Abstract
Autophagy, a process for catabolizing cytoplasmic components, has been implicated in the modulation of interactions between RNA viruses and their host. However, the mechanism underlying the functional role of autophagy in the viral life cycle still remains unclear. Hepatitis C virus (HCV) is a single-stranded, positive-sense, membrane-enveloped RNA virus that can cause chronic liver disease. Here we report that HCV induces the unfolded protein response (UPR), which in turn activates the autophagic pathway to promote HCV RNA replication in human hepatoma cells. Further analysis revealed that the entire autophagic process through to complete autolysosome maturation was required to promote HCV RNA replication and that it did so by suppressing innate antiviral immunity. Gene silencing or activation of the UPR-autophagy pathway activated or repressed, respectively, IFN-β activation mediated by an HCV-derived pathogen-associated molecular pattern (PAMP). Similar results were achieved with a PAMP derived from Dengue virus (DEV), indicating that HCV and DEV may both exploit the UPR-autophagy pathway to escape the innate immune response. Taken together, these results not only define the physiological significance of HCV-induced autophagy, but also shed light on the knowledge of host cellular responses upon HCV infection as well as on exploration of therapeutic targets for controlling HCV infection.
Introduction
Hepatitis C virus (HCV) is a major cause of chronic liver disease, with more than 170 million infected individuals worldwide (1, 2). In 50%–80% of infected patients, HCV establishes persistent infection, often leading to chronic liver disease (3). At present, HCV isolates can be classified into 6 major genotypes that differ in their nucleotide sequences by 30%–35%, and several subtypes can be defined within these genotypes (4). HCV is thought to be non-cytopathic in vivo, and the pathogenesis of hepatitis is assumed to reflect destruction of HCV-infected cells by cytotoxic CD8+ T cells (5, 6). Current therapy consists of a combination of pegylated IFN and ribavirin, but the success rate is limited, and the outcome of therapy is dependent on the genotype of the infecting virus (7).
HCV is an enveloped, single-stranded, positive-sense RNA virus of the genus Hepacivirus within the family Flaviviridae (8, 9). The RNA genome is of about 9.6 kb, and flanked at the 5′ and 3′ ends by untranslated regions (UTRs) (9, 10) (Supplemental Figure 1A, scheme 1; supplemental material available online with this article; doi: 10.1172/JCI41474DS1). The viral RNA encodes a single polypeptide precursor of about 3,000 amino acids, which is co- and post-translationally processed by a combination of cellular and viral proteases into at least 10 individual proteins, including 4 structural proteins (core, glycoproteins E1 and E2, and p7) and 6 nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Supplemental Figure 1, scheme 1). The structural proteins core, E1, and E2 are the major components of the viral particle, while the NS gene products participate in genome replication by organizing the replication complexes within a unique multi-vesiculated membrane structure, called membranous web (11, 12).
Viral infection often causes stress to the ER. The cellular response to ER stress, known as the unfolded protein response (UPR), is designed to allow the cell to recover by attenuating translation and upregulating the expressions of chaperone proteins and degradation factors to refold or eliminate misfolded proteins (13). Several viruses have been reported to induce UPR activation (14). For instance, herpes simplex virus type 1, human cytomegalovirus, and Epstein-Barr virus induce ER stress and activate the UPR signaling cascade to promote the assembly of infectious particles, thereby benefiting the establishment of infection (14). In the case of HCV, HCV utilizes the ER or ER-derived membrane structure as the primary site of envelope protein biogenesis, RNA replication, and viral particle assembly (10). Thus, it is conceivable that HCV-infected cells experience ER stress and the UPR. Although viral protein such as NS4B and expression of an HCV replicon were shown to induce UPR through transactivation of ER chaperons (15), another study showed that the inositol requiring–1α/X box–binding protein 1 (Ire1α/XBP1) pathway is inhibited in the HCV replicon cells (16). Despite this discrepancy, the functional significance of the UPR in the HCV life cycle is still poorly understood.
Autophagy is a highly evolutionarily conserved process in virtually all eukaryotic cells (17, 18). It involves the sequestration of regions of cytosol within double-membrane-bound compartments and delivery of the contents to lysosome for degradation (18). The process of autophagy initiates with steps including the nucleation and elongation of vesicles to form the phagophore. The edges of phagophore in turn fuse to assemble the autophagosome. Finally, the autophagosome fuses with a lysosome to form an autolysosome, where the captured cytosol component and the inner membrane are degraded (17, 18). Autophagy has been shown to be an important player in stresses such as nutrient starvation, damaged organelles, unfolded protein aggregation, and cell death (19). In addition, several studies have shown that autophagy is activated to act as a survival mechanism in cells exposed to ER stress, and that the UPR is specifically induced to trigger the initiation of autophagosome formation (20–22).
In mammalian cells, the autophagic process was reported to be exploited by many RNA viruses, such as mouse hepatitis virus, poliovirus, and rhinovirus, to promote their infections by serving as the membrane scaffold for RNA replication (23). In the case of HCV, the results concerning the involvement of autophagy in the HCV life cycle have been controversial. Ait-Goughoulte initially found that serial passage of HCV H77 (genotype 1a) in human immortalized hepatocytes reveals the characteristics of autophagy (24). Sir et al. reported that transfection of an HCV RNA into human hepatoma Huh7 cells induces an incomplete autophagic process that positively regulates viral RNA replication (25). On the other hand, Dreux et al. suggested that the autophagic machinery is required for the initiation of translation of the HCV incoming RNA, but not for the maintenance of ongoing RNA replication (26). In contrast, Tanida et al. recently reported that knockdown of autophagy-related gene 7 (Atg7) decreases the production of infectious HCV particles, with no apparent effects on the expressions of viral RNA and proteins (27). Nevertheless, the exact physiological role of autophagy in the HCV life cycle still remains unclear.
On the other hand, viral infection, including HCV, may trigger an immediate immune response, so-called innate immunity, to establish an antiviral state through retinoic acid–inducible gene I (RIG-I)–like receptor (RLR)–mediated activation of type I IFN (28, 29). The newly synthesized IFN is then secreted, binds to its cognate receptors on the surface of infected and uninfected neighboring cells, and stimulates a downstream cascade to turn on the production of a family of IFN-stimulated genes (ISGs); these ISGs then exert their inhibitory effects on viral replication in infected cells (30). Recently, autophagy has emerged as playing a regulatory role in controlling the innate and adaptive immune responses against intracellular pathogens (31–33). In the innate immunity, autophagy acts as a cell-autonomous defense, directly eliminating intracellular microbes or their products, including bacteria, viruses, and protozoa (32). Furthermore, autophagy can be an effector and regulator of pattern recognition receptor response to pathogen molecular pattern, such as DNA virus containing double-stranded DNA (34–36). Recently, the ATG5-ATG12 conjugate, a component of the autophagic machinery, was shown to associate directly with RIG-I, melanoma differentiation–associated gene 5 (MDA5), and mitochondrial antiviral signaling proteins (MAVS) through the caspase recruitment domains (CARDs) on these IFN induction–signaling molecules (37), resulting in the inhibition of type I IFN production and permitting the replication of vesicular stomatitis virus (VSV) in its host cells. Conversely, VSV infection was shown to induce autophagy, which in turn activates the antiviral response and inhibits virus replication in the model organism Drosophila (38). As a result, the exact physiological role of autophagy in controlling innate immunity remains enigmatic. More importantly, whether autophagy participates in innate immunity to control HCV replication has been an interesting open question.
Taking advantage of the genotype 2a JFH1 RNA–based cell culture-derived virus (HCVcc) infection approach (39–41), in the present study we studied the virus-cell interactions and cellular responses in a system that most likely mimics natural infection. We found that HCV induced complete autophagy through activation of the UPR and that the complete autophagic process is required for HCV RNA replication. Loss of autophagy signaling further upregulated HCV pathogen-associated molecular pattern–mediated (PAMP-mediated) cytoplasmic RIG-I signaling and IFN-β–mediated antiviral responses. And vice versa, enhanced UPR autophagic pathway led to dramatic downregulation of HCV and Dengue virus (DEV) PAMP–mediated activation of the IFNB promoter. Our results indicate for the first time to our knowledge that UPR-autophagy positively regulates HCV RNA replication through its inhibitory action on innate immunity. This study also provides better perspective for understanding HCV-induced pathogenic mechanisms as well as for exploring potential therapeutic applications.
Results
HCV infection induces complete UPR-autophagy.
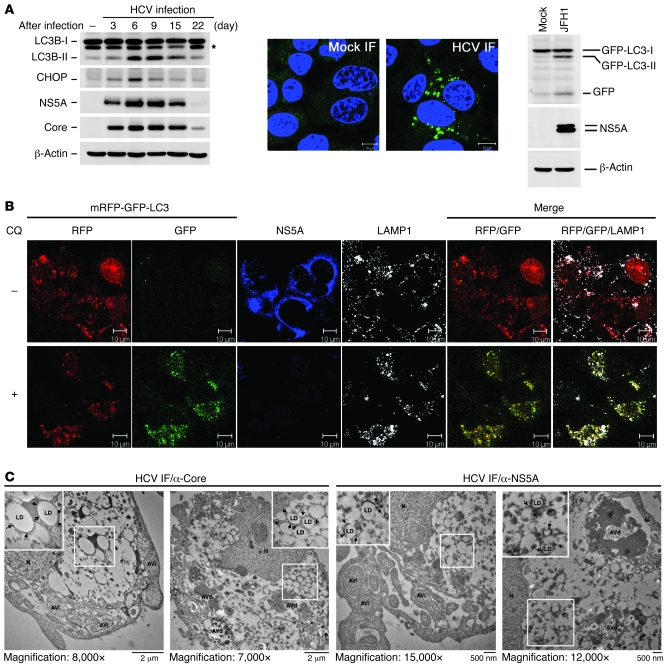
There is growing evidence showing that various host cellular responses, including autophagy, innate immunity, and apoptosis, are affected or activated by HCV in vitro (24, 25, 42–44). Here, we aimed to investigate the autophagic response upon HCV JFH1 infection and its regulatory role in virus replication in a condition that mimics natural viral infection. First, we found that the phosphatidylethanolamine-conjugated form of microtubule-associated protein 1 light chain 3 β (LC3B-II), a hallmark of autophagosome formation, was highly activated at the acute infection phase (6–9 days after infection) (Figure 1A, left panel). A UPR-activated transcriptional factor, CCAAT/enhancer binding protein (c/EBP) homologous protein (CHOP) was also greatly upregulated in these acutely HCV-infected cells (Figure 1A, left panel), suggesting that autophagosome formation in HCV infection was associated with ER stress and UPR. Further analysis of the long-term-cultured HCV-infected cells revealed that LC3B-II and CHOP levels started to diminish when the cells entered the chronic phase (15–22 days after infection) (Figure 1A, left panel). Similarly, HCV infection also led to puncta formation of GFP-LC3–labeled vacuoles in most of Huh7 cells stably expressing GFP-LC3 (Figure 1A, middle panels) and the formation of GFP-LC3-II (Figure 1A, right panel), confirming that HCV infection indeed induces the formation of autophagosomes. These results also showed a close correlation of HCV viral protein expression with CHOP induction and autophagosome formation (Figure 1A, left panel).
Figure 1. Induction of UPR and autophagy by HCV infection.
(A) Huh7 cells were inoculated with HCV at an MOI of 10, and then harvested at different times for Western blot analysis (left panel). The asterisk indicates the nonspecific band. HCV-infected Huh7/GFP-LC3 cells (MOI of 10) were fixed at 6 days after infection and analyzed by confocal microscopy (middle panels) and Western blotting (right panel). Scale bars: 10 μm. (B) HCV-infected Huh7/mRFP-GFP-LC3 cells (MOI of 10) were maintained for 6 days, treated with (+) or without (–) CQ (50 μM) for 6 hours, and assessed for subcellular localization of the indicated proteins. Scale bars: 10 μm. (C) HCV-infected cells as described in B were processed for immuno-TEM analysis. The larger boxed images represent enlargements of the smaller boxed insets. Another set of enlarged images is shown in Supplemental Figure 2A. The black arrows indicate LD-associated core and NS5A proteins, which were respectively immmunolabeled with 12-nm and 18-nm gold particles. N, nucleus; LD, lipid droplet.
