The cell nucleus is a highly compartmentalized organelle harboring a variety of dynamic membraneless nuclear bodies1–4. How these subnuclear domains are established and maintained is not well understood5–8. Here we investigated the molecular mechanism of how one nuclear body, the paraspeckle, is assembled and organized. Paraspeckles are discrete ribonucleoprotein bodies found in mammalian cells and implicated in nuclear retention of hyperedited mRNAs9–11. We developed a live-cell imaging system that allows for the inducible transcription of Men ε/β (also known as Neat112) noncoding (nc) RNAs and the direct visualization of the recruitment of paraspeckle proteins. Using this system, we demonstrate that Men ε/β ncRNAs are essential to initiate the de novo assembly of paraspeckles. These newly formed structures effectively harbor nuclear retained mRNAs confirming that they are bona fide functional paraspeckles. By three independent approaches, we show that it is the act of Men ε/β transcription, but not ncRNAs alone, that regulates paraspeckle maintenance. Finally, FRAP analyses supported a critical structural role of Men ε/β ncRNAs in paraspeckle organization. Together, this study establishes a model in which Men ε/β ncRNAs serve as a platform to recruit proteins to assemble paraspeckles.
Two distinct models of nuclear body formation have been proposed: (1) random self-organization and (2) ordered assembly pathway6–8. The random self-organization model posits that the recruitment of individual subunits from the soluble nucleoplasmic pool is self-organizing in a random and stochastic fashion. This has been supported by tethering experiments of Cajal body components to a specific genomic locus in living cells. Essentially, immobilization of any given key Cajal body protein was able to initiate the nucleation of a Cajal body13.
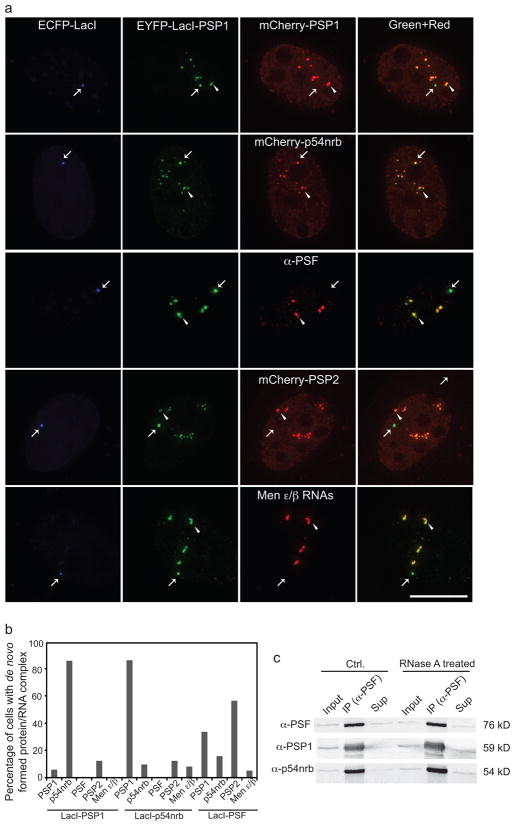
To access the possibility that such a model applies to paraspeckles, which are known to contain Men ε/β ncRNAs, core paraspeckle proteins PSP1, p54nrb, and PSF, and other proteins such as PSP214, we individually tethered multiple paraspeckle proteins fused with EYFP-Lac repressor (LacI) to a 256 repeat Lac operator (LacO) array stably integrated into the genome of C2C12 myoblast cells. These paraspeckle proteins were detected in the endogenous paraspeckles shown by either transient expression of multiple paraspeckle proteins or antibody labeling of an endogenous paraspeckle marker (Fig. 1a, arrowhead), indicating that they were correctly targeted. To distinguish the newly tethered foci from the endogenous paraspeckles, ECFP-LacI was coexpressed (Fig. 1a, arrow). Upon tethering of EYFP-LacI-PSP1, mCherry-p54nrb was recruited to the tethering site, but mCherry-PSP1, PSP2, endogenous PSF, or Men ε/β failed to be recruited (Fig. 1a).
