Abstract
AIM: To investigate the prognostic value of chromosome 18q microsatellite alterations (MA) in stage II colon cancer.
METHODS: One hundred and six patients with sporadic stage II colon cancer were enrolled in this study. DNA was extracted from formalin-fixed, paraffin-embedded tumor and adjacent normal mucosal tissue samples. MA, including loss of heterozygosity (LOH) and microsatellite instability (MSI), was analyzed by polymerase chain reaction, polyacrylamide gel-electrophoresis and DNA sequencing at 5 microsatellite loci on chromosome 18q (D18S474, D18S55, D18S58, D18S61 and D18S64).
RESULTS: Among the 102 patients eligible for MA information, the overall frequencies of LOH, high and low frequency MSI/microsatellite stable were 49.0%, 17.6% and 82.4%, respectively. The high frequency of 18q-LOH was significantly associated with the poor 5-year overall survival (OS) (P = 0.008) and disease free survival (P = 0.006). High levels of MSI were significantly associated with a longer 5-year OS (P = 0.045) while the higher frequency of 18q-LOH at the loci of D18S474 and D18S61 was significantly associated with a poorer 5-year OS (P = 0.010 and 0.005, respectively). But multivariate analysis showed that only the frequency of 18q-LOH was significantly associated with the prognosis of the disease.
CONCLUSION: High frequency of 18q-LOH is an independent prognostic factor indicating poor prognosis of the patients with stage II colon cancer.
Keywords: Chromosome 18q, Loss of heterozygosity, Microsatellite instability, Stage II colon cancer, Prognosis
INTRODUCTION
Colorectal cancer (CRC) is the third most common malignant cancer worldwide, with an estimate of one million new cases and a half million deaths annually[1]. Although the clinicopathological stage is currently the gold standard for prognosis, the molecular biological factors that determine the outcome of patients exhibiting the same clinicopathological stage are poorly understood, especially for stage II and III patients.
In recent years, multiple tumor-related biomarkers have been proposed as prognostic factors for CRC, but their predictive value has not been consistently demonstrated. Rapid advances in the molecular genetics of CRC have stimulated attempts to evaluate the prognostic significance of specific genetic alterations in this tumor type. The American Society of Clinical Oncology (ASCO) has published evidence-based clinical practice guidelines for the use of tumor markers in CRC bi-annually from 1996 to 2006[2]. Similarly, the European Group on Tumor Markers revised its recommendations for the use of tumor marker tests in the prevention, screening, treatment and surveillance of CRC in 2007[3]. These recommendations described new tumor markers, such as microsatellite instability (MSI), loss of heterozygosity (LOH) at chromosome 18q, as well as the traditional markers such as carcinoembryonic antigen (CEA), CA19-9, p53, Ras, and DNA ploidy.
It has been shown that CRC arises through at least two distinct pathways of genetic instability pathways of microsatellite alteration (MA): one involving chromosomal instability presenting as LOH, and the other involving MSI[4-6]. Although most studies have proven the prognostic value of LOH at chromosome 18q (18qLOH) in CRC, the outcomes are controversial. To further define the clinical effect of molecular genetic alterations on clinicopathological features and prognosis, we analyzed LOH and MSI at chromosome 18q in 106 patients retrospectively with stage II colon cancer.
MATERIALS AND METHODS
This study consisted of randomly selected 106 consecutive patients who had undergone curative colon resection for sporadic stage II colon cancer between 1996 and 2001. All cases were deemed sporadic, based on the absence of relevant family history in the initial patient interview records. None of the patients had additional synchronous colon cancers or other evidence of a heritable form of CRC.
A curative operation was defined as one in which no macroscopic tumor remained after surgery and histopathological examination showed radical resection margins were free of tumor at lines of the operative specimen. The methods used to exclude distant metastases at the time of resection included preoperative liver ultrasonography, abdominal and pelvic CT scan, chest X-ray, and intraoperative exploration. The right colon was defined as the large bowel proximal to the right half of the transverse colon; the left colon was defined as the large bowel between the right colon and the rectosigmoid junction. All specimens were histopathologically reviewed by a single gastrointestinal pathologist who was unaware of the results of molecular genetic testing. In accordance with the classification of tumors by the World Health Organization[7], we defined tumors as mucinous if 50% or more of the tumors displayed mucinous differentiation and as undifferentiated if features of tumor cell differentiation were absent. Other tumors were classified as adenocarcinomas.
Twelve lymph nodes on average were examined per case. Tumors were staged according to the TNM classification system and based on pathological findings after operation. Adjuvant chemotherapy was administered to patients with (1) clinicopathological factors with emergent presentation and associated with high risk of recurrence (i.e. bowel perforation or occlusion); (2) poorly differentiated tumors (histological grade); (3) deep tumor invasion and adjacent organ involvement (T4); (4) venous invasion; and (5) peritoneal involvement. The drug regimen for chemotherapy was 5-fluorouracil-based adjuvant chemotherapy for nearly 6 mo.
Patients were observed after the completion of therapy every 3 mo for 2 years, every 6 mo for the next 3 years, and then annually, or until relapse. History and physical examination, complete blood cell and platelet count, blood biochemistries, ultrasound, CEA and CA19-9 measurements were performed at each visit. The chest X-ray, colonoscopy, and CT scan were performed at least once a year.
The overall survival (OS) time was calculated from the date of surgery to the time of the last visit or death. The disease-free survival time was calculated from the date of resection to the time of relapse.