To verify whether the HCV-induced autophagosome fuses with lysosome, we made use of a tandem reporter construct, mRFP-GFP-LC3 (45). The green fluorescence of this tandem autophagosome reporter is attenuated in the acidic pH lysosomal environment by lysosomal hydrolysis, while the mRFP is not. Therefore, the green fluorescent component of the composite yellow fluorescence from this mRFP-GFP-LC3 reporter is lost upon autophagosome fusion with lysosome, whereas the red fluorescence remains detectable. In the absence of an acidification inhibitor of lysosome degradation, chloroquine (CQ), remarkable red fluorescence signals were detected in HCV-infected cells (Figure 1B, top row). And the RFP-LC3–labeled puncta structures were also colocalized with a lysosome marker, lysosome-associated membrane protein 1 (LAMP1) (Figure 1B, top row). In sharp contrast, treatment with CQ greatly restored the expression of GFP and resulted in yellow color–labeled autophagosomes in HCV-infected cells (Figure 1B, bottom row).
To confirm that HCV infection induced autophagic activation, we performed transmission electron microscopy–based (TEM-based) ultrastructural analysis. Both initial- and late-stage autophagic vacuoles (AVi and AVd, respectively) were observed in HCV-infected cells, which were judged by the detection of lipid droplets (LDs) surrounded by immunogold-labeled core or NS5A (Figure 1C and Supplemental Figure 2A), whereas no apparent AVi or AVd was detected in mock-infected cells (Supplemental Figure 2B). Moreover, the AVi and AVd detected in HCV-infected cells were, respectively, labeled with LC3B and LAMP1 antibodies (Supplemental Figure 2C, left and right panels), in agreement with the earlier studies showing that LC3B and LAMP1 are localized on the autophagosome and autolysosome, respectively, in the autophagic process (46–48). Our results confirm again that the HCV-induced autophagic process involves formation of different stages of autophagic vacuoles.
Next, we found that the HCV-induced LC3B-II formation was increased when infected cells were treated with lysosomal protease inhibitors (E64 and pepstatin A), CQ, and a vacuolar ATPase inhibitor (bafilomycin A1 [BAF-A1]) (Supplemental Figure 2D), which are well-known acidification inhibitors shown to block the activity of the pH-dependent lysosomal proteases and to bring about accumulation of immature autolysosome (49, 50). Further study showed that treatment with CQ and BAF-A1 led to more accumulation of LC3B-II in HCV-infected cells compared with that detected in mock-infected cells (Supplemental Figure 2E). It should be noted that interference with complete autolysosome fusion was also concomitant with reduced HCV expression (Supplemental Figure 2, D and E). In addition, the detection of free GFP fragment in HCV infection (Figure 1A, right panel) was indicative of the proceeding of autophagic vacuoles to autolysosome degradation (51). These results indicate that HCV induces the complete autophagic process through enhancing the autophagic flux.
UPR is required for HCV-induced CHOP expression and autophagosome formation.
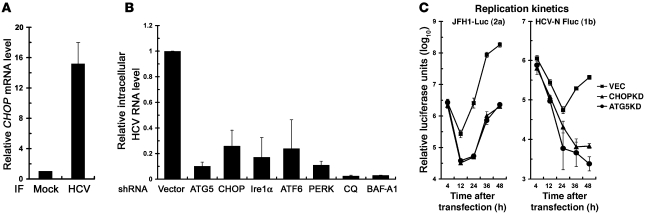
In parallel with increased CHOP expression, HCV infection also led to upregulation of the CHOP mRNA level by 16-fold compared with mock-infection (Figure 2A), suggesting that the accumulated CHOP protein level in infected cells resulted from elevated CHOP mRNA levels. Moreover, HCV infection also activated the 3 UPR modulators, as demonstrated by the cleavage of activating transcription factor 6 (ATF6) to yield p50ATF6; the splicing of XBP1, a downstream regulator of inositol requiring–1α (Ire1α); and phosphorylation of phosphorylated dsRNA-dependent protein kinase–like ER-localized eIF2α kinase (PERK) (Supplemental Figure 3A). Knockdown of CHOP by siRNA duplexes also greatly downregulated the expression of LC3B-II and HCV core expression (Supplemental Figure 3B). And siRNA-mediated transient knockdown of each of Ire1a, ATF6, and PERK specifically inhibited the HCV-triggered increase in CHOP as well as LC3B-II expression (Supplemental Figure 3C). Theses results imply that HCV infection triggers activation of UPR modulators to transactivate the CHOP expression, which in turn activates the autophagy process to enhance HCV viral protein expression.
Figure 2. Inhibition of HCV replication by knockdown of UPR and autophagy-related genes.
(A) HCV-infected Huh7 cells (MOI of 0.1) were maintained for 3 days and analyzed for CHOP mRNA levels. (B) HCV-infected Huh7 cells (MOI of 0.01) were maintained for 24 hours and transduced with the indicated shRNA lentiviruses or treated with CQ (50 μM) or BAF-A1 (100 nM) for 24 hours. Seventy-two hours after infection, cells were assessed for intracellular viral RNA levels. (C) VEC, ATG5KD, and CHOPKD cells were used for the assessment of the replication kinetics of JFH1-Luc and HCV-N-FLuc. Data represent mean ± SEM (n = 3).
HCV-induced UPR-autophagy pathway controls viral RNA replication.
Next, we examined whether HCV-induced UPR-autophagy is required for HCV RNA biogenesis. Individual knockdown of various genes involved in UPR-autophagy, such as ATG5, CHOP, and the 3 UPR regulators Ire1α, ATF6, and PERK, strikingly reduced the level of viral RNA in HCV-infected cells (Figure 2B) and also suppressed production of infectious virus into the culture medium (data not shown). Moreover, treatment with CQ or BAF-A1 reduced the level of intracellular viral RNA and infectious virus released into the culture medium to almost the background level (Figure 2B and data not shown).
Furthermore, we then determined whether the UPR-autophagy pathway functions in initial-stage viral RNA synthesis using CHOP- and ATG5-stable-knockdown Huh7 cells, i.e., CHOPKD and ATG5KD (Supplemental Figure 3D). A full-length (FL) bicistronic JFH1 carrying firefly luciferase, designated JFH-Luc (52) (Supplemental Figure 1, scheme 2), was examined. The replication kinetics of JFH1-Luc RNA were greatly reduced in ATG5- and CHOP-stable-knockdown Huh7 cells even at 12 hour after transfection, at which time viral assembly and release are unlikely to be attained, as compared with those observed in VEC cells, which harbored a stably transduced empty lentiviral vector (Figure 2C, left panel). Similarly, stable knockdown of ATG5 or CHOP also greatly reduced the replication kinetics of the genotype 1b HCV-N strain replicon (Figure 2C, right panel), indicating a strain-independent requirement of UPR-autophagy for HCV RNA replication. In addition, there were no apparent changes in HCVpp infection and HCV internal ribosome entry site (IRES)–mediated translation in ATG5KD and CHOPKD cells (data not shown). These results together indicate that the initial-stage RNA replication step is the primary target in the HCV life cycle most affected by the UPR-autophagy. Moreover, we found that transient knockdown of ATG5, CHOP, or LC3B by specific shRNAs in HCV-infected Huh7/RFP-LC3 cells not only disrupted RFP-LC3 puncta formation (Supplemental Figure 4A) but also greatly attenuated the HCV-induced formation of membranous web (Supplemental Figure 4B), known as the assembly site of HCV replication complex, as opposed to the obvious puncta structures and multivesicular membrane alterations seen in cells transduced with the empty lentivirus vector (Supplemental Figure 4, A and B, respectively). These results together reinforce the crucial role of the complete UPR-activated autophagy in HCV RNA replication.
Interference with UPR-autophagy activates the innate immune response.
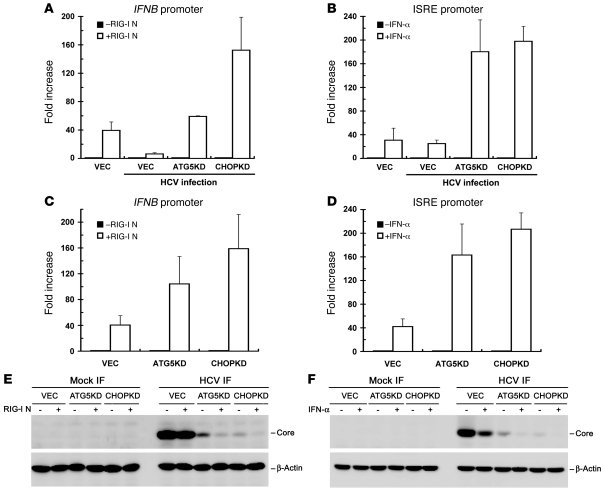
HCV escapes the innate immune defense and establishes chronic infection by NS3/4A-dependent proteolysis of MAVS, also called IFN-β promoter stimulator 1 (IPS1)/virus-induced signaling adaptor (VISA)/CARD adaptor–inducing IFN-β (Cardif), the downstream effector of pathogen recognition receptors, RIG-I, and MDA5 (53, 54). As the loss of UPR-autophagy activation by gene silencing led to inhibition of HCV RNA replication and infectious virus production (Figure 2B and data not shown), which was reminiscent of the antiviral state activated by the innate immune response, we wondered whether signaling of type I IFN is activated in these UPR-autophagy gene–knockdown cells. To test this hypothesis, we compared RLR signaling among VEC, ATG5KD, and CHOPKD cells. The activation of RLR signaling was determined by measurement of the IFNB promoter activity induced by ectopic expression of an N-terminal fragment of RIG-I (RIGI-N), which was shown to activate the IFNB promoter through the phosphorylation, dimerization, and translocation of IFN regulatory factor 3 (IRF3) into the nucleus (55). In VEC cells, HCV infection inhibited RIGI-N–mediated IFN-β activation, as opposed to uninfected VEC cells, in which RIGI-N–activated IFNB promoter increased 40-fold (Figure 3A), presumably due to the inactivation of MAVS signaling by NS3/4A protease activity in infected cells. In contrast, RIGI-N maintained a greater ability to activate the IFNB promoter in infected ATG5KD and CHOPKD cells compared with infected VEC cells (Figure 3A). HCV core expression was also greatly reduced in ATG5KD or CHOPKD cells compared with that observed in infected VEC cells (Figure 3E). And ectopically expressed RIGI-N in HCV-infected ATG5KD and CHOPKD cells also coincidently reduced the expression of core protein compared with that observed with HCV-infected VEC cells (Figure 3E). These results reveal a critical role of UPR-autophagy in modulation of innate immune response and HCV expression.
Figure 3. Activation of RIG-I–mediated IFN-β induction by knockdown of signaling molecules in UPR-autophagy.
(A) VEC, ATG5KD, and CHOPKD cells were infected with HCV at an MOI of 10. Three days after infection, infected cells along with uninfected VEC cells were cotransfected with pIFN-β/Fluc reporter and pCMV22-Rluc plasmids together with an empty vector (–) or a RIG-I N expression construct (+). Twenty-four hours after DNA transfection, cells were harvested for the dual luciferase assay. The Renilla luciferase unit was used to normalize the transfection efficiency. The results are expressed as fold increase in IFNB promoter activity in the presence of RIGI-N relative to the basal level of the empty vector. (B) HCV infection and reporter transfection were performed as described in A, except that the ISRE promoter luciferase reporter was used instead of pIFN-β/Fluc and RIG-I N expression constructs. Twenty-four hours after transfection, cells were treated with or without 100 U/ml IFN-α for 16 hours before the dual luciferase assay. The results represent the fold induction of ISRE promoter activity in the presence of IFN-α relative to the basal level of cells without IFN-α treatment. (C) The indicated cells were analyzed for RIGI-N–mediated IFNB promoter activation as described in B. (D) The indicated cells were assessed for IFN-α–stimulated ISRE promoter activity as described in B. (E and F) Cell samples obtained from A and C and from B and D were analyzed by Western blotting, and the results are shown in E and F, respectively. Data represent mean ± SEM (n = 3) (A–D).