Figure 1. Immobilization of protein components fails to assemble paraspeckles.
a, Tethering PSP1 protein to a specific locus (arrow) in the C2C12 nucleus recruited p54nrb, but not Men ε/β ncRNAs, PSP1, PSF, or PSP2 protein therefore failing to form paraspeckles. Scale bar, 5 μm. b, Quantification of protein tethering experiments shows that none of the immobilized paraspeckle proteins can initiate the formation of paraspeckles (n>50 cells in each condition). c, Co-IP experiments show that paraspeckle proteins can interact with each other in the absence of RNA.
Immobilization of PSP1 resulted in the recruitment of p54nrb in 87% of the cells and vice versa (Fig. 1b). Our results agree with previous studies indicating that PSP1 and p54nrb can form heterodimers in vivo and even in vitro15. However, the other essential components of paraspeckles were not recruited to the immobilized foci. Therefore, the efficient recruitment of one paraspeckle protein component is not an indication of bona fide paraspeckle formation, but rather due to direct protein:protein interaction. Quantification shows that immobilized PSP1 and p54nrb were unable to initiate paraspeckle formation (Fig. 1b). Tethered PSF was capable of recruiting some paraspeckle proteins (PSP1 in 35%, p54nrb in 17%, and PSP2 in 58% of the cells examined) in a relatively more efficient manner. However, PSF recruited Men ε/β ncRNAs in only 6% of the cells (Fig. 1b).
To address the question of how PSF recruits paraspeckle proteins but not RNAs, we performed co-immunoprecipitation (IP) experiments from C2C12 cells. Transiently expressed PSF was immunoprecipitated by anti-PSF antibody and both PSP1 and p54nrb were detected in the IP by immunoblotting (Fig. 1c). We then performed the co-IP experiment after RNase A treatment to abrogate paraspeckles10,15, and found that PSF still immunoprecipitated PSP1 and p54nrb (Fig. 1c), confirming that LacI-PSF did not initiate paraspeckle formation but rather induced protein:protein interactions. These results further suggested that the artificial scaffold created by protein tethering may not faithfully reflect bona fide formation of a nuclear body in vivo6.
None of the paraspeckle proteins examined was able to efficiently initiate paraspeckle assembly arguing that the random self-organization model does not seem to apply to paraspeckles. The alternative model is the hierarchical assembly pathway, in which single, some or all subunits are assembled in an ordered fashion around a central scaffolding component6–8. Men ε/β are nuclear ncRNAs that have been shown by us and other groups to play important roles in paraspeckle organization16–19. The depletion of Men ε/β ncRNAs in cells disrupts paraspeckles implicating that they act as structural RNAs in paraspeckle formation16–19. However, whether Men ε/β ncRNAs are the seeding molecules during paraspeckle assembly has not been directly demonstrated and if so, how this process occurs in vivo has not been investigated.
In order to characterize the de novo assembly of paraspeckles, we developed a live-cell imaging system that enables the control and visualization of the transcription of Men ε/β reporter ncRNAs with the capacity to directly observe the recruitment of mCherry-paraspeckle proteins (Fig. 2a). The reporter, modified from our previous studies20–22 and integrated at a single site in the C2C12 genome, is visualized by expression and binding of ECFP-LacI at the LacO array. Upon doxycycline (DOX) induction, a minimal CMV promoter drives expression of Men ε/β ncRNAs tagged with 24 MS2 RNA stem loop repeats23,24. Nascent Men ε/β transcripts are visualized by accumulation of EYFP-tagged MS2 viral coat protein, which binds the MS2 repeats within the transcripts. Using this system the Men ε/β ncRNAs transcription site, nascent Men ε/β ncRNAs transcripts, and protein recruitment can be simultaneously visualized in single living cells.
