Abstract
DNA damage is a well-known initiator of tumorigenesis. Studies have shown that most cancer cells rely on aerobic glycolysis for their bioenergetics. We sought to identify a molecular link between genomic mutations and metabolic alterations in neoplastic transformation. We took advantage of the intrinsic genomic instability arising in xeroderma pigmentosum C (XPC). The XPC protein plays a key role in recognizing DNA damage in nucleotide excision repair, and patients with XPC deficiency have increased incidence of skin cancer and other malignancies. In cultured human keratinocytes, we showed that lentivirus-mediated knockdown of XPC reduced mitochondrial oxidative phosphorylation and increased glycolysis, recapitulating cancer cell metabolism. Accumulation of unrepaired DNA following XPC silencing increased DNA-dependent protein kinase activity, which subsequently activated AKT1 and NADPH oxidase-1 (NOX1), resulting in ROS production and accumulation of specific deletions in mitochondrial DNA (mtDNA) over time. Subcutaneous injection of XPC-deficient keratinocytes into immunodeficient mice led to squamous cell carcinoma formation, demonstrating the tumorigenic potential of transduced cells. Conversely, simultaneous knockdown of either NOX1 or AKT1 blocked the neoplastic transformation induced by XPC silencing. Our results demonstrate that genomic instability resulting from XPC silencing results in activation of AKT1 and subsequently NOX1 to induce ROS generation, mtDNA deletions, and neoplastic transformation in human keratinocytes.
Introduction
Early studies of the metabolic changes that accompany the development of cancer led Otto Warburg to propose that a respiratory deficiency might drive neoplastic transformation (1), prompting many investigators to analyze the metabolism of tumor cells. These analyses revealed that a large number of cancer cell lines have a higher rate of glycolysis, an increased rate of glucose transport, increased pentose phosphate pathway (PPP) activity, decreased numbers of mitochondria, and a reduction in mitochondrial oxidative phosphorylation (OXPHOS) proteins and activities (2–4). These alterations in cancer cell energy metabolism could be related to somatic mutations in mitochondrial DNA (mtDNA); oxidative stress as a result of increased ROS level; adaptation to tissue hypoxia (4–7); the activation of oncogenes and/or inactivation of tumor suppressors (TP53, HIF-1A, c-MYC); as well as deregulation of PI3K/AKT, which influences the glycolytic flux through regulation of different factors (8).
However, the etiologic relationship among genomic mutations, the Warburg effect, and increased ROS levels in tumor induction remains unclear (3, 5, 9). Furthermore, there is no clear mechanism(s) linking genomic mutations and modified cellular bioenergetics. To understand the relationships between these factors, we speculated that cells with heightened predisposition to malignant transformation, or cells with the capacity to accumulate mutations, could be helpful in elucidating this mechanism. To this end, xeroderma pigmentosum (XP) cells are a useful tool since they manifest varying defects in nucleotide excision repair (NER), rendering them more susceptible to neoplastic transformation and cancer induction (10).
NER is the major pathway for repairing numerous types of DNA damage including helix-distorting lesions produced mostly by UV radiation and bulky lesions created by carcinogenic chemicals (11). The pivotal role of NER is demonstrated in 3 dermatological diseases that manifest skin photosensitivity — namely, XP, Cockayne syndrome, and trichothiodystrophy (11). Patients with XP exhibit extreme sensitivity to solar UV light and have a disproportionately high incidence of skin cancers (11, 12). XP patients are also shown to have a high incidence of developing several types of internal malignancies before the age of 20 (13). There are multiple subtypes of XP that have been defined on the basis of complementation groups (XPA-XPG). Among XP patients, XPC patients have proficient transcription-coupled repair and defective global genome repair. Therefore, in these patients, the cells accumulate mutations in nontranscribed parts of the genome, leading to neoplastic transformation (11). Notably, XPC-knockout mice have heightened predisposition to many types of UV-induced and spontaneous cancers (14, 15), indicating that XPC plays a role in the removal of non–UV-related mutations as well. Moreover, lymphocytes from Xpc–/– mice accumulate spontaneous lesions in the hypoxanthine guanine phosphoribosyl transferase (Hprt) gene (16). Interestingly, the most frequent mutations observed in older mice (i.e., >1 year of age) are G to T transversions, which also result from oxidative processes (16), suggesting a causative role for ROS. Here, we used XPC-deficient cells as a model system for assessing how the accumulation of DNA damage alters cellular energy metabolism in the context of cancer induction.
Results
XPC downregulation results in mitochondrial bioenergetics deficiency.
To determine whether DNA repair deficiency has an effect on cell bioenergetics, endogenous XPC protein expression was first inhibited using lentivirus-mediated expression of shRNA against XPC (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI40087DS1). To rule out “off target” effects of shXPC, all experiments were performed with 2 distinct shRNAs against XPC (Supplemental Figure 1). Both shXPCs stably inhibited more than 92% of XPC expression in keratinocytes (Supplemental Figure 1). Since both shRNAs had similar effects, hereafter only the results of shXPC1 have been shown.
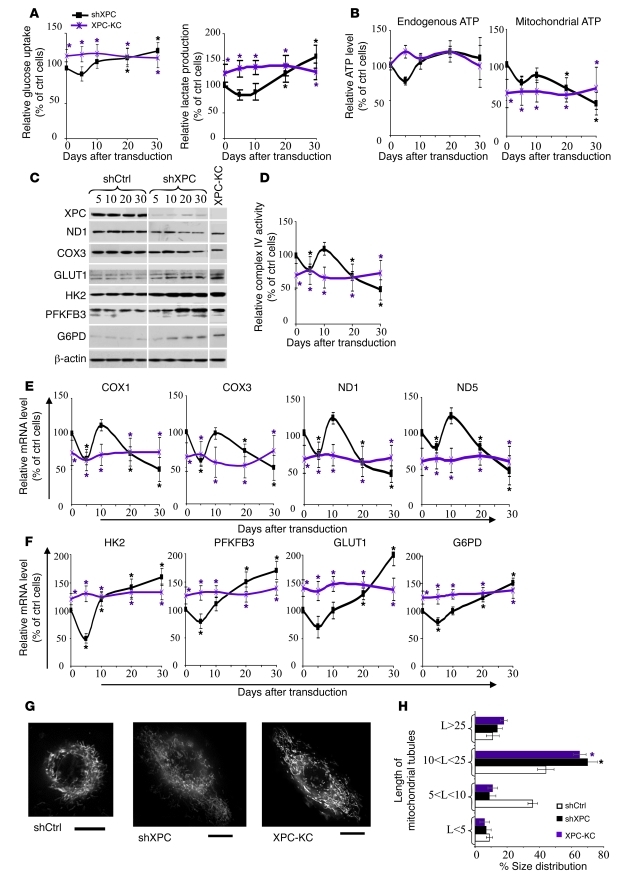
Measurement of glucose consumption and lactate production in shCtrl- and shXPC-transduced keratinocytes on different days after transduction revealed that XPCKD cells exhibited lactic acidosis at 20 and 30 days after transduction (Figure 1A), suggesting a deficiency in the mitochondrial metabolism of pyruvate through the Krebs cycle and OXPHOS system. Consumption of glucose and the production of lactate were higher in keratinocytes taken from XPC patients (XPC-KC) than in control keratinocytes over time (Figure 1A). ATP levels were similar in shCtrl- and shXPC-transduced keratinocytes on different days after transduction (Figure 1B), although a nonsignificant decrease in ATP production occurred 5 days after shXPC transduction. To assess whether XPCKD cells and XPC-KC are more dependent on glycolysis for ATP generation, the ATP level was measured in the presence of iodoacetate, an inhibitor of glycolysis. ATP levels were decreased to a greater extent in the iodoacetate-treated XPCKD cells as compared with control cells on days 20 and 30 after transduction (Figure 1B). Similarly, XPC-KC produced less mitochondrial ATP compared with control keratinocytes (Figure 1B).
Figure 1. Effects of XPC downregulation on mitochondrial metabolism and glycolysis.
Glucose consumption and lactate production (A) as well as the total endogenous ATP levels and ATP levels produced by mitochondria (B) were measured at the indicated time points after transduction. The glucose uptake, lactate production, and the total endogenous and mitochondrial ATP levels by shCtrl-transduced cells were set to 100% at each time point. The results were then compared with the shCtrl and are expressed as the average percentage of shCtrl ± SD of 3 independent experiments. *P < 0.05 (black) for shXPC-transduced cells versus shCtrl-transduced cells; *P < 0.05 (purple) for XPC-KC versus control keratinocytes at the indicated time points. (C) The protein expression levels of XPC, ND1, COX3, GLUT1, HK-2, PFKPB-3, and G6PD were determined by Western blot at the indicated days after transduction. β-actin was used as a loading control. (D) The relative activity of complex IV of the mitochondrial respiratory chain was assessed. The mRNA levels of COX1 and COX3, ND1 and ND5 (E), HK-2 and PFKFB3, and GLUT1 and G6PD (F) were quantified by qRT-PCR. The results are shown as the average percentage of control ± SD of 3 independent experiments. (G) The mitochondrial network morphology in XPC-KC, shCtrl-, and shXPC-transduced keratinocytes was determined by microscopy using MitoTracker. Scale bars: 10 μM. (H) Length of mitochondrial tubules was measured in 50 cells (25 mitochondrial tubules per cell) per condition. Results are expressed as average percentage of mitochondrial tubule size distribution ± SD of 3 independent experiments. *P < 0.05 (black) for shXPC cells versus shCtrl-transduced cells. shCtrl, keratinocytes transduced with control shRNA; shXPC, keratinocytes transduced with shXPC; XPC-KC, keratinocytes isolated from XPC patients.
The mitochondrial OXPHOS pathway includes the respiratory chain, which consists of about 85 individual subunits segregated into 5 main complexes (I to V). Among the mitochondrial protein subunits, 13 polypeptides are encoded by mtDNA (17). To determine whether OXPHOS subunits are affected by XPC downregulation, we assessed the levels of 2 mtDNA-encoded proteins: NADH dehydrogenase subunit 1 (ND1, from complex I) and cytochrome c oxidase subunit III (COX3, from complex IV). The levels of ND1 and COX3 decreased from around 20 days after XPC silencing (Figure 1C). Complex IV activity also diminished following XPC downregulation (Figure 1D). Both COX1 and COX3 mRNA expression decreased, correlating with the reduced activity of complex IV around 20 days after XPC downregulation (Figure 1E). XPCKD cells also showed decreased expression of ND1 and ND5 mRNA compared with shCtrl-transduced cells at approximately 20 days after transduction (Figure 1E).