To further assess the role of UPR-autophagy in IFN-mediated downstream signaling, we determined the promoter activity of IFN-stimulated gene-responsive element (ISRE) upon IFN stimulation. Infection of HCV did not greatly affect the IFN-α–stimulated activation of the ISRE promoter, as shown by the comparable ISRE-Fluc activities in VEC cells infected or not infected with HCV (Figure 3B). This phenomenon was consistent with a previous report that HCV infection does not block IFN-stimulated activation of the ISRE promoter (42). Interestingly, HCV-infected ATG5KD or CHOPKD cells still possessed a greater capacity for IFN-α–stimulated activation of the ISRE promoter, compared with infected VEC cells (Figure 3B). The IFN-α–triggered ISRE promoter activation also led to inhibition of HCV core protein expression in VEC, ATG5KD, and CHOPKD cells (Figure 3F). Similarly, in the absence of HCV infection, ATG5KD and CHOPKD cells still showed a higher potential for RIGI-N–mediated activation of the IFNB promoter as well as IFN-α–stimulated activation of the ISRE promoter than VEC cells (Figure 3, C and D, respectively), indicating that the incremental increase in innate immune signaling triggered by the loss of UPR-autophagy does not necessarily result from a decrease in NS3/4A expression due to the reduced HCV RNA replication, but may also proceed in an HCV-independent manner.
Interference with UPR-autophagy enhances HCV PAMP-triggered innate immune activation.
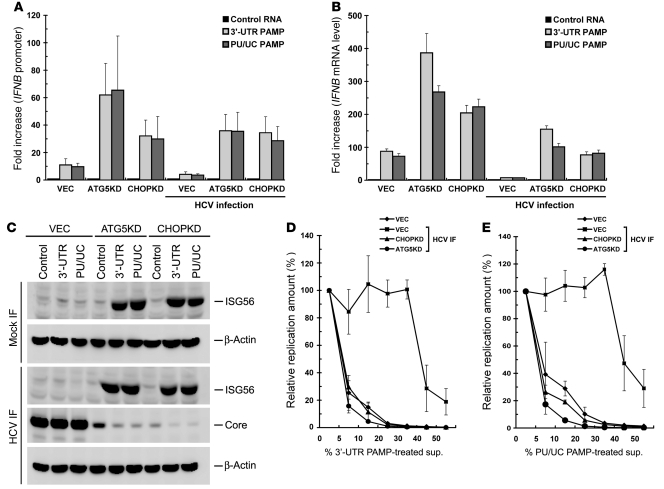
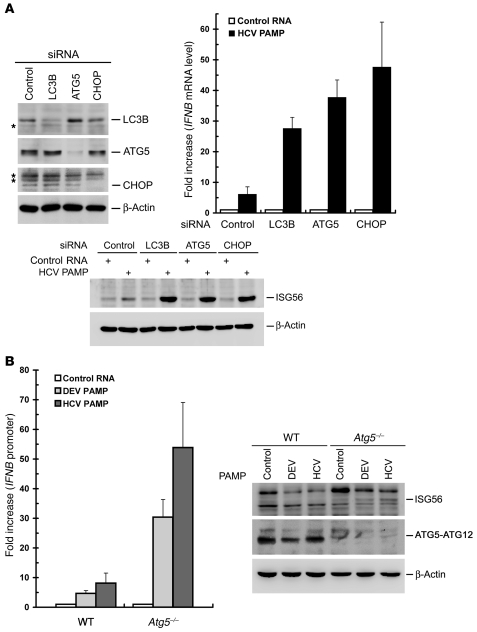
The homopolymeric uridine and cytidine (poly-U/UC; hereafter abbreviated as PU/UC) sequence located within the 3′-UTR of the genomes of various HCV genotypes has been identified as an HCV PAMP motif, mainly for its ability to activate RIG-I–triggered innate immunity (56, 57). It was imperative to examine whether stable silencing of ATG5 or CHOP also increases HCV PAMP-mediated activation of type I IFN, since ATG5 and CHOP function in the inhibition of RLR signaling (Figure 3). First, the HCV JFH1 3′-UTR and PU/UC PAMP RNAs were synthesized in vitro and checked for their authenticity (Supplemental Figure 5), and then used to test this hypothesis. As expected, both HCV 3′-UTR and PU/UC PAMPs efficiently increased IFNB promoter activity (~10-fold) and stimulated IFNB mRNA expression (~80-fold) in uninfected VEC cells (Figure 4, A and B, respectively). Stable knockdown of ATG5 or CHOP further upregulated HCV PAMP–triggered IFNB promoter activity and IFNB mRNA level in uninfected VEC cells (Figure 4, A and B, respectively). HCV infection inhibited both HCV PAMP–triggered IFNB promoter activation and IFNB mRNA expression in VEC cells (Figure 4, A and B, respectively). In contrast, 3′-UTR and PU/UC PAMP each still greatly stimulated IFNB promoter activation as well as IFNB mRNA production in HCV-infected ATG5KD and CHOPKD cells (Figure 4, A and B, respectively). These results confirm again that silencing of ATG5 or CHOP enhances HCV PAMP–mediated IFNB promoter activation even in the context of HCV infection.
Figure 4. Activation of HCV PAMP–mediated IFN responses by knockdown of UPR-autophagy signaling molecules.
(A) The indicated cells were infected with or without HCV (MOI of 10). Three days after infection, cells were transfected with pIFN-β/Fluc promoter reporter and pCMV22-Rluc plasmids. Twenty-four hours after transfection, cells were transfected with HCV 5′-UTR (Control), 3′-UTR, or PU/UC PAMP RNAs, followed by dual luciferase assay 24 hours after PAMP transfection. The fold increase is calculated by normalization to the basal level of control RNA transfection. (B) Virus infection and PAMP RNA transfection were performed as described in A but without DNA transfection. Cells were harvested for determination of the IFNB mRNA level, and fold increase was determined by normalization to the basal level of control RNA transfection. (C) Cell samples from B were assessed for the indicated proteins by Western blotting. (D) The indicated cells were infected (MOI of 10), and 3 days after infection, infected cells along with uninfected VEC cells were transfected with the 3′-UTR PAMP RNA motif for 24 hours. Culture supernatants were harvested, filtered, and used as conditioned medium. For inhibition experiments, Huh7 cells were transfected with JFH1-Luc RNA for 4 hours and incubated for an additional 48 hours with the indicated percentages of conditioned medium collected from different treatments as indicated in the total culture medium, which are expressed as percent 3′-UTR PAMP–treated supernatant (sup.) or percent PU/UC PAMP–treated supernatant. Replication of JFH1-Luc was monitored by the firefly luciferase activity. (E) Generation of PU/UC-treated conditioned medium and JFH1-Luc inhibition assay were performed as described in D. Data represent mean ± SEM (n = 3) (A, B, D, and E).
UPR-autophagy–modulated activation of HCV PAMP–triggered innate immunity correlates with its paracrine antiviral activity.
Since stable knockdown of ATG5 or CHOP upregulated IFNB promoter activation and IFN-α–stimulated ISRE promoter activation in infected cells (Figure 3, A and B, and Figure 4, A and B), we then determined whether interference with UPR-autophagy can trigger ISG expression. HCV 3′-UTR and PU/UC each were able to induce the expression of IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), also named ISG56 (58, 59), in HCV-infected ATG5KD and CHOPKD cells, but not in HCV-infected VEC cells (Figure 4C). Next, we examined whether stable knockdown of ATG5 or CHOP would exert a paracrine antiviral action on neighboring cells through secreted IFN-β. The supernatants collected from VEC cells transfected with 3′-UTR or PU/UC RNA specifically inhibited expression of JFH1-Luc reporter virus in a dose-dependent manner (Figure 4, D and E, respectively). As expected, HCV infection greatly inhibited the antiviral activities in the supernatants of 3′-UTR– and PU/UC RNA–transfected VEC cells on JFH-Luc replication, as shown in HCV-infected VEC cells (VEC HCV IF in Figure 4, D and E, respectively). Remarkably, the supernatants from 3′-UTR and PU/UC RNA–transfected, HCV-infected ATG5KD and CHOPKD cells possessed mediated a more potent inhibitory effect on HCV replication than the supernatants obtained from HCV-infected VEC cells (Figure 4, D and E). These results together highlight the notion that interference with UPR-autophagy not only amplifies HCV PAMP–mediated IFN-β activation but also elevates the downstream innate immune response to inhibit HCV replication in a paracrine fashion.
UPR-autophagy suppressing antiviral innate immunity can occur independently of HCV infection.
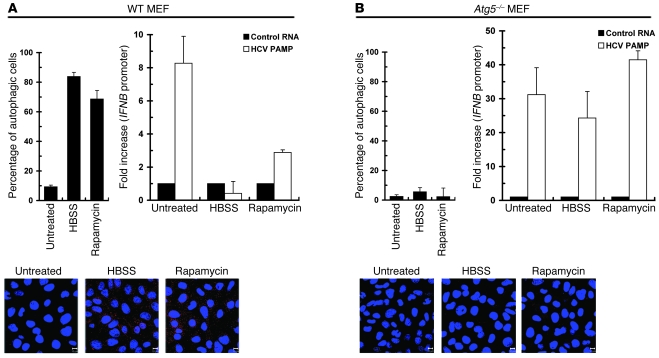
In addition, we examined the effects of silencing of UPR-autophagy on upregulation of the HCV PAMP–mediated innate immune response in the context of no viral infection. Knockdown of LC3B, ATG5, or CHOP by siRNA duplexes was attained by transient transfection of siRNA duplexes (Figure 5A, top left panel). HCV 3′-UTR PAMP–mediated IFNB mRNA induction and IFN-triggered ISG56 expression were greatly amplified in Huh7 cells in which LC3B, ATG5, and CHOP were individually knocked down (Figure 5A, top right and bottom panels, respectively). Also, IFNB mRNA and ISG56 protein expression were further increased in HeLa cells transiently knocked down for ATG5 or CHOP, albeit with a smaller effect than those observed with Huh7 cells (Supplemental Figure 6). Additionally, as shown in Figure 5B, left and right panels, respectively, HCV PAMP RNA was able to trigger further induction of IFNB promoter and ISG56 expression in ATG5-knockout, i.e., Atg5–/–, mouse embryonic fibroblasts (MEFs) (60).
Figure 5. Enhancement of HCV PAMP–mediated IFN response by disruption of UPR-autophagy activation.
(A) Huh7 cells were first transfected with 400 pmol each of control, LC3B, ATG5, or CHOP siRNA duplexes for 72 hours and then harvested for analysis of LC3B, ATG5, CHOP, and β-actin expression (top left panel). The asterisks indicate nonspecific background signals. An uncropped image of A is shown in Supplemental Figure 9, left panel. A portion of siRNA-transfected cells was then transfected with the control HCV 5′-UTR RNA or HCV 3′-UTR PAMP RNA 36 hours after siRNA transfection. Twenty-four hours after PAMP RNA transfection, the cells were harvested for analyses of the IFNB mRNA level (top right) and ISG56 expression (bottom). The fold increase in IFNB mRNA level was determined by normalization to the basal level of control RNA transfection. (B) WT and Atg5–/– MEFs were transfected with pIFN-β/Fluc promoter reporter and pCMV22-Rluc plasmids. Twenty-four hours later, cells were transfected with control HCV 5′-UTR RNA (Control), HCV 3′-UTR PAMP RNA, or DEV PAMP RNA. An additional 24 hours after PAMP RNA transfection, cells were harvested and dual luciferase activity determined (left panel). The fold increase was calculated by normalization to the basal level of control RNA transfection. In parallel, the cells were also analyzed for ISG56, ATG5-ATG12, and β-actin expression by Western blotting (right panel). Data represent mean ± SEM (n = 3) (A, middle panel, and B).