Figure 2. Men ε/β transcription induces de novo formation of functional paraspeckles.
a, Schematic representation of an inducible system to visualize expression of the Men ε/β reporter ncRNAs and recruitment of paraspeckle proteins. A LacO array, a tetracycline response element (TRE) array, and 24 MS2 stem loop repeats are placed upstream of the Men ε/β gene allowing visualization of the locus by expression of ECFP-LacI. Transcription is initiated by the addition of DOX and nascent transcripts are visualized through binding of EYFP-MS2 to the MS2 stem loops. Not drawn to scale. b, Live-cell imaging shows that transcriptional induction of Men ε/β ncRNAs initiated de novo formation of paraspeckles labeled by PSP1 (arrow). Scale bars, 5 μm. c, De novo formed paraspeckles (arrow) can retain inverted repeat-containing mRNAs, similar to the endogenous paraspeckles (arrowhead), confirming that they are bona fide functional paraspeckles. d, Quantification shows that Men ε/β transcription initiated the recruitment of different paraspeckle proteins, but not SC35 and SF2/ASF, nuclear speckle RNA binding proteins; and resulted in the retention of different inverted repeat containing mRNAs, but not mCat2 mRNA (n>50 cells in each condition).
DOX-induced transcription in cells transfected with Tet-On led to a strong local accumulation of nascent transcripts (EYFP-MS2), whereas little to no transcript was detected in cells without DOX induction (Fig. 2b). Consistently, quantitative RT-PCR (qRT-PCR) analysis revealed that Men ε/β ncRNAs were expressed at approximately one fold over the endogenous level and Men ε ncRNA was expressed at approximately 8-fold over Men β ncRNA, which is similar to their endogenous ratio without induction (Supplementary Information, Fig. S1). These data suggest that the reporter generates physiological levels of Men ε/β ncRNAs after DOX induction.
We next tested the hypothesis that transcription of Men ε/β ncRNAs directly initiates the de novo assembly of paraspeckles by monitoring paraspeckle protein dynamics using live-cell imaging. We found that upon DOX induction, transcription of Men ε/β ncRNAs was sufficient to trigger the recruitment of mCherry-PSP1 to the newly formed paraspeckles (Fig. 2b and Supplementary Information, Movies 1–3). Quantification showed that more than 95% of the newly formed bodies recruited all four paraspeckle proteins examined, including transiently expressed or endogenous PSP1, p54nrb, PSF, and PSP2 (Fig. 2d and Supplementary Information, Figs. S2, S3 and Movies 4, 5), but not SC35 and SF2/ASF, two major RNA binding proteins enriched in nuclear speckles and not found in endogenous paraspeckles (Fig. 2d and Supplementary Information, Fig. S4). Thus, Men ε/β ncRNAs are the initial nucleating factor providing a seed to recruit other building blocks during paraspeckle assembly.
To ascertain whether de novo formed paraspeckles were functional, we cotransfected cells with constructs encoding mRNAs containing SINE repeats to test if the induced paraspeckles could retain these mRNAs as has been shown for endogenous paraspckles9. The 3′UTR of mouse cationic amino acid transporter 2 (mCat2) transcribed nuclear RNA (Ctn RNA) contains repeat elements essential for adenosine-to-inosine editing and has been shown to be retained in nuclear paraspeckles10,11. Indeed, by RNA FISH we found that newly formed paraspeckles (Fig. 2c, upper row arrow) efficiently retained Ctn RNA as did endogenous paraspeckles (Fig. 2c, upper row arrowhead). In contrast, the structures induced by tethering individual paraspeckle proteins did not retain Ctn RNA (Supplementary Information, Fig. S5). We also tested constructs encoding reporter mRNAs containing inverted repeats of human Alu (IRAlus) sequences9,16 and found that the newly assembled paraspeckles were capable of retaining these RNAs as well (Fig. 2c, lower row). Quantification revealed that all hyperedited mRNAs tested, but not mCat2 mRNA, which does not contain repeats and is not enriched in nuclei10, were harbored in de novo assembled paraspeckles, confirming that these induced paraspeckles were functional (Fig. 2d). This functional study and subsequent kinetic studies strongly indicate that bona fide paraspeckles can be de novo assembled in living cells upon Men ε/β ncRNAs transcription using our inducible system.