DNA extraction, LOH and MSI analysis
DNA extraction: Three to four 8 μm-thick sections were obtained from archival blocks of formalin-fixed, paraffin-embedded tumor and adjacent normal mucosal tissues. One 8 μm-thick section of each tumor sample was stained with HE, and the percentage of tumor cells was estimated by visual examination by a pathologist. Representative tumor samples contained a minimum of 80% of tumor cells. Microdissection of regions representing tumor and matching normal tissues was performed. Samples were selected and collected into Eppendorf tubes. The tissue samples were de-paraffinized with xylene, and the DNA was extracted by incubating the samples overnight at 56°C in lysis buffer [10 mmol/L Tris-Cl (pH 8.0), 2.5 mmol/L MgCl2, 50 mmol/L KCl, 0.5% Tween 20] containing 0.2 mg/mL proteinase K (Invitrogen Inc., Carlsbad, CA). Digested products were boiled for 8 min at 95°C to inactivate enzymes, centrifuged, and cooled on ice. The supernatant was used directly for polymerase chain reaction (PCR).
PCR: A panel of five polymorphic microsatellite markers located in chromosomal regions potentially involved in colon carcinoma development and progression were interrogated. These regions were D18S474 (18q21.1), D18S55 (18q23), D18S58 (18q23), D18S61 (18q22.2), and D18S64 (18q21.3). The primer sets were obtained from published sequences and purchased from Invitrogen Inc. (Carlsbad, CA). The target sequences in normal and tumor DNA were amplified by PCR in a 50 μL reaction mixture containing 5 μL of DNA sample, 10 × buffer (10 mmol/L Tris-HCl pH 9.0, 50 mmol/L KCl, 0.1% Triton X-100), 3 mmol/L MgCl2, 5 U Taq Polymerase, 10 mmol/L of each dNTP, and 10 mmol/L of each primer. Each forward primer was coupled with the fluorescent dye TET (4,7,2’,7’-tetrachloro-6-carboxyfluorescein; Invitrogen Inc., Carlsbad, CA). The reactions were submitted to 35 cycles of amplification in a PTC-100™ thermal cycler (Bio-Rad Inc., America) at the following annealing temperatures: 55°C for D18S474, 57°C for D18S55, 61°C for D18S58, 55°C for D18S61, and 59°C for D18S64. The presence and correct size of all amplicons were determined using 2% agarose gel electrophoresis (Biowest Inc., CA).
Fragment analysis: Amplicons were separated in a 6% polyacrylamide denaturing gel (Bio-Rad Inc., CA) using the ABI 377 DNA Sequencer (ABI Inc., CA). A total of 0.5 μL PCR product was mixed with 0.5 μL blue dextran loading solution (Bio-Rad Inc., CA). After 5 min of denaturation at 95°C, the sample was loaded onto the gel and subsequently run for two h at 2750 V, 125 W, and 50°C. After electrophoresis, the fluorescent signals were automatically collected by a gene scanner. Analysis of these signals with the Genotyper (Version 2.0) image analysis software (ABI Inc., CA) generated electropherograms, which displayed alleles as peaks. The height and area of the peaks calculated by the software were proportional to the concentration of the alleles in the sample.
LOH: The presence of two distinctly sized alleles in normal tissues was the necessary condition for evaluation of allelic losses. Cases in which the normal DNA sample was homozygous were classified as non-informative. The ratio of allele peak areas calculated for each tumor sample was divided by the allele peak area ratio of the normal matching control. If this quotient was greater than 1.00, the quotient was converted to give a result ranging from 0.00 to 1.00. Ratios below 0.6, indicating at least a 40% reduction of a tumor allele, were indicative of LOH[8,9].
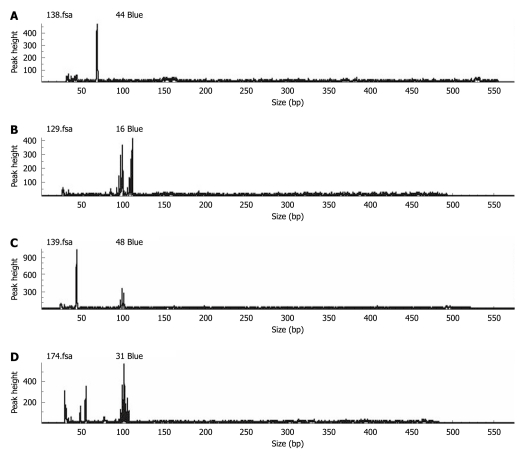
MSI: The unique appearance of one or more alleles in the tumor DNA but not in its paired normal DNA, such as new peaks in the electropherogram, indicated the MSI[10]. To ensure the reproducibility of the results, DNA analysis of the first 20 samples was repeated and 99% concordance was recorded. Tumors were classified as demonstrating a high frequency of MSI (MSI-H) if instability was detected in more than two loci of the five interpretable microsatellite markers investigated. Tumors were deemed as low frequency MSI (MSI-L) if instability was found in one of the five markers according to the international criteria. Tumors without MSI were considered to be microsatellite stable (MSS)[11]. In our study, MSI-L and MSS tumors were considered as a single group in comparison with MSI-H tumors. However, further studies of the significance of MSI-L[12] are needed, which may represent a biologically and clinically distinct entity[13,14] (Figure 1).
Figure 1.
Representative examples showing homozygosity and heterozygosity in normal tissues and loss of heterozygosity and microsatellite instability in tumor tissues. A: Normal sample shows homozygosity; B: Normal sample shows heterozygosity; C: Tumor sample shows loss of heterozygosity; D: Tumor sample shows microsatellite instability.