The above results indicate that XPCKD cells generate most of their ATP through glycolysis. To further examine this hypothesis, we measured the mRNA and protein levels of 4 enzymes that are involved in the regulation of the glycolytic flux. The hexokinase (HK) isoenzymes catalyze the first step of glucose metabolism following glucose uptake into cells. Among the 4 HK subtypes, HK-2 is the most highly expressed in tumors (18) and the XPCKD cells displayed increased expression of HK-2 mRNA (Figure 1F) and protein (Figure 1C). The bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB) catalyzes the phosphorylation of fructose-6-phosphate to fructose-2,6-bisphosphate. The PFKFB3 isozyme has the highest kinase/phosphatase activity ratio of all the PFKFB isoforms and is overexpressed in many types of neoplastic cells (19). Our data showed that XPCKD cells manifest elevated PFKFB3 as compared with control cells (Figure 1, C and F). The enhanced rate of glycolysis in tumors is usually accompanied not only by a marked increase in HK activity, but also by an upregulation of GLUT proteins that facilitate its cellular uptake (3). Therefore, we quantified GLUT1 mRNA and protein levels and showed that they were increased after XPC silencing (Figure 1, C and F). Cancer cells are known to have increased PPP activity, which is the only known pathway for the direct synthesis of ribose from glucose and the main pathway for NADPH synthesis. Glucose-6-phosphate dehydrogenase (G6PD), the first enzyme of this pathway, is known to be upregulated in cancer cells (20), and it was increased in XPCKD cells (Figure 1, C and F). Consistently, XPC-KC also showed a reduction in the expression of mitochondrial OXPHOS subunits and increased expression of HK-2, PFKFB3, GLUT-1, and G6PD (Figure 1, C–F).
Mitochondrial morphology is critically linked to bioenergetics, and changes in mitochondrial energy state cause structural alterations of the mitochondrial network (21). To determine whether XPC downregulation alters mitochondrial network morphology, cells were stained with a MitoTracker probe. Normal keratinocytes demonstrated an interconnected network located mostly around the nucleus, while XPCKD cells and XPC-KC exhibited a larger tubular mitochondrial network more dispersed throughout the cytoplasm (Figure 1, G and H). Moreover, XPCKD cells were substantially larger than control cells (Figure 1G and Supplemental Figure 2).
Taken together, our results demonstrate that XPC silencing in normal human keratinocytes leads to altered metabolism, which resembles the metabolic signature of most cancer cells.
XPC downregulation elevates intracellular ROS levels and results in mtDNA oxidation through the activation of NADPH oxidase.
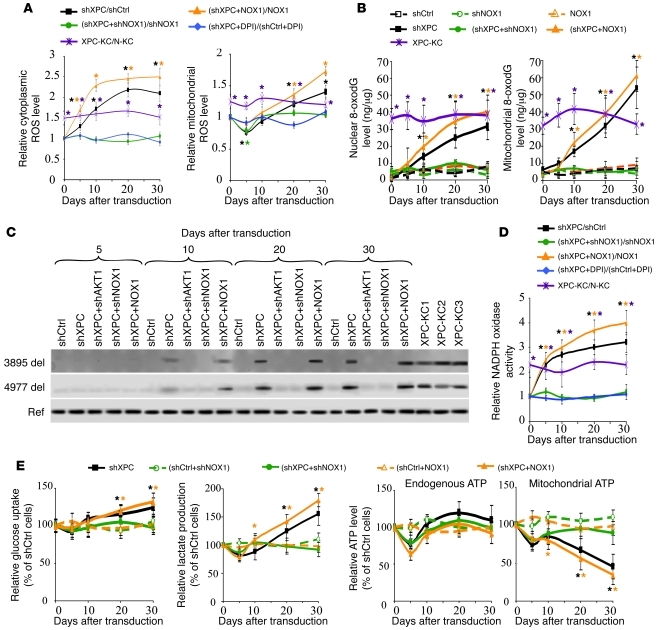
The mitochondrial respiratory chain is the major source of ROS generation in cells, and altered OXPHOS activity is associated with variations in ROS levels (7, 9). To evaluate ROS following XPC knockdown, we measured the cytoplasmic and mitochondrial steady-state levels of ROS using CM-H2DCF-DA and MitoSOX probes, respectively (Figure 2A and Supplemental Figure 3). A significant increase in cytoplasmic ROS levels occurred in XPCKD cells over time as compared with control cells. In contrast, the mitochondrial ROS showed a moderate but significant (P < 0.01, n = 3) decrease 5 days after XPC downregulation, followed by a gradual increase that peaked 30 days after transduction. Consistently, cytoplasmic and mitochondrial ROS levels were higher in XPC-KC than normal keratinocytes (Figure 2A).
Figure 2. XPC silencing–induced ROS production leads to mtDNA deletions.
(A) ROS levels in different cells were measured by flow cytometry using cytoplasmic- and mitochondrial-specific probes at the indicated days after transduction. The ROS level in the shCtrl-transduced cells was arbitrarily set to 1. Results are then assessed as shown at the top of the panels and expressed as the mean ± SD of 3 independent experiments. (B) Genomic and mtDNA oxidation were assessed by quantification of 8-oxodG levels in nuclear genome and mtDNA of different cells. Results are expressed as ng of 8-oxodG per μg DNA. (C) mtDNA was extracted and subjected to PCR, amplifying either reference fragments (Ref) representing total mitochondrial genome or 2 known deletions (3895 del and 4977 del). (D) NADPH oxidase activity was assessed as shown at the top of the panels and expressed as the mean ± SD of 3 independent experiments. (E) Glucose consumption and lactate production as well as the total endogenous ATP levels and ATP levels produced by mitochondria were measured in the different transduced cells. *P < 0.05 for different cells versus shCtrl-transduced cells at the indicated time points.
Since ROS can cause oxidative DNA damage, we determined the level of genomic and mtDNA oxidation using an antibody directed against 7,8-dihydro-8-oxoguanine (8-oxodG) (Figure 2B). 8-oxodG increased in the nuclear genome and mtDNA after XPC silencing in a time-dependent manner that was accompanied by increased cytoplasmic and mitochondrial ROS levels (Figure 2B), indicating that XPC silencing results in increased oxidative stress in both the nuclear and mitochondrial compartments.
Since increased oxidative stress is associated with mtDNA deletions and subsequent mitochondrial dysfunction (22, 23), we next assessed whether ROS generated in XPCKD cells led to mtDNA deletions and OXPHOS deficiency. To this end, we initially looked for 2 characteristic mtDNA deletions: (a) the 3895-bp deletion, which is known to occur more frequently in sun-exposed skin and is present in nonmelanoma skin cancer (24), and (b) the 4977-bp deletion, which is known to be involved in neurodegenerative diseases and the processes of aging and photoaging (22, 23). As shown in Figure 2C, XPCKD cells accumulated both types of mtDNA deletions in a time-dependent manner.
We further investigated the role of ROS in mtDNA deletions using XPCKD cells overexpressing the antioxidant enzymes catalase, CuZnSOD, or MnSOD, as well as treating these cells with diphenylene iodonium (DPI) (2.5 μM), which is an inhibitor of cytoplasmic NADPH oxidase (Supplemental Figures 1 and 3). Overexpression of catalase, CuZnSOD, and MnSOD significantly decreased steady-state levels of endogenous ROS but had no effect on XPC downregulation-induced enhancement of ROS generation. In contrast, DPI treatment eliminated the effects of XPC silencing on cytoplasmic and mitochondrial ROS alterations over time (Figure 2A and Supplemental Figure 3), suggesting that cytoplasmic NADPH oxidase is crucial for the metabolic alterations observed in XPCKD cells. NADPH oxidase consists of 6 heterosubunits that, when activated, associate to form an active enzyme complex generating O2– from oxygen using NADPH as an electron donor (25). The catalytic subunit of this complex, NOX, has 7 homologous members, of which NADPH oxidase-1 (NOX1) and NOX2 are expressed in keratinocytes (26, 27). NOX proteins are involved in host defense, posttranslational processing of proteins, cellular signaling, regulation of gene expression, and cellular differentiation (25). We further characterized the role of NADPH oxidase in ROS production in XPCKD cells by overexpressing or downregulating NOX1 and NOX2 (Supplemental Figure 1). Our results indicated that NOX2 has little or no involvement in ROS generation in XPCKD cells (data not shown). In contrast, NOX1 downregulation completely blocked both cytoplasmic and mitochondrial ROS generation in XPCKD cells (Figure 2A and Supplemental Figure 3). The ROS level was higher in the XPCKD cells that overexpressed NOX1 than in normal cells overexpressing NOX1 (Figure 2A and Supplemental Figure 3). These results suggest that ROS generation associated with XPC downregulation might be dependent on NADPH oxidase activation. To assess this, NADPH oxidase activity was measured. Results indicated that XPC silencing resulted in increased NADPH oxidase activity and that NOX1 downregulation or DPI treatment of XPCKD cells completely abrogated the modification in NADPH oxidase activity (Figure 2D). Overexpression of NOX1 enhanced NADPH oxidase activity in XPCKD cells, indicating a synergistic effect between XPC deficiency and NOX1 overexpression (Figure 2D). Our data further indicated that downregulation of NOX1 blocked the oxidative effects of XPC silencing on genomic and mtDNA (Figure 2B) and inhibited mtDNA deletions (Figure 2C). In contrast, NOX1 overexpression in XPCKD cells increased the nuclear and mitochondrial levels of 8-oxodG (Figure 2B) and the degree of the 2 mtDNA deletions (Figure 2C). Interestingly, NOX1 overexpression alone had no effect on DNA oxidation (Figure 2B) and mtDNA deletions (data not shown).
We next determined whether variations in glycolysis and mitochondrial metabolism following XPC downregulation are dependent on upregulation of NADPH oxidase. We found that downregulation of NOX1 in XPCKD cells restored glucose consumption and lactate production (Figure 2E); mitochondrial ATP production (Figure 2E); expression levels of ND1, ND5, COX1, and COX3; and expression levels of HK-2, PFKFB3, GLUT1, and G6PD to normal (Supplemental Figure 4, A–C). In contrast, NOX1 overexpression in XPCKD cells increased the rate of metabolic alterations (Figure 2E and Supplemental Figure 4, A–C). Taken together, our results demonstrate that NADPH oxidase activation plays an essential role in the XPC-silencing–induced metabolism remodeling.
XPC deficiency increases the proliferative capacity of normal keratinocytes.