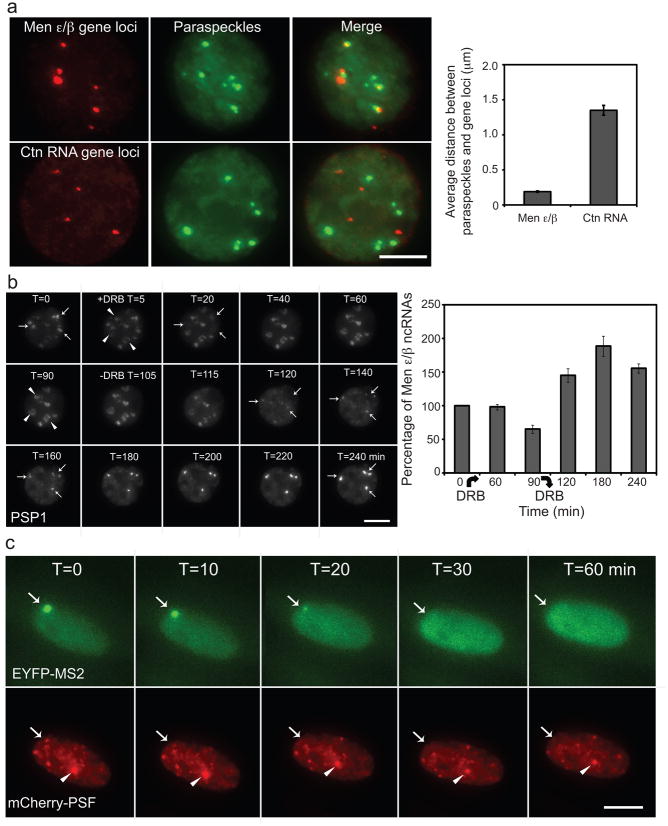
All new paraspeckles were assembled co-transcriptionally at the Men ε/β ncRNAs transcription sites and rarely left the sites after they were formed (Fig. 2b and Supplementary Information, Movies 1–5). We then examined endogenous paraspeckles to see if they were also localized close to the endogenous Men ε/β gene loci by DNA FISH. We found that paraspeckles were generally localized near the endogenous Men ε/β gene loci as shown previously17 and sometimes a cluster of paraspeckles could be found around one Men ε/β gene locus (Fig. 3a). Quantification indicated that the average distance between paraspeckles and the closest adjacent Men ε/β gene loci is 0.19 μm, while the distance to Ctn RNA gene loci, which serve as a negative control, is 1.35 μm (n=100 paraspeckles) (Fig. 3a). Similar results were also obtained in NIH 3T3 cells (Supplementary Information, Fig. S6). Together, these data support a model whose central premise is the requirement of transcription of Men ε/β ncRNAs to initiate paraspeckle formation.
Figure 3. Maintenance of paraspeckles depends on active Men ε/β transcription.
a, Left, paraspeckles, labeled by PSP1 antibody, were often found localized adjacent to Men ε/β gene loci, but not to Ctn RNA gene loci, in interphase nuclei of C2C12 cells. Scale bars, 5 μm. Right, quantification of the distance between paraspeckles and Men ε/β gene and Ctn RNA gene loci (n=100 paraspeckles in 20 cells each, mean±s.e.m.). b, Left, paraspeckle protein PSP1 redistributed to perinucleolar caps (arrowhead) upon transcriptional inhibition by DRB treatment and paraspeckles reassembled at the same location (arrow) after DRB wash-out. Right, qRT-PCR analysis shows Men ε/β transcript level upon DRB treatment (n=4, mean±s.e.m.). Note that 60-min DRB treatment did not affect the Men ε/β transcript level, but paraspeckles already disassembled at this time point. c, Withdrawing DOX to switch off Men ε/β transcription disassembled de novo formed paraspeckle (arrow) but had no effect on endogenous paraspeckles (arrowhead) demonstrating that the maintenance of paraspeckles is coupled with Men ε/β transcription.
