Abstract
Although skeletal muscle fiber types are often defined as belonging to discrete categories, many muscles possess fibers with intermediate phenotypes. These hybrid fiber types can be identified by their expression of two or more myosin heavy chain (MHC) isoforms within the same single fiber. In mouse muscles, the most common hybrid fibers are those coexpressing the IIX and IIB MHC isoforms. In the present study, we focused on these IIX/IIB fibers from normal mouse muscles to determine the relative proportions of MHC isoforms at both the protein and mRNA levels and to examine the longitudinal distribution of isoforms within single fibers. We found that IIX/IIB hybrids represent ∼25 and 50% of the fibers in the mouse tibialis anterior and brachioradialis, respectively. The relative proportion of the IIX and IIB isoforms in these fibers spans a continuum, from predominantly IIB-like hybrids to IIX-like hybrids. Quantitative assessment of mRNA levels using real-time PCR from single fibers indicated that IIB expression dominated over IIX expression in most fibers and that a general correlation existed between mRNA isoform levels and MHC protein content. However, the match between mRNA levels and protein content was not precise. Finally, we measured MHC isoform proportions in adjacent fiber segments and discovered that ∼30% of hybrids possessed significant differences in isoform content along their length. In some instances, the muscle fiber type as defined by MHC content changed completely along the length of a fiber. This pattern of asymmetrical MHC isoform content along the length of single fibers suggests that the multiple myonuclei of a muscle fiber may express distinct myofibrillar isoforms in an uncoordinated fashion.
Keywords: fiber type, hybrid fibers, skeletal muscle
whole skeletal muscles are comprised of hundreds to thousands of individual muscle cells, or fibers, which represent specialized units of muscle contraction. Each individual fiber possesses a limited range of physiological properties including activation rate, contractile speed, force production, fatigue resistance, and other properties that depend on the specialized cellular and molecular organization of the fiber (51). Whole muscles are capable of generating a continuous range of mechanical output by recruiting different populations of specific fiber types. Specialized fiber types are essential for the production of the vast array of mechanical output generated by skeletal muscles, from slow and steady efficient locomotion to explosive bursts of power (50, 51). Because of this important functional role, it is critical to understand how many different specialized fiber types exist, and in what proportions they are represented in different muscles. The most physiologically relevant system for classifying skeletal muscle fiber types is based on identification of myosin heavy chain (MHC) isoform(s) within single fibers. MHC is the motor protein that powers muscle contraction, and the specific type of the molecule directly determines contractile speed, efficiency, and power output (54). The skeletal muscles of most animals express multiple isoforms of MHC, which are generally encoded by distinct genes. In mammalian skeletal muscles, nine genes are expressed that encode alternate isoforms of the MHC protein (54). In adults, four of these are commonly found within the limb muscles: I, IIA, IIX, and IIB (54). These alternate isoforms provide different rates of ATP hydrolysis and muscle shortening, thereby imparting individual muscle fibers with specific physiological properties (54).
Shortly after the advent of techniques used to identify MHC isoforms within single fibers, researchers reported that some fibers coexpressed two or more different isoforms (hybrid fibers) (3, 55, 57–59, 67). Since skeletal muscles exhibit a high degree of phenotypic plasticity, these observations were frequently discussed within the context of fibers in the process of switching from one type to another. Indeed, a variety of experimental interventions driving a shift in muscle fiber type lead to changes in the relative number of hybrid fibers. Mechanical load (13, 15), denervation (41, 63–65), spinal cord transection (63–65), hormonal manipulation (13, 15, 19), and chronic electrical stimulation (47) are each capable of producing major changes in the hybrid fiber content of skeletal muscles. The influence of more natural interventions, like aging and exercise, on hybrid skeletal muscle fibers generally appears to be much less dramatic (7, 25, 27, 29, 68, 69). As data from skeletal muscles under a variety of conditions have accumulated, it has become clear that hybrid fibers often represent stable phenotypes and are common components of normal muscles (16, 60). The relative proportion of hybrid fibers varies significantly from muscle to muscle, and even among anatomical regions within the same muscle. While some muscles have only a small content of hybrid fibers (∼1–2%), others are comprised of up to ∼75% hybrid fibers (1, 16, 25). The common occurrence of hybrid fibers in diverse animal groups (32, 34, 43, 60) suggests that the coexpression of multiple myofibrillar protein isoforms within single fibers is a pattern common to all skeletal muscles, and it is therefore essential to understand what role these fibers play in skeletal muscle diversity and plasticity.