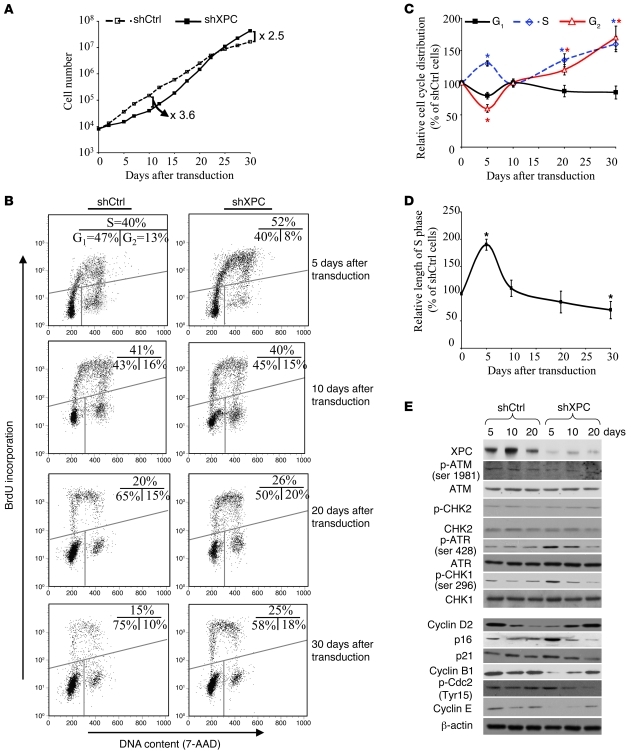
To examine the effects of altered metabolism on the proliferative capacity of XPCKD cells, we performed serial cell counts on different days following XPC downregulation. An initial period of growth retardation was followed by increased proliferation of XPCKD cells as compared with control cells (Figure 3A). Cell-cycle analysis indicated cells in S phase increased within 5 days of XPC downregulation. The number of S phase cells then declined over the next few days, and a second persistent increase occurred 15 days after transduction (Figure 3, B and C). Measurement of DNA synthesis time demonstrated a prolongation of S phase in XPCKD cells 5 days after transduction that was followed by a gradual decrease that peaked around day 30 (Figure 3D). Western blotting revealed that XPC knockdown resulted in immediate ataxia telangiectasia–mutated and Rad3-related protein–mediated (ATR-mediated) Chk1 phosphorylation, which then declined over time (Figure 3E). XPC downregulation also resulted in an initial reduction of cyclin B1, D2, and E protein expression 5 days after transduction. Thereafter, the levels of these cell-cycle proteins increased over time (Figure 3E). Concomitantly, the expression of inhibitors of cell-cycle progression, namely p16, p21WAF1, and p-Cdc2, initially increased shortly after XPC downregulation and then declined in a time-dependent manner (Figure 3E). Taken together, these results indicate that XPCKD cells acquire increased proliferative capacity shortly after ATR-mediated cell-cycle delay.
Figure 3. XPCKD cells display increased proliferative capacity associated with an increased fraction of S phase cells and decreased length of S phase following an initial stalled phase.
(A) The proliferation capacities of shCtrl- or shXPC-transduced keratinocytes were measured by serial cell counts on different days after transduction. (B) The distribution of cells in the G1, S, and G2 phases was measured at the indicated time intervals after transduction using 7-AAD and BrdU staining. (C) Graphic representation of the distribution of cells in the G1, S, and G2 phases. The percentage of cells in each phase for shCtl-transduced keratinocytes was considered to be 100%. The results were then compared with the shCtrl and are expressed as the mean ± SD of 3 independent experiments. (D) DNA synthesis time was measured by a BrdU pulse assay. Results are shown as the average percentages of shCtrl ± SD of 3 independent experiments. *P < 0.05 for shXPC-transduced cells versus shCtrl-transduced cells at the indicated time points. (E) Total protein extracts were assessed for cell-cycle regulators by Western blot analysis. β-actin was used as a loading control.
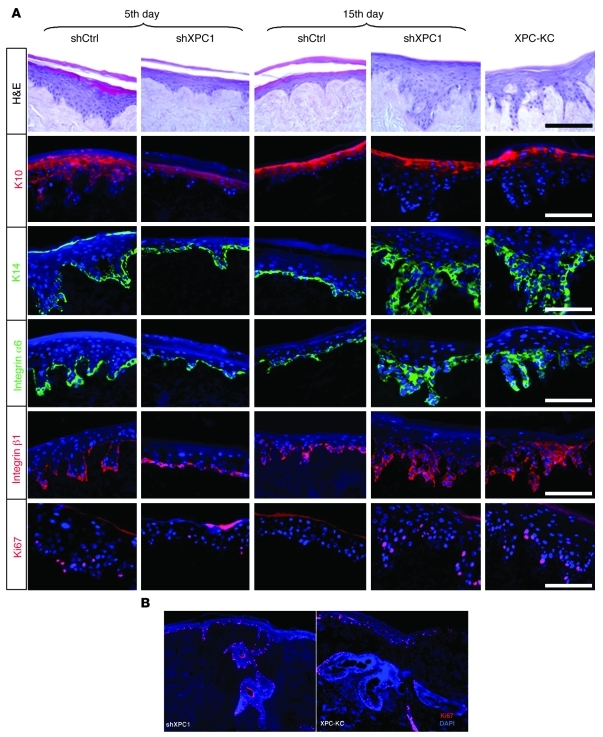
To assess the chronic effect of XPC downregulation on keratinocyte proliferation and differentiation, XPCKD cells were seeded either 5 or 15 days after transduction on dead dermis. Epidermis reconstructed with XPCKD cells at 5 days after transduction showed a marked reduction in thickness and also in the number of Ki67-positive cells, compared with control cells (Figure 4A). In contrast, epidermal thickness, the number of Ki67-positive cells, and levels of basal cell layer markers (keratin 14, α6, and β1 integrin) were substantially higher in epidermis reconstructed with XPCKD cells 15 days after transduction compared with control constructs (Figure 4A). It is important to emphasize that the epidermis reconstructed with XPC-KC manifested the same characteristics as the epidermis reconstructed with XPCKD cells 15 days after transduction, i.e., higher proliferation, increased levels of keratin 14, α6, and β1 integrin, and limited expression of keratin 10 to the upper epidermis (Figure 4A). Furthermore, both epidermis reconstructed with XPCKD cells at 15 days after transduction and XPC-KC manifested large and frequent extension of rete pegs (Figure 4, A and B) compared with control epidermis. Altogether, these results show that knockdown of XPC results in epithelial hyperplasia.
Figure 4. Knockdown of XPC results in epithelial hyperplasia.
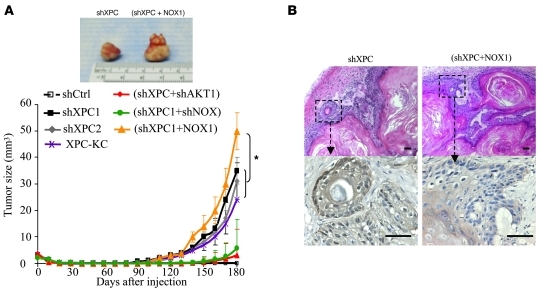
Epidermis reconstructed with XPC-KC, shCtrl-, or shXPC-transduced keratinocytes on days 5 and 15 after transduction. (A) Architectures of epidermis were evaluated with H&E staining. Proliferation and differentiation status of epidermis were assessed using immunofluorescence staining of K10, K14, integrin α6, integrin β1, and Ki67. The nuclei were marked in blue with DAPI. (B) High proliferation detected with Ki67 staining in a large extension of rete pegs in epidermis reconstructed with XPCKD cells and XPC-KC. Scale bars: 200 μm.
NOX1 downregulation inhibits neoplastic transformation of XPCKD cells.
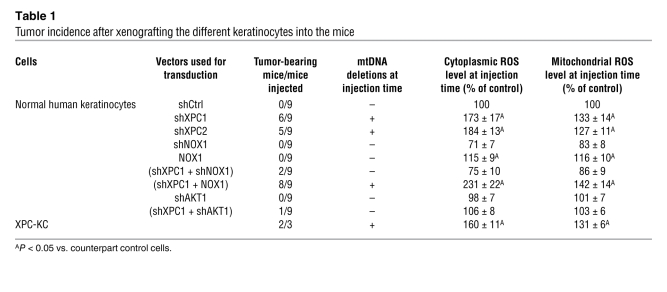
To determine whether XPCKD cells have tumorigenic potency, cells were injected subcutaneously into immunodeficient NOD/SCID mice 15 days after transduction. Around 17 weeks after xenografting, well-differentiated squamous cell carcinomas (SCCs) developed in 11 of the 18 injected mice (6 of 9 mice xenografted with shXPC1-transduced keratinocytes and 5 of 11 mice xenografted with shXPC2-transduced keratinocytes) and grew over time (Table 1 and Figure 5). XPC-KC, when injected subcutaneously into NOD/SCID mice, also developed differentiated SCCs around 13 weeks after xenografting (Table 1 and Figure 5A).
Table 1 .
Tumor incidence after xenografting the different keratinocytes into the mice
Figure 5. XPC silencing drives neoplastic transformation of human keratinocytes via NOX1.
Keratinocytes were transduced with indicated lentivirus. On the 15th day after transduction, cells were injected subcutaneously into NOD/SCID mice. (A) The in vivo tumor growth rate of XPCKD cells injected in mice is enhanced by overexpression of NOX1. Results are presented as mean ± SD. (B) H&E staining (upper panels) shows that tumors are well-differentiated SCCs with keratin pearl formation. Expression of GFP in tumors was confirmed using immunostaining with an anti-GFP antibody (lower panels). Scale bar: 100 μm.
To investigate the role of NADPH oxidase activity on tumorigenic potency of XPCKD cells, the double-knockdown cells (XPCKD/NOX1KD) and XPCKD cells overexpressing NOX1 were injected into mice. While XPCKD/NOX1KD cells failed to form tumors (2 of 9 injected mice; Table 1), the XPCKD cells overexpressing NOX1 formed tumors (8 of 9 injected mice; Table 1 and Figure 5) of larger size (1.6-fold) compared with the tumors formed by XPCKD cells alone (Figure 5A), suggesting that ROS level may be an essential factor for tumor initiation.
To further investigate the role of ROS in XPC-silencing induced tumorigenic transformation of keratinocytes, we looked at the p53 status in tumors developed by xenografted XPCKD cells. Results showed that the majority of tumors (7 of 11) had G to T transversion mutations in p53 (our unpublished observations), suggesting that XPC silencing initially causes ROS generation, which then leads to mutations in oncogenes and tumor suppressors.