To test this possibility, we monitored the dynamics of paraspeckles after treating cells with 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), a reversible RNA Pol II inhibitor. Consistent with previous findings15,17–19, paraspeckle protein PSP1 began to relocate to the perinucleolar caps (Fig. 3b, arrowhead) from paraspeckles at 5 minutes after DRB treatment and paraspeckles were completely disassembled within 40 minutes. Remarkably, after washing away DRB to allow cells to recover from transcription inhibition paraspeckles were usually reformed at the sites where they were disassembled (Fig. 3b, arrow and Supplementary Information, Movie 6). DRB-induced paraspeckle disassembly was previously explained to result from the decreased RNA level of Men ε/β transcripts by transcription inhibition15,25. By qRT-PCR analysis, we found this was not the case because within 60 minutes of DRB treatment, there was no change in transcript level of Men ε/β RNAs (Fig. 3b, right). Therefore, we speculate that the transcription of Men ε/β RNAs per se instead of Men ε/β transcripts is required for paraspeckle maintenance.
Our live-cell system provides a means to control the transcription of Men ε/β reporter RNAs without affecting other important transcriptional events in the cells, and thereby allowing us to characterize the transcription requirement of paraspeckle organization. When we withdrew DOX to shut down the transcription of Men ε/β reporter RNAs, we found that Men ε/β RNAs were no longer concentrated at the transcription sites and newly formed paraspeckles were disassembled accordingly, while endogenous paraspeckles were not affected (Fig. 3c and Supplementary Information, Movie 7). This clearly demonstrated that paraspeckle maintenance is coupled with Men ε/β transcription.
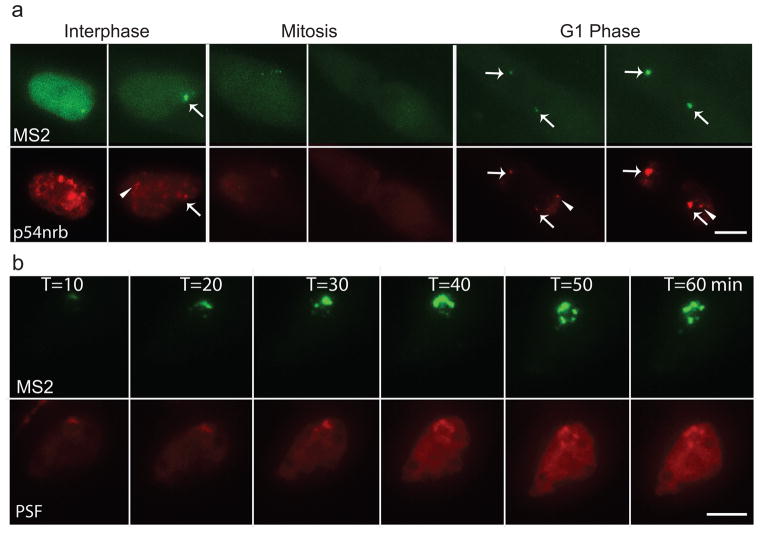
We then tested if the arrest of Men ε/β reporter gene transcription during mitosis would also result in the disassembly of paraspeckles. Indeed, we found that the reporter paraspeckles disassembled during mitosis and reassembled in the next G1 phase (Fig. 4a, arrow) like the endogenous paraspeckles (Fig. 4a, arrowhead and Supplementary Information, Movie 8). Imaging of the essential processes of paraspeckle dynamics, including their initiation, disassembly, and reassembly, by three different approaches (± DRB, ± DOX, and entry and exit from mitosis), demonstrated that paraspeckle formation/maintenance is dependent on the active transcription of Men ε/β ncRNAs. Interestingly, the nucleolus does form in the absence of RNA Pol I transcription. However, its structural organization is altered26.
