Abstract
Autoimmune diseases develop in approximately 5% of humans. They can arise when self-tolerance checkpoints of the immune system are bypassed as a consequence of inherited mutations of key genes involved in lymphocyte activation, survival, or death. For example, autoimmune lymphoproliferative syndrome (ALPS) results from defects in self-tolerance checkpoints as a consequence of mutations in the death receptor–encoding gene TNF receptor superfamily, member 6 (TNFRSF6; also known as FAS). However, some mutation carriers remain asymptomatic throughout life. We have now demonstrated in 7 ALPS patients that the disease develops as a consequence of an inherited TNFRSF6 heterozygous mutation combined with a somatic genetic event in the second TNFRSF6 allele. Analysis of the patients’ CD4–CD8– (double negative) T cells — accumulation of which is a hallmark of ALPS — revealed that in these cells, 3 patients had somatic mutations in their second TNFRSF6 allele, while 4 patients had loss of heterozygosity by telomeric uniparental disomy of chromosome 10. This observation provides the molecular bases of a nonmalignant autoimmune disease development in humans and may shed light on the mechanism underlying the occurrence of other autoimmune diseases.
Introduction
Autoimmune lymphoproliferative syndrome (ALPS, also known as lymphoproliferative syndrome with autoimmunity or Canale-Smith syndrome) (1) is a human disease characterized by benign lymphoproliferation (splenomegaly and/or diffuse lymphadenopathy), and elevated serum immunoglobulins (IgG and IgA), plasma IL-10 levels (2, 3), and FAS-L levels. (4), accumulation in the blood and lymphoid organs of CD4–CD8–TCRαβ+ T cells (also referred to as double-negative [DN] T cells), susceptibility to malignant conditions (5), and autoimmune manifestations (6, 7).
Mutations in the TNF receptor superfamily, member 6 (TNFRSF6) gene (encoding FAS, also known as the CD95 receptor) are the most common cause of ALPS (8, 9). Patients carrying a homozygous TNFRSF6 mutation have an early-onset severe phenotype, whereas subjects with heterozygous TNFRSF6 mutations present ALPS of variable intensity. T cells from patients with homozygous or heterozygous TNFRSF6 mutations, respectively, exhibit a complete or partial functional impairment in an in vitro FAS-induced apoptosis assay. Moreover, somatic heterozygous mutations in TNFRSF6 in patients with a clinical ALPS phenotype were recently described (10, 11). In our experimental conditions, activated T cells showed normal sensitivity to FAS-induced apoptosis in vitro (11). The inability to detect an apoptosis defect was due to spontaneous in vitro apoptosis of the mutant T cells. However, more than 80% of these patients’ DN T cells were mutated. These patients displayed mosaic expression of a somatic TNFRSF6 mutation, which provided the affected cells a selective advantage and accounted for the observed lymphoproliferation and autoimmunity.
However, heterozygous germline TNFRSF6 mutations are not always associated with clinical expression (partial clinical penetrance), since some mutation-positive relatives (MPRs) remain asymptomatic despite an in vitro FAS-induced apoptosis impairment on their T cells. This observation led us to postulate that a second event is necessary for disease expression in patients with TNFRSF6 mutations when an incomplete penetrance is observed. Here, we report that the combination of a germline TNFRSF6 mutation and a somatic event impairing the second TNFRSF6 allele can account for the onset of clinical phenotype in ALPS.
Results
Biological characteristics of the 7 ALPS patients and their asymptomatic relatives.