AKT downregulation inhibits NOX1 activation and neoplastic transformation of XPCKD cells.
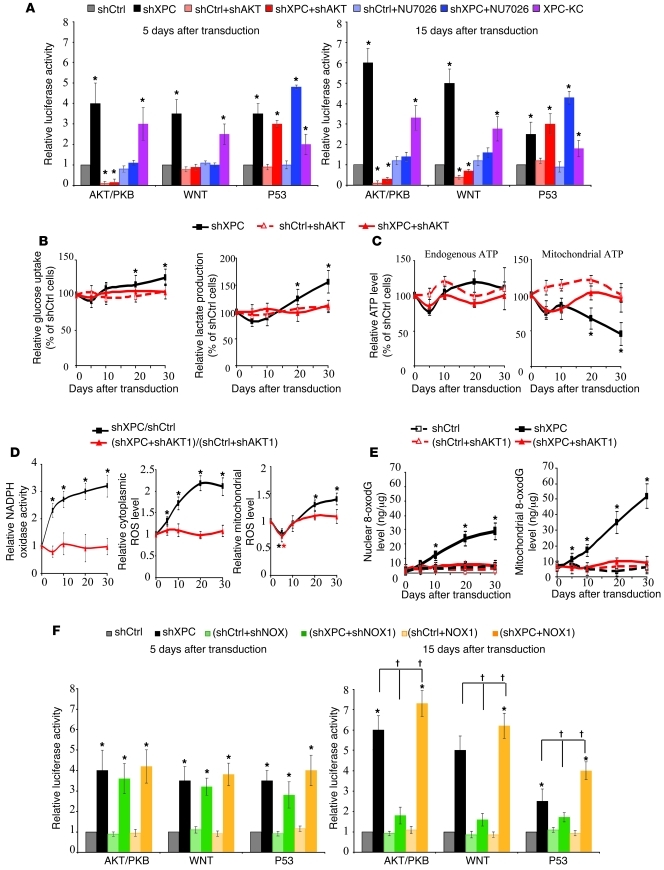
To define the signaling pathways resulting in tumorigenic transformation of XPCKD cells, we assessed the activation profile of 10 “cancer” pathways (AKT, WNT, p53, TGFB, pRB-E2F, NFKB, MYC, hypoxia, MAPK/ERK, and MAPK/JNK) using cancer reporter arrays. Among them, WNT–β-catenin, AKT/PKB, and p53 were upregulated in XPCKD cells (Figure 6A). Interestingly, knockdown of AKT1 — using lentivirus-mediated expression of shRNA directed against AKT1 (Supplemental Figure 1) — in XPCKD cells repressed WNT activation (Figure 6A). This is consistent with the previous data indicating that AKT could positively regulate Wnt–β-catenin signaling through GSK-3β phosphorylation (28). We also found an increase in phosphorylation of GSK-3β in XPCKD cells; this increase was blocked in the absence of AKT (Supplemental Figure 5A), suggesting that AKT activation is the principal event following XPC downregulation.
Figure 6. AKT activation in XPCKD cells triggers NADPH oxidase activation and metabolic alteration.
(A) To assess activation of different cancer pathways, luciferase-reporter lentivirus was used to transduce the keratinocytes in the indicated conditions at days 5 and 15. For each reporter, the mean ± SD luciferase activity is presented as the relative value to the activity in shCtrl-transduced cells (n = 3). (B and C) Glucose consumption and lactate production (B) as well as the total ATP levels and ATP levels produced by mitochondria (C) were measured in the different cells. The results were then compared with the shCtrl and are expressed as the average percentage of shCtrl ± SD of 3 independent experiments. (D) NADPH oxidase activity and cytoplasmic and mitochondrial ROS levels were measured in different transduced cells. (E) The effect of AKT on XPC silencing–induced genomic and mtDNA oxidation were assessed by quantification of 8-oxodG levels in nuclear and mtDNA. (F) To assess the effect of NOX1 on activation of AKT, WNT, and P53 pathways following XPC downregulation, luciferase reporter lentivirus was used as described in A. *P < 0.05 for indicated cells versus shCtrl; †P < 0.05 for indicated cells versus shXPC-transduced cells. Results are presented as mean ± SD.
AKT is shown to regulate multiple steps in glycolysis via posttranscriptional mechanisms that include localization of the glucose transporter to the cell surface, maintenance of HK function in the absence of extrinsic factors, and phosphorylation of some proteins that influence glycolytic flux (29, 30). Therefore, we next determined whether variations in bioenergetics following XPC downregulation are dependent on AKT upregulation. We found that downregulation of AKT1 in XPCKD cells restored glucose consumption and lactate production; mitochondrial ATP production (Figure 6, B and C); expression levels of ND1, ND5, COX1, and COX1; and expression levels of HK2, PFKFB3, GLUT1, and G6PD to normal (Supplemental Figure 5, B–E). That downregulation also blocked XPC knockdown–induced NADPH oxidase activation; cytoplasmic and mitochondrial ROS generation (Figure 6D); genomic and mtDNA oxidation (Figure 6E); and mtDNA deletions (Figure 2C). AKT1 downregulation also blocked the changes in mitochondrial network morphology (Supplemental Figure 5E) and in cell size (Supplemental Figure 2). As AKT downregulation had no effect on early decreased mRNA expression of all examined genes (i.e., COX1, COX3, etc.), their biphasic change observed in XPCKD cells could be related to stalled cell cycle and the selection of cell subsets with overactivation of AKT.
The above results prompted us to determine whether AKT1 downregulation blocks tumorigenic transformation of XPCKD cells. We initially reconstructed epidermis with the AKT1KD cells and double-knockdown (XPC+AKT1) cells (XPCKD/AKT1KD cells) (Supplemental Figure 6). The epidermis reconstructed with XPCKD/AKT1KD cells 5 days after transduction showed the same characteristic as XPCKD cells, i.e., a marked reduction in the epidermal thickness and in the number of Ki67-positive cells compared with control cells (see Supplemental Figure 6, and compare with Figure 4A). The thickness of epidermis, number of Ki67-positive cells, and levels of basal cell layer markers in epidermis reconstructed with XPCKD/AKT1KD cells 15 days after transduction resembled control epidermis (see Supplemental Figure 6, and compare with Figure 4A). Interestingly, we did not observe any hyperplasia, especially large extension of rete pegs found in epidermis reconstructed with XPCKD cells or XPC-KC. To assess the tumorigenic potency in vivo, the XPCKD/AKT1KD cells were subcutaneously injected into immunodeficient mice at 15 days after transduction. These mice were followed for up to 6 months, during which only 1 of the 9 injected mice developed well-differentiated SCCs (Table 1).
Concerning the relation between AKT and NOX, our data indicated that AKT downregulation in XPCKD cells blocked both increased NADPH oxidase activity and increased cytoplasmic and mitochondrial ROS levels observed in XPCKD cells (Figure 6D), suggesting that AKT activation is upstream of NOX1 activation. We further found that NOX downregulation had no effect on AKT activity at 5 days after transduction but abrogated XPC-silencing–induced AKT activation over the following days (Figure 6F and Supplemental Figure 7A). Interestingly, an absence of AKT activation in the double-knockdown (XPC+NOX) cells (XPCKD/NOXKD cells) after day 5 inhibited XPC silencing–induced metabolic alteration (Supplemental Figure 4) and resulted in increased apoptosis (Supplemental Figure 7B). These results suggest a crosstalk between NADPH oxidase activation and AKT activity during tumoral transformation of XPCKD cells.
DNA-PKcs activation results in AKT activation in XPCKD cells.
We then attempted to determine the mechanism by which AKT is activated in XPCKD cells. Since a considerable decrease in cell numbers (not just a plateau) was apparent with cell proliferation curves (see Figure 3A), it is reasonable to speculate that the majority of cells harboring stalled cell-cycle progression undergo apoptosis and only subsets of cells with some modification will be selected. A marked increase in the percentage of apoptotic cells 5 days after transduction that was followed by a rapid decrease in the following days confirmed this concept (Figure 7A). Furthermore, Western blotting revealed that knockdown of XPC resulted in an imbalance between proapoptotic (BAX and BAD) and antiapoptotic (BCL-XL and BCL-2) proteins in favor of the former 5 days after transduction, leading to induction of apoptosis (Figure 7B). Decreased BAX and Bad levels and increased BCL-2 and BCL-XL levels could account for the reduction in apoptotic cell death in XPCKD cells (Figure 7B).
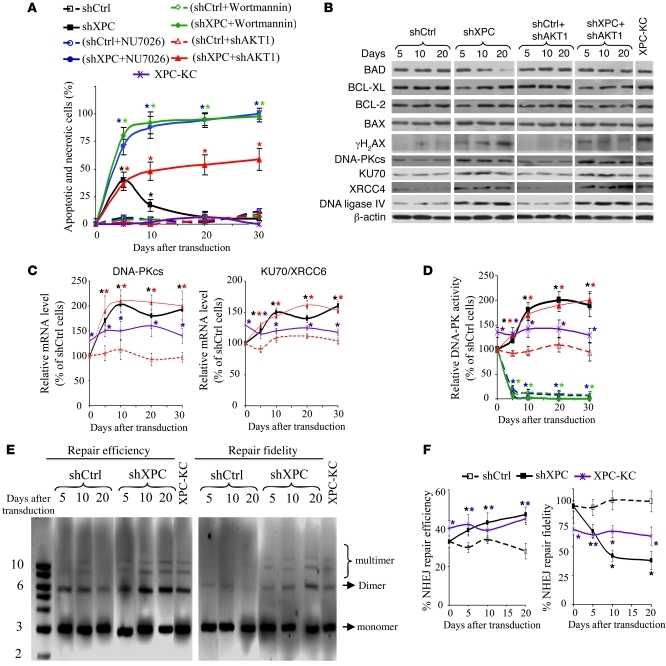
Figure 7. NHEJ activation in XPCKD cells results in overcoming stalled cell-cycle progression and AKT activation.
