Abstract
Mitochondrial ATP synthase has been recently detected at the surface of different cell types, where it is a high affinity receptor for apoA-I, the major protein component in high density lipoproteins (HDL). Cell surface ATP synthase (namely ecto-F1-ATPase) expression is related to different biological effects, such as regulation of HDL uptake by hepatocytes, endothelial cell proliferation or antitumor activity of Vγ9/Vδ2 T lymphocytes. This paper reviews the recently discovered functions and regulations of ecto-F1-ATPase. Particularly, the role of the F1-ATPase pathway(s) in HDL-cholesterol uptake and apoA-I-mediated endothelial protection suggests its potential importance in reverse cholesterol transport and its regulation might represent a potential therapeutic target for HDL-related therapy for cardiovascular diseases. Therefore, it is timely for us to better understand how this ecto-enzyme and downstream pathways are regulated and to develop pharmacologic interventions.
Keywords: F1Fo ATP synthase, High density lipoproteins receptor, Apolipoprotein A-I, Purinergic receptor P2Y13, Adenylate kinase, Nucleotide, Endothelium, Antitumor immunity
INTRODUCTION
F1Fo ATP synthase, the terminal enzyme of the oxidative phosphorylation pathway, is a complex molecular motor responsible for the large majority of ATP synthesis in all living beings, except in archaea which use a related enzyme to produce ATP, the A1Ao ATP synthase[1]. It is located in the inner membrane of mitochondria, in the thylakoid membrane of chloroplasts in plants, and in the plasma membrane (PM) of certain bacteria. ATP synthase is highly concentrated in inner membrane cristae, where it is part of the so-called “ATP synthasome”, in association with an inorganic phosphate (Pi) carrier as well as the adenine nucleotide translocase (ANT) which exchanges ADP and ATP[2]. Experimental evidence also suggests that ATP synthase is organized in dimers[3], or even oligomers[4]. This supramolecular organization is involved in the generation of inner membrane cristae and depends on the e and g subunits[5].
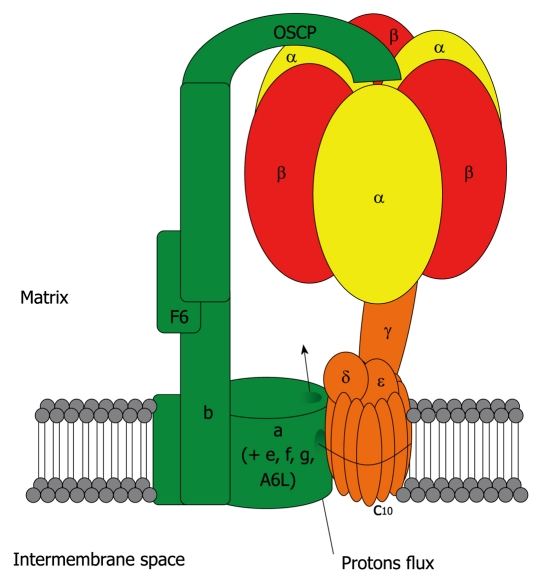
Despite a few differences in subunit composition, the general structure of ATP synthase is highly conserved throughout evolution. In eukaryotic cells, it is composed of 16 unique subunits (α, β and c being present in multiple copies in the whole complex). It comprises the activity-bearing subunits (three αβ heterodimers), a rotor (subunits γ, δ, ε and the c ring) and a stator (subunits a, e, f, g, A6L, b, F6, d and OSCP) which holds the αβ heterodimers. It is organized into two major domains: a soluble F1 domain (α, β, γ, δ, ε) and a membrane-associated Fo domain (Figure 1). The F1 domain bears the catalytic activity while the Fo domain is a proton channel. The biogenesis of ATP synthase follows a complex and timed mechanism[6] which involves at least five assembly factors: Atp10p, Atp11p, Atp12p, Atp22p and Fmc1p[7-10]. So far only the human orthologs of Atp11p and Atp12p have been identified[11].
Figure 1.
Mitochondrial ATP synthase. In eukaryotic cells, F1FO ATP synthase is a complex molecular motor composed of 16 unique subunits (α, β and c being present in multiple copies in the whole complex). It comprises the activity-bearing subunits (three αβ heterodimers), a rotor (in orange: subunits γ, δ, ε and the c ring) and a stator (in green: subunits a, e, f, g, A6L, b, F6, d and OSCP) which holds the αβ heterodimers. It is organized into two major domains: a soluble F1 domain (α, β, γ, δ, ε) and a membrane-associated FO domain. The F1 domain bears the catalytic activity while the FO domain is a proton channel. The FO domain uses the proton gradient created by the four other complexes of the respiratory chain to drive the rotation of the central stalk, which will alternatively change the conformation of the three αβ heterodimers, leading to the synthesis of ATP from ADP and Pi. In the case of the eukaryotic ATP synthase, the translocation of 10 protons through the c ring will lead to a full turn, and the synthesis of 3 ATP molecules. In the absence of a proton gradient, or if the F1 domain is isolated from the FO domain, the enzyme behaves as an ATP hydrolase.
The Fo domain uses the proton gradient created by the four other complexes of the respiratory chain to drive the rotation of the central stalk, which will alternatively change the conformation of the three αβ heterodimers, leading to the synthesis of ATP from ADP and Pi. The rotation of ATP synthase was demonstrated by a remarkable experiment in which the αβ heterodimers were immobilized on a solid support while the γ chain was coupled to a fluorochrome-labeled actin filament, and visualized under a microscope after addition of ATP[12]. ATP synthesis and hydrolysis involve a binding-change mechanism[13,14] in which catalytic sites undergo conformational changes driven by the interactions with the γ chain. In the case of the eukaryotic ATP synthase, the translocation of 10 protons through the c ring will lead to a full turn, and the synthesis of 3 ATP molecules. However, the exact mechanism of catalysis is still debated, as experimental data support both a bi-site mechanism, in which one of the catalytic sites is empty at any given moment[15], or a tri-site mechanism where all sites are filled with nucleotides[16]. The F1 domain also bears three non-catalytic sites which have been proposed to enhance ATP synthesis/hydrolysis upon binding of nucleotides[17].
In the absence of a proton gradient, or if the F1 domain is isolated from the FO domain, the enzyme behaves as an ATP hydrolase. This potential energy loss is tightly regulated by the inhibitor factor 1 (IF1) protein when the proton gradient collapses, for instance in the case of hypoxia as the respiratory chain stops functioning. Under normal conditions, IF1 exists as a homotetramer which turns into a homodimer when the pH in the matrix compartment decreases[18]. This allows it to tightly bind ATP synthase by interacting with both the αβ dimers and the central stalk, thereby blocking the rotation of the latter and preventing ATP hydrolysis[19]. This inhibitory mechanism has recently been shown to be highly dependent on the catalytic activity of the enzyme[20]. For extensive reviews on the structure and rotary mechanism of ATP synthase[21-23].
Given its essential role in ATP synthesis, defects in ATP synthase are involved in extremely severe and often lethal diseases because of the marked reduction in ATP production and concomitant accumulation of reactive oxygen species (ROS)[24,25]. Such pathologies are mostly linked to mutations in ATP synthase subunits, the most frequently described being mapped to the mitochondrial ATP6 gene (encoding for subunit a) which is responsible for the maternal-inherited NARP syndrome (Neurogenic muscle weakness, Ataxia, Retinitis Pigmentosa) or Leigh syndrome (severe and fatal encephalopathy), depending on the extent of heteroplasmic mutations. Defects in ATP synthase biogenesis, due to mutations in genes encoding its assembly factors for instance, have also been described[26]. Finally, abnormal accumulations of ATP synthase subunits have been found in various neurodegenerative diseases, such as subunit c accumulation in neuronal ceroid lipofuscinosis[27,28], or subunit α accumulation in Alzheimer’s disease[29].