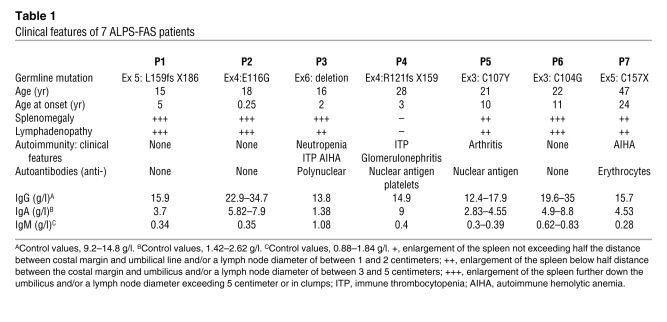
Within the cohort of ALPS patients with heterozygous TNFRSF6 mutation studied in our institution (n = 87), 16% (n = 14) of the germline mutations affected the extracellular domain (ECD) of TNFRSF6. The clinical penetrance has been found to be much lower for heterozygous ECD mutations (30% of carriers developed ALPS) than for heterozygous intracellular domain (ICD) mutations (80% of carriers developed ALPS) (12, 13). This observation suggests that additional events are necessary for ALPS disease expression in carriers of TNFRSF6 mutations with incomplete clinical penetrance. In order to search for these additional events, we analyzed the available T cells from 7 patients with the typical clinical symptoms of ALPS associated with monoallelic mutations affecting the ECD of TNFRSF6 (Table 1) and compared with asymptomatic family carriers in 4 cases. We first quantified ALPS markers: the percentage of DN T cells, plasma FAS-L, and IL-10 concentrations (2, 4, 14) (Figure 1A). In strict agreement with the clinical phenotype, all 7 patients presented markedly elevated numbers of DN T cells and plasma FAS-L and IL-10 concentrations, whereas their asymptomatic MPRs carrying the same TNFRSF6 mutations exhibited normal DN T cell percentages and plasma IL-10 concentration and little or no increase in the plasma FAS-L concentration.
Table 1 .
Clinical features of 7 ALPS-FAS patients
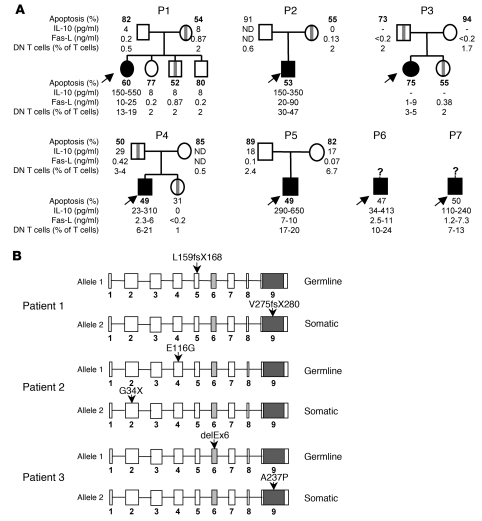
Figure 1. Family tree of 7 ALPS patients with germline heterozygous TNFSFR6 mutations and additional somatic mutations in patients 1, 2, and 3.
(A) Family trees of 7 ALPS patients with a germline ECD TNFRSF6 mutation. Asymptomatic mutated relatives are indicated by vertical gray bars. Subjects with ALPS are marked in black (arrows). The range of values determined for the 4 ALPS markers, i.e., apoptosis of activated T cells (control > 75%), plasma IL-10 concentration (control < 20 pg/ml), plasma FAS-L concentration (control < 0.2 ng/ml), and the percentage of DN T cells (< 2% of TCRαβ cells) are shown for these patients and their relatives. Apoptosis values depicted in bold were obtained in an assay performed at the same time for patients’ cells and relatives’ cells. P1, patient 1. (B) Localization of the 2 mutations in the TNFRSF6 alleles (9 exons) for patients 1, 2, and 3. The oblique gray bars in exon 6 and the dark section in exon 9 represent the transmembrane domain and the death domain, respectively.
For all 7 patients, FAS-mediated T cell apoptosis was lower than for controls (Figure 1A). When simultaneously tested, we found that FAS-mediated apoptosis of patients’ T cells and their asymptomatic MPRs had similarly low apoptosis levels; this contrasted with the results for nonmutated relatives (Figure 1A). Since the magnitude of the FAS-mediated apoptosis defect was similar in symptomatic and asymptomatic carriers of TNFRSF6 mutations, we postulated that any additional causative events did not directly have an impact on the FAS-induced apoptosis as assayed in vitro. This result was reminiscent of ALPS cases carrying somatic TNFRSF6 mutations but which were associated with normal FAS-mediated T cell apoptosis in vitro (11).
Somatic mutations of TNFRSF6 in 3 patients with heterozygous germline mutations.
Based on the literature description of ALPS sufferers carrying a somatic TNFRSF6 mutation, which was predominantly found in DN T cells, we sequenced the TNFRSF6 gene in sorted DN T cells from each of the 7 patients. In addition to the previously identified germline TNFRSF6 mutation, we found a second heterozygous TNFRSF6 mutation in the DN T cells from patients 1, 2, and 3 (Figure 1B). In patient 1, a base insertion in exon 9 had created a premature stop codon at position 280. In patient 2, a nonsense mutation was found in exon 2 (G34X). In patient 3, a missence mutation was found in exon 9 (A237P). These somatic heterozygous TNFRSF6 mutations were present in the second allele, since the cloned PCR products from cDNA amplified from these patients’ DN T cells exhibited only 1 of the 2 TNFRSF6 mutations (data not shown).