Figure 4. Dynamic behavior of paraspeckles by live-cell imaging.
a, Dynamics of paraspeckles through the cell cycle: both endogenous (arrowhead) and de novo formed paraspeckles (arrow) disassembled during mitosis, and re-assembled at the Men ε/β loci in the daughter nuclei during next G1 phase. Scale bars, 5 μm. b, Budding and splitting of paraspeckles during interphase: a paraspeckle assembled upon DOX induction at the Men ε/β locus, increased in size and formed clusters of paraspeckles.
We further characterized the dynamic behavior of paraspeckles during interphase and we were able to capture the budding or splitting of paraspeckles during interphase in about 15% of the cells. We found that when the transcription of Men ε/β RNAs continues, more Men ε/β transcripts were generated and de novo assembled paraspeckles increased in size accordingly and eventually bud or split into a cluster of paraspeckles around the transcription sites (Fig. 4b and Supplementary Information, Movies 9–11). The average distance between these paraspeckles in the cluster to the Men ε/β reporter loci (0.22 μm, n=50 paraspeckles) is consistent to that of endogenous paraspeckles. This explains why cells sometimes have more paraspeckles in number than Men ε/β gene loci and why they are often found in clusters17–19.
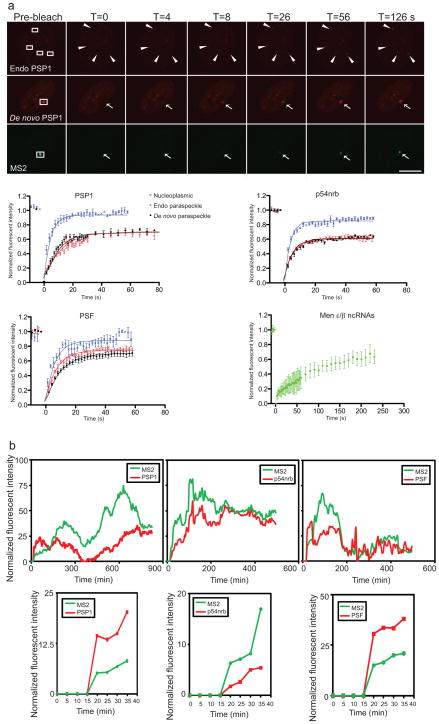
To determine the kinetics of paraspeckle components, we performed fluorescence recovery after photobleaching (FRAP) analyses on newly formed and endogenous paraspeckles. All three core proteins in de novo formed and in endogenous paraspeckles exhibited similar recovery profiles (Fig. 5a and Supplementary Information, Table S1). Approximately 30–40% of paraspeckle proteins are immobile in de novo formed and endogenous paraspeckles. The t1/2 of PSP1 measured here (t1/2 = 6.42 s) is smaller than that shown by photoactivated GFP-PSP1 in HeLa cells (t1/2 = 31 s)15. The methodological differences may contribute to this discrepancy. The rapid exchange rate detected here was corroborated by a FRAP analysis of CFIm (t1/2 ≈ 5–10 s), another paraspeckle component27. Nevertheless, the similar dissociation kinetics of paraspeckle proteins in de novo formed as in endogenous paraspeckles further confirmed that the newly induced structures in our system exhibit the same protein kinetics as endogenous paraspeckles.
