Abstract
We analyzed the natural genetic variation between Landsburg erecta (Ler) and Cape Verde Islands (Cvi) accessions by studying 105 recombinant inbred lines to search for players in the regulation of sensitivity to light signals perceived by phytochromes in etiolated seedlings of Arabidopsis. In seedlings grown under hourly pulses of far-red (FR) light, we identified three quantitative trait loci (QTLs; VLF3, VLF4, and VLF5) for hypocotyl growth inhibition and three different QTLs (VLF6, VLF7, and VLF1) for cotyledon unfolding. This indicates that different physiological outputs have selective regulation of sensitivity during de-etiolation. Ler alleles, compared with Cvi alleles, of VLF3, VLF4, VLF5, VLF7, and VLF1 enhanced, whereas the Ler allele of VLF6 reduced, the response to pulses of FR. We confirmed and narrowed down the position of some QTLs by using near-isogenic lines. VLF6 mapped close to the CRY2 (cryptochrome 2) gene. Transgenic Ler seedlings expressing the Cvi allele of CRY2 showed enhanced cotyledon unfolding under hourly pulses of FR compared with the wild type or transgenics expressing the CRY2-Ler allele. This response required phytochrome A. The cry1 cry2 double mutant lacking both cryptochromes showed reduced cotyledon unfolding under FR pulses. Because the CRY2-Cvi is a gain-of-function allele compared with CRY2-Ler, cryptochrome activity correlates positively with cotyledon unfolding under FR pulses. We conclude that the blue light photoreceptor cryptochrome 2 can modulate seedling photomorphogenesis in the absence of blue light. In addition to the nuclear loci, we identified cytoplasmic effects on seedling de-etiolation.
Seedlings grown in darkness show elongated shoot axis and rudimentary development of the foliage and of the photosynthetic machinery (Nemhauser and Chory, 2002). This etiolated body form is considered to facilitate penetration through the soil and seedling emergence. Light induces the transition from this so-called skotomorphogenic pattern to the photomorphogenic pattern of development. The seedling becomes de-etiolated as rapid axis growth ceases the leaves or cotyledons unfold, expand, and become green, whereas the cellular and biochemical components of the photosynthetic machinery are established. De-etiolation is initiated by red (R) and farred (FR) light perceived by phytochromes (Quail, 2002), and blue light perceived primarily by cryptochromes (Cashmore et al., 1999) and secondarily by phototropins (Parks and Spalding, 1999; Briggs and Christie, 2002) and even by phytochromes. These photoreceptors form an interconnected signaling network (Casal, 2000). There are five phytochromes (phyA–phyE) in Arabidopsis (Mathews and Sharrock, 1997). De-etiolation induced by R requires phyB (Reed et al., 1994) and phyA (Mazzella et al., 1997), whereas de-etiolation under FR requires phyA (Whitelam et al., 1993).
Three photobiologically discrete components of the phytochrome-mediated de-etiolation response can be distinguished in Arabidopsis and other species based on the effects of different R/FR treatments and different frequencies of excitation (Casal et al., 2003). The very low-fluence response (VLFR) is mediated by phyA and saturates with very low fluences of R or FR. Long-term effects on hypocotyl growth inhibition or cotyledon unfolding require these pulses to be repeated. The low-fluence response (LFR) of phyB is induced by R and not by FR, which is actually able to revert the effect of R. The high-irradiance response (HIR) is also mediated by phyA and requires sustained excitation with FR. HIR and VLFR can be dissected genetically and molecularly at the level of phyA domains (Casal et al., 2002; Yanovsky et al., 2002) and cis-acting regions of target gene promoters (Cerdán et al., 2000). The VLFR pathway would initiate de-etiolation when seedling emergence from the soil is impending. The LFR of phyB mediates deetiolation under the high R to FR ratios of open places, and the HIR of phyA does the same under the low R to FR ratios of dense canopies (Yanovsky et al., 1995; Smith and Whitelam, 1997). The contribution of each of these pathways differs with the species and the specific process under phytochrome control.
A source of significant genetic variation in deetiolation responses can be found among naturally occurring populations of a given species. For example, in a set of 97 accessions of Arabidopsis, the promotion of hypocotyl growth by low R to FR ratios typical of dense canopies compared with high R to FR ratios of open places ranged from 50% to 600% (Botto and Smith, 2002). Compared with other accessions, the Lm-2 has reduced HIR caused by a single amino acid change affecting the biochemical properties of phyA (Maloof et al., 2001). Recombinant inbred lines (RILs) are available and allow a detailed investigation of variation between two parental strains (Alonso-Blanco and Koornneef, 2000; Maloof, 2003). This variability provides an excellent opportunity to identify loci involved in the fine regulation of VLFR, LFR, and HIR, particularly because the contribution of each one of these signaling pathways can be estimated by simultaneous analysis of different samples of each line under various light/dark conditions. The later is not possible in a mutant screening protocol. The first genetic dissection between VLFR and HIR was obtained by using RILs between Landsberg erecta (Ler) and Columbia (Col) to identify two quantitative trait loci (QTLs; VLF1 and VLF2) controlling the VLFR of seed germination, hypcotyl growth inhibition, and cotyledon opening (Yanovsky et al., 1997). In addition, several QTLs for hypocotyl length were mapped based on the genetic variation between Ler and the Cape Verde Islands (Cvi) accessions under different light/hormone environments (Borevitz et al., 2002). Here, we uncover new loci that selectively regulate subsets of phytochrome-mediated deetiolation responses by using Cvi/Ler RILs and nearisogenic lines (NILs). The identification of one of the genes involved in QTL analysis demonstrates a previously unsuspected role for a known player in photomorphogenesis. In addition to several nuclear loci, we identified cytoplasmic effects on seedling de-etiolation.