Statistical analysis
Statistical analysis was performed using the SPSS 12.0 package. The main factors compared in this study were frequency of MA and survival. Contingency tables and the χ2 test were used to evaluate differences between percentages. The association of disease-free and OS with prognostic factors was evaluated by means of multivariate logistic regression.
Disease-free intervals in patients with recurrence were measured as the interval between the date of resection and the date of diagnosis of recurrence. Duration of survival was measured from the date of resection until the date of death from any cause or until the censoring date of June 30, 2008. In the survival analysis, deaths caused by postoperative complications within 30 d were excluded. Survival curves were drawn according to the Kaplan-Meier method, and differences in disease-free and OS were evaluated by means of log-rank test. The simultaneous effects of more than one prognostic factors were estimated by the Cox’s proportional hazards regression model. Mortality rate ratios were used to assess the difference in deaths caused by colon cancer. The significance level was set at 0.05; the confidence interval was 95%; and all P values reported were two-sided.
RESULTS
We investigated 66 male and 40 female patients. The mean age of this group was 57 years (range, 19-84 years). Of the 106 primary tumors, 47 were located in the right colon and 59 were located in the left colon. A total of 88 (83.0%) patients with the high recurrent clinicopathological factors mentioned above underwent adjuvant chemotherapy (Table 1).
Table 1.
Clinical features of patients and survival status after surgery n (%)
| Characteristics | Patients | Survival (mo) (mean ± SE) | 3-yr overall survival (%) | 5-yr overall survival (%) | P value |
| Gender | 0.753 | ||||
| Male | 66 (62.3) | 104 ± 3 | 90.7 | 83.2 | |
| Female | 40 (37.7) | 100 ± 3 | 91.6 | 84.1 | |
| Age (yr) | 0.011 | ||||
| ≤ 60 | 60 (56.6) | 107 ± 2 | 93.5 | 89.7 | |
| > 60 | 46 (43.4) | 97 ± 4 | 88.0 | 77.4 | |
| Tumor site | 0.467 | ||||
| Right colon | 47 (44.3) | 100 ± 3 | 89.5 | 82.9 | |
| Left colon | 59 (55.7) | 104 ± 3 | 92.2 | 84.8 | |
| Tumor size (cm) | 0.213 | ||||
| < 5 | 48 (45.3) | 108 ± 3 | 93.8 | 91.8 | |
| ≥ 5 | 58 (54.7) | 101 ± 3 | 90.1 | 80.6 | |
| Grade | 0.220 | ||||
| Well/moderately differentiated | 81 (76.4) | 103 ± 2 | 92.0 | 85.3 | |
| Poorly differentiated | 25 (23.6) | 104 ± 4 | 90.7 | 82.7 | |
| Histologic features | 0.246 | ||||
| Adenocarcinoma | 79 (74.5) | 104 ± 2 | 91.6 | 83.9 | |
| Mucinous | 27 (25.5) | 101 ± 4 | 87.6 | 78.3 | |
| pT stage | 0.090 | ||||
| T3 | 63 (59.4) | 106 ± 2 | 93.9 | 85.8 | |
| T4 | 43 (40.6) | 96 ± 4 | 87.0 | 78.5 | |
| Chemotherapy | 0.114 | ||||
| No | 18 (17.0) | 99 ± 3 | 83.3 | 69.8 | |
| Yes | 88 (83.0) | 103 ± 4 | 90.9 | 84.8 |
Microsatellite alteration
The number of tumors that could be evaluated for microsatellites (MA) varied in region because the polymorphic markers were non-informative for some patients. 18qLOH was observed in 50 (49%) of the 102 patients. The incidence of 18qLOH was 30.2% (26 of 86), 23.4% (18 of 77), 28.6% (20 of 70), 35.0% (28 of 80), and 20.8% (15 of 72) at D18S474, D18S55, D18S58, D18S61 and D18S64, respectively. Of the 102 tumor specimens evaluated for MSI, 42 (41.2%) cases were identified as MSI in at least one locus. Furthermore, seven cases were MSI at three loci, 11 cases at two loci, and 24 cases at one locus. According to the criteria mentioned above, 18 cases (17.6%) that showed MSI in at least two loci were defined as MSI-H. The remaining 84 cases (82.4%) were defined as MSI-L/MSS (Table 2).
Table 2.
Informative cases, loss of heterozygosity and microsatellite instability (n = 106) n (%)
| Marker | Location | Informative cases | LOH (+) | MSI |
| D18S474 | 18q21.1 | 86 | 26 (30.2) | 15 (17.4) |
| D18S55 | 18q23 | 77 | 18 (23.4) | 11 (14.3) |
| D18S58 | 18q23 | 70 | 20 (28.6) | 18 (25.7) |
| D18S61 | 18q22.2 | 80 | 28 (35.0) | 13 (16.3) |
| D18S64 | 18q21.3 | 72 | 15 (20.8) | 10 (13.9) |
| Overall | 18q21-23 | 102 | 50 (49.0) | 42 (41.2) |
LOH: Loss of heterozygosity; MSI: Microsatellite instability.
Poorly differentiated (P = 0.023) nonmucinous (P = 0.005) tumors located in the left colon (P = 0.023) more likely displayed chromosome 18qLOH. No association was found between 18qLOH, age, and gender. MSI-H was more likely to be displayed by well/moderately differentiated (P = 0.047) mucinous (P = 0.004) tumors located in the right colon (P < 0.001). No association was found between MSI-H, age and gender (Table 3).
Table 3.