Figure 5. Differential kinetics of Men ε/β ncRNAs and paraspeckle proteins.
a, FRAP analyses of paraspeckles. Paraspeckle proteins exhibit a rapid exchange rate, while Men ε/β ncRNAs exhibit a much slower exchange. Top, cells expressing mCherry-PSP1 and EYFP-MS2 labeling Men ε/β ncRNAs were imaged before and after bleaching of endogenous (arrowhead) or de novo formed (arrow) paraspeckles. Scale bar, 5 μm. Bottom, kinetics of recovery of paraspeckle proteins after bleaching in nucleoplasm (blue cross), endogenous (open red circle) or de novo formed paraspeckles (solid black circle), and of Men ε/β ncRNAs (square) (n=8 for PSP1 and p54nrb, n=5 for PSF, and n=12 for Men ε/β ncRNAs, mean±s.e.m.). Intensity was normalized as such that the first time point pre-bleach was set as 1 and the first time point post-bleach was set as 0. Note that the scale of the X-axis of Men ε/β ncRNAs recovery profile is different from those of paraspeckle proteins. b, Protein dynamics during paraspeckle formation. Top, the intensity of EYFP-MS2 and mCherry-fused paraspeckle proteins at the transcription site was quantified and normalized over time. Bottom, kinetics of first 40 minutes after DOX induction is shown. Within the temporal resolution (5 min), all paraspeckle proteins examined were found to be recruited to newly formed paraspeckles instantly upon the detection of Men ε/β transcription.
Conversely, the recovery of Men ε/β ncRNAs was significantly delayed compared to those of paraspeckle proteins (Fig. 5a). Their t1/2 could not be accurately measured because Men ε/β ncRNAs recovery never reached a plateau, but a reasonable estimation suggests a t1/2 > 60–90 s. The markedly long residence time of Men ε/β ncRNAs in paraspeckles is consistent with mechanistic insights of Men ε/β ncRNAs being the structural seeding molecules to nucleate and coordinate protein recruitment to accomplish paraspeckle assembly and maintenance.
The rapid exchange of paraspeckle proteins with the nucleoplasmic pool is consistent with the model that proteins are able to diffuse freely through the nuclear space allowing them to scan the nucleus for specific binding sites. Indeed, all three paraspeckle core proteins in the nucleoplasm showed very fast recovery and a small immobile fraction (Fig. 5a and Supplementary Information, Table S1). When they encounter Men ε/β ncRNAs at the transcription sites, proteins associate and assemble paraspeckles. This process could be accomplished by either stochastic interactions of single subunits or small preformed subcomplexes6–8. Previous studies and our data revealed that some paraspeckles proteins associate with each other in a RNA-independent manner15,28. In fact, all three paraspeckle proteins examined (PSP1, p54nrb, and PSF) showed similar kinetics during de novo paraspeckle assembly upon DOX induction (Fig. 5b). Although individual cells transcribe Men ε/β ncRNAs with different kinetics due to different transcription initiation rates21,29, protein recruitment always closely followed Men ε/β transcription (Fig. 5b, top row). More importantly, at least within our temporal resolution (5 min interval of image acquisition), paraspeckle proteins were immediately recruited to the Men ε/β transcription sites once the nascent transcripts were synthesized (Fig. 5b, bottom row). Thus, neither a solely stochastic nor hierarchical model could explain paraspeckle formation. Rather, our results are consistent with a seeding model in which Men ε/β ncRNAs serve as seeding molecules to provide platforms to recruit proteins, which self-organize randomly and stochastically around the Men ε/β ncRNAs, to assemble paraspeckles at the Men ε/β gene loci.
In conclusion, we developed a live-cell imaging system to allow direct visualization of the de novo assembly of a nuclear body via the transcription of ncRNAs. We provided evidence to demonstrate that de novo assembled paraspeckles are bona fide functional nuclear bodies that harbor nuclear-retained mRNAs. Using this system, we demonstrated that paraspeckle formation and maintenance are coupled to Men ε/β transcription. Our study has provided mechanistic insights into paraspeckle assembly via the transcription of Men ε/β ncRNAs and how proteins are kinetically coordinated in this process. These data established a seeding model for nuclear body formation in which Men ε/β ncRNAs were identified as pivotal nucleating molecules, driven by transcription, to recruit proteins to assemble paraspeckles.