We also examined whether knockdown of UPR modulators upregulates the HCV PAMP RNA–mediated innate immune response, as these UPR modulators controlled the HCV-induced expression of CHOP (Supplemental Figure 3). Stable silencing of each of Ire1α, ATF6, and PERK greatly increased the fold increases in HCV 3′-UTR– and PU/UC PAMPs–mediated IFNB promoter activation (Supplemental Figure 7A).
UPR-autophagy is also required for DEV PAMP–mediated suppression of innate immunity.
Given that several RNA viruses such as DEV and coxsackievirus may activate the UPR and autophagic pathway in their life cycle to benefit their infection (61–63), we then determined whether UPR-autophagy is also required for suppression of other RNA viral PAMP–mediated innate antiviral immunity. The PAMPs derived from DEV, rabies virus (RV), and Ebola virus were examined, since poly-U sequences are also present in the genomes of these 3 RNA viruses, which are known to trigger RIG-I–mediated innate immune signaling (56, 57, 64). As observed in HCV, stable knockdown of ATG5 or CHOP resulted in a greater capacity to activate the IFNB promoter in response to PAMP RNA motifs derived from these 3 viruses (Supplemental Figure 5 and Supplemental Figure 7B). In particular, the DEV PAMP RNA, just like the HCV PAMP, also enhanced IFNB promoter activation and ISG56 expression in Atg5–/– MEFs (Figure 5B). Collectively, these results indicate that HCV and, perhaps, DEV and its flaviviral relatives require the UPR-autophagy pathway to repress innate antiviral immunity.
Enhanced UPR-autophagy by chemical inducers inhibits IFN-β activation.
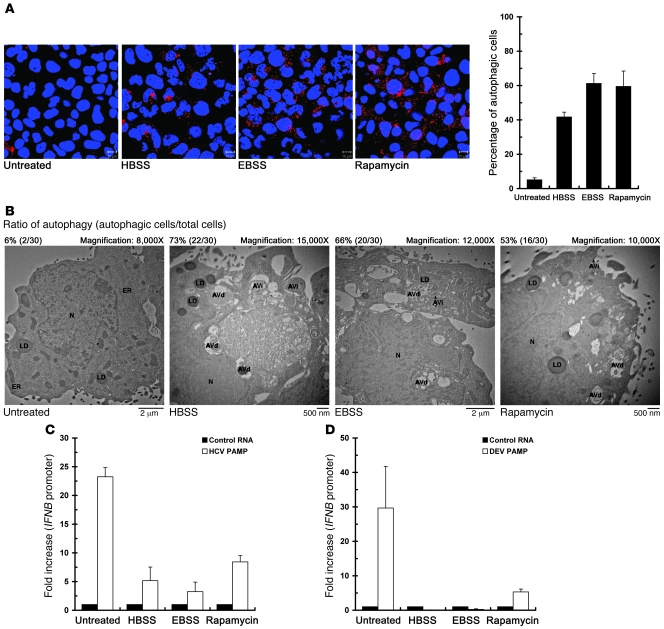
To ascertain the role of UPR-autophagy in modulation of the innate immune response, we then investigated whether induction of the UPR-autophagy signaling may directly downregulate the HCV PAMP RNA–mediated activation of innate immunity. Because nutrient starvation and rapamycin have been shown to induce the autophagic pathway through interference with the insulin receptor and mammalian target of rapamycin (mTOR) signaling cascades (17, 18), we treated Huh7/RFP-LC3 cells with Earle’s balanced salt solution (EBSS), HBSS, or fresh medium containing the mTOR inhibitor rapamycin, and cells were then quantified for the formation of autophagic vacuoles. A large portion of EBSS-, HBSS-, and rapamycin-treated RFP-LC3 reporter cells displayed RFP-LC3–labeled puncta structures (Figure 6A). Consistent with this finding, a higher percentage of autophagic cells containing AVi and AVd structures were detected in treated cells compared with untreated cells (Figure 6B), indicating that nutrient starvation and rapamycin can activate the complete autophagic process in a manner similar to that observed in HCV infection. In these autophagy-activated cells, the HCV- and DEV-PAMP RNA–mediated IFNB promoter activation was greatly reduced (Figure 6, C and D, respectively).
Figure 6. Downregulation of HCV- and DEV-PAMP RNA–triggered IFN-β activation by autophagy inducers.
(A) Huh7/RFP-LC3 cells were treated with EBSS, HBSS, or with fresh medium containing 4 mM rapamycin for 6 hours. Cells were fixed and analyzed for the formation of RFP-LC3B–labeled autophagic vacuoles by confocal microscopy (scale bars: 10 μm). A set of confocal images is shown (left panel). The degree of cells forming autophagic vacuoles was also quantified (right panel). (B) Huh7/RFP-LC3 cells were starved or treated with rapamycin as described in A, and cells were fixed and subjected to TEM analysis. The ratio of autophagy (autophagic cells/total cells) was determined by counting the number of cells containing autophagic vacuoles among the total 30 randomly selected cells. (C) Huh7/RFP-LC3 cells were transfected with the pIFN-β/Fluc promoter reporter and cultured for 24 hours. Cells were then transfected with control HCV 5′-UTR RNA or 3′-UTR PAMP RNA, and maintained for an additional 12 hours. The transfected cells were then treated with chemicals as described in A prior to determination of IFNB promoter activation. The fold increase in IFNB promoter of the PAMP RNA–transfected cells was determined by normalization to the basal level of the control RNA–transfected cells. (D) The effects of EBSS, HBSS, and rapamycin on DEV PAMP RNA–induced IFNB promoter reporter activation were determined as described in C. Data represent mean ± SEM (n = 3) (A, C, and D).
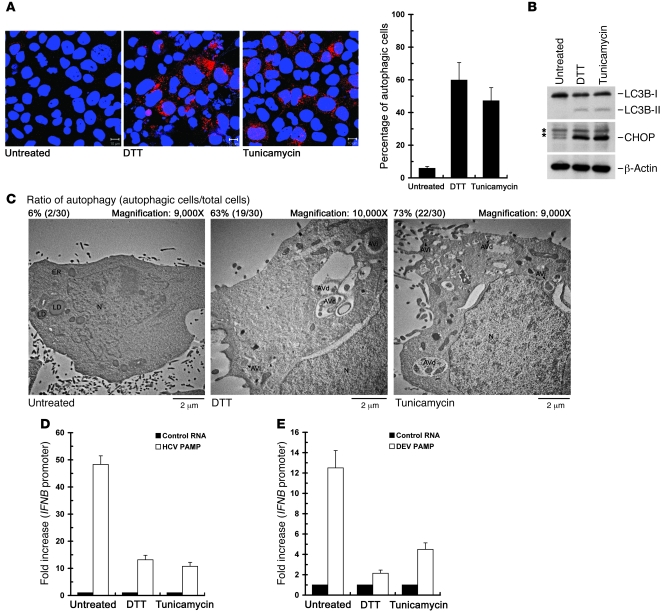
Moreover, we examined whether activation of UPR may lead to inhibition of HCV PAMP RNA–mediated IFN-β activation through the activated autophagic process. DTT and tunicamycin are two well-known inducers of UPR (20–22, 65) and have been reported to trigger the activation of autophagy through the UPR signaling (21, 25). Treatment with either DTT or tunicamycin led to more accumulation of RFP-LC3–labeled puncta structures (Figure 7A), upregulated expression of both CHOP and LC3B-II (Figure 7B), and a higher percentage of cells containing autophagic vacuoles (Figure 7C) in the Huh7/RFP-LC3 cells, indicating their ability to trigger the UPR-autophagy pathway in Huh7 cells. Meanwhile, DTT and tunicamycin reduced HCV- and DEV-PAMP RNA–mediated IFNB promoter activation (Figure 7, D and E, respectively). Likewise, the repressing effects of activated UPR-autophagy by these autophagy and UPR inducers on HCV- and DEV-PAMP RNA–mediated IFNB promoter induction were also observed in HeLa cells (Supplemental Figure 8). Taken together, these results clearly demonstrate that activated UPR-autophagy suppresses the Flaviviridae PAMP–mediated innate immune response.
Figure 7. Suppression of the HCV- and DEV-PAMP RNA–induced IFNB promoter activation by UPR inducers.
(A–C) Huh7/RFP-LC3 cells were left untreated or treated with 2 mM DTT or 4 μg/ml tunicamycin for 6 hours, and then subjected to quantification of RFP-LC3–labeled puncta structure (scale bars: 10 μm) (A), Western blot analysis of the expressions of indicated proteins (B), or TEM analysis of autophagic vacuoles (C). The asterisk in B indicates the nonspecific background signal. An uncropped image of B is shown in Supplemental Figure 9, right panel. The ratio of autophagy in C was determined as described in Figure 6B. (D) Huh7/RFP-LC3 cells were transfected with the pIFN-β/Fluc promoter, followed by transfection with HCV PAMP RNA. The transfected cells were then treated with DTT or tunicamycin as described in A prior to assessment of IFNB promoter activation. The fold increase in IFNB promoter of the PAMP RNA–transfected cells was determined by normalization to the basal level of the control RNA–transfected cells. (E) The effects of DTT and tunicamycin on DEV PAMP RNA–mediated IFNB promoter reporter induction were determined as described in D. Data represent mean ± SEM (n = 3) (A, D, and E).
Deficiency in UPR/autophagy-related genes disrupts chemical inducer–mediated downregulated antiviral innate immunity.
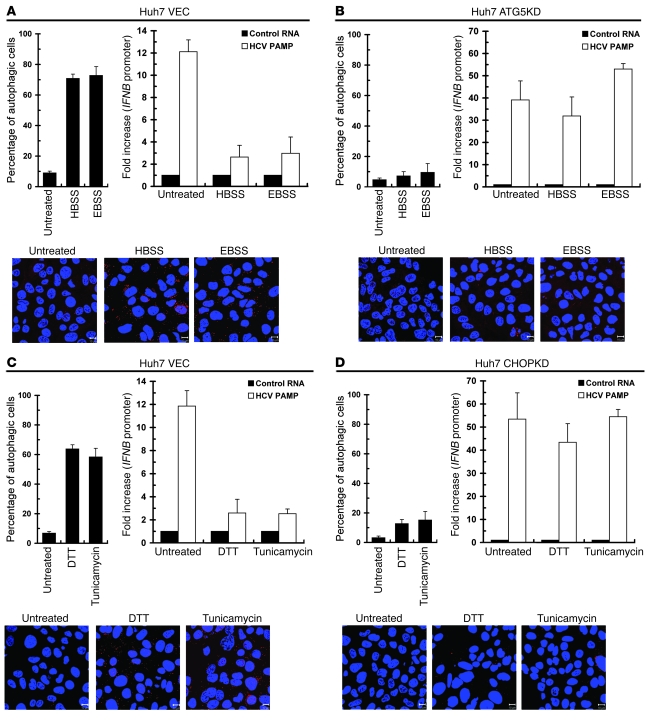
Next, we explored whether EBSS- and HBSS-mediated suppression of IFNB promoter activity truly relies on the activated autophagic process. First, a large number of endogenous LC3B-labeled puncta structures were detected in EBSS- and HBSS-treated VEC cells, but not in the ATG5KD cells treated in parallel (Figure 8, A and B, top left and bottom panels), indicating that nutrient starvation–triggered activation of autophagy is lost in ATG5KD cells. In contrast to the VEC cells, which exhibited reduced HCV PAMP–triggered IFNB promoter activation upon HEBS or EBSS treatment (Figure 8A, top right panel), the HBSS- and EBSS-treated ATG5KD cells still retained a high capacity for IFNB activation in a fashion similar to that of the untreated cells (Figure 8B, top right panel).