Over the past few years, various reports by many independent groups have described the presence, at the cell surface of mammalian cells, of various subunits of ATP synthase, which will also be referred to as ecto-F1-ATPase throughout this review. The exact structure of this ectopically-expressed ATP synthase and whether it is exactly the same enzyme as the mitochondrial complex have not been definitely determined. Nevertheless, a number of pieces of experimental evidence suggest that it is a closely related entity. We will review here the various reports describing the unexpected cell surface expression of ATP synthase and its possible functions.
IS ECTO-F1-ATPASE THE SAME ENTITY AS MITOCHONDRIAL ATP SYNTHASE?
Over the past few years, a growing number of reports, including our work[30,31], have described the presence of ATP synthase subunits at the surface of a wide variety of tumor as well as normal cells by fluorocytometry.
While most of these reports focused on the α and β subunits against which monoclonal antibodies are available[30-32], a few have described the presence of other subunits at the cell surface by using in-house manufactured polyclonal reagents analyzed by cytometry or confocal microscopy. Vantourout et al[33] have shown that high cell surface expression of major histocompatibility complex class I (MHC-I) antigens interferes with the detection of the α and β subunits using some monoclonal reagents, probably through molecular hindrance. In a recent work, Mangiullo et al[34] described the presence of FOI-PVP (b subunit), γ and OSCP subunits at the cell surface of rat hepatocytes. The inhibitory protein IF1 has also been shown to be present at the cell surface[35]. Recently, d and OSCP subunits were shown to be expressed on the surface of osteosarcoma cells[36]. In a number of studies, biochemical approaches have been used to confirm the presence of F1-ATPase/ATP synthase in plasma membranes. Bae et al[37] have shown via a proteomics approach that multiple ATP synthase subunits are found in the lipid rafts of rat hepatocytes and HepG2 cells, a human hepatocarcinoma cell line. F1-ATPase components have been coprecipitated with cell surface proteins from hemopoietic tumors by Vantourout et al[33]. One could argue that pure plasma membrane preparations are extremely difficult to obtain and easily contaminated by membranes originating from different sub-cellular compartments. A very low amount of mitochondrial material could explain the presence of ATP synthase subunits in membrane preparations. Nevertheless, in some works using biochemical approaches[33,34] the absence of other mitochondrial proteins in samples has been carefully checked, ruling out a contamination by mitochondrial material. The observation of multiple components of F1-ATPase by cytometry, and the identification of some of them by mass spectrometry and peptide sequencing[30,37,38], make very unlikely the hypothesis that ecto-F1-ATPase detection could be due to cross-reactivity with other proteins bearing sequence similarities.
The presence of subunits other than α and β is also not completely unexpected. Indeed, since both these subunits do not have any membrane-anchoring domain, the cell surface expression of other subunits, especially from the FO domain, would be required to allow such a localization of the complex.
To summarize, while definitive proof may still be lacking, it seems now highly probable that ecto-F1-ATPase is a cell surface complex identical or closely related to the mitochondrial ATP synthase. The wide range of cellular types that have been shown to express ecto-F1-ATPase (Table 1) certainly suggest that this ectopic expression is more likely to be a ubiquitous phenomenon rather than restricted to a few cell lines. Among all samples that we have analyzed for ecto-F1-ATPase expression using different approaches, so far only one, namely 721.221 (an EBV-immortalized B cell line), seemed to not express ecto-F1-ATPase, or at extremely low levels that are not detectable[33,46].
Table 1.
Ecto-F1-ATPase subunit detection in various cell types
| Cells/tissues | Subunits | Methods | Ref. |
| K562, A549, Raji | β | B, FC, S | [38] |
| HUVEC | α, β, IF1 | B, FC, CM | [35,39,40] |
| HepG2, IHH, Human hepatocytes | α, β | SPR, FC, CM | [30] |
| 3T3-L1 | α, β | B, CM | [41] |
| Jurkat | β | RP + MS | [42] |
| Rat hepatocytes | α, β, δ, γ, b, d, e, F6, OSCP | CM | [37] |
| HaCat, Human skin | β | RP + MS, CM | [43] |
| Daudi, RPMI-8226, U937, SK-NEP, G401, G402 | α and/or β | FC | [31] |
| Awells, ST-Emo, HeLa, fibroblasts, Rma-S | α, β | B, CM, FC | [33] |
| Rat tonsils | β | PM | [44] |
| Neurons, neuroblastoma, astrocytoma | α | B, CM | [45] |
| Osteosarcoma | α, β, d | CM | [36] |
Studies reporting a cell surface expression of ATP synthase subunits are summarized. The experimental approaches used in these studies are mentioned. B: Cell surface biotinylation; FC: Flow cytometry; S: Sequencing; CM: Confocal microscopy; SPR: Surface plasmon resonance (Biacore); RP + MS: Rafts purification and mass spectrometry; PM: Plasma membrane extraction. Cell lines/tissues are of human origin unless otherwise noted. K562: Erythroleukemia; A549: Lung adenocarcinoma; RPMI-8226: B myeloma; Raji, Daudi: Burkitt lymphomas; Jurkat: T lymphoma; HUVEC: Human umbilical vein endothelial cells; HepG2: Hepatocarcinoma; IHH: Immortalized human hepatocytes; 3T3-L1: Murine fibroblasts (differentiated into adipocytes); HaCat: Immortalized human keratinocytes; U937: Monocytic leukemia; SK-NEP, G401, G402: Renal cell carcinomas; Awells, ST-EMO: EBV-immortalized B cells (ST-EMO was derived from a major histocompatibility complex classI deficient patient); HeLa: Cervix carcinoma; Rma-S: Murine T lymphoma.
AN UNEXPECTED AND SO FAR UNEXPLAINED LOCALIZATION
While many proteins are now known to have multiple sub-cellular localizations, a cell surface expression of mitochondrial proteins is quite unusual and puzzling. To our knowledge, there is so far no experimental work precisely deciphering the mechanism used by ATP synthase to reach the plasma membrane (PM).
In our view, two main hypotheses are conceivable: (1) ATP synthase subunits are routed to the PM instead of the mitochondria. The concept of multiple localization of a given protein is quite established now and several mechanisms can be involved (see Karniely et al[47] for an extensive review). The first is that different mRNA isoforms of ATP synthase subunits could hold a cell-surface signal peptide rather than mitochondrial targeting signal. Bovine subunit c has been reported to have two isoforms (encoded by two different genes) with slightly different signal peptides[48]. However, to our knowledge, this is an isolated case so far, and there is no experimental evidence suggesting that these different peptides lead to different localizations. Another explanation is that the known signal peptides of ATP synthase subunits are ambiguous and could lead to a mitochondrial or cell surface expression. We have analyzed some of ATP synthase subunit leader peptides with the SignalP software[49] and some leader peptides of ATP synthase subunits may share some predicted similarities with PM-targeting sequences. Finally, some proteins use two targeting signals on the same polypeptide to reach different destinations, such as Catalase A in yeast[50]. This latter possibility seems unlikely since no PM signal can be identified in the sequences of ATP synthase subunits. Even if this first hypothesis cannot be definitely ruled out yet, it is in our opinion the less likely for a number of reasons. Firstly, it would imply that all subunits display ambiguous signals, or have different isoforms, and so far there is no formal proof of this. Secondly, two subunits (namely A6L and a) are encoded by the mitochondrial genome, and it seems unlikely that they would be targeted to the cell surface instead of staying in the mitochondria. Finally, the ATP synthase assembly factors would also have to be expressed elsewhere [such as the endoplasmic reticulum (ER)] to allow the expression of a complete and correctly assembled complex at the cell surface; and (2) The simpler and thus more tempting hypothesis is that once assembled into the mitochondria, ATP synthase then reaches the cell surface. As a direct rerouting of the whole complex via transporters is improbable in this case, given the size of ATP synthase (around 600 kDa and a dimension of 19 nm × 11 nm), it is suggested that the complex reaches the cell surface via vesicular transport or fusion of mitochondrial membranes with the PM.