The proportion of cells carrying the somatic mutation was established in several lineages for patients 1 and 2. In patient 1, the second mutation was found only in DN T cells and not in either single-positive T cells or epithelial cells (sensitivity threshold: 2.5% of cells) (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI43752DS1). It was therefore impossible to determine at which stage of lymphocyte differentiation the mutation had occurred. In patient 2, the mutation was found in 7%–35% of T cells, B cells, NK cells, and monocytes but also in buccal epithelial cells — indicating that the somatic mutation had occurred very early in embryogenesis. In the other 4 patients, TNFRSF6 sequencing did not highlight any second heterozygous mutation but revealed only the mutated allele in DN T cells (not shown), suggesting a somatic loss of the wild-type allele.
Loss of the wild-type TNFRSF6 allele in 4 patients.
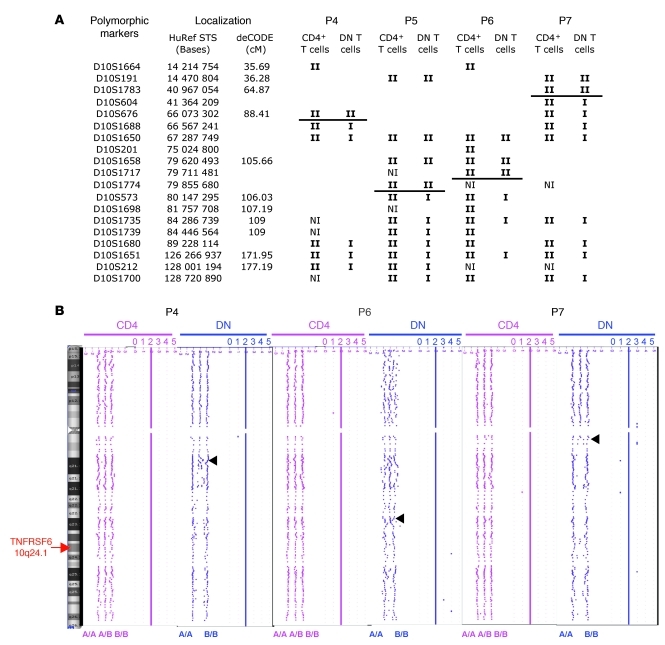
In order to identify the molecular basis of the loss of the wild-type allele in DN T cells from patients 4 to 7, we checked for single-nucleotide polymorphisms at the TNFRSF6 locus in DNA from DN T cells and from CD4+ T cells. In all 4 patients, DNA from the CD4+ T cells displayed heterozygous polymorphic microsatellite markers, whereas DNA from the DN T cells displayed homozygous markers (Figure 2A). We then used serial microsatellite typing along chromosome 10q in CD4+ T cell and DN T cell DNA to determine the location of the mutations. For patients 5 and 6, we honed in on a centromeric breakpoint region at about 1.3 kb upstream of the TNFRSF6 locus (Figure 2A). Loss of heterozygosity (LOH) was observed down to the more distal telomeric marker, indicating that about 45 Mb of parental DNA was missing. The conserved heterozygosity observed in the CD4+ T cells suggested that very few (if any) of these cells carried the same chromosome aberration. Our data indicated that DN T cells exhibited telomeric uniparental disomy (UPD) or partial loss of the long arm of chromosome 10. The same analysis performed on the DN T cells from patient 4 and patient 7 also revealed a mosaic LOH encompassing 60 Mb or more, respectively (Figure 2A). This phenomenon was not observed in DN T cells from 3 ALPS patients carrying ICD mutations of TNFRSF6 with incomplete clinical penetrance or from healthy carriers from families 1 to 4 (data not shown). Array-based comparative genomic hybridization in patients 4, 6, and 7 (Figure 2B) and FISH in patients 6 and 7 (data not shown) confirmed the LOH and conservation of the same gene copy number. The size of the chromosomal segment affected by the LOH in patient 4 was about 75 Mb. These results demonstrate that (a) the wild-type allele had been lost and (b) the mutated allele had been duplicated from the estimated breakpoint down to the end of chromosome 10.