Correlation between loss of heterozygosity at chromosome 18q, microsatellite instability and clinicopathologic variables of patients n (%)
| Characteristics | Cases | LOH (+) | LOH (-) | P value | MSI-H (%) | MSI-L/MSS (%) | P value |
| Gender | 0.662 | 0.272 | |||||
| Male | 63 (61.8) | 27 | 36 | 9 | 54 | ||
| Female | 39 (38.2) | 23 | 16 | 9 | 30 | ||
| Age (yr) | 0.435 | 0.111 | |||||
| ≤ 60 | 58 (56.9) | 32 | 26 | 11 | 47 | ||
| > 60 | 44 (43.1) | 18 | 26 | 7 | 37 | ||
| Tumor site | 0.023 | < 0.001 | |||||
| Right colon | 46 (45.1) | 13 | 33 | 15 | 31 | ||
| Left colon | 56 (54.9) | 37 | 19 | 3 | 53 | ||
| Tumor size (cm) | 0.232 | 0.327 | |||||
| < 5 | 48 (47.1) | 28 | 20 | 8 | 40 | ||
| ≥ 5 | 54 (52.9) | 22 | 32 | 10 | 44 | ||
| Grade | 0.016 | 0.047 | |||||
| Well/moderately differentiated | 78 (76.5) | 34 | 44 | 15 | 63 | ||
| Poorly differentiated | 24 (23.5) | 16 | 8 | 3 | 21 | ||
| Histologic features | 0.005 | 0.004 | |||||
| Adenocarcinoma | 77 (75.5) | 44 | 33 | 5 | 72 | ||
| Mucinous | 25 (24.5) | 6 | 19 | 13 | 12 | ||
| pT stage | 0.126 | 0.272 | |||||
| T3 | 61 (59.8) | 33 | 28 | 11 | 50 | ||
| T4 | 41 (40.2) | 17 | 24 | 7 | 34 | ||
| Chemotherapy | 0.650 | 0.782 | |||||
| No | 17 (16.7) | 10 | 7 | 4 | 13 | ||
| Yes | 85 (83.3) | 40 | 45 | 14 | 71 |
LOH: Loss of heterozygosity; MSI-L: Low frequency microsatellite instability; MSI-H: High frequency of microsatellite instability; MSS: Microsatellite stable.
Survival analysis
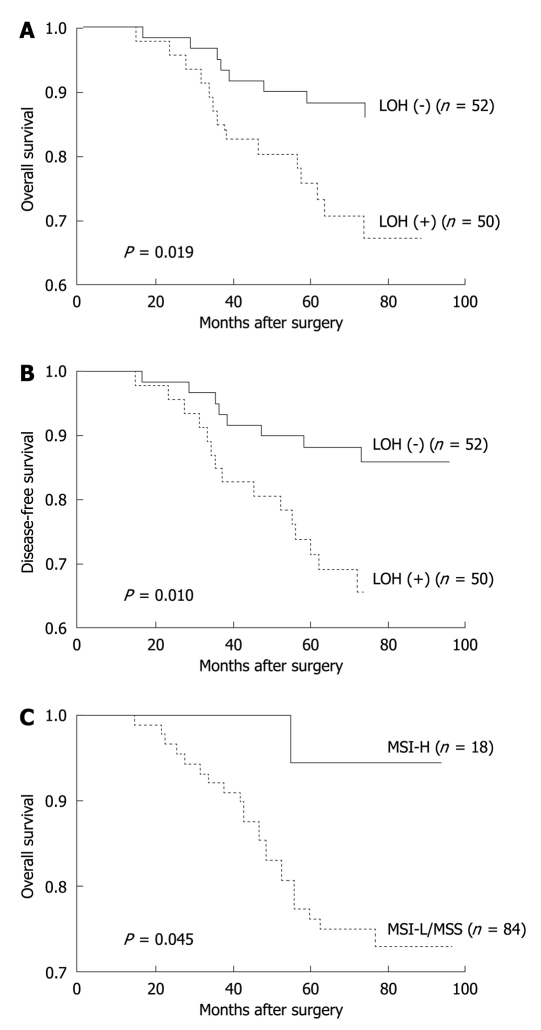
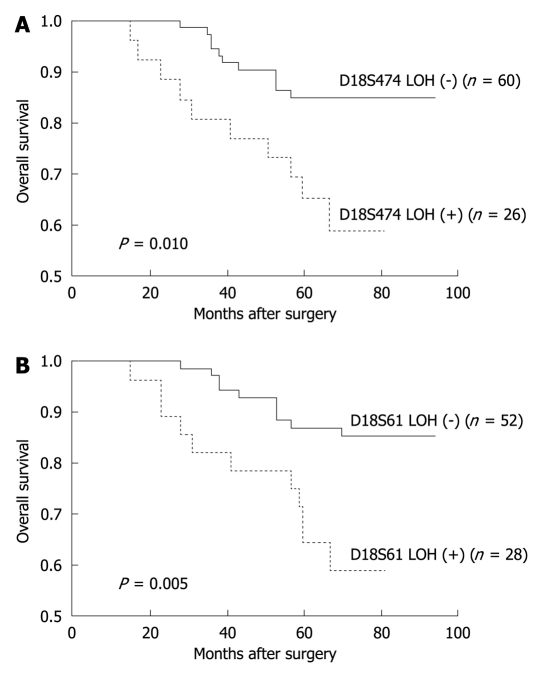
Of the molecular markers tested, 18qLOH was significantly associated with a reduced 5-year OS (P = 0.019) and a disease-free survival (P = 0.010) during the follow-up period. High levels of MSI were moderately associated with an improved 5-year OS during the follow-up period (P = 0.045). However, the rates of disease-free survival were not significantly different between patients with MSI-H and MSI-L/MSS tumors (Table 4, Figure 2). When the status of chromosome 18q was evaluated according to each microsatellite locus, the D18S474 and D18S61 loci were associated with a poor 5-year OS (P = 0.010 and 0.005, respectively) (Table 5, Figure 3).