Pette and Staron (45, 46) developed the concept of nearest-neighbor expression of MHC isoforms. This idea is that fiber phenotype falls along a continuum, with hybrid fiber types (bold) bridging the gaps between pure fiber types (italics): I ↔ I/IIA↔ IIA ↔ IIA/IIX ↔ IIX ↔ IIX/IIB ↔ IIB. Most changes in fiber type are thought to take place gradually from one MHC combination to another, although some experimental interventions lead to mismatched fibers containing MHCs that are not nearest neighbor isoforms (5a, 15, 19, 64). The most prominent hybrid fiber types in rat and mouse muscles are the IIX/IIB hybrids (16, 25), which, according to the above scheme, are intermediate between the IIX and IIB fiber types. Compared with rat muscles, mouse fibers are proportionately shifted toward the IIB end of the MHC spectrum, which is apparently a consequence of their smaller body size and faster muscle requirements (25). In rats, several limb muscles are composed of ∼70% IIX/IIB hybrids (16), while in mice the most hybrid-rich muscles contain ∼25–50% IIX/IIB hybrids (25).
The goal of this study was to use the IIX/IIB hybrids to answer several basic questions about the organization of MHC isoforms within single hybrid fibers. We focused on IIX/IIB hybrids from the mouse tibialis anterior (TA) and the brachioradialis (BR) muscles because we have found these to possess large proportions of these hybrid fibers (25). Our study focused on three essential questions. First, what are the relative proportions of the IIX and IIB MHC isoforms within single fibers? It is possible that some of these are more IIB-like in their proportions, while in others the IIX isoform is dominant. Second, how closely do the mRNA levels expressed for these two isoforms match the fiber type as determined by MHC protein isoforms present within a fiber? Previous studies have shown that the relationship between transcript levels and fiber type at the protein level can be complex (6, 12, 48). Finally, we sought to determine whether asymmetries in the distribution of the two MHC isoforms exist along the length of single fibers and how common these differences in MHC isoform content might be. Our findings have important implications for understanding how MHC isoform content is organized within hybrid fibers from normal muscles, and how this organization relates to skeletal muscle diversity and plasticity.
MATERIALS AND METHODS
Animals.
Adult male and female mice (C57BL/6J) were purchased from Jackson Laboratory (Bar Harbor, ME) and maintained in accordance with an approved IACUC protocol (BIO12075N) at the University at Buffalo. The ∼30 mice used in the study ranged in age from just over 3 wk to 6 mo. They were housed in cages with a 12:12-h light-dark cycle and were provided with food and water ad libitum.
Collection of muscle tissues.
Mice were euthanized by exposure to CO2, and muscles were collected from freshly killed animals. Whole muscles were dissected from mice and either processed for sectioning and histochemical staining or for the isolation of single fibers. For histochemistry, whole muscles were coated in OCT compound (Sakura Finetek, Torrance, CA) and mounted on balsa wood platforms. Muscles were rapidly frozen in isopentane cooled in liquid N2 for 15 to 30 s and then quickly transferred to −20°C, where they were stored prior to sectioning. For single-fiber isolation, muscles were transferred to a relaxing solution (50% glycerol, 2 mM EGTA, 1 mM MgCl2, 4 mM ATP, 10 mM imidazole and 100 mM KCl) and stored at −20°C. For fibers used for RNA isolation, the relaxing solution was made with RNase-free components (diethyl pyrocarbonate-treated H2O) and fibers were taken from freshly harvested muscle tissues.
Histochemistry.
Histochemical analyses were used to assess the general characteristics of the muscles used as a source of single fibers. ATP histochemistry was used to determine muscle fiber types present and to assess the regionalization of fiber types within the whole muscles. NADH diaphorase staining was used as a measure of the mitochondrial content, and aerobic capacity of the fibers. Serial sections of muscles (8–10 μm) were cut on a cryostat and mounted on glass slides. Before staining, sections were air dried for ∼30 min.