Figure 2. LOH on chromosome 10 of DN T cells from 4 patients carrying germline heterozygous TNFRSF6 mutations.
(A) Polymorphic markers used for LOH characterization. Each allele is represented by the symbol I. Horizontal bars indicate the upstream LOH region in DN T cells. NI, not informative. (B) An array comparative genome hybridization profile of chromosome 10, showing the LOH (phenotype A/A or B/B but no heterozygosity A/B) and the stable allele copy number (the numbers above the schema depicted the allele copy number: 2 copies along the entire chromosome) that are characteristic of UPD (duplication of the mutant allele with loss of the wild-type allele) in DN T cells of patients 4, 6, and 7. The arrowhead indicates the upstream site of LOH.
Measurement of FAS expression in cells with 2 disrupted TNFRSF6 alleles.
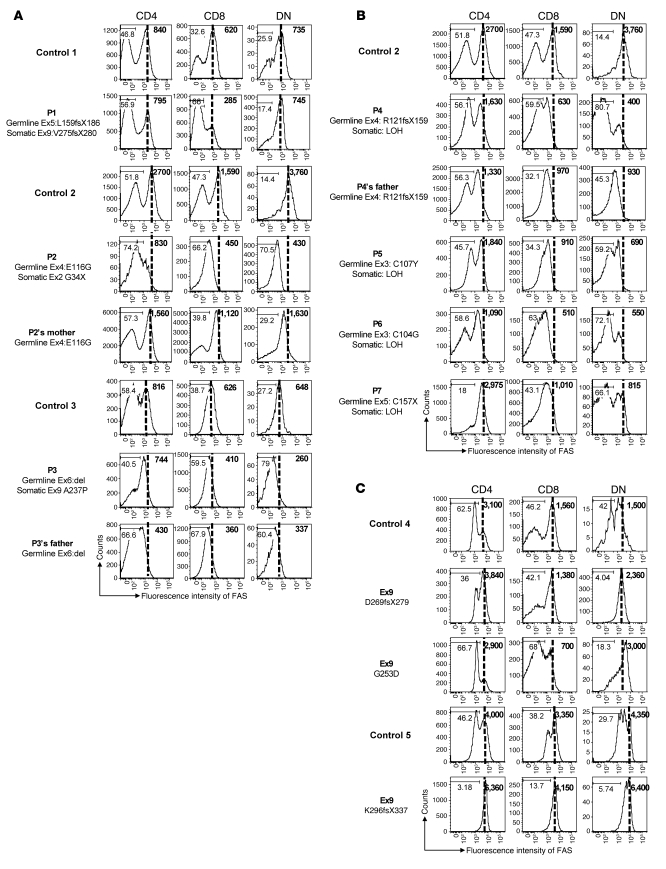
In order to determine the impact of these additional TNFRSF6 mutations or chromosome aberration, we analyzed FAS protein expression in CD4, CD8, and DN T cells from the 7 patients, 3 asymptomatic MPRs (patient 2’s mother, patient 3’s father, and patient 4’s father), and 5 healthy controls (Figure 3). First, we found that FAS expression in single-positive (CD4 or CD8) T cells was slightly lower than control values in all patients or relatives carrying a low-penetrance ECD TNFRSF6 mutation (except for patient 1); this suggests that the germline TNFRSF6 mutation in allele 1 led to defective expression in patients 2 to 7. Further analysis indicated that the great majority of these patients’ DN T cells did not express FAS. Interestingly, DN T cells from patients 2, 3, and 4 expressed less FAS than those from patient 2’s mother, patient 3’s father, and patient 4’s father, respectively (Figure 3, A and B). The low FAS expression in these cells was not due to an activation defect, since they expressed high amounts of human leukocyte antigen class II molecules (Supplemental Figure 1). Strikingly, no decrease of FAS expression was observed in 3 patients carrying germline ICD TNFRSF6 mutations associated with low penetrance. Accordingly, no somatic mutation has been detected in these patients. These results indicated that the somatic event in DN T cells is mostly associated with reduced FAS expression via loss of the wild-type TNFRSF6 allele.
Figure 3. Impact of TNFRSF6 mutations on FAS protein expression by lymphocyte subsets.