Figure 8. The effect of UPR-autophagy gene silencing on chemical inducer–mediated suppression of IFN-β activation.
(A and B) VEC (A) and ATG5KD (B) Huh7 cells were transfected with the IFNB promoter reporter and cultured for 24 hours. The cells were then transfected with HCV control HCV 5′-UTR RNA or 3′-UTR PAMP RNA and maintained for an additional 12 hours. Transfected cells were left untreated, or treated with EBSS or HBSS for 6 hours, and then IFNB promoter reporter activation determined (upper right panels). Another set of cells without DNA and RNA transfections was handled in parallel prior to quantification for endogenous LC3B-labeled puncta structures by immunostaining (upper left and bottom panels, scale bars: 10 μm). (C and D) VEC (C) and CHOPKD (D) Huh7 cells were transfected with the IFNB promoter reporter and HCV 3′-UTR PAMP RNA and then treated with 2 mM DTT or 4 μg/ml tunicamycin for 6 hours before assessment of IFNB promoter reporter activation (upper right panels). Another set of cells without DNA and RNA transfections was handled in parallel and then quantified for LC3B-labeled puncta structures (upper left and bottom panels; scale bars: 10 μm). Data represent mean ± SEM (n = 3).
Moreover, DTT and tunicamycin failed to induce expression of CHOP (data not shown) and formation of endogenous LC3B puncta structures in CHOPKD cells, as compared with those observed in VEC cells (Figure 8, C and D, top left and bottom panels), supporting again the vital role of CHOP in the UPR-induced autophagic process. Intriguingly, the DTT- and tunicamycin-mediated reduction in HCV PAMP RNA–triggered IFNB activation observed in VEC cells was strikingly alleviated in CHOPKD cells (Figure 8, C and D, top right panels). These results together indicate that the downregulation of antiviral innate immunity triggered by chemical inducers requires UPR-autophagy.
Similarly, despite enhancing formation of endogenous LC3B puncta structures and reducing HCV PAMP RNA–mediated IFNB promoter activation in control MEFs (Figure 9A), HBSS and rapamycin lost their inhibitory effect on innate antiviral immunity in Atg5–/– MEFs, which did not show the activated autophagic LC3B punta vacuoles (Figure 9B). These observations are in line with the earlier report that Atg5–/– MEFs are more susceptible to RLR signaling (37). The results indicate that activated autophagy represses viral PAMP–induced innate immunity in a broad spectrum of cells derived from different species.
Figure 9. Effect of Atg5 gene knockout in MEFs on chemical inducer–mediated autophagic activation and suppression of IFN-β activation.
WT (A) and Atg5–/– (B) MEFs were examined for the effects of HBSS and rapamycin on HCV 3′-UTR PAMP RNA–mediated IFNB promoter (upper right panels). Another parallel set of cells without DNA and RNA transfections was quantified for LC3B-labeled puncta structures (upper left and bottom panels, scale bars: 10 μm). Data represent mean ± SEM (n = 3).
CQ ablates rapamycin- and DTT-mediated suppression of antiviral innate immunity.
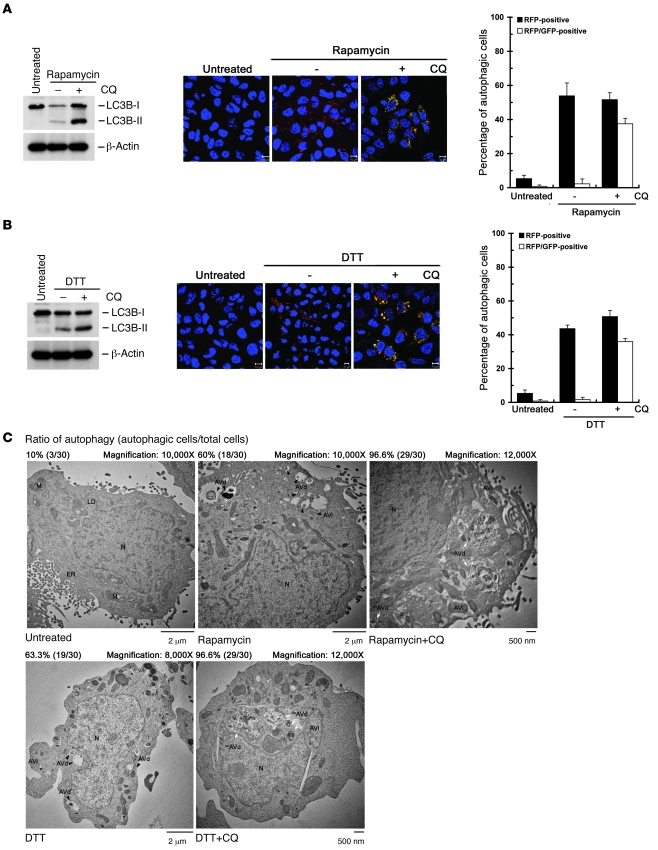
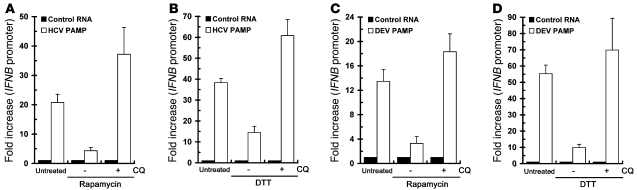
Since HCV-induced complete autolysosome formation was critical for HCV RNA replication (Figure 2B) as well as viral protein expression (Supplemental Figure 2, D and E), we then examined whether the complete autolysosome formation is analogously necessary for the repression of antiviral innate immunity by UPR-autophagy. CQ was shown to block the maturation of autolysosomes and lead to accumulated autophagic vacuoles in cells (49, 66). Cotreatment of rapamycin- and DTT-treated Huh7/mRFP-GFP-LC3 cells with CQ led to the restoration of yellow-colored autophagic vesicles and accumulation of LC3B-II expression, as opposed to rapamycin- and DTT-treated dual reporter cells without CQ administration (Figure 10, A and B, respectively). Similarly, administration of rapamycin- and DTT-treated cells with CQ showed an even higher percentage of autophagic cells harboring large, partial, or even nondegraded AVd (unAVd), which contained a majority of undigested organelles, as opposed to the large population of cells containing only AVi and AVd structures in rapamycin and DTT treatment (Figure 10C), in agreement with a previous study showing that CQ impairs the digestion of autolysosome content (66). Meanwhile, CQ hindered rapamycin- and DTT-mediated repression of HCV PAMP RNA–mediated IFN-β activation (Figure 11, A and B). Rapamycin- and DTT-mediated repression of DEV PAMP RNA–triggered IFN-β activation was also palliated by CQ treatment (Figure 11, C and D). Moreover, pretreatment of CQ and BAF-A1 augmented the HCV PAMP RNA–mediated induction of IFNB mRNA and ISG56 expression (data not shown).
Figure 10. Effect of CQ on rapamycin- and DTT-triggered autophagic process.
(A–C) Huh7/mRFP-GFP-LC3 cells were left untreated or treated with 4 mM rapamycin or 4 mM rapamycin in the presence (+) or absence (–) of 100 μM CQ. Six hours after treatment, cells were analyzed by Western blot analysis (A and B, left panels) and quantified for RFP-LC3–labeled (RFP-positive) and RFP-GFP-LC3–labeled (RFP-GFP-positive, yellow-colored) puncta structures by confocal microscopy (A and B, middle and right panels; scale bars: 10 μm). Another set of reporter cells was treated with drugs according to the procedure described above and fixed and analyzed by TEM for formation of autophagic vacuoles (C). The black arrowheads indicate the AVi or AVd, and the white arrows unAVd. Data represent mean ± SEM (n = 3) (A and B).
Figure 11. Interference with the UPR-autophagy inducer–triggered repression of IFN-β activation by CQ.
(A and B) Huh7/mRFP-GFP-LC3 cells were transfected with the pIFN-β/Fluc promoter reporter and cultured for 24 hours. Cells were then transfected with control HCV 5′-UTR RNA or 3′-UTR PAMP RNA and then maintained for an additional 12 hours. Transfected cells were left untreated or treated with rapamycin (A) or DTT (B) in the presence or absence of CQ as described in the legend to Figure 10, A and B. The fold increase in the IFNB promoter activity of viral PAMP RNA–transfected cells was determined by normalization to the basal level of the control RNA–transfected cells. (C and D) The effect of CQ on rapamycin-induced (C) or DTT-induced (C) suppression of DEV PAMP–mediated IFNB promoter activation was assessed as described in A and B. Data represent mean ± SEM (n = 3).
Knockdown of LAMP2 and Rab7 relieves EBSS- and tunicamycin-induced repression of antiviral innate immunity.
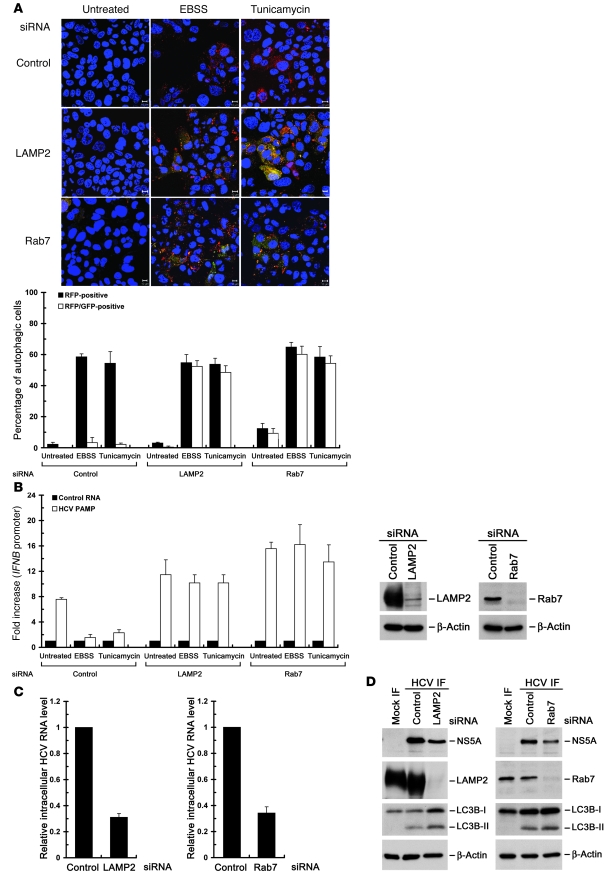
Various cellular proteins such as lysosome-associated membrane protein 2 (LAMP2) and the small GTP-binding protein Rab7 are essential for lysosome biogenesis and participate in the maturation step of autolysosomes (45–47). Disruption of LAMP2 expression and interference with the biological function of Rab7 were shown to result in accumulated autophagosomes and/or partially undigested autolysosomes due to blocking of the final maturation step of late autophagic vacuoles (45–47). We first found that individual silencing of LAMP2 and Rab7 resulted in the rescued GFP signals in EBSS- or tunicamycin-treated Huh7/mRFP-GFP-LC3 cells (Figure 12A), indicating that individual knockdown of LAMP2 and Rab7 specifically inhibits the maturation of autolysosomes. Meanwhile, EBSS- and tunicamycin-activated autophagy lost its ability to repress HCV PAMP RNA–mediated IFNB promoter activation in LAMP2- and Rab7-knockdown cells (Figure 12B). In parallel, knockdown of LAMP2 and Rab7 also reduced the intracellular viral RNA level and NS5A expression in HCV-infected cells (Figure 12, C and D, respectively). Collectively, these results demonstrate that the complete autophagic process is indispensable not only for suppression of the innate immune response, but also for productive HCV replication.