Table 4.
Five-year overall survival and disease-free survival after surgery in relation to microsatellite alteration in patients with stage II colon cancer
| Parameter | Cases | 5-yr overall survival (%) | P value | 5-yr disease-free survival (%) | P value |
| 18q-LOH | 0.019 | 0.010 | |||
| (+) | 50 | 71.3 | 70.5 | ||
| (-) | 52 | 88.3 | 87.1 | ||
| MSI | 0.045 | 0.155 | |||
| MSI-H | 18 | 94.4 | 88.5 | ||
| MSI-L/MSS | 84 | 77.8 | 76.7 |
18q-LOH: Loss of heterozygosity at chromosome 18q; MSI: Microsatellite instability; MSI-L: Low frequency MSI; MSI-H: High frequency of MSI; MSS: Microsatellite stable.
Figure 2.
Kaplan-Meier estimates of overall survival and disease-free survival among patients with stage II colon cancer according to microsatellite alteration. In our study, 18qLOH was significantly associated with reduced 5-year overall survival (P = 0.019) (A) and disease-free survival (P = 0.010) (B). High levels of microsatellite instability was moderately associated with improved 5-year overall survival during the follow-up period (P = 0.045) (C). LOH: Loss of heterozygosity; MSI-L: Low frequency microsatellite instability; MSI-H: High frequency of microsatellite instability; MSS: Microsatellite stable.
Table 5.
Five-year overall survival after surgery in relation to each microsatellite locus
| Marker | Cases | 5-yr overall survival (%) | P value |
| D18S474 | 86 | 0.010 | |
| LOH (+) | 26 | 65.2 | |
| LOH (-) | 60 | 84.9 | |
| D18S55 | 77 | 0.504 | |
| LOH (+) | 18 | 77.8 | |
| LOH (-) | 59 | 81.6 | |
| D18S58 | 70 | 0.293 | |
| LOH (+) | 20 | 71.4 | |
| LOH (-) | 50 | 83.3 | |
| D18S61 | 80 | 0.005 | |
| LOH (+) | 28 | 64.3 | |
| LOH (-) | 52 | 87.0 | |
| D18S64 | 72 | 0.076 | |
| LOH (+) | 15 | 69.0 | |
| LOH (-) | 57 | 86.3 |
LOH: Loss of heterozygosity.
Figure 3.
Kaplan-Meier estimates of overall survival among patients with stage II colon cancer at microsatellite loci, D18S474 and D18S61. When the status of chromosome 18q was evaluated according to each microsatellite loci, the D18S474 (A) and D18S61 (B) loci were associated with worse 5-year overall survival (P = 0.010 and 0.005, respectively). LOH: Loss of heterozygosity.
Cox’s proportional hazards regression model was adjusted for gender, age, T stage and chemotherapy, and multiple markers were analyzed. Only 18qLOH was independently associated with the OS rate (RR = 2.679, P = 0.021). However, when 18qLOH was excluded from the proportional hazards regression model, MSI-H status was found to be associated with the OS (RR = 3.149, P = 0.039) (Tables 6 and 7).
Table 6.
Univariate analysis of prognostic factors for 5-year survival according to Cox’s proportional hazards regression model in patients with stage II colon cancer
| Variables | B | SE | Wald value | P value | RR |
| Age | -0.837 | 0.404 | 4.290 | 0.038 | 0.433 |
| pT stage | 0.667 | 0.395 | 2.859 | 0.091 | 1.949 |
| Chemotherapy | -0.727 | 0.443 | 2.698 | 0.100 | 0.483 |
| 18q-LOH | 0.903 | 0.425 | 4.509 | 0.014 | 2.467 |
| MSI | 1.147 | 0.736 | 2.428 | 0.039 | 3.149 |
18q-LOH: Loss of heterozygosity at chromosome 18q; MSI: Microsatellite instability.
Table 7.
Multivariate analysis of prognostic factors for 5-year survival according to Cox's proportional hazards regression model in patients with stage II colon cancer
| Variables | B | SE | Wald value | P value | RR |
| 18q-LOH | 0.985 | 0.429 | 5.286 | 0.021 | 2.679 |
18q-LOH: Loss of heterozygosity at chromosome 18q.
DISCUSSION
In 2006, the ASCO published evidence-based clinical practice guidelines for the use of tumor markers in CRC to update the recommendations for the use of tumor marker tests in the prevention, screening, treatment, and surveillance of gastrointestinal cancers. As part of the update, 18q-LOH and MSI were mentioned in CRC for the first time[2]. In this publication, there were 16 series focusing on the LOH at 18q in the prognosis of early-stage CRC[15-30]. Of the 16 studies, 8 found that CRC patients with harbored LOH at 18q had a significantly lower survival than those who were heterozygous[15,19,20,22,25,28-30]. Four of the eight positive series also found that 18qLOH tumors had a significantly worse prognosis in a multivariate analysis with hazard ratios for death of 2.0, 2.75 and 7.30[19,22,25], or for recurrence of 9.60[15]. Three reports did not find 18qLOH to be independently prognostic[20,28,30] and one did not perform multivariate analysis[29]. In two studies where 18qLOH was not found to be prognostic in a univariate or multivariate analysis[20,21], LOH at 18q did yield a poor prognosis in stage II disease.