FAS expression by CD4, CD8, and DN T cells was determined by FACS analysis (A) for patients 1 to 3 carrying a germline TNFRSF6 mutation and a somatic TNFRSF6 mutation in the second allele; (B) for patients 4 to 7 carrying a germline heterozygous TNFRSF6 mutation and exhibiting LOH on the second allele; and (C) for 3 ALPS patients carrying germline ICD TNFRSF6 mutations. Analysis was also performed for 3 MPRs (patient 2’s mother, patient 3’s father, and patient 4’s father). Patients 2 and patient 4 to 6 and their respective MPRs were analyzed in parallel in the same experiment, using an ARIA II cytometer. Patient 1, patient 3, and all patients with ICD mutations were processed in distinct experiments in parallel with different healthy controls, using an ARIA II cytometer. Numbers on histograms indicate the percentage of cells in the depicted FAS-negative gates, and numbers in bold type indicate the mean fluorescence of FAS intensity in the whole population.
Discussion
Severe ALPS always occurs when both TNFRSF6 alleles are mutated in the germline in humans (9, 15, 16) or mice (17). In contrast, if the FAS defect is partial (as in the case of a heterozygous mutation, for example), the human disease develops in 70% of carriers (13). The partial clinical penetrance of heterozygous TNFRSF6 mutations in ALPS suggests that in addition to the germline TNFRSF6 mutation, a “second hit” is required for disease onset. Here, we identified somatic genetic events affecting the second TNFRSF6 allele in 7 patients carrying a germline heterozygous mutation with low clinical penetrance. In these patients, a small proportion of lymphocytes lacked functional FAS — leading to uncontrolled proliferation and autoimmunity. Missence or nonsense somatic mutations were identified in 3 patients, whereas somatic UPD (duplication of the mutant allele with loss of the wild-type allele) were found in the other 4 patients.
Interestingly, we identified somatic events in 7 patients carrying germline heterozygous TNFRSF6 mutations affecting the ECD of FAS. The latter are known to frequently induce a loss of FAS expression at the cell membrane and therefore haploinsufficiency (18). This was confirmed by analyzing cells from the MPRs, in which the only germline mutation was present. Expression of half the normal level of FAS receptor is sufficient to trigger apoptosis and might preclude the onset of ALPS and account for the low clinical penetrance seen with ECD TNFRSF6 mutations. The additional somatic event led to a clear, profound expression defect in a small proportion of single-positive T cells and in the majority of DN T cells from all but 1 patient. In this latter case, patient 1 carried 2 missence mutations (one germline and one somatic), which were predicted to be nonfunctional. The subsequent in vivo functional defect could thus be the consequence of either the lack of FAS expression at the cell membrane or the lack of apoptosis signal. This could result from a lack of multimerization of the receptor or the lack of death-inducing signaling complex (DISC) recruitment by the residual FAS molecules. Consequently, the double-mutant lymphocytes could not be killed by FAS-L. A similar situation has been observed in patients carrying inherited homozygous or compound heterozygous mutations (9, 15). The lymphoproliferation observed in the latter patients indicated that a complete FAS defect led to uncontrolled proliferation of T and (to a lesser extent) B cells. In contrast, most missence mutations affecting the ICD of TNFRSF6 are known to exert a transdominant negative effect, since the mutant protein is likely to trimerize with wild-type FAS. Assuming that efficient DISC formation and subsequent apoptosis triggering require wild-type FAS trimers only, it was estimated that 1 out of 8 trimers would be functional. The ensuing apoptosis defect may be sufficient to account for the onset of ALPS. Accordingly, the clinical penetrance of the ICD missence TNFRSF6 mutation was complete in most families (19). Moreover, no additional defect in the TNFRSF6 gene could be found in 3 tested patients with ICD mutations. It is noteworthy that previously described cases of ALPS caused by somatic heterozygous mutations (10, 11) were mostly associated with ICD TNFRSF6 mutations. This observation suggested that somatic ICD TNFRSF6 mutations markedly impair FAS function and provide the mutant cells with a selective advantage in vivo. Moreover, the patients carrying somatic ICD mutations developed clinical disease, despite the low proportion (10%–20%) of mutant lymphocytes. A similar picture was seen in our current cases, since the somatic events were seen in a minor fraction of single-positive T cells and in a major fraction of DN T cells. This observation supports the concept whereby the second hit (somatic mutation or UPD) together with the germline ECD-mutated TNFRSF6 gene, profoundly impair FAS function and provide the double-mutant T cells with a selective advantage in vivo. The double-mutant cells died spontaneously in vitro and so could not be tested. Consequently, in vitro expansion of the patients’ T cells led to accumulation of the germline mutants only. Accordingly, the magnitude of the FAS-induced apoptosis defect was similar in T cells from the patients and their asymptomatic mutated relatives, respectively.