(A) The percentage of apoptotic keratinocytes was evaluated by flow cytometry. Results are expressed as the average percentage of apoptotic and necrotic cells ± SD of 3 independent experiments. *P < 0.05 for different cells versus shCtrl-transduced cells at the indicated time points. (B) The effects of XPC and/or AKT downregulation on the protein expression level of pro- and antiapoptotic proteins and the major components of NHEJ repair were determined by Western blot. (C) The mRNA levels of DNA-PKcs and KU70 were quantified by qRT-PCR. (D) DNA-PK kinase activity was measured in nuclear extracts of various cells. The results were then compared with the shCtrl and are expressed as the mean ± SD of 3 independent experiments. (E) Nuclear extract of cells transduced with shCtrl or shXPC were used in in vitro end-joining assays. The pUC19 plasmid digested with EcoRI was used as monomer DNA substrate. The amount of ligated pUC19 following incubation with various nuclear extracts shows repair efficiency (left side of photomicrograph). Fidelity of repair was assessed by redigestion of end-jointed products with the EcoRI (right side of photomicrograph). (F) Graphic representation of the end-joining experiment in E. Data are expressed as mean ± SD of the 3 independent experiments.
To determine the mechanism whereby some cells escape the stalled cell-cycle–induced apoptosis, we assessed the expression profile of 84 genes with established roles in DNA damage signaling using a DNA damage PCR array kit. This revealed a 2.8 ± 0.7–fold increase in DNA-dependent protein kinase catalytic subunit (DNA-PKcs) mRNA and a 2.2 ± 0.6–fold increase in KU70/XRCC6 mRNA — the main component of the nonhomologous end-joining (NHEJ) repair pathway— in XPCKD cells, compared with control cells. NHEJ, as well as homologous recombination (HR), is important for repairing double-stranded breaks (DSBs) that are induced by ionizing radiation, collapsed replication forks, or oxidative damage. HR is an error-free pathway, whereas NHEJ is error prone, which can provoke genomic changes (31, 32). The results of PCR arrays were then validated with quantitative real-time RT-PCR (qRT-PCR) (Figure 7C). Western blotting revealed that the expression levels of DNA-PKcs and KU70/XRCC6 as well as those of XRCC4 and DNA-ligase IV (other components of NHEJ repair) were also increased in XPCKD cells and XPC-KC (Figure 7B). Both cell types further displayed a marked increase in DNA-PK kinase activity compared with control cells over time (Figure 7D). We then performed in vitro NHEJ assay to measure efficiency and fidelity of NHEJ in cell extracts obtained from XPCKD, XPC-KC, and control cells. The conversion of monomeric linear plasmid into multimer products, an indicator of end-joining efficiency, increased significantly when cell extracts from XPCKD cells and XPC-KC were used (Figure 7, E and F). Repair fidelity was then assessed by comparing the percentage of end-joined products that could be cut again with the same restriction enzyme initially used to generate the linear DNA substrate. Interestingly, extracts from XPCKD cells and XPC-KC showed a substantial reduction in correct end joining compared with that of control cells (Figure 7, E and F).
To evaluate the effect of NHEJ repair activation on cell survival, XPCKD cells were treated with 2 DNA-PKcs inhibitors: (a) NU7026, which at low concentrations inhibits DNA-PKcs but not PI-3 kinase, ATM, or ATR; and (b) wortmannin, which inhibits all PI-3 and PI-3-like kinases, including DNA-PKcs. Treatment of XPCKD cells with both inhibitors resulted in a major increase in apoptotic cell death (Figure 7A), indicating that DNA-PK activity in XPCKD cells is essential to bypass stalled cell-cycle progression.
It has been shown that DNA-PK can phosphorylate AKT on Ser473, resulting subsequently in the activation of the AKT signaling pathway (33, 34). Our results showed that XPC silencing–induced phosphorylation of AKT on Ser473 (Supplemental Figure 7A) and that inhibition of DNA-PK upon treatment with NU7026 blocked the AKT activation (Figure 6A). In contrast, knockdown of AKT1 in XPCKD cells had no effect on the expression and activity of DNA-PK (Figure 7, B–D). Overall, these results indicate that XPC downregulation results in AKT activation through DNA-PK activation.
Discussion
Cancer cells are now known to utilize complex mechanisms to remodelize their bioenergetics. The factors contributing to the shift from classic OXPHOS to glycolysis in tumor cells include mitochondrial dysfunction, redox stress, oncogene activation, loss of tumor suppressor genes, and environmental hypoxia adaptation. We exploited the intrinsic genomic instability of XPC to investigate the relationship among the accumulation of genomic mutations, bioenergetic remodeling, and ROS in driving tumorigenesis.
Effects of XPC silencing on NHEJ and AKT activation.
Structural DNA damage evokes the recruitment of DNA damage signaling proteins, including DNA-PK, ATM, or ATR to the sites of damaged bases. They amplify and convey the signal from the damaged DNA to DNA-repair, cell-cycle, and apoptotic cell-death machineries (31, 35). The ultimate fate of cells with damaged DNA depends on the type and extent of damage. It has been postulated that arrested replication due to damaged bases on the nontranscribed portion of the genome (in contrast with the transcribed part) does not result in apoptotic cell death. Instead, cells escape replicative arrest utilizing DNA damage tolerance mechanisms that include error-free and error-prone DNA repair systems (36). We and others have shown that UV radiation–induced apoptosis is decreased in XPC–/– cells compared with XPC+/+ wild-type cells (37–39). Our results show that the XPCKD cells overcome the early stalled cell cycle and replicate their DNA efficiently, albeit with greater infidelity in the days following overactivation of DNA-PK and the NHEJ pathway (Figure 7). The high incidence of spontaneous cancers in Xpc–/– mice (14, 15) and the increased risk of developing internal cancers in XPC patients (13) support this concept.
Increased formation of DSBs, as detected by γH2AX (Figure 7B), could be the signal that activates DNA-PK in XPCKD cells. DSBs, in turn, could result from stalled DNA replication observed 5 days after XPC downregulation (Figure 3) as well as from subsequent enhanced ROS-induced oxidative DNA damage (Figure 2B). The decrease in 8-oxodG level and γH2AX following downregulation of NOX1 or AKT1 in XPCKD cells (see Figure 2B, Figure 6E, Figure 7B, and Supplemental Figure 7A) supports this notion.
AKT activation has been revealed to be sufficient to induce the Warburg effect in either nontransformed cells or cancer cells (40, 41). Our results indicate that XPC silencing–induced DNA-PK activation triggers phosphorylation and subsequently activation of the AKT pathway, which in turn results in a characteristic increase in aerobic glycolysis and finally tumorigenic transformation. Interestingly, AKT inhibition blocks both events. AKT signaling is also an essential mediator of epidermal morphogenesis and contributes to epidermal homeostasis by modulating keratinocyte proliferation and apoptosis. Transgenic mice with overexpressed Akt targeted to the epidermal basal cell layer develop spontaneous epithelial tumors in multiple organs with age and are also susceptible to skin carcinogenesis (42).
Effects of XPC-silencing–induced NADPH oxidase activation on bioenergetics.
One of the mechanisms explaining the Warburg effect is the occurrence of a deficiency in mitochondrial respiration during cell growth due to somatic mutations in mtDNA or increased oxidative stress (4, 5, 7). mtDNA mutations are known to correlate with the 8-oxodG content of mtDNA (43). Increased ROS in response to environmental insults (i.e., UV irradiation) or oxidative stress can cause mtDNA deletions in vitro and in vivo (23, 44). It is likely that NOX activation–induced increased cytoplasmic ROS leads to enhanced oxidation of nuclear and mtDNA, followed by the induction of mtDNA deletions and alterations in mitochondrial bioenergetics. Interestingly, inhibition of NOX1 activation (by AKT1 or NOX1 downregulation or through treatment with DPI) inhibits the Warburg effect in XPCKD cells, indicating that NOX activation–induced ROS generation is the cause of the Warburg effect in XPCKD cells.
According to our data, which supports the supposition that oxidative damage contributes to the tumorigenicity of XPCKD cells, it is known that XPC-deficient cells are more sensitive to oxidative stress (45, 46). Spontaneous mutation frequencies in the Hprt gene are 30-fold higher in the spleen of Xpc–/– mice as compared with normal mice and the majority of these mutations result from oxidative damage (16). Also, the frequency of spontaneous lung tumors and hepatocellular adenomas is enhanced in Xpc–/– mice (47). Consistent with the contribution of NOX1-induced ROS in tumorigenicity, it has been shown that NOX overexpression enhances the tumorigenicity of NIH3T3 cells implanted in athymic nude mice (48).
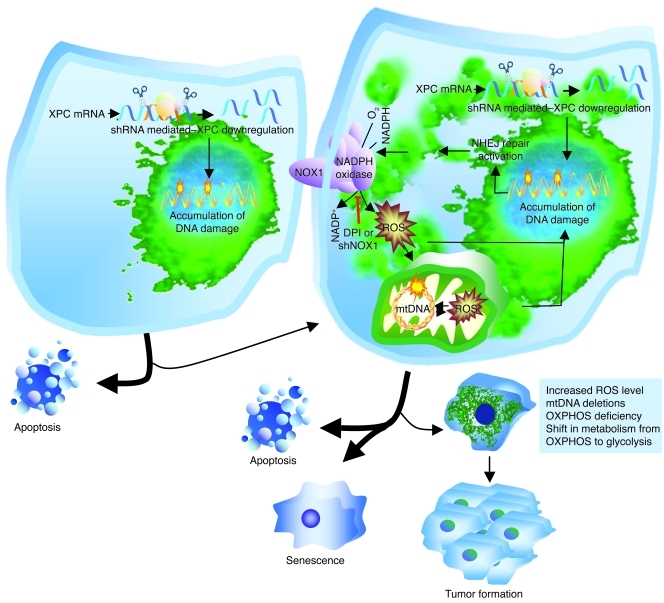
A model outlining cellular responses to XPC downregulation.
The connection between XPC downregulation, ROS, mtDNA deletion, and tumorigenic transformation is depicted in the following scheme (Figure 8). Accumulation of unrepaired damaged DNA bases in XPCKD cells triggers apoptotic death. However, in some cells, the unrepaired bases are recognized by other repair systems such as NHEJ repair, thereby enhancing infidelity of damage repair. DNA-PK activation subsequently promotes AKT activation that in turn mediates induction of NADPH oxidase. Cytoplasmic ROS generation will consequently increase, leading to enhanced oxidation of nuclear and mtDNA, followed by the induction of mtDNA deletions in an amplification loop by which DNA damage results in ROS generation and vice versa. This amplification loop might reduce the chance that cells with unrepaired DNA escape senescence and death through bypass repair systems. This could diminish the risk of cancer. Consistent with this assertion, it is known that mtDNA mutations result in premature aging (49) or sensitize cells to mitochondrial-dependent apoptosis (50). Meanwhile, increased ROS can result in mutations of oncogenes and/or tumor suppressor genes, leading to the deregulation of cell-cycle progression and proliferation, the escape of some cells from apoptosis, and, finally, formation of cancer cells.