RESULTS
Hypocotyl Growth Inhibition and Cotyledon Unfolding in Ler, Cvi, and RILs
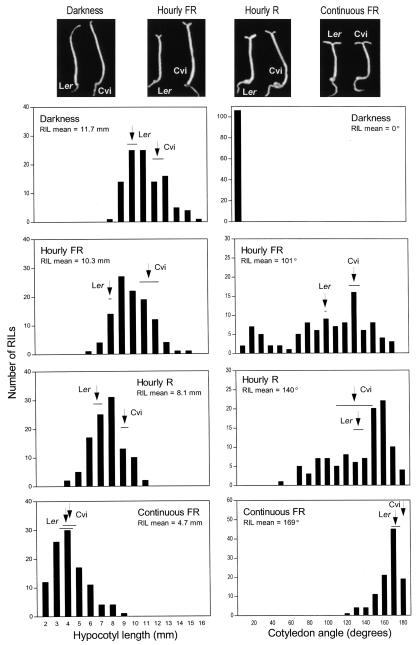
We measured hypocotyl length and the angle between cotyledons in seedlings of Ler and Cvi accessions and of 105 RILs grown in darkness or under hourly pulses of FR, hourly pulses of R, or continuous FR. In dark-grown seedlings, the hypocotyl was shorter in Ler compared with Cvi parental accessions. The difference was even larger under hourly pulses of FR and R, but it was not significant under continuous FR (Fig. 1). The cotyledons were closed in darkness. Cotyledon unfolding was enhanced in Cvi compared with Ler under hourly pulses of FR (Fig. 1). However, the differences were not significant under hourly pulses of R or continuous FR. Actually, R pulses failed to enhance cotyledon unfolding compared with FR pulses in Cvi (Fig. 1).
Figure 1.
Phenotypes of Ler, Cvi, and Cvi/Ler RILs. Histograms show the distributions of mean hypocotyl length and mean cotyledon angle in different dark/light environments. The mean (arrows) and sds (horizontal lines) of parental accessions are indicated. Photographs of representative seedlings of both parental accessions grown in different conditions are shown at the top.
The frequency distribution of RILs was very different for each physiological output. For hypocotyl length, the range of variation was large (7–8 mm compared with an average length between 11.7 mm in darkness and 4.7 mm under continuous FR) and similar for each light condition (Fig. 1). For cotyledon unfolding, the RILs showed a full-scale variation (7°–178°) under hourly pulses of FR, a narrow range under continuous FR (128°–180°) and an intermediate variation under hourly pulses of R (Fig. 1; Supplemental Table I).
The RILs showed transgressive variation in both directions compared with the parental lines for both traits in almost all conditions, with the exception of cotyledon angle in darkness (no unfolding) and under continuous FR (parental lines showed saturated response; Fig. 1). This suggests that parental lines contain a balance of different alleles at several loci with contrasting effects on a given physiological output. In addition, cotyledon angle and hypocotyl length values of the RIL population showed no obvious correlation under any light condition (Supplemental Fig. 1). This suggests that different loci are involved in the control of each physiological output, an expectation that was met by the results of QTL mapping described below.
Different QTLs Control Hypocotyl Growth and Cotyledon Unfolding Responses
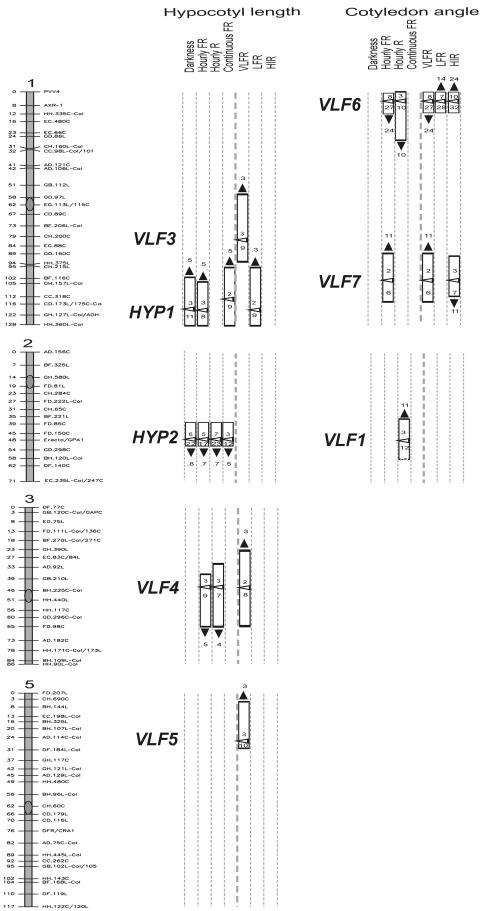
For mapping purposes, we used two complementary approaches based either on: (a) the absolute values of hypocotyl length and cotyledon angle in darkness, pulses of FR and R, and continuous FR; or (b), the differences between darkness and pulses of FR, between pulses of FR and pulses of R, and between pulses of FR and continuous FR. These differences calculate the contribution of phyA acting in the VLFR mode, phyB mediating the LFR, and phyA acting in the HIR mode, respectively (Casal et al., 1998). Conservatively, our analysis detected five QTLs controlling hypocotyl growth and three QTLs controlling cotyledon unfolding (Fig. 2; Supplemental Figs. 2–5; Supplemental Tables II and III). The total additive effects of these QTLs accounted for a maximum of 41% of the phenotypic variation of a given trait (Fig. 2; Supplemental Tables II and III). No significant two-way QTL × QTL interaction among the various nuclear QTLs identified was detected (P > 0.001).
Figure 2.