The combination of germline and somatic mutations in the same gene has been reported in cancers with a hereditary predisposition, such as retinoblastoma (20). Similarly, missence mutations of TNFRSF6 outside the death domain are associated with LOH in non-Hodgkin lymphomas (21). The LOH observed in 4 of our patients with germline TNFRSF6 ECD mutations resulted from loss of the wild-type allele and concomitant duplication of the mutant one, i.e., telomeric UPD. This type of mechanism is usually observed in genetic disorders of development, where it can be involved in trisomy or monosomy rescue during embryonic development (22). The telomeric UPD observed in our ALPS cases might reflect a recombination hotspot on the 10q chromosome (as described in the 10q syndrome; ref. 23), but more likely reflect the selective advantage provided by the resulting homozygous TNFRSF6 mutations. These events may occur during embryogenesis (patient 2) or later in life (since the onset of clinical manifestations was delayed in patients 5, 6, and 7). Implication of modifier genes in the onset of ALPS was previously postulated. The present findings highlight the role of somatic events affecting the second TNFRSF6 allele as modifier in ALPS expression. However, we could not strictly exclude that these somatic FAS events result from other, yet undefined, germline mutation in the actual “ modifier” genes.
Lymphocyte homeostasis and self tolerance are maintained by several checkpoints, including antigen receptor editing, modulation, anergy, and central or peripheral clonal deletion of self-reactive lymphocytes (24). Variants of proteins involved in cell activation, survival, proliferation, and death may contribute to the emergence of autoimmunity (25). For instance, variants of the genes for HLA molecules (selecting for self-reactive lymphocytes), PTPN22 (regulating Antigen receptor signaling), and CTLA-4 (inhibiting T cell proliferation) are risk factors in the occurrence of various autoimmune diseases. It has been postulated that in view of the moderate functional impact of these variants, additional events are required to bypass the tolerance checkpoints. This model accounts for the relatively long time to onset of autoimmune disease in most patients. Our identification of a combined germline TNFRSF6 mutation (with incomplete clinical penetrance) and a somatic event affecting the second TNFRSF6 allele now demonstrates that a hit on each of the 2 alleles of the same gene can lead to lymphoproliferation and autoimmunity. On the basis of this observation, one can postulate that combinations of gene variants and somatic mutations in genes involved in cell death (such as Bim, Bid, or caspases) or in the regulation of cell proliferation (such as PD-1 or CTLA-4) could be involved in the emergence of autoimmune disease via the selection of self-reactive T and B lymphocytes. This hypothesis could be tested in models of autoimmunity in which self-reactive T and/or B cell clones can be identified. Somatic mutations occur relatively frequently (i.e., 10–6 per gene per cell division) (26). Consequently, in the context of the high-rate cell division seen in both lymphoid precursors and mature lymphocytes, a role for somatic mutations in triggering autoimmune disease onset should be considered.
In conclusion, our present findings validate the “2-hit” hypothesis in the setting of a nonmalignant disease and highlight the possible impact of somatic mutations on the onset of other autoimmune diseases in humans.
Methods
Study population.
Seven patients presenting with typical ALPS manifestations (including autoimmune manifestations in 3 cases) and carrying germline heterozygous TNFRSF6 mutations affecting the ECD of the protein were enrolled in this study (Table 1). Relatives of patients 1 to 5 (including 7 asymptomatic MPRs) as well as 3 patients carrying a germline heterozygous ICD TNFRSF6 mutation (D269fsX279; G253D; K296fsX279) were also investigated as controls. The study protocol was approved by the local investigational review board (CPP ile de France II, Ministry of Research, Paris France). Informed consent was obtained from the patients or their families prior to study entry.
Immunofluorescence and cell sorting.