Figure 12. Abrogation of the UPR-autophagy–mediated suppression of IFN-β activation by knockdown of LAMP2 and Rab7.
(A) Huh7/mRFP-GFP-LC3 cells were transfected with 400 pmol each of control, LAMP2, or Rab7 siRNA duplexes for 48 hours, and cells were quantified for RFP-LC3– (RFP-positive) and RFP-GFP-LC3–labeled (RFP-GFP-positive) puncta structures by confocal microscopy (bottom panel). The yellow-colored image represents the RFP-GFP-LC3–labeled puncta structure. A set of representative confocal images is also shown (top panel; scale bars: 10 μm). (B) Huh7/mRFP-GFP-LC3 cells were transfected with 400 pmol each of the indicated siRNA duplexes. Forty-eight hours later, cells were transfected with the pIFN-β/Fluc promoter reporter and cultured for 24 hours. The cells were then transfected with control HCV 5′-UTR RNA or 3′-UTR PAMP RNA and maintained for an additional 12 hours. The cells were left untreated or treated with EBSS or tunicamycin for 6 hours. Cells were then analyzed for IFNB promoter reporter activation (left panel). A set of cells 48 hours after siRNA transfection was analyzed by the Western blotting (right panel). (C and D) Huh7/mRFP-GFP-LC3 cells were infected with HCV at an MOI of 10 and cultured for 6 days. The cells were then transfected with the indicated siRNA duplexes as described in A. Seventy-two hours after transfection, cells were harvested and analyzed for the intracellular HCV RNA level (C) and analyzed for the indicated proteins by Western blotting (D). Data represent mean ± SEM (n = 3) (A–C).
Discussion
Research into the essential role of autophagy in pathogen-host immune responses has been mainly focused on its elimination of pathogens and the activation of the adaptive immune defense for MHC II antigen presentation (31–33). In this study, we demonstrate that infection of HCV specifically activates the UPR as well as the downstream autophagic pathway throughout complete autolysosome formation. We also identify that the activated UPR-autophagy to the stage of autolysosome maturation, conversely, regulates HCV RNA replication by repressing the PAMP-mediated innate immune response.
HCV-induced UPR-autophagy is required for HCV RNA biogenesis.
Recently, challenge of homozygous urokinase plasminogen activator–SCID mice (uPA-SCID) with HCV was shown to activate oxidative and ER stresses in infected hepatocytes (67). Consistent with this finding, we first showed here that infection with HCV activates UPR modulators and CHOP upregulation, which in turn modulate the autophagic response (Figure 2A and Supplemental Figure 3). The observation that CHOP expression coincided with the lipidation of LC3B (Figure 1A) is in line with the recent findings that UPR-triggered CHOP promotes the activation of autophagic process through the transcriptional control of ATG5 and LC3B (68, 69). On the other hand, we showed that HCV-induced UPR-autophagy is conversely required for HCV RNA replication. First, lentiviral shRNA-mediated knockdown of each of the signaling molecules involved in UPR-autophagy resulted in a reduction in viral RNA replication (Figure 2B). Second, disruption in UPR-autophagy neither affected HCV entry or HCV IRES-mediated translation nor altered the translation of incoming viral RNAs (data not shown). Instead, it greatly affected early-stage viral RNA biogenesis (Figure 2C).
HCV-activated autolysosome formation is required for viral RNA replication.
Moreover, we showed that complete autophagy is necessary for HCV RNA replication. Several lines of evidence supporting this proposition include (a) fusion of autophagosome with lysosome by confocal microscopy, and immunogold-TEM (immuno-TEM) analyses of AVi and AVd in HCV-infected cells (Figure 1, B and C, and Supplemental Figure 2, A and C); (b) accumulation of HCV-induced LC3B-II upon treatment with lysosomal inhibitors (E64 and PepA) and acidification inhibitors (CQ and BAF-A1) (Supplemental Figure 2, D and E); and (c) reduction of HCV RNA replication and infectious virus production by CQ and BAF-A1 (Figure 2B and data not shown). Most importantly, the observations that individual knockdown of LAMP2 and Rab7, which impaired complete autolysosome maturation (Figure 12A), inhibited both HCV RNA replication and viral protein expression (Figure 12, C and D) are also in accordance with this viewpoint.
Activation of UPR and complete autolysosome fusion promotes HCV replication through suppression of innate immunity.
In this study, we also showed that several important UPR-autophagy molecules, such as CHOP, ATG5, LC3B, and the 3 UPR modulators, play a negative role in regulation of HCV PAMP–triggered innate immune responses, including the IFNB mRNA level, IFNB promoter activation, the paracrine antiviral activity of IFN, as well as the ISG56 expression (Figures 3–5, Supplemental Figure 6, and Supplemental Figure 7A). These results imply that suppression of innate immunity upon HCV infection relies on the entire UPR-activated autophagy pathway, not simply on individual ATG5 or CHOP gene expression. HCV PAMP–induced IFN-β activation could be amplified even in the context of no viral infection in Huh7, HeLa, and MEFs in which UPR-autophagy–related genes were knocked down or knocked out (Figure 4, A and B, Figure 5, A and B, and Supplemental Figure 6). Indeed, stable knockdown of ATG5 and CHOP in Huh7 cells and knockout of ATG5 in MEFs interfered with the formation of autophagic vacuoles induced by UPR-autophagy inducers (Figures 8 and 9). The UPR-autophagy preactivation approach by chemical inducers further assured the suppressive effect of UPR-autophagy on HCV PAMP–mediated IFN-β activation (Figures 6 and 7 and Supplemental Figure 8, A and B). Interestingly, stable knockdown or knockout of UPR-autophagy genes and CQ treatment counteracted the negative role of UPR-autophagy inducers in innate immune response (Figures 8–11). Accompanied by ablation with chemical inducer–mediated suppression of antiviral innate immunity (Figure 11), CQ led to accumulated unAVd structures in the majority of rapamycin- and DTT-treated cells (Figure 10C), supporting the notion that the function of autolysosomes is crucial for UPR-autophagy–mediated suppression of innate immunity. More importantly, separate knockdown of LAMP2 and Rab7 abolished the counteracting effect of UPR-autophagy inducers on downregulated innate immunity, as well as impairing HCV RNA replication and viral protein expression (Figure 12). All these results convincingly underscore the pivotal roles of the induction of UPR-autophagy and complete autolysosome formation in the repression of the innate immune response as well as the promotion of HCV RNA replication.
The UPR-autophagic pathway is a crucial alternative for HCV to escape innate immunity.
Cheng et al. reported that ectopic expression of RIG-I restores dsRNA signaling and IFN-β activation in HCV-infected cells, indicating that excess RIG-I may protect MAVS from proteolytic cleavage by NS3/4A (42). Under more physiological conditions, inoculation of HCV into chimpanzees induces the liver-specific overexpression of RIG-I, which can circumvent the NS3/4A-mediated cleavage of MAVS and thus trigger the innate antiviral response to restrict HCV replication and pathogenecity (70). In addition to the NS3/4A-mediated suppression of innate immunity, Cheng et al. also found that dsRNA-induced IFNB promoter activity in HCV-infected cells cannot be restored by an NS3 protease inhibitor, BILN2061 (42), suggesting the existence of NS3/4A-independent signaling to counteract HCV-mediated innate immunity, in particular when NS3/4A is present at an insufficient level to shut down MAVS signaling. In accordance with this hypothesis, our findings here show that HCV can pirate the cellular UPR-autophagy pathway to escape the innate immune response in an NS3/4A-independent fashion.
The current study revise the model of HCV-cell interaction.
Autophagy has been implicated in various steps in the HCV life cycle, such as HCV RNA replication, translation of incoming viral RNA, or viral particle production (25). In the present study, we provide evidence to demonstrate the tightly associated effect of activated UPR-autophagy on downregulation of the innate immune response and enhancement of HCV RNA replication. Our results thus reveal a paradigm of HCV–host cell interaction, i.e., HCV RNA replication can be finely tuned by the interplay between UPR-autophagy and innate immunity.
What causes the disparities among these previous studies and our finding is presently unclear. Since individual genes involved in autophagy may regulate vesicle trafficking and protein translocation as well (35, 71), it is likely that silencing of different autophagy-related genes may exert an effect on the cellular function other than modulation of the autophagic pathway. The highly heterogeneous nature of Huh7 cells (72); the different Huh7 and derivative clones, such as the Huh7.5 clone, which harbors a defective mutation in the RIG-I gene locus (73); and/or the different strategies of HCV expression, such as RNA transfection versus infection, used in different laboratories may account for the different conclusions obtained, since activation of autophagic process varies among different cells types and also depends on the cellular context (51).
As seen in HCV, knockdown of ATG5 or CHOP increased the IFN-β activation mediated by other viral RNA PAMP motifs (Supplemental Figure 7B). The enhancement in UPR-autophagy by inducers also reduced DEV PAMP RNA–mediated IFN-β activation (Figure 6D, Figure 7E, and Supplemental Figure 8, C and D), and this reduction was relieved by inhibition of autolysosome formation, such as with CQ (Figure 11, C and D). Thus, our present study may conceptualize a new mode of host cell–virus interactions, i.e., many membrane-enveloped RNA viruses, particularly, the flaviviruses, may exploit UPR-autophagy signaling as a general mechanism to evade the innate immune defense, thereby promoting their infections and growth.
Attenuation in the UPR-autophagic pathway provides a potential therapeutic application.
The results presented here corroborate the notion that UPR-autophagy can serve as an ideal target for the development of anti-HCV agents and/or intervening approaches. CQ and BAF-A1 were reported to have an inhibitory role in HCV entry (74, 75), possibly through their perturbing role in the endocytic pathway. CQ is a drug widely used in the treatment of malaria (76) and a potential anti-HIV agent, as it interferes with the glycosylation of envelope glycoprotein 120 (gp120) in the trans-Golgi apparatus, which requires a low pH (77). Nevertheless, our present results provide a mechanistic basis for the therapeutic application of CQ and BAF-A1 in order to intervene in HCV replication via inhibition of complete autolysosome formation. Most importantly, inhibition of autolysosome maturation by CQ also counteracted the UPR-autophagy–mediated repression of viral PAMP–mediated antiviral innate immunity (Figures 10 and 11). Our results together indicate that interference with the UPR-autophagy pathway could amplify the cellular capacity of immune response to RLR signaling. Along this line of thinking, small molecule inducers of the cytostatic effects of rapamycin have been shown to enhance the autophagic clearance of A35T α-synuclein and reduce the toxicity of aggregated proteins in a Huntington disease model (78), implying that small molecule modulators of autophagy, including enhancers or inhibitors, will have practical applications in the treatment of neurodegenerative diseases, and possibly infectious diseases as well.
Perspectives.
Herein, we report what we believe to be a novel regulatory role of UPR-autophagy in HCV RNA replication through its suppression of innate immunity. Our findings suggest that UPR-autophagy at the basal level may sense incoming HCV, inhibiting the HCV PAMP–triggered innate immune response, thereby supporting initial HCV RNA synthesis during the early stage of HCV infection, at which time the level of NS3/4A expression may be insufficient to downregulate the inhibitory effect of innate immunity on HCV RNA replication. When the HCV RNA replicates more vigorously, the expression of HCV further induces UPR-autophagy to suppress the antiviral response and to promote viral RNA replication. Nevertheless, as viral protein expression proceeds in a time-dependent manner upon HCV infection (Figure 1A), this implies that HCV infection may also trigger an as-yet-unidentified cellular mechanism to curtail the promoting effect of HCV-activated UPR-autophagy on virus replication and/or expression in order to maintain virus cell homeostasis. Our findings thus contribute to the understanding of the physiological significance of UPR-autophagy in HCV replication, but also provide a framework for the rational design of feasible anti-HCV drugs or intervening applications.