QTL mapping of hypocotyl growth and cotyledon angle for the Cvi/Ler RIL population grown under different light/dark environments. Mapping was based either on absolute values of hypocotyl length and cotyledon angle in darkness, hourly pulses of FR, hourly pulses of R, and continuous FR, or on the specific photobiological response (VLFR, LFR, and HIR). VLFR, Difference between darkness and pulses of FR. LFR, Difference between pulses of FR and pulses of R. HIR, Difference between pulsed and continuous FR. Rectangles, Two-log of the odds (LOD) support interval. The white arrowhead points to the position of the highest LOD score. The numbers above the arrowhead indicate the LOD score. The numbers below the white arrowhead indicate the percentage of explained variance for each QTL. Additive allele effects of each QTL are shown close to the 2-LOD support interval. The black arrowheads point upwards when the value of a given trait is increased by Ler compared with Cvi alleles and downwards when the value of a given trait is increased by Cvi compared with Ler alleles. Note that for hypocotyl growth, higher values of VLFR, LFR, or HIR mean shorter hypocotyls.
HYP1 and HYP2 Control Hypocotyl Growth in Darkness
Two loci mapping to chromosomes 1 and 2 were detected in darkness and almost under every light condition (Fig. 2). Based on Borevitz et al. (2002), who observed loci with pleiotropic effects in darkness and/or continuous light environments (i.e. white, blue, R, and FR) at similar positions using the Cvi/Ler RIL population, the loci were named HYP1 (chromosome 1) and HYP2 (chromosome 2).
The Ler allele of HYP1 increased hypocotyl length compared with the Cvi allele, and the opposite was true for HYP2 (Fig. 2). HYP1 was not significant for hypocotyl length under hourly pulses of R. This situation reflects the fact that the Ler allele of HYP1 increased hypocotyl length in darkness and under FR, and it also increased the inhibition of hypocotyl growth caused by R compared with FR pulses (i.e. LFR; Fig. 2). The effect on the LFR seemed rather specific because neither the VLFR nor the HIR were affected by HYP1. HYP2 mapped close to the ER (ERECTA) marker (Fig. 2), but because Ler and La(ER+) seedlings showed no differences in hypocotyl growth (mean ± se, darkness, 11.9 ± 0.4 and 12.3 ± 1.3 mm; hourly FR pulses, 8.4 ± 0.7 and 8.5 ± 0.9 mm; hourly R pulses, 6.9 ± 0.3 and 6.2 ± 0.5 mm, respectively), we conclude that HYP2 is not the ERECTA locus.
VLF3, VLF4, and VLF5 Control the VLFR of Hypocotyl Growth
Three QTLs mapping to chromosomes 1, 3, and 5 affected the VLFR of hypocotyl growth (Fig. 2). Following the terminology used by Yanovsky et al. (1997), who identified VLF1 and VLF2 in Ler/Col RILs, the loci at novel positions were named VLF3, VLF4, and VLF5, respectively. The three VLF QTLs together accounted for 26.3% of the total phenotypic variation observed in the VLFR. For all three loci, Ler alleles increased the VLFR compared with Cvi alleles (Fig. 2).
VLF4 was also detected for hypocotyl length under hourly pulses of FR or R, and this indicates that the magnitude of the effect on the VLFR was enough to modify the absolute hypocotyl length under these conditions. To confirm VLF4, we used NIL DOG6 carrying a small Cvi fragment between 40 and 65 cM in chromosome 3. NIL DOG6 showed reduced VLFR of hypocotyl growth compared with Ler (12% ± 3% and 27% ± 4% inhibition, respectively), thus confirming the presence of VLF4.
VLF5 was localized toward the top of chromosome 5 close to VLF2 (maximum LOD score at 25 and 7 cM, respectively). However, VLF2 affected primarily the VLFR of cotyledon unfolding and secondarily the VLFR of hypocotyl growth, whereas VLF5 only affected hypocotyl growth. VLF5 also mapped close to the flowering repressor FLC (flowering locus C), but flc mutants had no obvious VLFR phenotype suggesting that VLF5 and FLC are two different loci (data not shown). To confirm the position of VLF5, we used NIL 130 carrying an introgressed Cvi fragment of 30 cM at the top of the chromosome 5. As expected, NIL 130 showed reduced VLFR of hypocotyl growth compared with Ler (16% ± 2% and 27% ± 4% inhibition, respectively).
VLF1, VLF6, and VLF7 Control the VLFR of Cotyledon Unfolding
Three QTLs, VLF6 and VLF7 mapping to chromosome 1 and VLF1 mapping to chromosome 2, affected the VLFR of cotyledon unfolding (Fig. 2). The VLFR of cotyledon unfolding was enhanced by the Cvi allele of VLF6 and the Ler alleles of VLF7 and VLF1. Interestingly, the Cvi allele of VLF6 increased the VLFR but reduced the LFR compared with the Ler allele (Fig. 2). A similar feature has been observed for other loci affecting the VLFR (e.g. EVE1; Luccioni et al., 2002) and has been interpreted as the result of an early element of the VLFR signaling pathway negatively regulating the LFR of phyB (Cerdán et al., 1999). The Cvi allele of VLF7 also negatively affected the HIR, but it was not significant for the LFR (Fig. 2).