In healthy cells, mitochondria form a dynamic network and constantly divide and fuse, a process which is believed to maintain the integrity and quality of the mitochondrial function and implicates several recently identified proteins such as mitofusins or Drp1[51,52]. A direct fusion of mitochondrial membranes with the PM, resulting in a cell surface expression of ATP synthase, seems difficult to imagine mostly because mitochondria have two membranes and ATP synthase is located in the inner one. Unless the fusion occurs between PM and mitochondrial contact sites, which are distinct structures playing an important role in the mitochondrial metabolism and apoptosis[53], this hypothesis is unlikely.
However, there are now several lines of evidence indicating that mitochondria communicate with other cellular compartments. A recent report by de Brito et al[54] has shown that mitofusin 2 is implicated in the interaction between ER and mitochondria, allowing the exchange of Ca2+ between those organelles. In another recent report, Neuspiel et al[55] identified a new outer membrane mitochondria-anchored protein ligase named MAPL (mitochondria-anchored protein ligase) and characterized the sorting of MAPL into mitochondria-derived vesicles which then fuse with peroxisomes. Interestingly, Neuspiel et al[55] have also identified distinct vesicles containing another mitochondrial protein, Tom20 (translocase of outer membrane 20, a member of the complex importing proteins encoded by the nuclear genome into mitochondria). These Tom20-containing vesicles do not fuse with peroxisomes and their fate is currently under investigation. In addition, Soltys et al[56] have shown that Hsp60, a mitochondrial chaperone, can be found in vesicles that are distinct from major organelles and may communicate with the PM. Interestingly, Hsp60 has also been shown to be expressed at the cell surface[57,58]. In both cases, these uncharacterized vesicles could thus be part of a shuttle system between mitochondria and PM, allowing the cell surface expression of selected mitochondrial proteins. In this context, Rab32 or endophilin B1, involved in mitochondrial dynamics and morphology, respectively[59,60], might also be involved in the biogenesis of mitochondria-derived vesicles and the trafficking towards the cell surface, raising the question of the existence of a new intracellular pathway from mitochondria to cell surface responsible for the targeting of the F1-ATPase.
A recent report has shown that a deficiency in palmitoyl protein thioesterase 1 (Ppt1), responsible for the infantile neuronal ceroid lipofuscinosis syndrome, was linked with an increase in ecto-F1-ATPase expression in neurons[61]. This suggests that Ppt1 might be involved in the regulation of the subcellular localization of ATP synthase. In addition, Schmidt et al[45] have immunoprecipitated F1-ATPase α chain from the synaptic membranes of murine neural cells. Strikingly, they reported that this subunit was at least in part N-glycosylated and that this post-translational modification was abrogated by treating cells with the Golgi-disrupting drug brefeldin A. They also identified a unique N-glycosylation site which is absent in the human α-subunit sequence. This suggests that at least some components of the mitochondrial F1-ATPase can transit through the Golgi apparatus, although the mechanism is still enigmatic.
Another question regarding the cell surface expression of ATP synthase is its sub-localization at the PM. Some of its subunits, as well as a few other mitochondrial proteins, have often been found in lipid rafts in proteomic studies[37,42]. However, a recent report by Zheng et al[62] suggested that mitochondrial proteins, including ATP synthase subunits, were the best contaminants of raft preparations, mostly based on the fact that they were resistant to cholesterol depletion by methyl-β-cyclodextrin, whereas raft-resident proteins usually are sensitive to this treatment. Detection of ecto-F1-ATPase by confocal microscopy usually reveals a patchy pattern[30,33], which is reminiscent of several proteins residing in lipid rafts. We also have experimental evidence showing at least a partial co-localization of ecto-F1-ATPase subunits with the raft markers cholera toxin (unpublished data). This may indicate that the presence of ecto-F1-ATPase in lipid rafts is not exclusive. It may also vary with the cell line or conditions of culture. Similarly, its detection in isolated rafts may depend on raft purification procedures. Thus, more extensive studies are required to definitively answer this question.
A NEW LOCALIZATION FOR MANY NEW ROLES: ECTO-F1-ATPASE AS A MOONLIGHTING PROTEIN
The term “moonlighting protein” was introduced in 1999 by Jeffery[63] to describe proteins that could perform several unrelated roles. Their different functions can depend on different localizations, expression in different cell lines, ligand concentration, or on many other mechanisms.
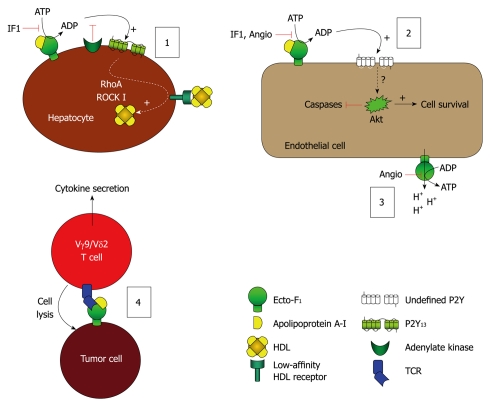
ATP synthase appears as a typical example of a moonlighting protein. Its α subunit, for instance, has been shown to be part of a complex importing cytosolic tRNAs into mitochondria in Leishmania[64]. Subunit F6 has also been shown to be present in plasma and involved in hypertension through its binding on ecto-F1-ATPase expressed on endothelial cells[65]. It is also now clear that the whole complex expressed at the cell surface is implicated in many physiological roles, depending on the ligand and the cell type (Table 2). So far, the most documented roles of ecto-F1-ATPase are its involvement in HDL endocytosis by hepatocytes, endothelial cell survival, and as an immunological ligand for a subset of non-conventional T lymphocytes (Figure 2).
Table 2.
Ligands and roles of Ecto-F1-ATPase
| Cells/tissues | Ligands | Proposed roles | Ref. |
| K562, A549, Raji | ? | NK/LAK-mediated tumor cell lysis | [38] |
| HUVEC | Angiostatin HDL/apoA-I | Cell survival through ATP production | [40,60,66] |
| FC6 | Cell survival through purinergic receptor activation | ||
| Control of blood pressure | |||
| Hepatocytes | HDL/apoA-I | HDL endocytosis | [30] |
| 3T3-L1, HaCat | ? | Cell survival | [41,43] |
| Tumor cells | Vγ9/Vδ2 TCR, apoA-I, MHC-I | Tumor cell recognition by Vγ9/Vδ2 T cells/presentation of phosphoantigens | [31,33,46,67] |
| Tonsils (mouse) | Enterostatin | Food intake regulation | [44] |
| Neurons | APP, Amyloid β-peptide | Synaptic plasticity regulation | [45] |
Major roles of Ecto-F1-ATPase described in the literature, as well as ligands involved, are summarized here. MHC-I: Major histocompatibility complex class I; NK: Natural killer; LAK: Lymphokine activated killer; HDL: High density lipoprotein; apoA-I: Apolipoprotein A-I; FC6: Coupling factor 6; ATP: Adenosine triphosphate; APP: Amyloid precursor protein. Cells/tissues are of human origin unless otherwise noted. K562: Erythroleukemia; A549: Lung adenocarcinoma; Raji: Burkitt’s lymphoma; HUVEC: Human umbilical vein endothelial cells; 3T3-L1: Murine fibroblasts (differentiated into adipocytes); HaCat: Immortalized human keratinocytes.
