite-modified DNA (5′-AGTGAGAGAGAGAGAAAGTGAGGAG-3′ and 5′-CACCCAAACCCAAAATCCTA-3′) and PCR products were cloned into TOPO TA Cloning Kit (Invitrogen Life Technologies). DNA was purified from 10 or 11 clones for each cell line and sequenced.
To determine the methylation status of each CpG site between −274 and +43, we analyzed DNA from three representative cell lines by direct bisulfite DNA sequencing. For each cell line, multiple DNA strand copies were analyzed by cloning the bisulfite PCR product and sequencing at least 10 individual clones. None of the 42 CpG sites was methylated in the 11 individual DNA isolates analyzed from HER4-expressing T47D cells (Figure 2d). In concordance with decreasing levels of HER4 expression, the extent of CpG site methylation increased within MDAMB468, and was even greater within BT20 cells (Figure 2d). Taken together, our data suggest that HER4 expression is suppressed in breast tumor cells through hypermethylation of multiple CpG sites within the HER4 promoter.
Reactivated expression of HER4 promotes breast tumor cell apoptosis
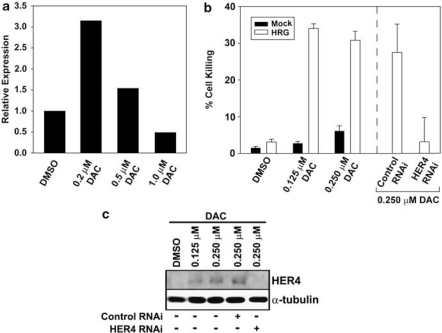
We have previously shown that HER4 functions as a pro-apoptotic BH3-only protein of the BCL-2 family and when ligand-activated HER4 induces apoptosis of multiple tumor cell lines (Vidal et al., 2005; Naresh et al., 2006). This apoptotic activity may not be unique to HER4. For example, HER2, another epidermal growth factor receptor-family member, undergoes caspase cleavage to generate a HER2 intracellular domain with pro-apoptotic BH3-only protein activity (Strohecker et al., 2008). Epigenetic silencing of HER4 expression may provide a mechanism for breast tumor cells to evade HER4-induced apoptosis. Indeed, evasion of apoptosis is one of the obligate hallmarks of tumorigenesis (Hanahan and Weinberg, 2000). To determine if reactivated HER4 expression can suppress tumorigenesis, we reactivated HER4 expression by treating BT20 cells with the demethylating agent 5-aza-2′-deoxycytidine (DAC). This cell line had the highest levels of HER4 promoter methylation and resembles triple-negative breast tumors. Patients with advanced triple-negative breast tumors have extremely poor prognosis and because these tumors lack targetable molecular markers therapeutic options for these patients are limited to cytotoxic therapy. DAC treatment of BT20 cells resulted in increased HER4 expression detectable at both the RNA and protein levels (Figures 3a and c). These results further confirm that HER4 expression is suppressed in these breast tumor cell lines through a reversible DNA methylation mechanism.
Figure 3.
Reactivation of HER4 expression results in tumor cell apoptosis. (a) The BT20 breast tumor cell line was treated with the indicated amount of DAC for 48 h. HER4 expression after DAC treatment relative to dimethylsulphoxide (DMSO) vehicle control was determined by quantitative reverse transcriptase (qRT)–PCR as described in Figure 2a. (b) BT20 cells were treated with the indicated amount of DAC in combination with 50 ng/ml of HRG and analyzed for apoptosis after 48 h. The percentage of cells undergoing apoptosis was determined using a combination of 4′,6-diamidoino-2-phenylindole (DAPI) staining for condensed chromatin and TUNEL assay as described elsewhere (Naresh et al., 2008). In some experiments, HER4 expression was suppressed using erbB-4/HER-4 small interfering RNA (siRNA) SMARTpool or a nonspecific negative control pool (Upstate Biotechnology, Lake Placid, NY, USA) exactly as described elsewhere (Naresh et al., 2008). BT20 cells were pretreated with HER4 siRNA 24 h before DAC and HRG treatments. Results represent the mean and standard error of at least three independent experiments. (c) Western blot analysis of HER4 expression in BT20 cells treated with DMSO or DAC in the presence or absence of control RNA interference (RNAi) or HER4 RNAi. Treatments were performed as described in panel (b). Total cell lysates were prepared and analyzed by western blot exactly as described elsewhere (Jones et al., 1999). Primary antibodies used for western blot analysis included ErbB4 E200 (Abcam, Cambridge, MA, USA) and α-tubulin #05829 (Upstate Biotechnology). Secondary antibodies were IRDye 680 goat anti-rabbit (Li-Cor Biosciences, Lincoln, NE, USA) or IRDye 800 goat anti-mouse (Li-Cor Biosciences) detected using an Odyssey Infrared Imaging System (Li-Cor Biosciences).