The locus VLF1 was identified in chromosome 2 with an LOD score significant under hourly pulses of R (Fig. 2) and incipient but below the threshold under hourly pulses of FR (LOD = 1.55). To confirm the presence of VLF1, we measured cotyledon unfolding in seedlings of NIL 43 carrying a 20-cM Cvi fragment around the ERECTA locus in the Ler background. The cotyledon unfolding of NIL 43 seedlings was significantly reduced under hourly pulses of FR (i.e. VLFR) compared with Ler (58° ± 8° and 99° ± 6°, P < 0.001, respectively), and the effect was not increased under hourly pulses of R (93° ± 9° and 127° ± 4°, P < 0.01, respectively). This indicates that this locus affects mainly the VLFR. In addition, since NIL 43 showed reduced VLFR of cotyledon unfolding but normal hypocotyl length compared with Ler in all light/dark conditions (e.g. darkness, 11.5 ± 1.3 and 11.6 ± 0.5 mm; hourly FR pulses, 8.4 ± 0.8 and 8.4 ± 0.7 mm, respectively). VLF1 and HYP2 appear to be different loci (i.e. NIL 43 retained the VLF1 but not the HYP2 phenotype). To further refine the position of VLF1, we analyzed the segregation of six markers in the F3 families from an F2 population of 40 individuals derived from a cross between NIL 43 and Ler. The LOD score was below the threshold for the three markers placed toward the top end of the insertion (F3P11, MSAT2.36, and PLS7), and this excludes the PHYB gene as a candidate for this locus. The LOD score was significant for F13B15, which showed no segregation with the bottom end markers MSAT241 and nga1126 (data not shown). The lines with Ler alleles for the markers F13B15 to nga1126 showed stronger cotyledon unfolding under FR pulses than the lines with Cvi alleles (85° ± 5° and 33° ± 10°; P = 0.003, respectively). Thus, VLF1 would be placed in the region between PLS7 and the bottom end of the insertion.
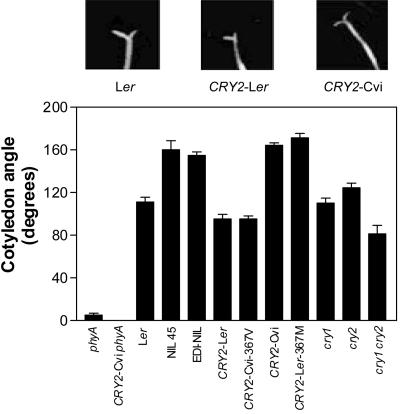
VLF6 Is the CRY2 Gene
To narrow down the position of VLF6, we used NIL 45- and EDI-NIL-containing Cvi regions introgressed into a Ler genetic background on the top of chromosome 1. Both NILs increased significantly cotyledon unfolding compared with Ler (Fig. 3), confirming the presence of VLF6 at the top of chromosome 1. Because this chromosomal region contains the CRY2 (cryptochrome 2) gene, which is known to be polymorphic between Ler and Cvi (El-Assal et al., 2001), we used transgenic seedlings in the Ler background carrying an additional copy of the CRY2 from Ler (CRY2-Ler) or from Cvi (CRY2-Cvi) to investigate whether this blue light photoreceptor gene was playing a role in the absence of blue light. Cotyledon unfolding was enhanced by the Cvi but not by the Ler transgene (Fig. 3). The natural substitution of Val (Ler) for Met (Cvi) at position 367 creates a dominant gain-of-function of CRY2 allele (El-Assal et al., 2001). Further support for a role of CRY2 in the control of the VLFR of cotyledon unfolding was provided by transgenic lines bearing as a transgene either the CRY2-Ler allele, where the Val-367 was changed to Met-367, or the CRY2-Cvi allele bearing the Met-367 to Val-367 replacement (El-Assal et al., 2001). The sole substitution of this amino acid was enough to modify the effect of the transgene on cotyledon unfolding in a manner consistent with a critical role of this residue in the whole CRY2 gene (Fig. 3). The single cry1 (cryptochrome 1) or cry2 loss-of-function mutants had no significant effects, but the cry1 cry2 double mutant showed reduced cotyledon unfolding under FR pulses (P < 0.01, Fig. 3). Because the Cvi is a gain-of-function allele compared with the Ler allele, the results of the loss-of function mutants indicate a positive correlation between cryptochrome activity and the VLFR of cotyledon unfolding. The phyA mutation was epistatic over the enhanced cotyledon unfolding under hourly FR (Fig. 3) but not on the flowering phenotype of the Cvi allele (data not shown; see also S.E.-D. El-Assal, C. Alonso-Blanco, A.J.M. Peeters, C. Wagemaker, J.L. Weller, and M. Koornneef, unpublished data).
Figure 3.
VLF6 is the CRY2 gene. Cotyledon angle in seedlings of Ler, NIL-EDI, NIL 45, transgenic lines carrying genomic alleles of CRY2 from Ler or Cvi (CRY2-Ler and CRY2-Cvi, respectively) or with the amino acid 367 replaced to Val (CRY2-Cvi-367V) or with the amino acid 367 replaced to Met (CRY2-Ler-367M), the cry1, cry2, and cry1 cry2 mutants grown under hourly pulses of FR. phyA-201 and CRY2-Cvi in the phyA-201 background are also included. Data are means ± se of three independent experiments with at least three replicate boxes each. Photographs of representative seedlings are shown at the top.
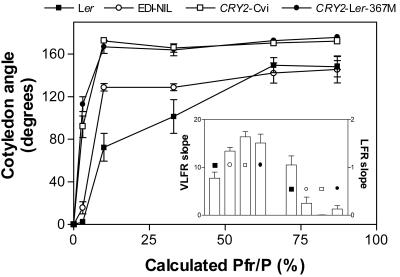
The analysis of RILs indicated that the Ler allele of VLF6 reduced the VLFR but increased the LFR of cotyledon unfolding (Fig. 2). This issue was investigated in further detail by characterizing the cotyledon unfolding response under hourly pulses of R, FR, or R-FR mixtures, providing a series of calculated proportions of phytochrome in the Pfr form (Fig. 4). Given the unexpected effects of cry2 in the absence of blue light, the first important conclusion from these experiments is that the Cvi allele of CRY2 enhances cotyledon unfolding even under hourly pulses of long-wavelength FR provided by a well-known filter (Schott RG9) that cuts off all visible light (Fig. 4, calculated Pfr/P = 3%; data not shown), indicating that the observed effects were not due to any contamination with blue light. The EDI-NIL showed enhanced VLFR but did not show LFR (Fig. 4, inset). This is a genuine negative effect on the LFR because these seedlings were well below the maximum attainable cotyledon unfolding (Fig. 4). The transgenic line overexpressing CRY2-Ler behaved like WT (calculated Pfr/P 10% = 60° ± 14° and calculated Pfr/P 87% = 139° ± 5°).