Figure 2.
Model of the leading hypotheses regarding the distinct cellular events mediated by ecto-F1-ATPase. (1) Model of F1-ATPase/P2Y13-mediated high density lipoproteins (HDL) endocytosis by hepatocytes. ApoA-I binding to ecto-F1-ATPase stimulates extracellular ATP hydrolysis, and the ADP generated selectively activates the nucleotide receptor P2Y13 and subsequent RhoA/ROCK I signaling that controls holo-HDL particle endocytosis. Conversely IF1 inhibitor which inhibits ecto-F1-ATPase activity or adenylate kinase (AK) activity which consumes ADP generated upon activation of ecto-F1-ATPase, downregulates holo-HDL particle endocytosis. Thus, HDL endocytosis depends on the balance between the AK activity that removes extracellular ADP and the ecto-F1-ATPase that synthesizes ADP. Activation of ADP synthesis by apoA-I unbalances the pathway and increases HDL endocytosis when required; (2), (3): Model of F1-ATPase-mediated endothelial cell protection. (2): Hydrolytic activity of ecto-F1-ATPase expressed on endothelial cells can be increased by apoA-I, stimulating cell proliferation. Conversely, inhibition by IF1, angiostatin increases apoptosis and blocks proliferation both at the basal level and after stimulation by apoA-I. The downstream receptors and intracellular signaling are still not completely characterized, but ADP-sensitive receptors could be involved; (3): The anti-angiogenic activity of angiostatin is attributed to its ability to inhibit ATP production by ecto-F1-ATPase, thus eliminating a significant source of energy and reducing proliferation. It is also proposed that this inhibition prevents the extrusion of protons by ecto-F1-ATPase, thus lowering intracellular pH and hampering cell survival; and (4) Model of F1-ATPase-mediated tumor cell recognition by Vγ9/Vδ2 T lymphocytes. Ecto-F1-ATPase is found at the surface of tumor cells and plays a role in the presentation of adenylated derivatives of small phosphoantigens, which are classical inducers of cell lysis by Vγ9/Vδ2 T lymphocytes. Recognition of ecto-F1-ATPase by the γ9/δ2 TCR is stimulated by apoA-I and requires a low expression of class I HLA-antigens, a situation commonly found in tumor cells.
Hepatic HDL-cholesterol uptake
Our work on high density lipoproteins (HDL) has identified ecto-F1-ATPase as the receptor of apolipoprotein A-I (apoA-I), the major protein component of HDL. Indeed, binding of HDL on hepatocyte membranes displayed two binding sites with high and low affinities[68]. A high-affinity receptor for lipid-free apoA-I was purified by Biacore and identified as the β-chain of ATP synthase. As binding of lipid-free apoA-I to ecto-F1-ATPase activated endocytosis of whole and fully lipidated HDL by the low-affinity binding sites, we have hypothesized that enzymatic activity of ecto-F1-ATPase might play a role in this stimulatory process. Indeed, direct addition of ADP increased HDL endocytosis. We showed that binding of apoA-I on ecto-F1-ATPase increased its hydrolase activity and led to an enhanced HDL endocytosis. Later, we identified the purinergic P2Y13 receptor as the target of ADP produced by ecto-F1-ATPase[69] and we have shown recently that the small GTPase RhoA and its effector ROCK I were activated downstream of P2Y13[70]. We could demonstrate that this F1-ATPase/P2Y13-mediated HDL endocytosis pathway is controlled through cell surface adenylate kinase (AK) activity which consumes ADP generated upon activation of ecto-F1-ATPase by apoA-I and therefore constitutively inhibits P2Y13 and subsequent hepatic HDL endocytosis[71]. In addition, niacin (vitamin B3), which is well-known to increase plasma HDL levels in humans, has recently been shown to inhibit cell surface expression of ecto-F1-ATPase in hepatocytes[72]. This study thus suggests the implication of ecto-F1-ATPase in HDL metabolism.
Our recent work has confirmed the role of the P2Y13 ADP-receptor in HDL-mediated reverse cholesterol transport (RCT) in vivo[73]. We showed that P2Y13-deficient mice exhibited a decrease in hepatic HDL cholesterol uptake, hepatic cholesterol content, and biliary cholesterol output, although their plasma HDL and other lipid levels were normal. These changes translated into a substantial decrease in the rate of macrophage-to-feces RCT. Therefore, hallmark features of RCT are impaired in P2Y13-deficient mice. Furthermore, cangrelor, a partial agonist of P2Y13, stimulated hepatic HDL uptake and biliary lipid secretions in normal mice and in mice with a targeted deletion of scavenger receptor class B type I (SR-BI) in liver [hypomSR-BI-knockout(liver)] but had no effect in P2Y13 knockout mice, which indicate that the P2Y13-mediated HDL uptake pathway is independent of SR-BI-mediated HDL selective cholesteryl ester uptake. Pharmacological activation of the P2Y13 receptor might thus represent a new therapeutic approach to improve HDL-cholesterol removal and/or turnover, which could be an alternative way to prevent and treat atherosclerosis.
Endothelial cell survival
Ecto-F1-ATPase has also been characterized by Moser et al[32] as the receptor of angiostatin, an inhibitor of angiogenesis originating from the proteolysis of plasminogen. Later work established that ecto-F1-ATPase was able to synthesize ATP at the cell surface of endothelial cells. This ATP production would facilitate the proliferation of these cells, especially under hypoxic conditions which are known to impair cell survival through the inhibition of oxidative phosphorylation. The anti-angiogenic activity of angiostatin is attributed to its ability to inhibit ATP production by ecto-F1-ATPase, thus eliminating a significant source of energy and reducing proliferation. It was also proposed that this inhibition prevents the extrusion of protons by ecto-F1-ATPase, thus lowering intracellular pH and hampering cell survival[39,66].
It is well known that HDL and apoA-I have a protective effect against the development of endothelial dysfunction, one of the first events in the pathogenesis of atherosclerosis, by enhancing endothelial cell survival. The mechanisms mediating this atheroprotection are not well understood, but interactions between HDL and apoA-I with cell surface receptors have been proposed to be essential contributors. Considering that both HDL and apoA-I bind ecto-F1-ATPase, we have evaluated the hypothesis that the protective effects of apoA-I on endothelial cells are dependent on ecto-F1-ATPase activity. This hypothesis is also supported by the anti-angiogenic effect of angiostatin.
As shown for hepatocytes[30], the hydrolytic activity of ecto-F1-ATPase expressed on endothelial cells can be increased by apoA-I, stimulating cell proliferation and inhibiting apoptosis in a dose-dependent manner[40]. Conversely, inhibition by IF1, angiostatin or antibodies directed against the β-chain of ecto-F1-ATPase increases apoptosis and blocks proliferation both at the basal level and after stimulation by apoA-I. These results are in favor of a direct effect of the ecto-F1-ATPase activity on endothelial cell survival through extracellular ADP production. The downstream receptors and intracellular signaling are still not completely characterized, but ADP-sensitive receptors could be involved.