ATP staining followed the general procedures described by Dubowitz and Sewry (20). Dried sections were incubated in a glycine-buffered saline solution (50 mM glycine, 50 mM NaCl, and 100 mM CaCl2, pH 9.4) containing 1 mM ATP for 30 min at 37°C. Sections were then washed 3 × 2 min in 2% CaCl2, followed by 2 × 1 min washes in 1% CoCl2. Sections were subsequently rinsed several times in deionized water (DI H2O) and then placed in a 2% ammonium sulfide solution for 30 s. Slides with sections were then rinsed several times in DI H2O, dehydrated through a graded ethanol series, cleared in xylenes, and then mounted with Permount (Fisher Scientific, Pittsburgh, PA). This staining reaction yields all fast fibers uniformly dark brown, while slow fibers remain light or unstained. To differentiate the fast isoforms, sections were incubated in 100 mM sodium acetate solution (pH 4.6) prior to incubation in the ATP solution (20). This preincubation changes the staining reaction so that fibers are differentiated on staining intensity: I > IIX > IIB > IIA (26).
NADH-diaphorase staining followed the procedures described by Dubowitz and Sewry (20). A solution containing nitro blue tetrazolium (0.8 mg/ml) and NADH (0.64 mg/ml) in 50 mM sodium phosphate (pH 7.3) was freshly prepared and used to cover the dried sections. Sections were incubated for 30 min at 22°C and then rinsed several times in DI H2O. Sections were dehydrated and mounted using the above procedure. Fibers with greater aerobic capacities stain an intense blue color, while those with less aerobic enzyme activity exhibit less staining.
Isolation of individual fibers for protein and mRNA analyses.
Fiber bundles were separated from whole muscles and placed in a glass petri dish chilled with ice. Fibers were dissected with the aid of a stereomicroscope using fine forceps, and a small volume (∼10 μl) of ice-cold relaxing solution was used to cover the fibers. To generate fiber segments, single fibers of ∼1–4 mm in length were divided using Vannas scissors (World Precision Instruments, Sarasota, FL). In some cases, we were able to cut single fibers into up to five segments of ∼0.5–1 mm in length. Isolated fibers and fiber segments were placed into ∼30 μl of urea sample buffer for protein analysis and into 100 μl of Trizol reagent (Invitrogen, Carlsbad, CA) for RNA isolation.
Determination of MHC isoforms within single fibers.
Single fibers were separated from larger bundles and placed into a 1.5-ml microcentrifuge tube containing ∼30 μl of sample buffer. The sample buffer contained 8 M urea, 2 M thiourea, 50 mM Tris base, 75 mM dithiothreitol, 3% SDS and 0.004% Bromophenol Blue, pH 6.8 (11). Samples were heated to 65°C for 15 min and 10 μl of each sample was applied to the gel. Samples taken from the soleus muscle containing both type IIA and type I isoforms were used as standards on each gel with samples from predominantly fast muscles. SDS-PAGE were used with a Hoeffer SE 600 to resolve individual isoforms of MHC. Resolving gels consisted of 9% acrylamide (200:1 acrylamide/methylene-bis-acrylamide), 12% glycerol, 0.675 M Tris base (pH 8.8) and 0.1% SDS. Stacking gels consisted of 4% acrylamide (20:1 acrylamide/methylene-bis-acrylamide), 0.125 M Tris base (pH 6.8) and 0.1% SDS. Running buffer contained 0.192 M glycine, 25 mM Tris base, 0.1% SDS, and 0.08% 2-mercaptoethanol. Gels were run with a constant current of 20 mA for ∼41 h at 8°C. At the end of the run, gels were fixed in 50% methanol with a trace of formaldehyde (0.037%) added to increase the sensitivity of the silver-staining procedure. Gels were allowed to fix for at least 3 h in this solution and then washed for 1 h in deionized water before staining with silver. Gels were then stained with an ammoniacal silver staining procedure (70).
Dried gels were scanned at high resolution, and the relative proportions of MHC isoforms in single fibers were determined from digital images using densitometry (Image J 1.42q, National Institutes of Health). Serial dilutions of standards containing known proportions of MHC isoforms were used to establish that concentrations were a linear function of sample density. To determine normal measurement error, replicates of samples containing two MHC isoforms were quantified.
RNA isolation and RT.