Figure 4.
The Cvi allele of CRY2 enhances the VLFR but reduces the LFR. Cotyledon angle in seedlings of Ler, NIL-EDI, CRY2-Cvi, and CRY2-Ler-367M grown under hourly pulses of FR, R, or R-FR mixtures providing a series of calculated Pfr/P (3%, 10%, 33%, 61%, and 87%). The standard FR source used in the rest of the experiments corresponds to a calculated Pfr/P = 10%. Inset, Slopes (Δ cotyledon angle/Δ Pfr/P) of the VLFR (0%–10% Pfr/P) and LFR (10%–87% Pfr/P). Data are means ± se of three independent experiments with at least three replicate boxes each.
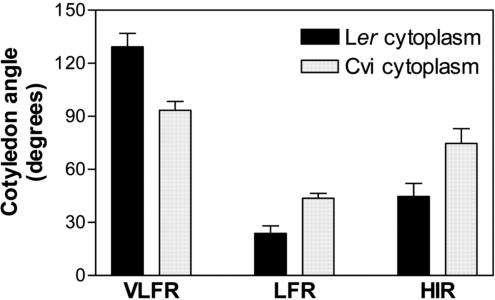
Cytoplasmic Effects on Cotyledon Unfolding
Because the RILs were obtained from reciprocal crosses (Alonso-Blanco et al., 1998a), cytoplasmic effects could be analyzed. Noteworthy, significant LOD scores were detected for VLFR and LFR (LOD = 2 and 2.6, respectively) No significant two-way interaction was found between the cytoplasm and the nuclear QTLs identified (P > 0.001). The RILs derived from crosses involving a Ler mother showed enhanced VLFR and reduced LFR when compared with the lines derived from a Cvi mother (Fig. 5).
Figure 5.
Cytoplasmic effects on cotyledon unfolding. Average VLFR, LFR, and HIR of RILs with an Ler or Cvi cytoplasm. Data are means ± se of 25 (Ler) or 81 (Cvi) RILs.
DISCUSSION
The analysis of VLFR, LFR, and HIR during deetiolation in RILs derived from the cross between Cvi and Ler has yielded three important conclusions: First, cryptochromes can control selective aspects of photomorphogenesis in the absence of blue light; second, multiple QTLs control specific features of the de-etiolation syndrome; and third, maternal extranuclear information controls some de-etiolation responses.
The existence of specific blue-light photoreceptors had been indicated first by elegant physiological experiments where the effects of blue light could not be accounted for by the action of phytochromes (Thomas and Dickinson, 1979) and by identification of the hy4 mutant of Arabidopsis affecting hypocotyl-growth responses to blue light but not to R or FR (Koornneef et al., 1980). Cloning of a T-DNA mutant allele of hy4 finally allowed the discovery of cry1 (Ahmad and Cashmore, 1993; Lin et al., 1995) followed shortly afterward by cry2, which affects deetiolation under very low fluences of blue light and the photoperiodic response of flowering (Lin et al., 1995, 1998). The Val residue at position 376 is highly conserved among cryptochrome genes, but it is substituted to Met in the Cvi allele of CRY2 (El-Assal et al., 2001). The EDI-NIL carrying CRY2-Cvi is hypersensitive to continuous blue light during deetiolation (El-Assal et al., 2001). Here, we show that an effect of cry2 on cotyledon unfolding can also be observed under hourly pulses of FR in the absence of any blue light. This is particularly obvious for the Cvi compared with the Ler allele of CRY2, but it is also detectable for the Ler compared with the cry2 mutant allele in the cry1 mutant background (Fig. 3). This observation cannot be accounted for by the absorption properties of cryptochromes measured in vitro (Lin et al., 1995; Ahmad et al., 2002). However, overexpression of the carboxy termini of cryptochromes can yield hypersensitivity to FR (Yang et al., 2000).
The phyA mutation is epistatic to the effects of CRY2-Cvi reported here (Fig. 3), indicating that cry2 is a positive regulator of phyA-mediated perception and/or signaling even in the absence of blue light. Genetic interaction between phyA and cry2 had been observed for hypocotyl growth inhibition in seedlings de-etiolating under white light, i.e. in the presence of blue light (Mazzella et al., 2001). In concordance, two detailed works studying the dynamics of the hypocotyl inhibition under blue light demonstrated that phyA fully impairs cry2 function between 30 and 120 min after the initiation of light treatment (Folta and Spalding, 2001a, 2001b).
In mammals, cryptochromes have a function as components of the clock observed even in darkness in addition to their function in the light input (van der Horst et al., 1999). In Arabidopsis, cry1 and cry2 are not key components of the clock (Devlin and Kay, 2000; Yanovsky et al., 2000a) but are involved in the light input to the clock (Yanovsky et al., 2001). In Arabidopsis, cry1 and cry2 mutants show an extended period of the circadian rhythm of expression of a light-harvesting chlorophyll-binding protein gene under R (Devlin and Kay, 2000). However, the latter effects are larger than those of the phyA mutation and can be observed in the phyA background (Devlin and Kay, 2000), indicating a fundamental difference between the effects of cryptochromes on clock period and cotyledon unfolding in the absence of blue light. El-Assal et al. (2001) have shown that CRY2-Cvi accelerates flowering under short photoperiods. The phyA mutation is not epistatic to the latter phenotype (data not shown; see also S.E.-D. El-Assal, C. Alonso-Blanco, A.J.M. Peeters, C. Wagemaker, J.L. Weller, and M. Koornneef, unpublished data), which, therefore, would be more directly mediated by CRY2-Cvi.