Figure 8. A model outlining cellular responses to XPC downregulation.
Lentivirus-mediated XPC silencing in normal human keratinocytes impairs DNA repair efficiency. To evade damaged base–induced cell-death apoptosis, some cells use bypass repair systems (such as NHEJ repair). Activation of the DNA-PK pathway results in upregulation of the AKT pathway, which in turn activates NADPH oxidase. Cytoplasmic ROS generation will consequently increase, leading to enhanced oxidation of nuclear and mitochondrial DNA, followed by the induction of mtDNA deletions, mitochondrial ROS, and alterations in mitochondrial bioenergetics. Meanwhile, it is likely that increased cytoplasmic ROS enhances the mutation of oncogenes and/or tumor suppressor genes, leading to aberrant cell-cycle progression, enhanced cell proliferation, and tumorigenesis.
G to T transversion mutations in P53 in the majority of tumors developed by xenografted XPCKD cells lend further credence to the proposed model. Consistent with our results, it has been reported that the predisposition to UV-induced skin cancer in Xpc–/– mice is enhanced if these animals are heterozygous additionally for Apex (51), a gene coding the apurinic/apyrimidinic endonuclease that is a key enzyme in the process of base excision repair, required for the repair of spontaneous base damage that arises as a result of oxidative damage to DNA. Interestingly, the kinetics of cancer induction in Xpc–/– mice that are heterozygous additionally for both p53 and Apex is indistinguishable from that in Xpc–/– mice heterozygous for just p53 (51), suggesting that the enhanced predisposition to skin cancer in Xpc–/–Apex+/– animals could result from oxidative stress–induced loss of p53 activity.
In conclusion, we demonstrate a mechanism whereby XPC silencing–induced ROS generation, through NOX1 activation, leads to neoplastic transformation of normal keratinocytes. Beyond skin cancer, since the activation of the NOX family has been shown in many cancer cells (5), blocking of NOX activation could offer a unique target for the prevention and treatment of cancers driven by the generation of ROS.
Methods
Source and culture of cells.
Normal human keratinocytes were isolated from skin of patients undergoing plastic surgery after informed consent as previously described (38, 52). Briefly, fresh skin fragments were cut into 5 × 5 mm pieces and treated with trypsin-EDTA overnight at 4°C, which allows for separation of the epidermis from the dermis. Keratinocytes were seeded at a concentration of 105 cells/cm2 in KGM medium, which included hydrocortisone, epidermal growth factor, insulin, and bovine pituitary extract (all from Lonza). The medium was changed 3 times a week. When the cultures reached 70%–80% confluence, the cells were detached with 10% trypsin-EDTA and then resuspended in KGM medium.
Three unrelated patients clinically classified as classical XP with no associated neurologic or extracutaneous findings were included in the study. Western blot analyses of XPC protein expression performed on total cellular extracts using anti-XPC and anti–β-actin antibodies showed that neither the full-length nor the truncated XPC protein was detectable in the keratinocytes from the 3 XPC patients (Supplemental Figure 8). XPC keratinocytes were isolated from punch biopsies obtained after informed consent from nonexposed body sites and seeded on mitomycin C–treated feeder cells originating from the mouse fibroblast J2-3T3 cell line in complete Green’s media as already described (38). Epidermis was reconstructed following seeding of keratinocytes on dead dermis as previously described (38, 52).
Lentiviral vectors constructs and keratinocyte transduction.
The different lentiviral vectors were constructed by inserting catalase, CuZnSOD, MnSOD (52), and NOX1 cDNAs into the multiple cloning site (MCS) of the TPW vector downstream of the human phosphoglycerate kinase (hPGK) promoter (Supplemental Figure 1A). TPW was also used as the backbone for the construction of the TEEHshXPCW, TEEHshNOX1W, and TEEHshRFPW by replacement of the hPGK promoter with the EF-1α promoter and insertion of a cassette containing the H1 promoter followed by shRNA sequences targeting either the XPC, AKT1, NOX1, or NOX2 mRNA or by a sequence targeting the red fluorescent protein mRNA (Supplemental Figure 1B). Lentiviral particles were produced by transient transfection of 293T cells using a calcium phosphate transfection technique as previously described (52). Determination of the titer of each viral supernatant was performed by assessing EGFP expression by flow cytometry and by enzyme-linked immunosorbent assays of p24. For transduction, keratinocytes (5 × 105 cells per T25 flasks) were incubated for 24 hours in complete medium. Prior to infection, the medium was removed, and the cells were incubated with viral supernatants for 24 hours at 37°C in the presence of 8 μg/ml of protamine sulfate. After 5 days, the percentage of GFP-positive cells was determined by cytofluorometry (52).
Western blotting procedure.
Western blotting was performed as previously described (52). Briefly, equal amounts of total protein were resolved by SDS-PAGE and electrophoretically transferred to PVDF membranes. The membranes were then incubated overnight at 4°C with a 1:1000 dilution of the anti-XPC (GeneTex), anti-ND1, anti-COX3, anti–cyclin D2, anti–cyclin B1, anti–p-Cdc2 p34, anti-p16, anti-p21, anti-MnSOD, anti-CuZnSOD, anti-AKT1, anti-G6PD, anti-KU70, anti-XRCC4, anti–DNA ligase IV (Santa Cruz Biotechnology Inc.), anti–DNA-PKcs (Thermo scientific), anti-ATM, anti-ATR, anti–phospho-ATR, anti–phospho-ATM, anti–phospho-ChK1, anti–phospho-ChK2, anti–phospho-AKT, anti–phospho-GSK3B, anti-HK2 (Cell Signaling), anti–PFKFB3 (Abnova; TEBU), and anti-catalase (Rockland) antibodies. After additional incubation with a 1:10,000 dilution of an anti-immunoglobin horseradish peroxidase–linked antibody (Vector Laboratories) for 1 hour, blots were developed using the chemiluminescence ECL reagent (Amersham).
Cell proliferation and apoptosis analysis.
Serial cultures of keratinocytes were performed in 24-well plates by seeding 8 × 103 cells in triplicate, refeeding the cells every second day, and subculturing every 5–6 days. The trypan blue exclusion test was used to obtain cell counts at different days after seeding.
For apoptosis analysis by flow cytometry, keratinocytes were incubated with 5 μmol/l FITC-VAD-FMK (Promega) for 30 minutes at 37°C in the dark and then with 2.5 μg/ml propidium iodide (PI) (Sigma-Aldrich) as already described (53, 54).
Cell-cycle analysis and S phase length calculation.
Cell-cycle analysis was performed using APC-linked anti-BrdU and 7-AAD according to the manufacturer’s protocols (BD Biosciences). Briefly, after 2 hours BrdU pulse labeling, the cells were trypsinized and fixed in Cytofix/Cytoperm buffer on ice for 20 minutes. The cells were then incubated with APC-linked anti-BrdU antibody overnight and then resuspended in 20 μl of 7-AAD solution for flow cytometric analysis.
The length of the S phase was assessed using the BrdU pulse-labeling method. Briefly, immediately after 2-hour pulse BrdU labeling and at 2-hour intervals thereafter, the cells were trypsinized, fixed, and stained with APC-conjugated anti-BrdU and 7-AAD. Changes in the 7-AAD intensity of S phase cells (relative movement [RM]) during the time relative to the 7-AAD positions of G1 and G2 were calculated according to Begg et al. (55) with the following formulas: RM(t) = (FL[t] – FG1[t])/(FG2/M[t] – FG1[t]), where FL is the mean red fluorescence (7-AAD) of BrdU-positive cells and FG1 and FG2/M are the mean 7-AAD fluorescence values for G1 and G2/M cells at time (t), respectively. The S phase duration was calculated as follows: Ts = (0.5/[RM – 0.5]) × t, where t is the postlabeling time. This procedure was repeated at different days after transduction.
Measurement of glucose consumption and lactate production.
Glucose consumption and lactate production were measured using colorimetric assay kits according to the manufacturer (both from BioVision).
Measurement of endogenous and mitochondrial ATP production.
The amount of intracellular ATP was measured by a luciferin/luciferase-based assay. In brief, the cells were trypsinized, rinsed with PBS, and lysed with 0.2 ml of cell lysis reagent. ATP concentrations in the cell lysates were quantified using an ATP bioluminescence assay kit HSII (Roche Applied Science) in accordance with the manufacturer’s instructions. A standard curve for ATP concentration was plotted using standard ATP solution. ATP levels were calculated and normalized to protein lysate concentrations. To assess the ATP levels produced by mitochondria, the cells were incubated with 1 mM of iodoacetate for 5 minutes before they were trypsinized. Iodoacetate is an inhibitor of glycolysis, as it inhibits glyceraldehyde 3-phosphate dehydrogenase. Therefore, any ATP generation by the cells following this treatment must be a consequence of OXPHOS activity. After treatment, the cells were subjected to the same procedures as the untreated cells.
Complex IV (cytochrome c–oxidase) activity.
Cytochrome c–oxidase activity was determined spectrophotometrically with cytochrome c (II) as the substrate. The oxidation of cytochrome c was monitored at 550 nm at 30°C using a double-wavelength Xenius spectrophotometer from SAFAS (Monaco) and standardized reproducible methods as described by Benard et al. (56).
RNA extraction and qRT-PCR.
Total cellular RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. Total cellular RNA (2 μg) was reverse transcribed at 42°C for 60 minutes using the First Strand cDNA synthesis kit (Roche Applied Science). Quantitative real-time PCR was carried out for ND1, ND5, Cox1, Cox3, GLUT1, HK2, PFKFB3, G6PD, DNA-PKcs, KU70, and tubulin using the SYBR Green method with an ABI instrument. Primer sequences are listed in Supplemental Table 1. The reactions were cycled 40 times after initial polymerase activation (50°C, 2 minutes) and initial denaturation (95°C, 15 minutes) using the following parameters: denaturation at 95°C for 15 seconds; and annealing and extension at 60°C for 1 minute. A final fusion cycle (95°C, 30 seconds; 60°C, 30 seconds; 95°C, 30 seconds) terminated these reactions. The standard curve demonstrated a linear relationship between the Ct values and the cDNA concentration. The relative expression of each gene was assessed by considering the Ct and efficiency values and normalized according to the tubulin expression level.
DNA damage signaling-pathway PCR array.