As for MSI, 17 series focusing on MSI in the prognosis of early stage CRC were considered in this review[15-17,19,31-43]. Six of the series were analyzed in patients on randomized therapeutic trials[16,17,19,31,40,41]. The remaining studies selected patients treated in defined time periods. Interestingly, 11 of the 17 series found that patients with MSI-H colon or rectal cancers had a significantly better survival than those with MSI-L/MSS[15,17,31,33-40,43]. Two series found no association of MSI-H with a better survival from randomized trials of adjuvant chemotherapy[16,19]. One study did find a statistically significant association of MSI-H status with a better disease-free survival[19]. Of the 11 positive series, six also found that MSI-H tumors had a significantly better prognosis in a multivariate analysis[17,33,37,38,40,43]. However, four did not find MSI-H to be independently prognostic[15,31,34,39]. Multivariate analysis was not done in one series[35].
Although there is evidence suggesting an association of 18qLOH and MSI with the prognosis of CRC, the ASCO expert panel determined that the data was insufficient and controversial to recommend using the two tumor markers as independent prognostic tests in the clinic.
Our study in patients with stage II colon cancer confirms some previous findings of a lower survival in patients with 18qLOH and a longer survival in patients with MSI-H to some extent. Our present study showed that stage II sporadic colon cancer patients with 18qLOH were associated with a significantly shorter OS and a disease-free survival as compared with 18qLOH-negative patients. This association was independent of other clinicopathological variables. According to other investigators, tumors with 18qLOH were more frequently located in the left colon. This observation, together with that of the worse prognosis of colon cancers with 18qLOH, may account for the findings that patients with tumors in the left side of the colon have a worse prognosis than those in the right side.
The reasons why some studies did not find that 18qLOH can predict survival in stage II CRC may have several explanations. First, different loci and number of 18q markers were used by various investigators. Second, variation in surgical techniques, regimens of chemotherapy or radiotherapy, and methods of histopathologic analysis of the tumor (i.e. electrophoresis or DNA sequencing directly) could create significant differences. Third, differences in TNM stage (stage II or stage III), T stage (T3 or T4), or tumor location (colon or rectum) could also contribute to the controversial results. For these reasons, our study selected a relatively small range of tumor locations (colon cancer) and TNM stage (stage II) to investigate the frequency of 18qLOH and its prognostic value.
In contrast to the unfavorable prognosis associated with 18qLOH, MSI-H is associated with a favorable prognosis. Patients with tumors exhibiting MSI-H had a better OS than patients with MSS tumors. The correlation between the MSI-H genotype and disease-free survival was not significant (P = 0.155). However, we hypothesize that this association lacks significance only as a result of the small sample size examined. The favorable effect of MSI on survival, however, resulted as being independent only when multivariate analysis did not consider 18qLOH. The survival advantage of patients with MSI-H tumors compared to those with MSS tumors is not unanimously accepted. Thus, the outcome referred to this phenotype is controversial. In our study, the National Institute of Health consensus definition of MSI has been used.
The chromosome 18q contains several genes with potential importance in colon cancer pathogenesis and progression. Deletion of portions of 18q has been implicated as an important step in the development of many CRCs. Among the genes located on 18q are the DCC gene that codes for a neutrin-1 receptor important in apoptosis, cell adhesion, and tumor suppression. SMAD-4 gene, which codes for a nuclear transcription factor in transforming growth factor-β1 signaling, is involved in tumor suppression and the SMAD22 gene is involved in endodermal differentiation[44].
MSI is a measure of the inability of the DNA nucleotide mismatch repair system to correct errors that commonly occur during the replication of DNA. It is characterized by the accumulation of single nucleotide mutations and length alterations in repetitive microsatellite nucleotide sequences. It is an alternative pathway to chromosomal instability with LOH in the pathogenesis of colon cancer[11].
In conclusion, the results of this study support the opinion that in colon cancer, genetic approaches may delineate subgroups of patients who would benefit most from specific treatment or other investigative therapies. We found that 18qLOH is a very informative prognostic genetic marker that could lead to the identification of patients who should be subjected to different adjuvant therapy plans. In addition, our study demonstrated that MSI-H in colon cancer is a potentially favorable prognostic genetic marker. This lends support to the hypothesis that MSI-H is an index of a less aggressive type of tumor growth and a heightened immunologic response. Although these results suggest that 18qLOH and MSI are prognostic genetic markers, the final verdict should depend on large-scale prospective randomized control trials.
COMMENTS
Background
It has been shown that colorectal cancer (CRC) arises through at least two distinct genetic pathways of microsatellite alteration (MA): one involving chromosome instability presented as loss of heterozygosity (LOH), and the other involving microsatellite instability (MSI). Although many studies have proven the prognostic value of MA at chromosome 18q in CRC, the outcomes are controversial.
Research frontiers
To further define the clinical effect of molecular genetic alterations on clinicopathological features and prognosis, the authors analyzed 106 patients retrospectively with stage II colon cancer for LOH and MSI at chromosome 18q.
Innovations and breakthroughs
In order to minimize the bias, the authors selected a relatively small range of tumor locations (colon cancer) and TNM stage (stage II) to analyze expression of LOH and MSI focusing on five microsatellite loci at chromosome 18q.
Applications
This study found LOH at chromosome 18q to be a very informative prognostic genetic marker that could identify patients who should be subjected to different adjuvant therapy plans.