We have identified two novel QTLs for the VLFR of cotyledon unfolding (VLF6 = CRY2 and VLF7) and three novel QTLs for the VLFR of hypocotyl growth (VLF3, VLF4, and VLF5). A third locus controlling cotyledon unfolding is tentatively equated to the VLF1 locus also polymorphic between Ler and Col (Yanovsky el al., 1997). Ler, compared with Cvi alleles of VLF1, VLF3, VLF4, VLF5, and VLF7, increased VLFR, whereas the opposite was true for VLF6. In addition, we have confirmed and extended the role of HYP1 and HYP2 in the control of hypocotyl growth (Borevitz el al., 2002). The Ler allele of HYP1 increased hypocotyl length in darkness, pulses of FR, or continuous FR, but not under pulses of R. This is due to enhanced LFR (phyB-mediated inhibition of hypocotyl growth caused by R compared with FR pulses; Fig. 2). Some of the QTLs affecting hypocotyl growth reported previously were not detected here, and this is likely to reflect differences in growth conditions. In particular, Borevitz et al. (2002) measured hypocotyl length after 7 d of growth in agar containing one-half-strength Murashige and Skoog salts; i.e. conditions that could enhance the impact of loci controlling seed carbohydrate reserves compared with the shorter experiments used here (3 d, plain agar). Actually, Borevitz et al. (2002) noted the coincidence between one of their loci and a previously identified seed-sized locus (Alonso-Blanco et al., 1999).
VLF6 = CRY2-Cvi enhanced the VLFR and reduced the LFR (Fig. 4). This is not surprising because the phyA or fhy1 mutations that eliminate or reduce VLFR also enhance the LFR (Mazzella et al., 1997), the overexpression of PHYA (Casal and Sánchez, 1994; Casal et al., 2002), and the eve1 (Luccioni et al., 2002) and spa1 (Baumgardt et al., 2002) mutations that enhance VLFR reduce the LFR. These observations have been interpreted as negative regulation of phyB-mediated LFR by some component of the VLFR signaling cascade (Cerdán et al., 1999). Other loci as VLF1, VLF2, VLF3, VLF5, and VLF7 affect VLFR but not LFR, suggesting that they would regulate VLFR downstream the point of the VLFR pathway controlling LFR signaling. The possibility of a negative regulation of HIR signaling by VLFR signaling also has been proposed (Luccioni et al., 2002). However, although CRY2-Cvi and VLF7-Ler reduced HIR, available information does not rule out a trivial explanation based on physical constraints for further cotyledon unfolding under HIR conditions (continuous FR), when the VLFR is already enhanced.
Most photomorphogenic mutants, including those with enhanced or reduced VLFR, have both altered hypocotyl growth and altered cotyledon unfolding responses to light (Baumgardt et al., 2002; Luccioni et al., 2002). In this report, we have demonstrated the power of QTL mapping to uncover novel loci that selectively affect one of the two physiological responses. This finding suggests a scenario where plants would bear a relatively large number of QTLs with developmental specific action, providing flexibility to the de-etiolation responses.
At least between Ler and Cvi, there is a clear imbalance in the variability for the different response modes. Six QTLs affect primarily the VLFR, but only one (HYP1) shows an effect on the LFR that cannot be accounted for on the basis of a primary action on VLFR (as discussed in the previous paragraph), and none affects selectively the HIR. This is also true for the variability between Ler and Col (Yanovsky et al., 1997). This might merely reflect the genetic information of Ler, present in both crosses, or a more general tendency for genetic variability in sensitivity to weak signals compared with maximum attainable response. It will be interesting to explore this issue as new RILs become available.
Because the RILs used here are derived from reciprocal crosses between the parental lines (Alonso-Blanco et al., 1998a), it is possible to evaluate the occurrence of cytoplasmic effects. The lines derived from a cross involving a Ler, compared with Cvi mother, showed enhanced VLFR and reduced LFR and HIR (Fig. 5). At least in principle, the effects on LFR and HIR could derive from those on VLFR as described in previous paragraphs for the effects of nuclear loci. The reports of cytoplasmic effects are scant (Corey et al., 1976). A similar analysis conducted for seed traits that are affected by the genotype of the mother plant concluded against the occurrence of a cytoplasmic control for those traits (Alonso-Blanco et al., 1999). Retrograde signals emanate from the chloroplast to control nuclear gene expression (Surpin et al., 2002). Mutations at a nuclear gene coding a soluble ATP-binding cassette protein that localizes at the chloroplast periphery impair phyA-mediated de-etiolation under continuous FR (Møller et al., 2001). Therefore, it is tempting to speculate that chloroplast loci polymorphic between Ler and Cvi could modulate retrograde signaling and, hence, modulate phytochrome-mediated responses.