We have previously shown that heregulin-β1 (HRG) stimulation of endogenous HER4 results in apoptosis of breast tumor cell lines (Naresh et al., 2006), however, non-malignant cells appear to be resistant to HER4 apoptotic activity (Vidal et al., 2007). In non-transformed cells, the HER4 intracellular domain and BH3-only protein, 4ICD, is sequestered to the nucleus in which it cooperates with STAT5A to regutle gene expression (Long et al., 2003; Williams et al., 2004). To determine if the apoptotic activity of reactivated HER4 can be induced in the BT20 cell line, we combined DAC treatment with HRG stimulation of the BT20 cell line. Treatment of BT20 cells with increasing concentrations of DAC resulted in an insignificant increase in cell death. Concurrent treatment with HRG, however, resulted in dramatic cell killing after 48 h of treatment (Figure 3b). Although DAC treatment typically affects expression of <1% of the human transcriptome, we cannot rule out the possibility that the DAC/HRG combination induces apoptosis independent of HER4. To confirm a role for reactivated HER4 in DAC/HRG-mediated cell killing, we suppressed reactivated HER4 expression by concurrent HER4 RNA interference treatment. The DAC/HRG combination failed to promote significant levels of tumor cell apoptosis when HER4 expression was suppressed thereby demonstrating an obligate role for HER4 expression in DAC/HRG-induced apoptosis (Figures 3b and c). Taken together, these data suggest that breast tumor cells selectively suppress HER4 expression through promoter hypermethylation to evade the cell-killing activity of HER4. Interestingly, the HRG gene is also epigenetically silenced in breast carcinomas (Chua et al., 2009; Fernandez et al., 2010). Our observation that HRG stimulation of reactivated HER4 results in breast tumor apoptosis serves as an important proof-of-principle for marshaling HER4 apoptotic activity as a therapeutic approach to breast cancer.
HER4 promoter hypermethylation in primary breast tumors is associated with decreased patient survival
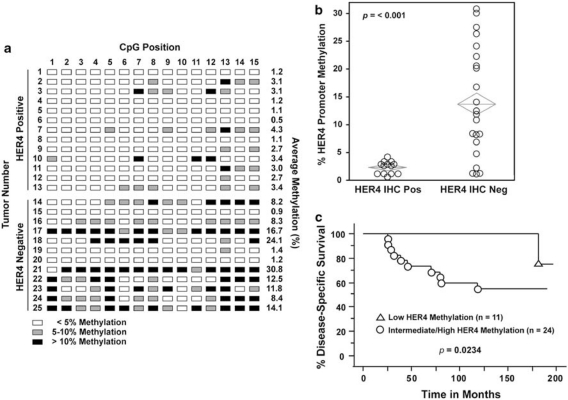
We next determined the levels of HER4 promoter methylation in a cohort of 25 primary breast tumors previously analyzed for HER4 expression by immunohistochemistry (Thor et al., 2009). In the 13 HER4-positive tumors, the levels of HER4 promoter methylation ranged from 0.5 to 4.3% with a mean of 2.4% (Figures 4a and b). HER4 promoter methylation in 12 HER4-negative tumors had a significantly higher mean of 11.5% (Figures 4a and b; P<0.001). These results implicate HER4 promoter hypermethylation as an important mechanism promoting suppression of HER4 expression in primary breast carcinomas. We expanded our cohort to 35 patients and determined the effect of HER4 promoter methylation on patient survival using a cutoff of 3% methylation. We found that patients whose tumors had HER4 promoter methylation levels ⩾3% had significantly shorter disease-specific survival (Figure 4c; P=0.0234). Strikingly, a failure in the low HER4 methylation population was not observed until nearly 15 years after diagnosis. Taken together, our results provide an important clinical correlate implicating HER4 promoter hypermethylation as an epigenetic event promoting breast tumor progression and reduced patient survival.