Using flow cytometry, the percentage of DN T cells was defined as the percentage of TCR-αβ–positive cells that lacked CD4 and CD8 expression. Thawed peripheral blood mononuclear cells were incubated with a cocktail of 2.5 μl of anti–TCR PAN α/β PC5 antibody (IP26A; Immunotech; Beckman Coulter), 5 μl of anti–CD4-APC antibody (SK3), 10 μl of anti–CD8-V500 antibody (RPAT8), 10 μl of anti–CD95-PE antibody (DX2), and 10 μl of anti–HLA-DP-DQ-DR-FITC (TU39) (BD Biosciences) per 4 million cells. Acquisition and cell sorting were performed with an ARIA I or ARIA II cytometer (BD Biosciences), and data were analyzed using Flowjo software (TreeStar Inc.).
Plasma IL-10 and FAS-ligand and apoptosis assays.
Plasma IL-10 and FAS-ligand levels and apoptosis were assayed as previously described (4).
Detection of TNFRSF6 mutations.
DNA was isolated by proteinase K digestion and phenol-chloroform extraction. Genomic DNA segments were amplified as previously described (11).
DNA cloning.
PCR products were cloned using TOPO TA cloning kit (Invitrogen) following the manufacturer’s instructions. The sequences of at least 80 clones were then determined by capillary electrophoresis using Applied Biosystems DNA sequencing kits and a 3130xl Genetic Analyzer (Applied Biosystems).
Genotype analysis.
All analyzed microsatellite markers flanked the TNFRSF6 locus or genes located on the 10q chromosome. Primer sequences were retrieved from the Genome Database ( http://www.ncbi.nlm.nih.gov). FISH experiments were performed with BAC RP11-399O19 (a probe spanning the TNFRSF6 locus) as previously described (27). Analysis of copy number variations and LOH was performed using Whole-Genome Cytogenetics 2.7M DNA chips (Affymetrix). The results were visualized using the Chromosome Analysis Suite (version 1.0.1; Affymetrix).
Supplementary Material
Acknowledgments
This work was funded by grants from INSERM, the Agence Nationale de la Recherche (ANR) (grant no. 08-GENO-015-01, E-rare 2007 [epinostics]), the Association pour la Recherche contre le Cancer (ARC), and the European STREP (autorome). A. Magerus-Chatinet was supported by a postdoctoral fellowship from ARC and the European STREP. B. Neven was supported by a doctoral fellowship from INSERM and the Fondation pour la Recherche Médicale. The authors thank Françoise Bourgue Vion-Dury, Corinne Cordier, Virginie Grandin, Chantal Harre, Corinne Jacques, Nathalie Lambert, Alexandra Léger, Stephanie Ndaga, Fabienne Poulain, Solenn Pruvost, and Magali Salmon; and M. Jerôme Megret for technical support.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(1):106–112. doi:10.1172/JCI43752.
See the related Commentary beginning on page 16.
References
- 1.Canale VC, Smith CH. Chronic lymphoadenopathy simulating malignant lymphoma. J Pediatr. 1967;70(6):891–899. doi: 10.1016/s0022-3476(67)80262-2. [DOI] [PubMed] [Google Scholar]
- 2.Lopatin U, et al. Increases in circulating and lymphoid tissue interleukin-10 in autoimmune lymphoproliferative syndrome are associated with disease expression. Blood. 2001;97(10):3161–3170. doi: 10.1182/blood.V97.10.3161. [DOI] [PubMed] [Google Scholar]
- 3.Ohga S, et al. Dominant expression of interleukin 10 but not interferon gamma in CD4(–) CD8(–) alphabetaT cells of autoimmune lymphoproliferative syndrome. Br J Haematol. 2002;119(2):535–538. doi: 10.1046/j.1365-2141.2002.03848.x. [DOI] [PubMed] [Google Scholar]
- 4.Magerus-Chatinet A, et al. FAS-L, IL-10, and double-negative CD4- CD8- TCR alpha/beta+ T cells are reliable markers of autoimmune lymphoproliferative syndrome (ALPS) associated with FAS loss of function. Blood. 2009;113(13):3027–3030. doi: 10.1182/blood-2008-09-179630. [DOI] [PubMed] [Google Scholar]
- 5.Straus SE, et al. The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood. 2001;98(1):194–200. doi: 10.1182/blood.V98.1.194. [DOI] [PubMed] [Google Scholar]
- 6.Rieux-Laucat F, Le Deist F, Fischer A. Autoimmune lymphoproliferative syndromes:genetic defects of apoptosis pathways. Cell Death and Differentiation. 2003;10(1):124–133. doi: 10.1038/sj.cdd.4401190. [DOI] [PubMed] [Google Scholar]
- 7.Sneller MC, et al. Clincal, immunologic, and genetic features of an autoimmune lymphoproliferative syndrome associated with abnormal lymphocyte apoptosis. Blood. 1997;89(4):1341–1348. [PubMed] [Google Scholar]