This pathway thus provides a new mechanism of action of apoA-I in endothelial protection distinct from other HDL-mediated protective effects previously described[74]. Thus, depending on the cell type, distinct pathways seem to be involved downstream of F1-ATPase activation by apoA-I and induce distinct cellular events such as stimulation of HDL uptake by hepatocytes or endothelial cell survival.
Tumor cell recognition
Vγ9/Vδ2 T lymphocytes are a non-conventional subpopulation of cytotoxic T cells unique to humans and higher primates. These cells are known to participate in the immune response against various intracellular pathogens as well as tumor cells, through the recognition of small alkyl diphosphate molecules termed phosphoantigens[75,76]. Experimental evidence suggests that recognition of these antigens requires a cell-cell contact, and thus may depend on a cell surface presentation molecule expressed on target cells but distinct from classical antigen presentation molecules such as major histocompatibility antigens and CD1[77]. This is confirmed by recent data showing that the recognition of phosphoantigens by a soluble recombinant Vγ9/Vδ2 T cell receptor (TCR) on the cell surface is dependent on a trypsin-sensitive structure[78]. We have initially identified apoA-I as an enhancer of the Vγ9/Vδ2 T cell response, and shown that these cells can actually recognize F1-ATPase on the surface of tumor cells through their TCR[31]. We have also shown that ecto-F1-ATPase interacts with MHC-I molecules on the cell surface of various cell lines, leading to a masking of epitopes which prevents the detection of ecto-F1-ATPase by commercially available antibodies[33]. This suggests that cell surface expression of ATP synthase cannot be formally ruled out simply because it is not detected by flow cytometry or confocal microscopy. It is also of particular interest for the physiology of Vγ9/Vδ2 cells because, like natural killer cells, they express inhibitory receptors which bind MHC antigens and modulate TCR signaling and lymphocyte activation[79].
Our latest work has focused on the study of a novel ATP analog, ApppI [triphosphoric acid 1-adenosin-5’-yl ester 3-(3-methylbut-3-enyl) ester] in which the γ-phosphate of ATP is esterified with isopentenol. This metabolite has recently been identified in cells treated with aminobisphosphonate drugs which are known to potently sensitize cells to Vγ9/Vδ2 T cell cytotoxicity. As it is an ATP analog and has a structure reminiscent of nucleotidic phosphoantigens naturally produced by some bacteria, we thus used this molecule to explore the potential role of ecto-F1-ATPase in phosphoantigen presentation to Vγ9/Vδ2 T cells. We found that classical targets of Vγ9/Vδ2 T cells can spontaneously produce ApppI[67]. Furthermore, it stably binds to purified ATP synthase and ApppI-loaded ATP-synthase efficiently activates Vγ9/Vδ2 T cells in vitro. Our current model proposes that phosphoantigens produced by tumor cells or intracellular pathogens are converted to nucleotidic derivatives, loaded onto ATP synthase, and presented to Vγ9/Vδ2 T cells when the complex is exported to the cell surface[46].
As ATP synthase is present at the cell surface of a number of normal cells, the potential self-reactivity of Vγ9/Vδ2 T cells is probably tightly controlled by the interaction between ecto-F1-ATPase and MHC-I, as well as by phosphoantigen production by the target cell. Moreover, these observations suggest another beneficial role for apoA-I; as an immunomodulator in anti-tumor responses.
Since ecto-F1-ATPase has already been detected in a wide variety of cells, with many different ligands, there is no doubt that it is probably implicated in many other cellular processes. In many instances, these roles seem to rely on the enzymatic activity of ATP synthase. While its hydrolase activity is undoubtedly possible since it does not need a proton flux nor requires a fully assembled F1FO complex, there are still conflicting reports regarding the ability of the cell surface complex to synthesize ATP.
TO SYNTHESIZE OR NOT TO SYNTHESIZE, THAT IS THE QUESTION
The first work showing that ecto-F1-ATPase can synthesize ATP from ADP and Pi was reported in endothelial cells[39] and showed that angiostatin was able to inhibit both the synthase and hydrolase activities of ATP synthase. This was shown by bioluminescence measurement of ATP and a radioactive thin layer chromatography (TLC) assay using [3H]ADP and 32Pi. In later work, the authors have shown that ATP production was enhanced when extracellular pH was low, hypothesizing that concomitantly with ATP synthesis, ecto-F1-ATPase would extrude protons from the cytoplasm. It was also proposed that this process improves the resistance of tumor and endothelial cells to low pH. Similarly, it would also improve cell survival in hypoxic conditions, a hallmark of tumoral micro-environment. Since then, a number of independent studies have reported a synthase activity in many different cell types, including keratinocytes, adipocytes, or hepatocytes, among others[34,43,41]. Most of these works have used a bioluminescence assay to measure the production of ATP after addition of ADP in the culture media and underlined the importance of ecto-F1-ATPase in this production through the use of oligomycin, a well known inhibitor of mitochondrial ATP synthase.
However, whether or not ecto-F1-ATPase can synthesize ATP is still debated, as there are several conflicting reports. Several studies by our group and others seem to completely rule out a potential contribution of ecto-F1-ATPase to extracellular conversion of ADP to ATP. Yegutkin et al[80] have shown that ATP production after addition of ADP to endothelial cells was not increased in the presence of Pi, which should be the case if ecto-F1-ATPase was able to synthesize ATP. They concluded that ADP phosphorylation was mostly due to cell surface adenylate kinase (AK, which produces ATP and AMP from two ADP molecules) and/or nucleotide diphosphokinase (NDPK, which transfers terminal phosphate from a nucleotide triphosphate to a nucleotide diphosphate) enzymatic activities. This is in agreement with our results in hepatocytes as well as in endothelial cells. Indeed, we have observed that addition of [3H]ADP and 32Pi to the cell medium does not lead to production of dual-labeled ATP, but only [3H]ATP[30,40,71], indicating that free inorganic phosphate is not used for ATP synthesis and excluding a role for ATP synthase in the process of ATP generation.
Most of the studies reporting a synthase activity have shown that oligomycin decreased extracellular ATP production after addition of ADP to culture media, thus suggesting that ecto-F1-ATPase was indeed able to synthesize ATP. However, in our hands, oligomycin had no effect on ATP synthesis on the cell surface of human hepatocytes after addition of ADP[71]. The reason for these discrepancies is not clear. One should note that oligomycin has been shown to inhibit other enzymes, such as the non-gastric H,K-ATPase[81]. Thus it does not appear to be a strictly specific inhibitor of ATP synthase/F1-ATPase and could act on other nucleotide-converting enzymes. One cannot exclude also that the activity of ecto-F1-ATPase depends on particular experimental settings (culture conditions, cell type) and on the metabolic status of the cells. Nevertheless, in mitochondria, ATP production by ATP synthase is strictly dependent on the electrochemical proton gradient between the two sides of the inner mitochondrial membrane. This gives the chemical energy required for ATP synthesis. Whether a strong proton flux can occur at the cell surface and lead to ATP synthesis by ecto-F1-ATPase is not certain and in the case of extracellular acidosis, the proton gradient would not be favorable for ATP synthesis. Mangiullo et al[34] have recently shown that ecto-F1-ATPase was active in ATP synthesis and that it was correlated to an oligomycin-sensitive proton flux from the cytoplasm to the extracellular medium in the case of intracellular acidosis. Although, as discussed above, oligomycin might inhibit other proton pumps, this suggests that synthesis might occur in particular metabolic settings. It remains to be demonstrated whether these experimental conditions are relevant to physiological situations. Thus, the actual role of ecto-F1-ATPase in proton transport across the PM remains arguable.