Peer review
The manuscript presented data of LOH and MSI assays on more than 100 cases of stage II colonic adenocarcinomas. The results showed the preferential occurrence of allelic imbalance on 18q in more aggressive lesions, indicating the clinical significance of these markers.
Footnotes
Peer reviewers: Dr. Lodewijk AA Brosens, MD, PhD, Department of Pathology, University Medical Center Utrecht, Postbus 85500, 3508 GA, Utrecht, The Netherlands; Qin Su, Professor, Department of Pathology, Cancer Hospital and Cancer Institute, Chinese Academy of Medical Sciences and Peking Medical College, PO Box 2258, Beijing 100021, China
S- Editor Tian L L- Editor Ma JY E- Editor Zheng XM
References
- 1.Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RL, Thun MJ. Global Cancer Facts & Figures 2007. Atlanta, GA: American Cancer Society; 2007. [Google Scholar]
- 2.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348–1360. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 4.Dutrillaux B. Pathways of chromosome alteration in human epithelial cancers. Adv Cancer Res. 1995;67:59–82. doi: 10.1016/s0065-230x(08)60710-1. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 6.Sweezy MA, Fishel R. Multiple pathways leading to genomic instability and tumorigenesis. Ann N Y Acad Sci. 1994;726:165–177. doi: 10.1111/j.1749-6632.1994.tb52810.x. [DOI] [PubMed] [Google Scholar]
- 7.Jass JR, Sobin LH, editors . World Health Organization international histological classification of tumours. Histological typing of intestinal tumours. 2nd ed. Berlin: Springer-Verlag; 1989. [Google Scholar]
- 8.Skotheim RI, Diep CB, Kraggerud SM, Jakobsen KS, Lothe RA. Evaluation of loss of heterozygosity/allelic imbalance scoring in tumor DNA. Cancer Genet Cytogenet. 2001;127:64–70. doi: 10.1016/s0165-4608(00)00433-7. [DOI] [PubMed] [Google Scholar]
- 9.Beckmann MW, Picard F, An HX, van Roeyen CR, Dominik SI, Mosny DS, Schnürch HG, Bender HG, Niederacher D. Clinical impact of detection of loss of heterozygosity of BRCA1 and BRCA2 markers in sporadic breast cancer. Br J Cancer. 1996;73:1220–1226. doi: 10.1038/bjc.1996.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller JD, Haegle N, Keller G, Mueller E, Saretzky G, Bethke B, Stolte M, Höfler H. Loss of heterozygosity and microsatellite instability in de novo versus ex-adenoma carcinomas of the colorectum. Am J Pathol. 1998;153:1977–1984. doi: 10.1016/S0002-9440(10)65711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 12.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright CM, Dent OF, Newland RC, Barker M, Chapuis PH, Bokey EL, Young JP, Leggett BA, Jass JR, Macdonald GA. Low level microsatellite instability may be associated with reduced cancer specific survival in sporadic stage C colorectal carcinoma. Gut. 2005;54:103–108. doi: 10.1136/gut.2003.034579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohonen-Corish MR, Daniel JJ, Chan C, Lin BP, Kwun SY, Dent OF, Dhillon VS, Trent RJ, Chapuis PH, Bokey EL. Low microsatellite instability is associated with poor prognosis in stage C colon cancer. J Clin Oncol. 2005;23:2318–2324. doi: 10.1200/JCO.2005.00.109. [DOI] [PubMed] [Google Scholar]
- 15.Pietra N, Sarli L, Costi R, Ouchemi C, Grattarola M, Peracchia A. Role of follow-up in management of local recurrences of colorectal cancer: a prospective, randomized study. Dis Colon Rectum. 1998;41:1127–1133. doi: 10.1007/BF02239434. [DOI] [PubMed] [Google Scholar]
- 16.Barratt PL, Seymour MT, Stenning SP, Georgiades I, Walker C, Birbeck K, Quirke P. DNA markers predicting benefit from adjuvant fluorouracil in patients with colon cancer: a molecular study. Lancet. 2002;360:1381–1391. doi: 10.1016/s0140-6736(02)11402-4. [DOI] [PubMed] [Google Scholar]
- 17.Halling KC, French AJ, McDonnell SK, Burgart LJ, Schaid DJ, Peterson BJ, Moon-Tasson L, Mahoney MR, Sargent DJ, O'Connell MJ, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–1303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Arbman G, Sun XF. Codon 201 polymorphism of DCC gene is a prognostic factor in patients with colorectal cancer. Cancer Detect Prev. 2003;27:216–221. doi: 10.1016/s0361-090x(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB 3rd, Hamilton SR. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diep CB, Thorstensen L, Meling GI, Skovlund E, Rognum TO, Lothe RA. Genetic tumor markers with prognostic impact in Dukes' stages B and C colorectal cancer patients. J Clin Oncol. 2003;21:820–829. doi: 10.1200/JCO.2003.05.190. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-López E, Abad A, Font A, Monzó M, Ojanguren I, Pifarré A, Sánchez JJ, Martín C, Rosell R. Allelic loss on chromosome 18q as a prognostic marker in stage II colorectal cancer. Gastroenterology. 1998;114:1180–1187. doi: 10.1016/s0016-5085(98)70423-8. [DOI] [PubMed] [Google Scholar]