Methods
Cell culture and drug treatments
C2C12 myoblasts were grown at 37 °C, 5% CO2 in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. C2C12 cells were transfected with pCC1BAC-LacO-MS2-Men ε/β and selected under 600 ng/ml Hygromycin B and colonies were picked, selected, and expanded. RNA Pol II was inhibited by DRB (33 μg/ml; Sigma). DOX (1 μg/ml; Clonetech) was used to induce the transcription of Men ε/β reporter ncRNAs.
Immunofluorescence labeling, RNA and DNA FISH, and qRT-PCR
Immunofluorescence staining, RNA and DNA-FISH, and qRT-PCR were performed as previously described10,19. Primary antibodies used were anti-PSF (FC23), anti-PSP1 (Ab2), and anti-p54nrb (9-99)19. BAC RP23-20N24 and RP24-164P20 served as Men ε/β gene locus probes and RP23-406L11 served as a Ctn RNA gene locus probe for DNA FISH.
Co-Immunoprecipitation
C2C12 cell lysate was prepared in RIPA buffer with or without RNase A (100 μg/ml, Sigma). Following lysate centrifugation, the supernatant was incubated with antibody and protein A beads. IP was subsequently analyzed by SDS-PAGE and immunoblotting.
Image acquisition and data analysis
Images were obtained using a DeltaVision system (Applied Precision) with a 100×/1.35 NA objective (Olympus) as stacks of 12 images taken with a Z step size of 1 μm. For intensity analyses, cells were randomly chosen and the average intensity of region of interest on unprocessed images was measured. For each cell, a separate region within the nucleus was measured for background subtraction. For distance measurement in 3D, ImageJ software was used.
Fluorescent recovery after photobleaching
Photobleaching and subsequent image acquisition (4 precleached images, and a sequence of post-bleach images at 30 images every 2 seconds and 36 images every 5 seconds) were performed on a spinning disk confocal microscope UltraViewVox (Perkin Elmer). Data were fitted in a one phase exponential association curve and t1/2 was measured by Volocity software (Perkin Elmer).
Supplementary Material
Acknowledgments
We thank Javier Caceres, Gordon G. Carmichael, Ling-ling Chen, Yasuyuki Kurihara, and Angus I. Lamond for reagents, Stephen Hearn and Zsolt Lazar for assistance in microscopy, Carmen Berasain, Megan Bodnar, Melanie Eckersley-Maslin, Michael Huebner, Ileng R. Kumaran, Jingjing Li, Eduardo Reis, Jeremy E. Wilusz, and Rui Zhao of the Spector laboratory for discussions and comments. Y.S.M. is supported by a National Cancer Center Postdoctoral Fellowship. B.Z. is supported by a Department of Defense Prostate Cancer Research Program Postdoctoral Fellowship. This work was supported by grants to D.L.S. from NIH (NIGMS 42694 and 5PO1CA013106-38).
Footnotes
Author Contributions
Y.S.M., H.S., and D.L.S. designed research; Y.S.M., H.S., and B.Z. performed experiments and analyzed data; and Y.S.M. and D.L.S. wrote the paper.
References
- 1.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. TrendsCell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 3.Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 4.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 9.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 12.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 14.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukamoto T, et al. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 23.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 24.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki YT, Hirose T. How to build a paraspeckle. Genome Biol. 2009;10:227. doi: 10.1186/gb-2009-10-7-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dousset T, et al. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardinale S, et al. Subnuclear localization and dynamics of the Pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol Biol Cell. 2007;18:1282–1292. doi: 10.1091/mbc.E06-09-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang WW, Zhang LX, Busch RK, Farres J, Busch H. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem J. 1993;290:267–272. doi: 10.1042/bj2900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shav-Tal Y, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.