Accordingly, the synthase activity of ecto-F1-ATPase is still an open issue, and there certainly is a need for reassessment of results obtained with the various techniques assessing ATP synthase activity as well as with the different inhibitors.
CONCLUSION
Over the past few years, data originating from many different groups have confirmed the cell surface expression of this mitochondrial enzyme. While this ectopic expression is now certainly widely accepted, there is still a need for a definitive characterization of its enzymatic activity, as well as new insights into the mechanism of its cell surface targeting.
In our opinion, the synthase activity of ecto-F1-ATPase is still heavily challenged on both experimental and theoretical aspects. It is an important issue that requires additional work and may depend on the development of novel techniques which would be more specific and sensitive than those currently used. Nevertheless, the monitoring of dual-labeled ATP production from [3H]ADP and 32Pi by high-performance liquid chromatography (HPLC) is probably so far the most reliable method to determine whether ecto-F1-ATPase is active in ATP synthesis, since ATP synthase is the only known enzyme able to generate ATP directly from ADP and inorganic free phosphate. Therefore, it should be the method of choice to study the enzymatic properties of ecto-F1-ATPase.
Understanding how ecto-F1-ATPase reaches the cell surface is a challenging but extremely important point. As the use of siRNAs targeting ATP synthase subunits is hardly conceivable since it will undoubtedly rapidly lead to cell death, inhibiting its cell surface expression might help to better characterize its roles. Further studies are definitely required to decipher this mechanism.
ATP synthase is a fascinating molecular machine and a perfect example of a nanomotor. It has been one of the most studied enzymes over the past 40 years and garnered Professor Boyer PD and Sir Walker JE the Nobel Prize in Chemistry in 1997 for their outstanding work on the comprehension of its structure and the ATP synthesis mechanism[82-84]. Its central role in energy production is fundamental, an active person synthesizing their own weight in ATP every day. This essential role is now enhanced by its new implication in various important cellular processes, achieved through this ectopic localization and interactions with diverse ligands. We are only beginning to decipher these new functions.
Footnotes
Supported by An INSERM Avenir Grant (Martinez LO), ANR (Martinez LO and Lichtenstein L, #GENO 102 01) and the French Association pour la Recherche sur le Cancer (Vantourout P and Champagne E, #3711-3913-4847); An INSERM young scientist fellowship (Pons V)
Peer reviewer: Dr. M H Ahmed, MD, PhD, Department of Chemical Pathology, Southampton University Hospital NHS trust, Mail point 6, Level D, South Academic Block, Southampton SO16 6YD, United Kingdom
S- Editor Sun H L- Editor Logan S E- Editor Zheng XM
References
- 1.Lewalter K, Müller V. Bioenergetics of archaea: ancient energy conserving mechanisms developed in the early history of life. Biochim Biophys Acta. 2006;1757:437–445. doi: 10.1016/j.bbabio.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 2.Ko YH, Delannoy M, Hullihen J, Chiu W, Pedersen PL. Mitochondrial ATP synthasome. Cristae-enriched membranes and a multiwell detergent screening assay yield dispersed single complexes containing the ATP synthase and carriers for Pi and ADP/ATP. J Biol Chem. 2003;278:12305–12309. doi: 10.1074/jbc.C200703200. [DOI] [PubMed] [Google Scholar]
- 3.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittig I, Schägger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- 5.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brèthes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nijtmans LG, Klement P, Houstĕk J, van den Bogert C. Assembly of mitochondrial ATP synthase in cultured human cells: implications for mitochondrial diseases. Biochim Biophys Acta. 1995;1272:190–198. doi: 10.1016/0925-4439(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 7.Ackerman SH, Tzagoloff A. Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci USA. 1990;87:4986–4990. doi: 10.1073/pnas.87.13.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackerman SH, Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990;265:9952–9959. [PubMed] [Google Scholar]
- 9.Helfenbein KG, Ellis TP, Dieckmann CL, Tzagoloff A. ATP22, a nuclear gene required for expression of the F0 sector of mitochondrial ATPase in Saccharomyces cerevisiae. J Biol Chem. 2003;278:19751–19756. doi: 10.1074/jbc.M301679200. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP, di Rago JP. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- 11.Wang ZG, White PS, Ackerman SH. Atp11p and Atp12p are assembly factors for the F(1)-ATPase in human mitochondria. J Biol Chem. 2001;276:30773–30778. doi: 10.1074/jbc.M104133200. [DOI] [PubMed] [Google Scholar]
- 12.Noji H, Yasuda R, Yoshida M, Kinosita K Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 13.Boyer PD. A model for conformational coupling of membrane potential and proton translocation to ATP synthesis and to active transport. FEBS Lett. 1975;58:1–6. doi: 10.1016/0014-5793(75)80212-2. [DOI] [PubMed] [Google Scholar]
- 14.Boyer PD. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 1989;3:2164–2178. doi: 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- 15.Milgrom YM, Cross RL. Rapid hydrolysis of ATP by mitochondrial F1-ATPase correlates with the filling of the second of three catalytic sites. Proc Natl Acad Sci USA. 2005;102:13831–13836. doi: 10.1073/pnas.0507139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber J, Senior AE. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 17.Harris DA. The 'non-exchangeable' nucleotides of F1-F0ATP synthase. Cofactors in hydrolysis? FEBS Lett. 1993;316:209–215. doi: 10.1016/0014-5793(93)81295-b. [DOI] [PubMed] [Google Scholar]
- 18.Cabezon E, Butler PJ, Runswick MJ, Walker JE. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J Biol Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- 19.Cabezón E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 20.Corvest V, Sigalat C, Haraux F. Insight into the bind-lock mechanism of the yeast mitochondrial ATP synthase inhibitory peptide. Biochemistry. 2007;46:8680–8688. doi: 10.1021/bi700522v. [DOI] [PubMed] [Google Scholar]
- 21.Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 22.Devenish RJ, Prescott M, Rodgers AJ. The structure and function of mitochondrial F1F0-ATP synthases. Int Rev Cell Mol Biol. 2008;267:1–58. doi: 10.1016/S1937-6448(08)00601-1. [DOI] [PubMed] [Google Scholar]
- 23.Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 24.Das AM. Regulation of the mitochondrial ATP-synthase in health and disease. Mol Genet Metab. 2003;79:71–82. doi: 10.1016/s1096-7192(03)00069-6. [DOI] [PubMed] [Google Scholar]