Figure 4.
Tumor hypermethylation of the HER4 promoter predicts breast cancer patient outcome. The patient population and HER4 expression analysis by immunohistochemistry (IHC) are described elsewhere (Thor et al., 2009). (a) Bisulfite-treated primary breast tumor DNA was analyzed by pyrosequencing as described in Figure 2d. DNA was isolated from 50 μm sections of formalin fixed and paraffin embedded (FFPE) primary breast tumors and bisulfite treated at EpigenDx. The position and extent of methylation for each site in 13 HER4-positive and 12 HER4-negative tumors is indicated. (b) Box plot of HER4 promoter methylation in HER4-positive (n=13) and HER4-negative (n=22) primary breast tumors (P<0.001). (c) Kaplan–Meier survival curves for disease-specific survival (DSS). HER4 methylation <3% (triangles; n=11) vs ⩾3% (circles; n=24) (P=0.0234) For Kaplan–Meier survival curves HER4 methylation determined by pyrosequencing was dichotomized at <3% vs ⩾3%. Outcomes included DSS defined as the number of months between the diagnosis date and the date of death from breast cancer.
In summary, we have identified a molecular mechanism to explain the selective suppression of HER4 expression observed clinically during breast tumorigenesis. Accordingly, we show that extinguished HER4 expression is associated with promoter hypermethylation in both breast tumor cell lines and primary carcinomas. Furthermore, breast tumor HER4 promoter methylation >3% was significantly associated with poor patient prognosis. In fact, all patients in our cohort with tumor HER4 promoter methylation <3% survived for at least 15 years after diagnosis. Significantly, reactivation of epigenetically silenced HER4 followed by HRG stimulation results in tumor cell apoptosis supporting a tumor suppressor function for HER4 in breast cancer. Our results, therefore, provide a proof-of-principle therapeutic rationale for marshaling HER4 tumor cell-killing activity in breast carcinomas associated with the worst patient prognosis.
Acknowledgments
We thank June Allison for excellent lab management and other members of the Jones lab for valuable input. This work is dedicated to the loving memory of June Allison who will no longer be subjected to the hardships of breast cancer. This work was supported by National Cancer Institute/National Institutes of Health Grants RO1CA95783 (FEJ) and RO1CA96717 (FEJ).
The authors declare no conflict of interest.
References
- Chua YL, Ito Y, Pole JC, Newman S, Chin SF, Stein RC, et al. The NRG1 gene is frequently silenced by methylation in breast cancers and is a strong candidate for the 8p tumour suppressor gene. Oncogene. 2009;28:4041–4052. doi: 10.1038/onc.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Fernandez SV, Snider KE, Wu YZ, Russo IH, Plass C, Russo J. DNA methylation changes in a human cell model of breast cancer progression. Mutat Res. 2010;688:28–35. doi: 10.1016/j.mrfmmm.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FE. HER4 intracellular domain (4ICD) activity in the developing mammary gland and breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:247–258. doi: 10.1007/s10911-008-9076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FE, Welte T, Fu X-Y, Stern DF. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J Cell Biol. 1999;147:77–90. doi: 10.1083/jcb.147.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreike B, Hart G, Bartelink H, van de Vijver MJ. Analysis of breast cancer related gene expression using natural splines and the Cox proportional hazard model to identify prognostic associations. Breast Cancer Res Treat. 2009. [DOI] [PubMed]
- Long W, Wagner K-U, Lloyd KCK, Binart N, Shillingford JM, Hennighausen L, et al. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development. 2003;130:5257–5268. doi: 10.1242/dev.00715. [DOI] [PubMed] [Google Scholar]