The human DNA damage signaling RT2 profiler PCR array (SABiosciences) was used to assess the expression of 84 genes with known roles in DNA damage response. Synthesis of complementary DNA, real-time PCR, and statistical analyses were done according to the manufacturer’s instructions. Five housekeeping genes were included as RNA content controls, and the 3 with the lowest SD across replicates were used in the analysis (GAPDH, ACTB, and B2M). In addition, a genomic DNA control primer set that can detect the presence of DNA contamination in each sample was included in each PCR run.
Cancer pathway lentiviral reporter arrays.
The cancer lenti reporter arrays (SABiosciences) and FOXO reporter (SABiosciences) were used to monitor the activation of 10 cancer-related signaling pathways and the AKT/PKB pathway, respectively, in accordance with the manufacturer’s instructions. Each lentiviral reporter construct contains a pathway-specific transcription factor–responsive element upstream of firefly luciferase.
Keratinocytes transduced by different shRNAs were seeded on 96-well plates (2 × 104 cells per well) at days 2 or 12 after transduction. One day after, the keratinocytes were incubated with the reporter lentivirus for 24 hours at 37°C. After 2 days (i.e., days 5 or 15 after transduction with shRNAs), keratinocytes were harvested by adding 20 μl of luciferase lysis buffer (Promega). The cell lysates were kept at –80°C and then subjected to luciferase activity assessment using the Luciferase Assay System (Promega) in accordance with the manufacturer’s instructions. The change in the activity of each signaling pathway was determined by measuring the activity of luciferase in transduced keratinocytes versus shCtrl-transduced cells. For each lenti reporter, at least 3 independent experiments were performed in duplicate.
Measurement of intracellular ROS.
The intracellular production of ROS was assessed using a CM-H2DCF-DA cytoplasmic probe or the MitoSOX red mitochondrial superoxide indicator (both from Molecular Probes; Invitrogen) (52). Briefly, after addition of CM-H2DCF-DA (5 μM) or MitoSOX (5 μM), the cells were incubated for 15 minutes at 37°C in the dark. While CM-H2DCF is oxidized by cytoplasmic ROS to the highly green fluorescent CM-DCF compound, MitoSOX is targeted to the mitochondria and oxidized by superoxide to a red fluorescence component. After 2 washes with PBS, the cells were detached by trypsin-EDTA and immediately analyzed by flow cytometry. Ten thousand individual data points were collected for each sample.
Determination of catalase, CuZnSOD, and MnSOD activities.
All these assays were performed as previously described (52) using the SOD Assay Kit-WST (Dojindo Molecular Technologies) and the Amplex Red Catalase Assay Kit (Molecular Probes; Invitrogen).
Measurement of NADPH oxidase activity in intact cells.
NADPH oxidase activity was assessed based on oxidation of luminol by NADPH-dependent O2– production. After trypsinization and washing in PBS, 1 × 106 keratinocytes were put in 50 μl of PBS supplemented with 0.9 mM CaCl2 and 0.5 mM MgCl2. After adding 200 μl of the solution containing 0.8 mM glucose, 2 mM luminol, and 500 U/ml horseradish peroxidase, a 96-well plate was incubated for 1 minute at 37°C. Following the addition of 10 ng/ml of phorbol myristate acetate (PMA), the RLU of chemiluminescence were recorded every 30 seconds for a total of 90 minutes at 37°C using a luminometer.
PCR analysis of mitochondrial deletions.
mtDNA was prepared using the mtDNA isolation kit (BioVision) in accordance with the manufacturer’s instructions. The PCR analysis was carried out on 50 ng of mtDNA using the AccuPrime Taq DNA Polymerase System (Invitrogen). Amplification of fragments representing 3895-bp and 4977-bp deletions were carried out using the primers designed to anneal outside the 3985 and 4977 deletions, respectively (the primer sequences and PCR conditions were determined as described previously in refs. 24 and 57; also see Supplemental Table 2). In brief, the A1/A2 and C1/C2 primers were designed to anneal outside the 3895-bp and 4977-bp deletions, respectively. To increase the sensitivity and specificity, a secondary nested PCR was performed using the B1/B2 and D1/D2 primers on the primary PCR products. To estimate the total mtDNA content, PCR was carried out with Ref1/2 primers that anneal to both wild-type and deleted mtDNA. The PCR conditions were 94°C for 10 minutes, 35 cycles of 94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 30 seconds, followed by a final extension of 10 minutes at 72°C. The short (30 seconds) polymerase extension time during DNA amplification permitted amplification of only the shorter and deleted mtDNA fragments. Amplification products were visualized in a 1% agarose gel stained with ethidium bromide.
DNA-PK activity assay.
DNA-PK activity was assessed using the SignaTECT DNA-dependent protein kinase assay kit (Promega) in accordance with the manufacturer’s instructions. Briefly, after removal of endogenous DNA from 100 μg nuclear extract cells using DEAE-Sepharose, samples were incubated with biotinylated peptide substrate in the presence of [γ-32P] ATP for 5 minutes at 30°C. 20 μl of reactant was spotted onto SAM2 membrane. The membrane was then washed, dried, and subjected to phosphorimaging system. The enzymatic activity of DNA-PK was calculated and expressed as pmol ATP min–1 μg–1 of protein.
In vitro DNA end-joining assay.
In vitro NHEJ repair assay was performed. 400 ng linear plasmid DNA (pUC19) obtained by restriction digestion using EcoRI (New England Biolabs) was incubated with 40 μg nuclear cell extracts at 25°C for 2 hours in 50 μl reaction buffer (50 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 75 mM KCl, 1 mM ATP, 1 mM DTT, 5% glycerol, and protease inhibitors). Reactions were performed in duplicate per set of experiments. The repair reactions were stopped by the addition of 1 μl 10% SDS followed by incubation at 65°C for 15 minutes. The DNA was recovered by Quick Purification Kit (QIAGEN). The second set of purified DNA products was cut again with EcoRI (10 U) in a 10-μl reaction buffer to investigate the fidelity of the end joining. The repair products were resolved in 1% agarose gels stained with ethidium bromide. Digital images of the gels were analyzed densitometrically. Repair efficiency was assessed as the conversion of monomeric linear plasmid into multimers products by the following formula: (total end-joined product [dimmers and multimers]/total DNA in the lane [end-joined products + monomer]). Repair fidelity was measured by comparing the percentages of end-joined products that could be recut with EcoRI using the following formula: ([% end-joined product before recutting – % end-joined product left after recutting with EcoRI]/% end-joined product before recutting).
Morphological studies of mitochondria and morphometry.
To observe the mitochondria network by fluorescence confocal microscopy, keratinocytes grown on 6-well plates were incubated with MitoTracker (Molecular Probes) at 150 nM for 20 minutes at 37°C for. After 2 washes in PBS, the cells were observed and photographed under a FluoView laser scanning inverted microscope (Nikon). The morphology of the mitochondrial network was studied by using MitoTracker Green (Invitrogen); the objective used was a Plan Apochromat 60.0X/1.4/0.21 oil spring loaded. The images were acquired using the EZ-C1 Gold Version 3.2 build 610, as follows (at 37°C): the pinhole was set at 33.3 μm and the image size was 1024 × 1024. The step size was 0.5 μm, and the acquisition time of the Z-series set at 10 frames per second. The number of images was adapted to the width of each cell determined individually. The reconstitution of the 3D images was performed using Imaris Software (Bitplane). Fifty cells per experimental condition were selected randomly on 5 different images, and the analysis of mitochondrial tubule length was performed with the morphometric software Morpho Pro by Explora Nova. It allows automatic selection of the region of interest (i.e., the mitochondrial tubule) and calculation of the length occupied by the selected pixels. In each cell, 25 tubules were measured. The tubules were divided in 4 categories based on their size: those less than 5 μm, those between 5 and 10 μm, those between 10 and 20 μm, and those greater than 20 μm. Results are expressed as average percentage of size distribution ± SD of 3 independent experiments.
Localization and quantification of 8-oxodG in nuclear and mtDNA.
8-oxodG adducts in genomic and mtDNA were quantified using an enzyme-linked immunosorbent assay kit (Trevigen) in accordance with the manufacturer’s instructions. Briefly, following nuclear and mtDNA isolation, 1 μg of DNA was incubated with anti–8-oxodG antibody overnight at 4°C. The samples were then added to 96-well microtiter plates containing bound albumin:8-oxodG adducts and incubated at room temperature for 2 hours. The plate was then washed and incubated with a peroxidase-coupled secondary antibody for 1 hour. After 6 times washing, tetramethylbenzidine substrate was added to each well and incubated for 15 minutes in the dark. After addition of a stop solution, absorbance read at 450 nm. Results were calculated based on a standard curve run simultaneously.
Xenografts of human keratinocytes.
Keratinocytes were transduced with different vectors. Fifteen days after transduction, keratinocytes were trypsinized, washed in DPBS, and resuspended in DPBS with 25% Matrigel (BD Biosciences). Under a protocol approved by the Committee on the Use and Care of Animals at Columbia University, 2 × 106 human keratinocytes were injected subcutaneously into 6- to 8-week-old NOD/SCID mice (Taconic Laboratories). Tumor formation was monitored at different time intervals. Each tumor was dissected, measured, fixed in 4% paraformaldehyde, embedded in paraffin, and processed for H&E staining. Histological assessment of tumor sections was performed by pathologists.
Immunohistochemical and immunofluorescence staining.
Following deparaffinization of formalin-fixed, paraffin-embedded tumor sections, antigen retrieval was performed using microwave oven treatment in a citric acid solution (Vector Laboratories). The sections were then incubated overnight at 4°C with anti-GFP antibody. After incubation with HRP-conjugated secondary antibody, revelation was performed using diaminobenzidine (Sigma-Aldrich). The nuclei were then counterstained with hematoxylin.
For immunofluorescence staining, following deparaffinization of formalin-fixed, paraffin-embedded reconstructed epidermis, antigen retrieval was performed using microwave oven treatment in a citric acid solution (Vector Laboratories). The sections were then incubated 1 hour at room temperature with antibodies against K14, K10, integrin α6 or integrin β1, or Ki67. After incubation with Alexa Fluor 488– or 555–conjugated secondary antibody, the nuclei were counterstained with DAPI.
Statistics.
Statistical analyses were performed using 1-way ANOVA tests, followed by post-hoc Tukey’s tests. P < 0.05 was considered significant. Results are presented as mean ± SD.