MATERIALS AND METHODS
Plant Material
The parental lines Ler and Cvi and a set of 105 RILs derived from reciprocal crosses between the two genotypes (Alonso-Blanco et al., 1998a) were obtained from Arabidopsis Biological Resource Center (Ohio State University, Columbus). The La(ER+), NIL 42, NIL 45, NIL-EDI (Alonso-Blanco et al., 1998b; Swarup et al., 1999), and NIL DOG6 (Alonso-Blanco et al., 2003) were described previously. NIL 43 carrying a single introgression of about 20 cM from physical positions 8.3 to 11.7 Mb of chromosome 2 from Cvi into a Ler background was kindly provide by Dr. Andrew Millar (University of Warwick, UK). NIL 130 carries a single introgression from the top of chromosome 5 of Cvi into a Ler genetic background, with a recombination breakpoint between markers AD129 and HH.480. The transgenic lines carrying CRY2-Ler or CRY2-Cvi transgenes or CRY2 transgenes mutated in a single amino acid at position 367 in the Ler background were described before (El-Assal et al., 2001) and kindly provide by Dr. Maarten Koornneef (University of Wageningen, The Netherlands). The phyA-201 allele in Ler (Reed et al., 1994) was used to produce CRY2-Cvi transgenic plants in the phyA background. F3 families from an F2 population of 40 individuals derived from a cross between NIL 43 and Ler were genotyped for six markers covering regularly the Cvi region introgressed in the NIL 43. The microsatellite markers MSAT2.36, MSAT2.41 (Loudet et al., 2002) and nga1126 (Bell and Ecker, 1994) and the INDELs F3P11, PLS7, and F13B15 (AtDB) were used.
Growth Conditions, Light Treatments, and Physiological Measurements
Twenty seeds were sown in clear plastic boxes (40- × 33-mm2 × 15-mm height) containing 3 mL of 0.8% (w/v) agar. The boxes were incubated at 4°C in darkness for 3 d and exposed to an R pulse followed by 24 h of darkness to promote seed germination. The seedlings were either kept in the dark or exposed to various light treatment for 3 d. Hypocotyl length was measured to the nearest 0.5 mm with a ruler, and, to eliminate defective seedlings, only the largest 10 seedlings of each box were averaged. The angle between the cotyledons was measured with a protractor.
Hourly pulses of R (3 min, 20 μmol m-2 s-1), hourly pulses of FR (3 min, 50 μmol m-2 s-1), continuous FR (2.5 μmol m-2 s-1), and the mixtures of R-FR (3 min, at least 11 μmol m-2 s-1) used to obtain intermediate calculated proportions of phytochrome in the Pfr form were provided by the sources described earlier (Yanovsky et al., 2000b). Long-wavelength FR (3 min, 40 μmol m-2 s-1) was provided by incandescent lamps in combination with a water filter and an RG9 filter (Schott, Maintz, Germany).
Analysis of QTLs and Cytoplasmic Effects
Each trait was analyzed separately by using the mean values from three independent experiments. All the lines were included in parallel in each experiment. Each experiment included 10 seedlings per RIL. Hypocotyl length data were normally distributed. Cotyledon angle data were transformed (arcsin √) to improve the normality of distributions, but none of the conclusions was changed when using the original data. Therefore, results are presented in figures with the original scale. A set of 99 markers covering nearly all of the Arabidopsis genetic map at average intervals of 5 cM was selected from the Cvi/Ler RIL map (Alonso-Blanco et al., 1998a). The computer software MapQTL version 4.0 (van Ooijen, 2000) was used to identify and locate QTLs on the linkage map by using interval mapping and multiple-QTL model mapping methods as described previously (Alonso-Blanco et al., 2003). LOD threshold values applied to declare the presence of a QTL were estimated with the permutation tests implemented in MapQTL version 4. The quantitative trait data of the RILs were permuted 1,000 times over the genotypes and empirical LOD thresholds corresponding to a genome wide significance α = 0.05 were estimated between 2.5 and 2.7 for the various data sets. Two-LOD support intervals were established as ≈95% QTL confidence intervals. The estimated additive genetic effect and the percentage of variance explained by each QTL and the total variance explained by all the QTLs affecting a trait were obtained with MapQTL in the final multiple-QTL model in which one cofactor marker was fixed per QTL. Additive genetic effects presented correspond to the differences between the estimated means of the two homozygous RIL genotypic groups, at each particular QTL, divided into two. Two-way interactions among the QTLs identified were tested by ANOVA using the corresponding two markers as factors. In addition, two-way interactions were searched for among all pair-wise combinations of the 99 nuclear markers and the cytoplasmic genotype, using the computer program EPISTAT (Chase et al., 1997) with log-likelihood ratio threshold corresponding to a significance P < 0.001. Because 81 of the RILs carry Ler cytoplasm and 25 carry Cvi cytoplasm, cytoplasmic genetics effects were analyzed in the RIL population using the cytoplasmic genotype as a factor in one-way ANOVA and in multiple factor linear models in combination with the nuclear QTL markers affecting each trait.
Supplementary Material
Acknowledgments
We thank Ana María Rodríguez for her technical assistance. We also thank Maarten Koornneef, Salah E.-D. El-Assal, and Leonie Bentsink for kindly providing the sees of CRY2 transgenic lines and NIL DOG6, Andrew Millar for the seeds of NIL 43, and the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the RILs.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.029546.
This work was supported by the University of Buenos Aires (grant no. G406 to J.F.B.), by the Agencia Nacional de Promoción Científica y Tecnológica (grant nos. BID 1201/OC–AR and PICT 06739 to J.J.C.), and by the Spanish Ministerio de Ciencia y Tecnología (salary contract “Ramón y Cajal” to C.A.-B.).
The online version of this article contains Web-only data.