- 25.Houstek J, Mrácek T, Vojtísková A, Zeman J. Mitochondrial diseases and ATPase defects of nuclear origin. Biochim Biophys Acta. 2004;1658:115–121. doi: 10.1016/j.bbabio.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 26.De Meirleir L, Seneca S, Lissens W, De Clercq I, Eyskens F, Gerlo E, Smet J, Van Coster R. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J Med Genet. 2004;41:120–124. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fearnley IM, Walker JE, Martinus RD, Jolly RD, Kirkland KB, Shaw GJ, Palmer DN. The sequence of the major protein stored in ovine ceroid lipofuscinosis is identical with that of the dicyclohexylcarbodiimide-reactive proteolipid of mitochondrial ATP synthase. Biochem J. 1990;268:751–758. doi: 10.1042/bj2680751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer DN, Fearnley IM, Medd SM, Walker JE, Martinus RD, Bayliss SL, Hall NA, Lake BD, Wolfe LS, Jolly RD. Lysosomal storage of the DCCD reactive proteolipid subunit of mitochondrial ATP synthase in human and ovine ceroid lipofuscinoses. Adv Exp Med Biol. 1989;266:211–222; discussion 223. doi: 10.1007/978-1-4899-5339-1_15. [DOI] [PubMed] [Google Scholar]
- 29.Sergeant N, Wattez A, Galván-valencia M, Ghestem A, David JP, Lemoine J, Sautiére PE, Dachary J, Mazat JP, Michalski JC, et al. Association of ATP synthase alpha-chain with neurofibrillary degeneration in Alzheimer's disease. Neuroscience. 2003;117:293–303. doi: 10.1016/s0306-4522(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 30.Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezón E, Champagne E, Pineau T, Georgeaud V, Walker JE, Tercé F, et al. Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature. 2003;421:75–79. doi: 10.1038/nature01250. [DOI] [PubMed] [Google Scholar]
- 31.Scotet E, Martinez LO, Grant E, Barbaras R, Jenö P, Guiraud M, Monsarrat B, Saulquin X, Maillet S, Estève JP, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity. 2005;22:71–80. doi: 10.1016/j.immuni.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Moser TL, Stack MS, Asplin I, Enghild JJ, Højrup P, Everitt L, Hubchak S, Schnaper HW, Pizzo SV. Angiostatin binds ATP synthase on the surface of human endothelial cells. Proc Natl Acad Sci USA. 1999;96:2811–2816. doi: 10.1073/pnas.96.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vantourout P, Martinez LO, Fabre A, Collet X, Champagne E. Ecto-F1-ATPase and MHC-class I close association on cell membranes. Mol Immunol. 2008;45:485–492. doi: 10.1016/j.molimm.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 34.Mangiullo R, Gnoni A, Leone A, Gnoni GV, Papa S, Zanotti F. Structural and functional characterization of F(o)F(1)-ATP synthase on the extracellular surface of rat hepatocytes. Biochim Biophys Acta. 2008;1777:1326–1335. doi: 10.1016/j.bbabio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Cortés-Hernández P, Domínguez-Ramírez L, Estrada-Bernal A, Montes-Sánchez DG, Zentella-Dehesa A, de Gómez-Puyou MT, Gómez-Puyou A, García JJ. The inhibitor protein of the F1F0-ATP synthase is associated to the external surface of endothelial cells. Biochem Biophys Res Commun. 2005;330:844–849. doi: 10.1016/j.bbrc.2005.03.064. [DOI] [PubMed] [Google Scholar]
- 36.Yonally SK, Capaldi RA. The F(1)F(0) ATP synthase and mitochondrial respiratory chain complexes are present on the plasma membrane of an osteosarcoma cell line: An immunocytochemical study. Mitochondrion. 2006;6:305–314. doi: 10.1016/j.mito.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Bae TJ, Kim MS, Kim JW, Kim BW, Choo HJ, Lee JW, Kim KB, Lee CS, Kim JH, Chang SY, et al. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004;4:3536–3548. doi: 10.1002/pmic.200400952. [DOI] [PubMed] [Google Scholar]
- 38.Das B, Mondragon MO, Sadeghian M, Hatcher VB, Norin AJ. A novel ligand in lymphocyte-mediated cytotoxicity: expression of the beta subunit of H+ transporting ATP synthase on the surface of tumor cell lines. J Exp Med. 1994;180:273–281. doi: 10.1084/jem.180.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ, Pizzo SV. Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin. Proc Natl Acad Sci USA. 2001;98:6656–6661. doi: 10.1073/pnas.131067798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radojkovic C, Genoux A, Pons V, Combes G, de Jonge H, Champagne E, Rolland C, Perret B, Collet X, Tercé F, et al. Stimulation of cell surface F1-ATPase activity by apolipoprotein A-I inhibits endothelial cell apoptosis and promotes proliferation. Arterioscler Thromb Vasc Biol. 2009;29:1125–1130. doi: 10.1161/ATVBAHA.109.187997. [DOI] [PubMed] [Google Scholar]
- 41.Kim BW, Choo HJ, Lee JW, Kim JH, Ko YG. Extracellular ATP is generated by ATP synthase complex in adipocyte lipid rafts. Exp Mol Med. 2004;36:476–485. doi: 10.1038/emm.2004.60. [DOI] [PubMed] [Google Scholar]
- 42.von Haller PD, Donohoe S, Goodlett DR, Aebersold R, Watts JD. Mass spectrometric characterization of proteins extracted from Jurkat T cell detergent-resistant membrane domains. Proteomics. 2001;1:1010–1021. doi: 10.1002/1615-9861(200108)1:8<1010::AID-PROT1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 43.Burrell HE, Wlodarski B, Foster BJ, Buckley KA, Sharpe GR, Quayle JM, Simpson AW, Gallagher JA. Human keratinocytes release ATP and utilize three mechanisms for nucleotide interconversion at the cell surface. J Biol Chem. 2005;280:29667–29676. doi: 10.1074/jbc.M505381200. [DOI] [PubMed] [Google Scholar]
- 44.Park M, Lin L, Thomas S, Braymer HD, Smith PM, Harrison DH, York DA. The F1-ATPase beta-subunit is the putative enterostatin receptor. Peptides. 2004;25:2127–2133. doi: 10.1016/j.peptides.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt C, Lepsverdize E, Chi SL, Das AM, Pizzo SV, Dityatev A, Schachner M. Amyloid precursor protein and amyloid beta-peptide bind to ATP synthase and regulate its activity at the surface of neural cells. Mol Psychiatry. 2008;13:953–969. doi: 10.1038/sj.mp.4002077. [DOI] [PubMed] [Google Scholar]
- 46.Mookerjee-Basu J, Vantourout P, Martinez LO, Perret B, Collet X, Périgaud C, Peyrottes S, Champagne E. F1-adenosine triphosphatase displays properties characteristic of an antigen presentation molecule for Vgamma9Vdelta2 T cells. J Immunol. 2010;184:6920–6928. doi: 10.4049/jimmunol.0904024. [DOI] [PubMed] [Google Scholar]
- 47.Karniely S, Pines O. Single translation--dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gay NJ, Walker JE. Two genes encoding the bovine mitochondrial ATP synthase proteolipid specify precursors with different import sequences and are expressed in a tissue-specific manner. EMBO J. 1985;4:3519–3524. doi: 10.1002/j.1460-2075.1985.tb04111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 50.Petrova VY, Drescher D, Kujumdzieva AV, Schmitt MJ. Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem J. 2004;380:393–400. doi: 10.1042/BJ20040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berman SB, Pineda FJ, Hardwick JM. Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ. 2008;15:1147–1152. doi: 10.1038/cdd.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brdiczka DG, Zorov DB, Sheu SS. Mitochondrial contact sites: their role in energy metabolism and apoptosis. Biochim Biophys Acta. 2006;1762:148–163. doi: 10.1016/j.bbadis.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 54.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 55.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 56.Soltys BJ, Gupta RS. Immunoelectron microscopic localization of the 60-kDa heat shock chaperonin protein (Hsp60) in mammalian cells. Exp Cell Res. 1996;222:16–27. doi: 10.1006/excr.1996.0003. [DOI] [PubMed] [Google Scholar]