Supplementary Material
Acknowledgments
We thank Yucui Zhu and Catherine Pain for technical assistance, and Aqeel Ahmed and Mireia Castillo Martin, Department of Pathology, Columbia University, for histological evaluation of the tumors. This work was supported by the French National Plan Maladies Rares and the GENESKIN European coordination action grant LSHM-CT-2005-512117 (to A. Taïeb), a grant from Enfants de la Lune (to H. de Verneuil and A. Taïeb), NIH grant R01 CA-097249-01 (to D.R. Bickers), the Irving Scholar Foundation (to A.L. Kim), and 5P30 ES009089.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(1):195–211. doi:10.1172/JCI40087.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of cancer. Expert Rev Mol Med. 2002;4(9):1–19. doi: 10.1017/S1462399402004453. [DOI] [PubMed] [Google Scholar]
- 3.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 4.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 5.Clerkin JS, Naughton R, Quiney C, Cotter TG. Mechanisms of ROS modulated cell survival during carcinogenesis. Cancer Lett. 2008;266(1):30–36. doi: 10.1016/j.canlet.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 7.Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA. Mitochondrial mutations contribute to HIF1alpha accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenease kinase 2 in head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15(2):476–484. doi: 10.1158/1078-0432.CCR-08-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci. 2008;65(24):3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelicano H, Carney D, Huang P. ROS stress in cancer cells and therapeutic implications. Drug Resist Updat. 2004;7(2):97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg EC. Rous-Whipple Award Lecture. Nucleotide excision repair and cancer predisposition: A journey from man to yeast to mice. Am J Pathol. 2000;157(3):693–701. doi: 10.1016/s0002-9440(10)64581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5(7):564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 12.van Steeg H, Kraemer KH. Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol Med Today. 1999;5(2):86–94. doi: 10.1016/s1357-4310(98)01394-x. [DOI] [PubMed] [Google Scholar]
- 13.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123(2):241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 14.Hollander MC, et al. Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc Natl Acad Sci U S A. 2005;102(37):13200–13205. doi: 10.1073/pnas.0503133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miccoli L, Burr KL, Hickenbotham P, Friedberg EC, Angulo JF, Dubrova YE. The combined effects of xeroderma pigmentosum C deficiency and mutagens on mutation rates in the mouse germ line. Cancer Res. 2007;67(10):4695–4699. doi: 10.1158/0008-5472.CAN-06-3844. [DOI] [PubMed] [Google Scholar]
- 16.Wijnhoven SW, et al. Age-dependent spontaneous mutagenesis in Xpc mice defective in nucleotide excision repair. Oncogene. 2000;19(43):5034–5037. doi: 10.1038/sj.onc.1203844. [DOI] [PubMed] [Google Scholar]
- 17.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathupala SP, Rempel A, Pedersen PL. Aberrant glycolytic metabolism of cancer cells: a remarkable coordination of genetic, transcriptional, post-translational, and mutational events that lead to a critical role for type II hexokinase. J Bioenerg Biomembr. 1997;29(4):339–343. doi: 10.1023/A:1022494613613. [DOI] [PubMed] [Google Scholar]
- 19.Bando H, et al. Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin Cancer Res. 2005;11(16):5784–5792. doi: 10.1158/1078-0432.CCR-05-0149. [DOI] [PubMed] [Google Scholar]
- 20.Frederiks WM, Vizan P, Bosch KS, Vreeling-Sindelarova H, Boren J, Cascante M. Elevated activity of the oxidative and non-oxidative pentose phosphate pathway in (pre)neoplastic lesions in rat liver. Int J Exp Pathol. 2008;89(4):232–240. doi: 10.1111/j.1365-2613.2008.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64(3):985–993. doi: 10.1158/0008-5472.CAN-03-1101. [DOI] [PubMed] [Google Scholar]
- 22.Berneburg M, Kamenisch Y, Krutmann J. Repair of mitochondrial DNA in aging and carcinogenesis. Photochem Photobiol Sci. 2006;5(2):190–198. doi: 10.1039/b507380d. [DOI] [PubMed] [Google Scholar]
- 23.Wei YH, Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp Biol Med (Maywood). 2002;227(9):671–682. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- 24.Harbottle A, Birch-Machin MA. Real-time PCR analysis of a 3895 bp mitochondrial DNA deletion in nonmelanoma skin cancer and its use as a quantitative marker for sunlight exposure in human skin. Br J Cancer. 2006;94(12):1887–1893. doi: 10.1038/sj.bjc.6603178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 26.Chamulitrat W, et al. A constitutive NADPH oxidase-like system containing gp91phox homologs in human keratinocytes. J Invest Dermatol. 2004;122(4):1000–1009. doi: 10.1111/j.0022-202X.2004.22410.x. [DOI] [PubMed] [Google Scholar]
- 27.Valencia A, Kochevar IE. Nox1-based NADPH oxidase is the major source of UVA-induced reactive oxygen species in human keratinocytes. J Invest Dermatol. 2008;128(1):214–222. doi: 10.1038/sj.jid.5700960. [DOI] [PubMed] [Google Scholar]
- 28.Takemaru K, Fischer V, Li FQ. Fine-tuning of nuclear-catenin by Chibby and 14-3-3. Cell Cycle. 2009;8(2):210–213. doi: 10.4161/cc.8.2.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robey RB, Hay N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19(1):25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9(4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 32.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531(1–2):231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Boehme KA, Kulikov R, Blattner C. p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci U S A. 2008;105(22):7785–7790. doi: 10.1073/pnas.0703423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30(2):203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 35.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297(5581):547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 36.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6(12):943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 37.Clement V, Dunand-Sauthier I, Clarkson SG. Suppression of UV-induced apoptosis by the human DNA repair protein XPG. Cell Death Differ. 2006;13(3):478–488. doi: 10.1038/sj.cdd.4401764. [DOI] [PubMed] [Google Scholar]
- 38.Rezvani HR, Ged C, Bouadjar B, de Verneuil H, Taieb A. Catalase overexpression reduces UVB-induced apoptosis in a human xeroderma pigmentosum reconstructed epidermis. Cancer Gene Ther. 2008;15(4):241–251. doi: 10.1038/sj.cgt.7701102. [DOI] [PubMed] [Google Scholar]
- 39.Ananthaswamy HN, et al. Persistence of p53 mutations and resistance of keratinocytes to apoptosis are associated with the increased susceptibility of mice lacking the XPC gene to UV carcinogenesis. Oncogene. 1999;18(51):7395–7398. doi: 10.1038/sj.onc.1203147. [DOI] [PubMed] [Google Scholar]
- 40.Elstrom RL, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64(11):3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 41.Karnauskas R, et al. Bcl-x(L) and Akt cooperate to promote leukemogenesis in vivo. Oncogene. 2003;22(5):688–698. doi: 10.1038/sj.onc.1206159. [DOI] [PubMed] [Google Scholar]
- 42.Segrelles C, et al. Deregulated activity of Akt in epithelial basal cells induces spontaneous tumors and heightened sensitivity to skin carcinogenesis. Cancer Res. 2007;67(22):10879–10888. doi: 10.1158/0008-5472.CAN-07-2564. [DOI] [PubMed] [Google Scholar]
- 43.Lu CY, Lee HC, Fahn HJ, Wei YH. Oxidative damage elicited by imbalance of free radical scavenging enzymes is associated with large-scale mtDNA deletions in aging human skin. Mutat Res. 1999;423(1–2):11–21. doi: 10.1016/s0027-5107(98)00220-6. [DOI] [PubMed] [Google Scholar]
- 44.Adachi K, et al. A deletion of mitochondrial DNA in murine doxorubicin-induced cardiotoxicity. Biochem Biophys Res Commun. 1993;195(2):945–951. doi: 10.1006/bbrc.1993.2135. [DOI] [PubMed] [Google Scholar]
- 45.D’Errico M, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25(18):4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melis JP, et al. Mouse models for xeroderma pigmentosum group A and group C show divergent cancer phenotypes. Cancer Res. 2008;68(5):1347–1353. doi: 10.1158/0008-5472.CAN-07-6067. [DOI] [PubMed] [Google Scholar]
- 47.Wijnhoven SW, Hoogervorst EM, de Waard H, van der Horst GT, van Steeg H. Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat Res. 2007;614(1–2):77–94. doi: 10.1016/j.mrfmmm.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Suh YA, et al. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401(6748):79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 49.Vermulst M, et al. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40(4):392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 50.Cai S, et al. Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res. 2005;65(8):3319–3327. doi: 10.1158/0008-5472.CAN-04-3335. [DOI] [PubMed] [Google Scholar]
- 51.Cheo DL, Meira LB, Burns DK, Reis AM, Issac T, Friedberg EC. Ultraviolet B radiation-induced skin cancer in mice defective in the Xpc, Trp53, and Apex (HAP1) genes: genotype-specific effects on cancer predisposition and pathology of tumors. Cancer Res. 2000;60(6):1580–1584. [PubMed] [Google Scholar]
- 52.Rezvani HR, Cario-Andre M, Pain C, Ged C, de Verneuil H, Taieb A. Protection of normal human reconstructed epidermis from UV by catalase overexpression. Cancer Gene Ther. 2007;14(2):174–186. doi: 10.1038/sj.cgt.7701000. [DOI] [PubMed] [Google Scholar]
- 53.Rezvani HR, et al. Hypoxia-inducible factor-1alpha, a key factor in the keratinocyte response to UVB exposure. J Biol Chem. 2007;282(22):16413–16422. doi: 10.1074/jbc.M611397200. [DOI] [PubMed] [Google Scholar]
- 54.Rezvani HR, et al. Protective effects of catalase overexpression on UVB-induced apoptosis in normal human keratinocytes. J Biol Chem. 2006;281(26):17999–18007. doi: 10.1074/jbc.M600536200. [DOI] [PubMed] [Google Scholar]
- 55.Begg AC, McNally NJ, Shrieve DC, Karcher H. A method to measure the duration of DNA synthesis and the potential doubling time from a single sample. Cytometry. 1985;6(6):620–626. doi: 10.1002/cyto.990060618. [DOI] [PubMed] [Google Scholar]
- 56.Benard G, et al. Physiological diversity of mitochondrial oxidative phosphorylation. Am J Physiol Cell Physiol. 2006;291(6):C1172–C1182. doi: 10.1152/ajpcell.00195.2006. [DOI] [PubMed] [Google Scholar]
- 57.Berneburg M, et al. Singlet oxygen mediates the UVA-induced generation of the photoaging-associated mitochondrial common deletion. J Biol Chem. 1999;274(22):15345–15349. doi: 10.1074/jbc.274.22.15345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.