Methods
Cells, antibodies, reagents, and siRNA oligonucleotides.
Huh7, HeLa, and 293T cells were cultured at 37°C in DMEM containing 10% FBS under a 5% CO2 atmosphere. In particular, nonessential amino acids were added to the Huh7 culture medium at a final concentration of 1%. WT and Atg5–/– MEFs were obtained and maintained as describe previously (60). Rabbit anti-core antibody was raised by immunization of New Zealand white rabbits with a purified recombinant core antigen (Genesis Biotech Inc., Hsin-Tien, Tapei, Taiwan) (79). The anti-NS5A mouse mAb (9E10) (39), mouse anti-core (2H9) mAb (80), and rabbit anti-NS5A (CL1) antibody (81) were as previously described in the references cited. A human anti-NS3 mAb was harvested from the cell culture supernatants of the CM3B6 hybridoma cell line. Mouse anti–β-actin mAb was obtained from Invitrogen. Rabbit anti-LC3B, human ATG5, and anti-CHOP mAb were obtained from Cell Signaling Technology. Mouse anti-PKR, anti–proliferation cell nuclear antigen (anti-PCNA), and anti-LAMP1 mAbs, and rabbit anti-ATF6, –XBP-1, -PERK, and –phospho-PERK polyclonal antibodies were all from Santa Cruz Biotechnology Inc. Rabbit polyclonal anti–mouse ATG5 antibody and anti-ISG56 antibody were purchased from MBL and Thermo Scientific, respectively. The Alexa Flour–conjugated secondary antibodies and DAPI were obtained from Invitrogen. IFN α-2a was purchased from Roche. Polybrene, E64, pepstatin A, CQ, bafilomycin A1, dithiothreitol, rapamycin, and tunicamycin were obtained from Sigma-Aldrich. HBSS and EBSS were obtained from Invitrogen and Sigma-Aldrich, respectively. The Bio-Rad protein assay kit was used. siRNA duplexes against human LC3B and a negative control siRNA that does not target any mammalian genes were synthesized and re-annealed as follows: LC3B (sense: 5′-CUAGAUAGUUACACACAUATT-3′ and anti-sense: 5′-UAUGUGUGUAACUAUCUAGTT-3′) and control (sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and anti-sense: 5′-ACGUGACACGUUCGGAGAATT-3′). The siRNA duplexes against human ATG5 (HSS114103 and HSS114104), CHOP (VHS40605 and VHS40607), Ire1α (HSS140846 and HSS140846), ATF6 (HSS177036 and HSS177037), PERK (HSS190343 and HSS190344), LAMP2 (HSS105968 and HSS105967), and Rab7 (HSS111821 and HSS187913) were purchased from Invitrogen Stealth RNAi collection.
Plasmids.
The pUC-JFH1 (40, 82) and pFL-JFH1-Luc (52) plasmids, pmRFP-LC3 (45), pmRFP-GFP-LC3 (45), and pGFP-LC3 (48), and the IFN-β/Fluc reporter and pEF-BOS-FLAG-RIGI-N constructs (55) were as previously described in the references cited. pHCV-N-Fluc is a firefly luciferase gene–encoding bicistronic HCV genotype 1b replicon. pT7-EMCV-IRES-RLuc is a plasmid encoding a Renilla luciferase gene driven by the encephalomyocarditis virus (EMCV) IRES, which is cloned downstream of the T7 promoter sequence. The pISRE-Fluc encodes the firefly luciferase gene driven by the ISRE promoter. pMD.G, pCMVΔR8.91, and pLKO.As1 were obtained from the National RNAi Core Facility, Academia Sinica. The detailed sequences of these shRNAs are listed in Supplemental Table 1.
Construction of plasmids.
To construct pcDNA3-T7-3′-UTR and pcDNA3-T7-PU/UC plasmids, the PCR-amplified 3′-UTR (nt 9389–9679) and PU/UC (nt 9436–9600) fragments flanked with HindIII and XbaI recognition sites at the 5′ and 3′ ends, respectively, were digested with the corresponding restriction enzymes and then subcloned at the same sites of the pcDNA3 vector (Invitrogen). To generate lentiviral shRNA constructs, the shRNA (sense-loop-antisense) sequences of target genes were cloned into the BsmBI site of the pLKO.As1 vector. The pCMV22-Rluc was constructed by insertion of the PCR-amplified fragment of the Renilla luciferase gene from pGL4.74 (Promega) into the HindIII and EcoRI sites of the pCMV22 vector (Sigma-Aldrich). The sequence of PCR-amplified fragment in each plasmid used in this study was confirmed by the automatic DNA sequencing method.
Generation of viral genomic and PAMP motif RNAs.
Generation of JFH1 and JFH1-Luc RNAs was performed as previously described (39–41). Briefly, pUC-JFH1 and pFL-JFH1-Luc (Supplemental Figure 1) were linearized by XbaI and MluI digestion, respectively, and treated with mung bean nuclease followed by purification and used as templates for in vitro RNA synthesis. HCV RNA was in vitro transcribed by T7 polymerase using a T7 RiboMAX Express kit (Promega), and the synthesized RNAs were purified by an RNeasy Mini Kit (QIAGEN). For synthesis of HCV-N-Fluc replicon RNA, the HCV-N-Fluc replicon was linearized by BamHI digestion, purified, and used as a template for in vitro RNA transcription. For synthesis of EMCV-IRES-Rluc, pT7-EMCV-IRES-Rluc (provided by Huey-Nan Wu, Academia Sinica) was linearized by MluI, purified, and used as a template for in vitro synthesis of RNA as described above.
The synthesis of 3′-UTR and PU/UC RNAs containing 5′-triphosphates was performed as previously described (56, 57). Briefly, the pcDNA3-T7-3′-UTR and pcDNA3-T7-PU/UC plasmids were linearized by XbaI digestion, purified, and used as the template for in vitro RNA synthesis. To generate the HCV 5′-UTR (nt 1–155) RNA motif, pUC-JFH1 was linearized by AgeI instead and used as a template for in vitro RNA transcription.
The generation of DEV PAMP RNA (3′-UTR) was performed as previously described (57). Briefly, The 3′-UTR (nt 10273–10723) of DEV2 (PL046 strain) (provided by Yi-Lin Lin, Academia Sinica) fragment flanked with HindIII and XbaI recognition sites at the 5′ and 3′ ends, respectively, was subcloned into the downstream of the T7 promoter sequence within pCR-BluntII using the pCR-Blunt II-TOPO kit (Invitrogen) to yield pCR-BluntII-DEV3′-UTR. The pCR-BluntII-DEV3′-UTR was then linearized by XbaI, purified, and used as a template for in vitro RNA synthesis. The rabies virus PAMP (leader RNA nt 11870–11925) and Ebola virus PAMP (nt 1–42) RNA motifs were synthesized according to the procedure described by Saito et al. (56). The sense and antisense primers containing the T7 promoter sequence and the corresponding nucleotide sequence of the viral genome were annealed and used as a template for in vitro transcription.
Viral RNA and siRNA duplex transfections.
Ten micrograms of purified HCV viral RNAs were transfected into 6 × 106 Huh7 cells by electroporation using the Neon MicroPorator MP-100 (Promega). Transfected cells were immediately transferred into one 15-cm culture dish containing 15 ml culture medium. The cell density of transfected cells was maintained at 50%–70% confluence by cell passage every 3 days. For siRNA duplex transfections and PAMP-triggered activation of IFNB promoter assays, 400 pmol of siRNA duplexes and 4 μg of PAMP RNA motifs, respectively, were transfected into Huh7 cells grown on 6-well plates using the Neon MicroPorator MP-100 and Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
Replication kinetic assay of HCV RNA.
VEC, ATG5KD, and CHOPKD cells were cotransfected with EMCV-IRES-Rluc RNA along with JFH1-Luc RNA or HCV-N-FLuc RNA by electroporation. The transfected cells were harvested at different times as indicated in Figure 2C and analyzed for firefly and Renilla luciferase activities by the Dual-Luciferase Reporter Assay System (Promega) with a Berthold Luminometer (Berthold Detection Systems). The luciferase activity of EMCV-IRES-Rluc was measured at 4 hours after transfection and used for normalization of the transfection efficiency.
DNA transfections.
For plasmid DNA transfections, Huh7 cells grown on 6-well plates, which were seeded at a density of 5 × 105 per well 18 hours prior to transfection, were transfected with a total of 4 μg of plasmid DNA using Lipofectamine 2000 reagent. To prepare shRNA lentiviral stocks, HEK 293T cells were seeded in a 6-cm dish at a density of 8 × 105 cells per well 18 hours prior to transfection. 293T cells were then transfected with 2.5 μg of pLKO.As1-shRNA plasmid DNA, 0.25 μg of pMD.G, a VSV envelope glycoprotein G–expressing plasmid, and 2.25 μg of pCMVΔR8.91 packaging plasmid by the calcium phosphate coprecipitation method.
For analysis of activation of the IFNB promoter by the N-terminal fragment of RIG-I, Huh7 cells grown on 6-well plates were cotransfected with 2 μg of pIFN-β/Fluc, 0.2 μg of pCMV22-Rluc, and 2 μg of the empty vector or pEF-BOS-FLAG-RIGI-N using the Lipofectamine 2000 transfection method. Twenty-four hours after DNA transfection, cells were harvested for analysis by the Dual-Luciferase Reporter Assay System. To determine the IFN-2α–induced stimulation of the ISRE promoter, Huh7 cells grown on 6-well plates were transfected with 2 μg of pISRE-Fluc and 0.2 μg of pCMV22-Rluc plasmids by Lipofectamine 2000 reagent. Twenty-four hours after DNA transfection, cells were treated with or without 100 units of IFN-α–2a for 16 hours before analysis by the Dual-Luciferase Reporter Assay. The Renilla luciferase unit was used to normalize the transfection efficiency.
HCVcc infection and titration of virus infectivity assays.
To harvest infectious HCVcc, the conditioned media from JFH1- and its derivative RNA–transfected cells were clarified by centrifugation at 3,000 g for 10 minutes and sterile-filtered through a 0.2-μm pore cellulose acetate disc filter. The culture supernatants containing HCVcc were aliquoted and stored at 4°C.
HCV infection was performed as described previously (39). Briefly, 2 × 105 Huh7 cells were seeded on a 10-cm dish 12 hours prior to infection. Then, the cells were inoculated with supernatants containing the indicated MOI of HCVcc, which were supplemented with 20 mM HEPES, pH 7.5, and 8 μg/ml Polybrene. Twelve hours after incubation, cells were washed and replenished with fresh medium.
shRNA lentiviral vector transduction analyses.
For shRNA lentiviral transduction assays, culture supernatants containing shRNA lentiviral vectors were collected over 48–60 hours after transfection, followed by brief centrifugation and filtration. Lentiviral shRNAs at an MOI of approximately 5 were used to infect Huh7 cells in the presence of 10 μg/ml polybrene. Four days after transduction, cells were harvested to analyze for proteins of target genes and HCV RNAs.
Generation of stable cells.
To generate Huh7/GFP-LC3, Huh7/RFP-LC3, and Huh7/mRFP-GFP-LC3 stable cells, Huh7 cells were transfected with pGFP-LC3, pmRFP-LC3, and pmRFP-GFP-LC3, respectively, using Lipofectamine 2000 and then selected with 1 mg/ml G418 for 2 weeks. The surviving cells were pooled and maintained in medium supplemented with 500 μg/ml G418. The expression of GFP-LC3, RFP-LC3, and RFP-GFP-LC3 proteins was examined by fluorescence microscopy. The ATG5-, CHOP-, Ire1α-, PERK-, and ATF6-stable-knockdown Huh7 cells were established by selection of the corresponding lentiviral shRNA–transduced cells with 5 μg/ml puromycin for 2 weeks, and the surviving cells were maintained in medium supplemented with 2 μg/ml puromycin.