- Muraoka-Cook RS, Caskey LS, Sandahl MA, Hunter DM, Husted C, Strunk KE, et al. Heregulin-dependent delay in mitotic progression requires HER4 and BRCA1. Mol Cell Biol. 2006a;26:6412–6424. doi: 10.1128/MCB.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook RS, Sandahl M, Husted C, Hunter D, Miraglia L, Feng SM, et al. The intracellular domain of ErbB4 induces differentiation of mammary epithelial cells. Mol Biol Cell. 2006b;17:4118–4129. doi: 10.1091/mbc.E06-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook RS, Sandahl MA, Strunk KE, Miraglia LC, Husted C, Hunter DM, et al. ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol Cell Biol. 2009;29:4935–4948. doi: 10.1128/MCB.01705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, et al. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66:6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- Naresh A, Thor AD, Edgerton SM, Torkko KC, Kumar R, Jones FE. The HER4/4ICD estrogen receptor coactivator and BH3-only protein is an effector of tamoxifen-induced apoptosis. Cancer Res. 2008;68:6387–6395. doi: 10.1158/0008-5472.CAN-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C, Liang G, Nguyen TT, Tsao-Wei D, Groshen S, Lubbert M, et al. Susceptibility of nonpromoter CpG islands to de novo methylation in normal and neoplastic cells. J Natl Cancer Inst. 2001;93:1465–1472. doi: 10.1093/jnci/93.19.1465. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Rokavec M, Justenhoven C, Schroth W, Istrate MA, Haas S, Fischer HP, et al. A novel polymorphism in the promoter region of ERBB4 is associated with breast and colorectal cancer risk. Clin Cancer Res. 2007;13:7506–7514. doi: 10.1158/1078-0432.CCR-07-0457. [DOI] [PubMed] [Google Scholar]
- Sassen A, Diermeier-Daucher S, Sieben M, Ortmann O, Hofstaedter F, Schwarz S, et al. Presence of HER4 associates with increased sensitivity to Herceptin in patients with metastatic breast cancer. Breast Cancer Res. 2009;11:R50. doi: 10.1186/bcr2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Wang YP, Jo KH, Moon SW, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer. 2006;118:1426–1429. doi: 10.1002/ijc.21507. [DOI] [PubMed] [Google Scholar]
- Strathdee G, Davies BR, Vass JK, Siddiqui N, Brown R. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis. 2004;25:693–701. doi: 10.1093/carcin/bgh066. [DOI] [PubMed] [Google Scholar]
- Strohecker AM, Yehiely F, Chen F, Cryns VL. Caspase cleavage of HER-2 releases a Bad-like cell death effector. J Biol Chem. 2008;283:18269–18282. doi: 10.1074/jbc.M802156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K. Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- Thor AD, Edgerton SM, Jones FE. Subcellular localization of the HER4 intracellular domain, 4ICD, identifies distinct prognostic outcomes for breast cancer patients. Am J Pathol. 2009;175:1802–1809. doi: 10.2353/ajpath.2009.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvorogov D, Sundvall M, Kurppa K, Hollmen M, Repo S, Johnson MS, et al. Somatic mutations of ErbB4: selective loss-of-function phenotype affecting signal transduction pathways in cancer. J Biol Chem. 2008;284:5582–5591. doi: 10.1074/jbc.M805438200. [DOI] [PubMed] [Google Scholar]
- Vidal GA, Clark DE, Marrero L, Jones FE. A constitutively active ERBB4/HER4 allele with enhanced transcriptional coactivation and cell-killing activities. Oncogene. 2007;26:462–466. doi: 10.1038/sj.onc.1209794. [DOI] [PubMed] [Google Scholar]
- Vidal GA, Naresh A, Marrero L, Jones FE. Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J Biol Chem. 2005;280:19777–19783. doi: 10.1074/jbc.M412457200. [DOI] [PubMed] [Google Scholar]
- Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, et al. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol. 2004;167:469–478. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Sullivan LL, Nair SS, Williams CC, Pandey A, Marrero L, et al. Coregulation of estrogen receptor by ERBB4/HER4 establishes a growth-promoting autocrine signal in breast tumor cells. Cancer Res. 2006;66:7991–7998. doi: 10.1158/0008-5472.CAN-05-4397. [DOI] [PubMed] [Google Scholar]