- 8.Fisher GH, et al. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81(6):935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 9.Rieux-Laucat F, et al. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268(5215):1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 10.Dowdell KC, et al. Somatic FAS mutations are common in patients with genetically undefined autoimmune lymphoproliferative syndrome. Blood. 2010;115(25):5164–5169. doi: 10.1182/blood-2010-01-263145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holzelova E, et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N Engl J Med. 2004;351(14):1409–1418. doi: 10.1056/NEJMoa040036. [DOI] [PubMed] [Google Scholar]
- 12.Vaishnaw AK, Orlinick JR, Chu JL, Krammer PH, Chao MV, Elkon KB. The molecular basis for apoptotic defects in patients with CD95 (Fas/Apo-1) mutations. J Clin Invest. 1999;103(3):355–363. doi: 10.1172/JCI5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieux-Laucat F, et al. Lymphoproliferative syndrome with autoimmunity: A possible genetic basis for dominant expression of the clinical manifestations. Blood. 1999;94(8):2575–2582. [PubMed] [Google Scholar]
- 14.Bleesing JJ, et al. TcR-alpha/beta(+) CD4(–)CD8(–) T cells in humans with the autoimmune lymphoproliferative syndrome express a novel CD45 isoform that is analogous to murine B220 and represents a marker of altered O-glycan biosynthesis. Clin Immunol. 2001;100(3):314–324. doi: 10.1006/clim.2001.5069. [DOI] [PubMed] [Google Scholar]
- 15.Bettinardi A, et al. Missence mutations in the Fas gene resulting in autoimmune lymphoproliferative syndrome: a molecular and immunological analysis. Blood. 1997;89(3):902–909. [PubMed] [Google Scholar]
- 16.Kasahara Y, et al. Novel Fas (CD95/APO-1) mutations in infants with a lymphoproliferative disorder. Int Immunol. 1998;10(2):195–202. doi: 10.1093/intimm/10.2.195. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe D, Suda T, Hashimoto H, Nagata S. Constitutive activation of the Fas ligand gene in mouse lymphoproliferative disorders. EMBO J. 1995;14(1):12–18. doi: 10.1002/j.1460-2075.1995.tb06970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roesler J, et al. Haploinsufficiency, rather than the effect of an excessive production of soluble CD95 (CD95{Delta}TM), is the basis for ALPS Ia in a family with duplicated 3′ splice site AG in CD95 intron 5 on one allele. Blood. 2005;106(5):1652–1659. doi: 10.1182/blood-2004-08-3104. [DOI] [PubMed] [Google Scholar]
- 19.Jackson CE, et al. Autoimmune lymphoproliferative syndrome with defective Fas: genotype influences penetrance. Am J Hum Genet. 1999;64(4):1002–1014. doi: 10.1086/302333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rushlow D, et al. Detection of mosaic RB1 mutations in families with retinoblastoma. Hum Mutat. 2009;30(5):842–851. doi: 10.1002/humu.20940. [DOI] [PubMed] [Google Scholar]
- 21.Gronbaek K, et al. Somatic Fas mutations in non-Hodgkin’s lymphoma: association with extranodal disease and autoimmunity. Blood. 1998;92(9):3018–3024. [PubMed] [Google Scholar]
- 22.Conlin LK, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19(7):1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsenko SA, et al. Identification of critical regions for clinical features of distal 10q deletion syndrome. Clin Genet. 2009;76(1):54–62. doi: 10.1111/j.1399-0004.2008.01115.x. [DOI] [PubMed] [Google Scholar]
- 24.Goodnow CC. Multistep pathogenesis of autoimmune disease. Cell. 2007;130(1):25–35. doi: 10.1016/j.cell.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 25.Zenewicz LA, Abraham C, Flavell RA, Cho JH. Unraveling the genetics of autoimmunity. Cell. 2010;140(6):791–797. doi: 10.1016/j.cell.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araten DJ, et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65(18):8111–8117. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- 27.Romana SP, Le Coniat M, Berger R. t(12;21): a new recurrent translocation in acute lymphoblastic leukemia. Genes Chromosomes Cancer. 1994;9(3):186–191. doi: 10.1002/gcc.2870090307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.