References
- Ahmad M, Cashmore AR (1993) HY4 gene of Arabidopsis thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162-166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Grancher N, Heil M, Black RC, Giovani B, Galland P, Lardemer D (2002) Action spectrum for cryptochrome-dependent hypocotyl growth inhibition in Arabidopsis. Plant Physiol 129: 1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-deVries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711-729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Blankestijn-deVries H, Hanhart CJ, Koornneef M (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710-4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE-D, Coupland G, Koornneef M (1998b) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde islands ecotypes of Arabidopsis thaliana. Genetics 149: 749-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22-29 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van der Bosch N, Pot J, Kuiper MTR (1998a) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259-271 [DOI] [PubMed] [Google Scholar]
- Baumgardt RL, Oliverio KA, Casal JJ, Hoecker U (2002) SPA1, a component of phytochrome A signal transduction, regulates the light signaling current. Planta 21: 745-753 [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137-144 [DOI] [PubMed] [Google Scholar]
- Botto JF, Smith H (2002) Differential genetic variation in adaptative strategies to a common environmental signal in Arabidopsis accessions: phytochrome-mediated shade avoidance. Plant Cell Environ 25: 53-64 [Google Scholar]
- Borevitz JO, Maloof JN, Lutes J, Dabi T, Redfern JL, Trainer GT, Werner JD, Asami T, Berry CC, Weigel D, Chory J et al. (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204-210 [DOI] [PubMed] [Google Scholar]
- Casal JJ (2000) Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol 71: 1-11 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Davis SJ, Kirchenbauer D, Viczian A, Yanovsky MJ et al (2002) The serine-rich N-terminal domain of oat phytochrome A helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol 129: 1127-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Luccioni LG, Oliverio KA, Boccalandro HE (2003) Light, phytochrome signalling and photomorphogenesis in Arabidopsis. Photochem Photobiol Sci 2: 625-636 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Sánchez RA (1994) Overexpression of oat phytochrome A gene differentially affects stem growth responses to red/far-red ratio signals characteristic of sparse or dense canopies. Plant Cell Environ 17: 407-419 [Google Scholar]
- Casal JJ, Sánchez RA, Botto JF (1998) Modes of action of phytochromes. J Exp Bot 49: 127-138 [Google Scholar]
- Cashmore AR, Jarillo JA, Wu Y-J, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760-765 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Staneloni RJ, Ortega J, Bunge MM, Rodríguez-Batiller J, Sánchez RA, Casal JJ (2000) Sustained but not transient phytochrome A signaling targets a region of a Lhcb1*2 promoter that is not necessary for phytochrome B action. Plant Cell 12: 1203-1211 [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Yanovsky MJ, Reymundo FC, Nagatani A, Staneloni RJ, Whitelam GC, Casal JJ (1999) Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J 18: 499-507 [DOI] [PubMed] [Google Scholar]
- Chase K, Adler FR, Lerk KG (1997) EPISTAT: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theor Appl Genet 242: 81-89 [Google Scholar]
- Corey LA, Matzinger DF, Cockerham CC (1976) Maternal and reciprocal effects on seedling characters in Arabidopsis thaliana (L.) HEYNH. Genetics 82: 677-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499-2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal SE-D, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of. CRY2 Nat Genet 29: 435-439 [DOI] [PubMed] [Google Scholar]
- Folta KM, Spalding EP (2001a) Opposing roles of phytochrome A and phytochrome B in early cryptochrome-mediated growth inhibition. Plant J 28: 333-340 [DOI] [PubMed] [Google Scholar]
- Folta KM, Spalding EP (2001b) Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J 26: 471-478 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147-160 [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968-970 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 93: 15491-15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Goubely C, Depeiges A, Picard G (2002) Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173-1184 [DOI] [PubMed] [Google Scholar]
- Luccioni LG, Oliverio KA, Yanovsky MJ, Boccalandro H, Casal JJ (2002) Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol 128: 173-181 [PMC free article] [PubMed] [Google Scholar]
- Maloof JN (2003) QTL for plant growth and morphology. Curr Opin Plant Biol 6: 85-90 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Babi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC et al. (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441-446 [DOI] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20: 666-671 [Google Scholar]
- Mazzella MA, Alconada Magliano TM, Casal JJ (1997) Dual effect of phytochrome A on hypocotyl growth under continuous red light. Plant Cell Environ 20: 261-267 [Google Scholar]
- Mazzella MA, Cerdán PD, Staneloni RJ, Casal JJ (2001) Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development 128: 2291-2299 [DOI] [PubMed] [Google Scholar]
- Møller SG, Kunkel T, Chua NH (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15: 90-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J, Chory J (2002) In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0054, http://www.aspb.org/publications/arabidopsis
- Parks BM, Spalding EP (1999) Sequential and coordinated action of phytochrome A and B during Arabidopsis stem growth revealed by kinetics analysis. Proc Natl Acad Sci USA 96: 14142-14146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signaling networks. Nat Rev 3: 85-93 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Whitelam GC (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20: 840-844 [Google Scholar]
- Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell 14: S327-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar A (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67-77 [DOI] [PubMed] [Google Scholar]
- Thomas B, Dickinson HG (1979) Evidence for two photoreceptors controlling growth in de-etiolated seedlings. Planta 146: 545-550 [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, Dewit J, Verkerk A, Eker APM, Van Leenen D et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 627-630 [DOI] [PubMed] [Google Scholar]
- van Ooijen (2000) MapQTL (r) Version 4.0: User Friendly Power in QTL Mapping: Addendum to the Manual of Version 3.0. Plant Research International, Wageningen, The Netherlands.
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757-768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815-827 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Luppi JP (1997) The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J 12: 101-109 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Whitelam GC (1995) Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ 18: 788-794 [Google Scholar]
- Yanovsky MJ, Luppi JP, Kirchbauer D, Ogorodnikova OB, Sineshchekov VA, Adam E, Kircher S, Staneloni RJ, Schäfer E, Nagy F, Casal JJ (2002) Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell 14: 1591-1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazzella MA, Casal JJ (2000a) A quadruple photoreceptor mutant still keeps track of time. Curr Biol 10: 1013-1015 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazella MA, Whitelam GC, Casal JJ (2001) Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J Biol Rhythms 16: 523-530 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Whitelam GC, Casal JJ (2000b) fhy3-1 retains inductive responses of phytochrome A. Plant Physiol 123: 235-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.