- 57.Soltys BJ, Gupta RS. Cell surface localization of the 60 kDa heat shock chaperonin protein (hsp60) in mammalian cells. Cell Biol Int. 1997;21:315–320. doi: 10.1006/cbir.1997.0144. [DOI] [PubMed] [Google Scholar]
- 58.Fisch P, Malkovsky M, Kovats S, Sturm E, Braakman E, Klein BS, Voss SD, Morrissey LW, DeMars R, Welch WJ. Recognition by human V gamma 9/V delta 2 T cells of a GroEL homolog on Daudi Burkitt's lymphoma cells. Science. 1990;250:1269–1273. doi: 10.1126/science.1978758. [DOI] [PubMed] [Google Scholar]
- 59.Alto NM, Soderling J, Scott JD. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J Cell Biol. 2002;158:659–668. doi: 10.1083/jcb.200204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyly A, Marjavaara SK, Kyttälä A, Uusi-Rauva K, Luiro K, Kopra O, Martinez LO, Tanhuanpää K, Kalkkinen N, Suomalainen A, et al. Deficiency of the INCL protein Ppt1 results in changes in ectopic F1-ATP synthase and altered cholesterol metabolism. Hum Mol Genet. 2008;17:1406–1417. doi: 10.1093/hmg/ddn028. [DOI] [PubMed] [Google Scholar]
- 62.Zheng YZ, Berg KB, Foster LJ. Mitochondria do not contain lipid rafts, and lipid rafts do not contain mitochondrial proteins. J Lipid Res. 2009;50:988–998. doi: 10.1194/jlr.M800658-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 64.Goswami S, Dhar G, Mukherjee S, Mahata B, Chatterjee S, Home P, Adhya S. A bifunctional tRNA import receptor from Leishmania mitochondria. Proc Natl Acad Sci USA. 2006;103:8354–8359. doi: 10.1073/pnas.0510869103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Osanai T, Magota K, Tanaka M, Shimada M, Murakami R, Sasaki S, Tomita H, Maeda N, Okumura K. Intracellular signaling for vasoconstrictor coupling factor 6: novel function of beta-subunit of ATP synthase as receptor. Hypertension. 2005;46:1140–1146. doi: 10.1161/01.HYP.0000186483.86750.85. [DOI] [PubMed] [Google Scholar]
- 66.Chi SL, Pizzo SV. Angiostatin is directly cytotoxic to tumor cells at low extracellular pH: a mechanism dependent on cell surface-associated ATP synthase. Cancer Res. 2006;66:875–882. doi: 10.1158/0008-5472.CAN-05-2806. [DOI] [PubMed] [Google Scholar]
- 67.Vantourout P, Mookerjee-Basu J, Rolland C, Pont F, Martin H, Davrinche C, Martinez LO, Perret B, Collet X, Périgaud C, et al. Specific requirements for Vgamma9Vdelta2 T cell stimulation by a natural adenylated phosphoantigen. J Immunol. 2009;183:3848–3857. doi: 10.4049/jimmunol.0901085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez LO, Georgeaud V, Rolland C, Collet X, Tercé F, Perret B, Barbaras R. Characterization of two high-density lipoprotein binding sites on porcine hepatocyte plasma membranes: contribution of scavenger receptor class B type I (SR-BI) to the low-affinity component. Biochemistry. 2000;39:1076–1082. doi: 10.1021/bi991971y. [DOI] [PubMed] [Google Scholar]
- 69.Jacquet S, Malaval C, Martinez LO, Sak K, Rolland C, Perez C, Nauze M, Champagne E, Tercé F, Gachet C, et al. The nucleotide receptor P2Y13 is a key regulator of hepatic high-density lipoprotein (HDL) endocytosis. Cell Mol Life Sci. 2005;62:2508–2515. doi: 10.1007/s00018-005-5194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malaval C, Laffargue M, Barbaras R, Rolland C, Peres C, Champagne E, Perret B, Tercé F, Collet X, Martinez LO. RhoA/ROCK I signalling downstream of the P2Y13 ADP-receptor controls HDL endocytosis in human hepatocytes. Cell Signal. 2009;21:120–127. doi: 10.1016/j.cellsig.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 71.Fabre AC, Vantourout P, Champagne E, Tercé F, Rolland C, Perret B, Collet X, Barbaras R, Martinez LO. Cell surface adenylate kinase activity regulates the F(1)-ATPase/P2Y (13)-mediated HDL endocytosis pathway on human hepatocytes. Cell Mol Life Sci. 2006;63:2829–2837. doi: 10.1007/s00018-006-6325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang LH, Kamanna VS, Zhang MC, Kashyap ML. Niacin inhibits surface expression of ATP synthase beta chain in HepG2 cells: implications for raising HDL. J Lipid Res. 2008;49:1195–1201. doi: 10.1194/jlr.M700426-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Fabre AC, Malaval C, Ben Addi A, Verdier C, Pons V, Serhan N, Lichtenstein L, Combes G, Huby T, Briand F, et al. P2Y13 receptor is critical for reverse cholesterol transport. Hepatology. 2010;52:1477–1483. doi: 10.1002/hep.23897. [DOI] [PubMed] [Google Scholar]
- 74.Barter PJ, Rye KA. Relationship between the concentration and antiatherogenic activity of high-density lipoproteins. Curr Opin Lipidol. 2006;17:399–403. doi: 10.1097/01.mol.0000236365.40969.af. [DOI] [PubMed] [Google Scholar]
- 75.Constant P, Davodeau F, Peyrat MA, Poquet Y, Puzo G, Bonneville M, Fournié JJ. Stimulation of human gamma delta T cells by nonpeptidic mycobacterial ligands. Science. 1994;264:267–270. doi: 10.1126/science.8146660. [DOI] [PubMed] [Google Scholar]
- 76.Gober HJ, Kistowska M, Angman L, Jenö P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. 2003;197:163–168. doi: 10.1084/jem.20021500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morita CT, Beckman EM, Bukowski JF, Tanaka Y, Band H, Bloom BR, Golan DE, Brenner MB. Direct presentation of nonpeptide prenyl pyrophosphate antigens to human gamma delta T cells. Immunity. 1995;3:495–507. doi: 10.1016/1074-7613(95)90178-7. [DOI] [PubMed] [Google Scholar]
- 78.Wei H, Huang D, Lai X, Chen M, Zhong W, Wang R, Chen ZW. Definition of APC presentation of phosphoantigen (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate to Vgamma2Vdelta 2 TCR. J Immunol. 2008;181:4798–4806. doi: 10.4049/jimmunol.181.7.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yegutkin GG, Henttinen T, Jalkanen S. Extracellular ATP formation on vascular endothelial cells is mediated by ecto-nucleotide kinase activities via phosphotransfer reactions. FASEB J. 2001;15:251–260. doi: 10.1096/fj.00-0268com. [DOI] [PubMed] [Google Scholar]
- 81.Swarts HG, Koenderink JB, Willems PH, De Pont JJ. The non-gastric H,K-ATPase is oligomycin-sensitive and can function as an H+,NH4(+)-ATPase. J Biol Chem. 2005;280:33115–33122. doi: 10.1074/jbc.M504535200. [DOI] [PubMed] [Google Scholar]
- 82.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 83.Boyer PD. A research journey with ATP synthase. J Biol Chem. 2002;277:39045–39061. doi: 10.1074/jbc.X200001200. [DOI] [PubMed] [Google Scholar]
- 84.Surridge C. Nobel prizes honour biologists' work on protein energy converters. Nature. 1997;389:771. doi: 10.1038/39674. [DOI] [PubMed] [Google Scholar]