- 22.Ogunbiyi OA, Goodfellow PJ, Herfarth K, Gagliardi G, Swanson PE, Birnbaum EH, Read TE, Fleshman JW, Kodner IJ, Moley JF. Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J Clin Oncol. 1998;16:427–433. doi: 10.1200/JCO.1998.16.2.427. [DOI] [PubMed] [Google Scholar]
- 23.Carethers JM, Hawn MT, Greenson JK, Hitchcock CL, Boland CR. Prognostic significance of allelic lost at chromosome 18q21 for stage II colorectal cancer. Gastroenterology. 1998;114:1188–1195. doi: 10.1016/s0016-5085(98)70424-x. [DOI] [PubMed] [Google Scholar]
- 24.Font A, Abad A, Monzó M, Sanchez JJ, Guillot M, Manzano JL, Piñol M, Ojanguren I, Rosell R. Prognostic value of K-ras mutations and allelic imbalance on chromosome 18q in patients with resected colorectal cancer. Dis Colon Rectum. 2001;44:549–557. doi: 10.1007/BF02234328. [DOI] [PubMed] [Google Scholar]
- 25.Lanza G, Matteuzzi M, Gafá R, Orvieto E, Maestri I, Santini A, del Senno L. Chromosome 18q allelic loss and prognosis in stage II and III colon cancer. Int J Cancer. 1998;79:390–395. doi: 10.1002/(sici)1097-0215(19980821)79:4<390::aid-ijc14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Laurent-Puig P, Olschwang S, Delattre O, Remvikos Y, Asselain B, Melot T, Validire P, Muleris M, Girodet J, Salmon RJ. Survival and acquired genetic alterations in colorectal cancer. Gastroenterology. 1992;102:1136–1141. [PubMed] [Google Scholar]
- 27.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Järvinen H, Mecklin JP, Hemminki A, Schwartz S Jr, Aaltonen LA, Arango D. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11:2606–2611. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- 28.Bisgaard ML, Jäger AC, Dalgaard P, Søndergaard JO, Rehfeld JF, Nielsen FC. Allelic loss of chromosome 2p21-16.3 is associated with reduced survival in sporadic colorectal cancer. Scand J Gastroenterol. 2001;36:405–409. doi: 10.1080/003655201300051252. [DOI] [PubMed] [Google Scholar]
- 29.Chang SC, Lin JK, Lin TC, Liang WY. Loss of heterozygosity: an independent prognostic factor of colorectal cancer. World J Gastroenterol. 2005;11:778–784. doi: 10.3748/wjg.v11.i6.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi SW, Lee KJ, Bae YA, Min KO, Kwon MS, Kim KM, Rhyu MG. Genetic classification of colorectal cancer based on chromosomal loss and microsatellite instability predicts survival. Clin Cancer Res. 2002;8:2311–2322. [PubMed] [Google Scholar]
- 31.Westra JL, Plukker JT, Buys CH, Hofstra RM. Genetic alterations in locally advanced stage II/III colon cancer: a search for prognostic markers. Clin Colorectal Cancer. 2004;4:252–259. doi: 10.3816/ccc.2004.n.024. [DOI] [PubMed] [Google Scholar]
- 32.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 33.González-García I, Moreno V, Navarro M, Martí-Ragué J, Marcuello E, Benasco C, Campos O, Capellà G, Peinado MA. Standardized approach for microsatellite instability detection in colorectal carcinomas. J Natl Cancer Inst. 2000;92:544–549. doi: 10.1093/jnci/92.7.544. [DOI] [PubMed] [Google Scholar]
- 34.Lothe RA, Peltomäki P, Meling GI, Aaltonen LA, Nyström-Lahti M, Pylkkänen L, Heimdal K, Andersen TI, Møller P, Rognum TO. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 35.Cawkwell L, Gray S, Murgatroyd H, Sutherland F, Haine L, Longfellow M, O'Loughlin S, Cross D, Kronborg O, Fenger C, et al. Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut. 1999;45:409–415. doi: 10.1136/gut.45.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curran B, Lenehan K, Mulcahy H, Tighe O, Bennett MA, Kay EW, O'Donoghue DP, Leader M, Croke DT. Replication error phenotype, clinicopathological variables, and patient outcome in Dukes' B stage II (T3,N0,M0) colorectal cancer. Gut. 2000;46:200–204. doi: 10.1136/gut.46.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gafà R, Maestri I, Matteuzzi M, Santini A, Ferretti S, Cavazzini L, Lanza G. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89:2025–2037. [PubMed] [Google Scholar]
- 38.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 39.Johannsdottir JT, Bergthorsson JT, Gretarsdottir S, Kristjansson AK, Ragnarsson G, Jonasson JG, Egilsson V, Ingvarsson S. Replication error in colorectal carcinoma: association with loss of heterozygosity at mismatch repair loci and clinicopathological variables. Anticancer Res. 1999;19:1821–1826. [PubMed] [Google Scholar]
- 40.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storojeva I, Boulay JL, Heinimann K, Ballabeni P, Terracciano L, Laffer U, Mild G, Herrmann R, Rochlitz C. Prognostic and predictive relevance of microsatellite instability in colorectal cancer. Oncol Rep. 2005;14:241–249. [PubMed] [Google Scholar]
- 42.Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 43.Wright CM, Dent OF, Barker M, Newland RC, Chapuis PH, Bokey EL, Young JP, Leggett BA, Jass JR, Macdonald GA. Prognostic significance of extensive microsatellite instability in sporadic clinicopathological stage C colorectal cancer. Br J Surg. 2000;87:1197–1202. doi: 10.1046/j.1365-2168.2000.01508.x. [DOI] [PubMed] [Google Scholar]
- 44.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]