Immunofluorescence and confocal microscopy.
Cells were fixed with 4% paraformaldehyde prepared in PBS for 30 minutes at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 10 minutes. After 3 washes with PBS, cells were incubated with the specific primary antibodies (LAMP1 Ab at a dilution of 1:100; NS5A Ab at a dilution of 1:5,000), followed by incubation with the cognate Alexa Fluor–conjugated secondary antibodies, respectively. For analysis of endogenous LC3B subcellular localization, cells were fixed by ice-cold methanol for 20 minutes and then permeabilized with 0.05% saponin prepared in PBS containing 2% FBS for 30 minutes. After blocking with 2% FBS for 30 minutes, cells were incubated with LC3B antibody (Sigma-Aldrich) at a dilution of 1:1,000 for 2 hours and then incubated with Alexa Fluor 555–conjugated anti-rabbit secondary antibody and DAPI (blue), which stains the nucleus, for 1 hour. The images were analyzed by LSM510 META Laser Scanning Confocal Microscopy and its associated software (Zeiss).
TEM and immuno-TEM.
For TEM analysis, cells were harvested and washed with PBS 3 times, each time for 5 minutes. Cells were then fixed with fixative I (2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.2) for 2 hours at room temperature and then incubated with fixative II (1% osmium tetroxide in 0.1 M sodium cacodylate buffer, pH 7.2) for 1 hour at 4°C. Then, the cells were dehydrated in a graded series of ethanol and embedded in Epon 812. Ultrathin (80-nm) cell sections were stained with saturated uranyl acetate and Reynolds lead citrate solution. For immuno-TEM analysis, cells were fixed, prepared for ultrathin cryosectioning, and labeled with immunogold in accordance with a previously described protocol (83). Briefly, cells were fixed by 2% formaldehyde and 1% acrolein for 1 hour and then washed once in PBS/0.02 M glycine. The washed cells were then scraped into and embedded in 12% gelatin in PBS. The cell-gelatin was cut into 1-mm blocks, infiltrated by 2.3 M sucrose, mounted on an aluminum pin, and frozen in liquid N2, after which the sample was ultrathin-sectioned and picked up in a mixture of 50% sucrose and 50% methylcellulose. Then samples were incubated with mouse anti-core (1:100) or rabbit anti-NS5A (1:500) and the cognate anti-mouse and anti-rabbit secondary antibodies, which were, respectively, labeled with 12-nm and 18-nm gold particles (Jackson ImmunoResearch Laboratories Inc.). For the double-immunogold labeling in Supplemental Figure 2, samples were incubated with a combination of mouse anti-core (1:100) and rabbit anti-LC3B (1:200), or rabbit anti-NS5A (1:500) and mouse anti-LAMP1 (1:20), followed by incubation with the corresponding protein A–gold secondary antibodies. Electron micrographs were obtained with a Hitachi H-7000 Transmission Electron Microscope.
UPR-autophagy activation and inhibition of autolysosome maturation.
For induction of autophagy, Huh7/RFP-LC3 stable cells were treated with EBSS, HBSS, or complete medium supplemented with 4 mM rapamycin for 6 hours. To activate UPR and autophagy, Huh7/RFP-LC3 stable cells were treated with 2 mM DTT or 4 μg/ml tunicamycin for 6 hours. For the inhibition of autolysosome maturation by CQ and knockdown of LAMP2 or Rab7, Huh7m/RFP-GFP-LC3 cells were treated with chemical inducer with or without CQ, or first transfected with siRNA duplexes targeted to the desired genes, followed by treatment with or without chemical inducers as indicated in the legend to each corresponding figure.
Quantification of autophagic cells.
The percentages of autophagic cells were determined by calculating the numbers of cells containing more than 5 dot-like structures of RFP-LC3, RFP-GFP-LC3, or endogenous LC3B from 5 fields containing more than 100 randomly selected cells in the microscopy-captured images.
SDS-PAGE and Western blotting.
Cells were lysed in RIPA buffer containing a protease inhibitor cocktail (Roche). For preparation of the nuclear extracts, the protocol of David Ron’s laboratory at New York University (84) was followed. Protein concentration was determined based on the Bradford method using the Bio-Rad protein assay kit. Equal amounts of protein were separated by SDS-PAGE and electrophoretically transferred onto a nitrocellulose membrane (Millipore). After blocking with 3% nonfat milk in Tris-buffered saline containing 0.2% Tween-20, the membrane was incubated with specific primary antibodies, followed by incubation with appropriate horseradish peroxidase–conjugated secondary antibodies. Signals were detected using an enhanced chemiluminescence system (Millipore) according to the manufacturer’s instructions.
Real-time RT-PCR.
For real-time RT-PCR analysis, total cellular RNA was extracted with Trizol reagent. Two micrograms of total cellular RNA was reverse-transcribed into cDNA by a high-capacity cDNA reverse transcription kit (Applied Biosystems). The synthesized cDNAs were immediately applied into quantitative real-time PCR using the TaqMan gene expression system and detected with the ABI Prism 7700 sequence detection system (Applied Biosystems). The relative amounts of HCV and IFN-β RNAs were calculated with the comparative Ct method (ΔΔCt) and normalized to the endogenous levels of GAPDH. The HCV forward and reverse primers were 5′-GCGGAACCGGTGAGTACAC-3′ and 5′-GGCATAGAGTGGGTTTATCCAAGAA-3′, respectively, and the probe sequence was 6FAM-5′-TTCCCGGCAATTCC-3′-MGBNFQ. The primers and probes for human CHOP (Hs99999172_m1), IFN-β (Hs00277188_s1), and GAPDH (Hs99999905_m1) were obtained from the TaqMan gene expression assay (Applied Biosystems).
Supplementary Material
Acknowledgments
We are indebted to the following individuals for providing the plasmids and antibodies used in this study: Takaji Wakita (pUC-JFH1; National Institute of Infectious Diseases, Tokyo, Japan), Ralf Bartenschlager (pFL-JFH1-Luc; University of Heidelberg, Heidelberg, Germany), Noboru Mizushima (pGFP-LC3, WT and Atg5–/– MEFs; Tokyo Medical and Dental University, Tokyo, Japan), Tamotsu Yoshimori (pmRFP-LC3 and pmRFP-GFP-LC3; Osaka University, Osaka, Japan), Michael Lai (HCV-N-Fluc replicon; Academia Sinica), Takashi Fujita (pIFN-β/Fluc and pEF-BOS-Flag-RIGI-N; Kyoto University, Kyoto, Japan), Yi-Lin Lin (pISRE-Fluc and DEV2 PL046 genome construct; Academia Sinica), Huey-Nan Wu (pT7-EMCV-IRES-Rluc; Academia Sinica), Charles Rice (9E10 mouse anti-NS5A MAb; The Rockefeller University, New York, New York, USA), Kunitada Shimotohno (CL1 rabbit anti-NS5A antibody; Chiba Institute of Technology, Chiba, Japan), and Tetsuro Suzuki (2H9 mouse anti-core antibody; National Institute of Infectious Diseases, Tokyo, Japan). The CM3B6 hybridoma cell line was obtained from Winand Habets (University of Pavia Medical School, Policlinico San Matteo, Pavia, Italy) through the NIH AIDS Research and Reference Reagent Program. We acknowledge the National RNAi Core Facility (Taiwan) for providing lentiviral shRNA expression plasmids and pLKO.As1, pMD.G, and pCMVΔR8.91. We also thank the core facilities of the Institute of Biomedical Sciences and Tzu-Han Hsu at the Institute of Cellular and Organismic Biology at Academia Sinica for technical assistance in confocal microscopy and TEM analyses, respectively. This study was supported by Foresight Research Grants (AS97FP-L15-1, AS98FP-L15-1, and AS99FP-L15-1) from Academia Sinica and a research grant (NSC95-2320-B-001-032-MY3) from the National Science Council, Taipei, Taiwan.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(1):37–56. doi:10.1172/JCI41474.
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5 suppl 1):S21–S29. doi: 10.1002/hep.1840360704. [DOI] [PubMed] [Google Scholar]
- 3.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 4.Simmonds P, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 5.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 6.Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 7.Gao B, Hong F, Radaeva S. Host factors and failure of interferon-α treatment in hepatitis C virus. Hepatology. 2004;39(4):880–890. doi: 10.1002/hep.20139. [DOI] [PubMed] [Google Scholar]
- 8.Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436(7053):930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 9.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436(7053):933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 10.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5(6):453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 11.Egger D, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76(12):5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosert R, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77(9):5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 14.Lee DY, Lee J, Sugden B. The unfolded protein response and autophagy: herpesviruses rule! J Virol. 2009;83(3):1168–1172. doi: 10.1128/JVI.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, et al. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J Microbiol. 2005;43(6):529–536. [PubMed] [Google Scholar]
- 16.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem. 2004;279(17):17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 17.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 21.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65(6):862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wileman T. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu Rev Microbiol. 2007;61:149–167. doi: 10.1146/annurev.micro.57.030502.090836. [DOI] [PubMed] [Google Scholar]
- 24.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82(5):2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48(4):1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106(33):14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5(7):937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 28.Katze MG, Fornek JL, Palermo RE, Walters KA, Korth MJ. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat Rev Immunol. 2008;8(8):644–654. doi: 10.1038/nri2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 30.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5(6):527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 33.Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16(1):57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27(7):1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106(49):20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 37.Jounai N, et al. The Atg5-Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 40.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102(26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng G, Zhong J, Chisari FV. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2006;103(22):8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loo YM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103(15):6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H, et al. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133(5):1649–1659. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 45.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 46.Eskelinen EL, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13(9):3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jager S, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117(pt 20):4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 48.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mousavi SA, Kjeken R, Berg TO, Seglen PO, Berg T, Brech A. Effects of inhibitors of the vacuolar proton pump on hepatic heterophagy and autophagy. Biochim Biophys Acta. 2001;1510(1–2):243–257. doi: 10.1016/s0005-2736(00)00354-0. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23(1):33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koutsoudakis G, et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80(11):5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foy E, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102(49):17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 56.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83(9):4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grandvaux N, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76(11):5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281(45):34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 60.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 61.Khakpoor A, Panyasrivanit M, Wikan N, Smith DR. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J Gen Virol. 2009;90(pt 5):1093–1103. doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- 62.Lee YR, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374(2):240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoon SY, et al. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82(23):11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14(9):1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 66.Gray RH, Sokol M, Brabec RK, Brabec MJ. Characterization of chloroquine-induced autophagic vacuoles isolated from rat liver. Exp Mol Pathol. 1981;34(1):72–86. doi: 10.1016/0014-4800(81)90037-X. [DOI] [PubMed] [Google Scholar]
- 67.Joyce MA, et al. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS Pathog. 2009;5(2):e1000291. doi: 10.1371/journal.ppat.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rouschop KM, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120(1):127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salazar M, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119(5):1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bigger CB, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78(24):13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16(7):956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 72.Akazawa D, et al. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81(10):5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76(24):13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blanchard E, et al. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80(14):6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80(4):1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hall AP. Preventing deaths from malaria. Br Med J. 1978;2(6141):877–879. doi: 10.1136/bmj.2.6141.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6(2):67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3(6):331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ai LS, Lee YW, Chen SS. Characterization of hepatitis C virus core protein multimerization and membrane envelopment: revelation of a cascade of core-membrane interactions. J Virol. 2009;83(19):9923–9939. doi: 10.1128/JVI.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masaki T, et al. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol. 2008;82(16):7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9(9):1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 82.Kato T, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125(6):1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 83.Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2(10):2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- 84. Ron Lab Home Page: Lab Protocols. Heather Harding and Huiquing Zeng’s protocol for isolating both cytoplasmic and nuclear extracts. http://saturn.med.nyu.edu/research/mp/ronlab/protocols/NucCyto.html . Updated October 29, 2002. Accessed October 27, 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.