Total RNA was isolated from single-fiber segments using Trizol reagent (Invitrogen, Carlsbad, CA). Segments were placed in 100 μl of Trizol and then stored at −80°C until the time of extraction. Samples were thawed and then homogenized using a hand-held pestle that fit directly into the microcentrifuge tube. RNA was then isolated per the manufacturer's protocol. Due to the small quantities of RNA to be isolated, 10 μg of molecular grade glycogen (Invitrogen) was added to each sample as a carrier before precipitation of dissolved RNA with isopropanol. Isolated RNA pellets were allowed to air dry for 10 min and were then resuspended in 10 μl of RNase-free water and heated at 59°C for 10 min to ensure solubility. Samples were then treated with DNase (Invitrogen) for 15 min at room temperature to remove any trace amounts of genomic DNA. The entire sample of RNA was subsequently used for reverse transcription.
First-strand synthesis of cDNA from RNA template was performed using SS III Reverse Transcriptase (Invitrogen, Carlsbad, CA). The 20-ml reaction contained 2.5 mg oligo(dT) 20 mer, 2.5 mmol/l dNTP, 1× first-strand buffer, 5 mmol/l DTT, 2.5 units of RNase inhibitor, total isolated RNA, and 200 units of SS III RT. The reaction mixture was incubated at 50°C for 45 min and then inactivated by incubation at 70°C for 15 min. The synthesized cDNA solutions were used for subsequent end-point and real-time PCR reactions.
Quantitative real-time PCR to measure mRNA levels from single fibers.
Before designing specific primers and optimizing PCR conditions, portions of the IIX and IIB MHC sequences were obtained, cloned into plasmid, and transfected into Escherichia coli cells. These MHC clones were then used as controls to optimize PCR conditions and to construct standard curves for real-time PCR. To obtain these clones, total RNA was first isolated from mouse muscles. The RNA was then used as template for a RT reaction, where a universal amplification linker [5′- GGCCACGCGTCGACTAGTACT(17)- 3′] was used to prime the poly-A tail sequences. The resulting cDNA was used as template for a PCR reaction using a reverse primer targeted to the universal amplification sequence (5′-GGCCACGCGTCGACTAGTAC-3′) and a forward primer to a conserved region of the MHC gene (5′-AACAGATCCAGAAACTGGAGGCCA-3′). Specific products were isolated from agarose gels and purified with gel spin columns (Qiagen, Valencia, CA) and then ligated into a plasmid using the TOPO TA cloning kit (Invitrogen). Multiple clones containing plasmids with DNA inserts were identified through blue/white screening, and the isolated clones were grown overnight. Plasmids from several clones were then purified and used for sequencing (Roswell Park Cancer Center DNA Sequencing Lab, Buffalo, NY). Sequence comparisons confirmed that some of these clones corresponded to the mouse IIX MHC sequence (Myh1; GenBank accession no. NM_030679.1), while others matched the mouse IIB sequence (Myh4; GenBank accession no. NM_010855.2).
PCR primers were designed to specifically amplify sequences spanning the end of the 3′ terminal sequence of the MHC gene and part of the 3′ untranslated region. Although the coding sequences of MHC isoforms share a high level of sequence identity, the 3′ untranslated regions of different isoforms are divergent and provide specificity for PCR reactions. Primers for the IIX isoform (forward: 5′-AGACCGCAAGAATGTTCTCAGGCT-3′; reverse: 5′-AGGAGGCTGAGGAACAATCCAACA-3′) amplified a 266-bp sequence, while primers for the IIB isoform (forward: 5′-AGTGAAAGCCTACAAGAGACAGGC-3′; reverse: 5′-CAGGACAGTGACAAAGAACGTCAC-3′) amplified a 252-bp sequence. For standard end point 25 μl PCR reactions contained 2.5 mM of dNTP, 1 μM forward primer, 1 μM reverse primer, 5 μl of cDNA template from RT reaction, and 1 unit of ExTaq DNA polymerase (Takara Bio, Madison, WI). Reaction conditions included 4 min at 94°C, followed by 30–35 cycles of 94°C (30 s), 60°C (30 s), and 72°C (30 s). PCR products were then separated on 1% agarose gels and stained with ethidium bromide.
Real-time PCR was used to quantify the expression levels of the IIX and IIB MHC genes from RNA isolated from single fibers. A SYBR green PCR master mix (Platinum SYBR Green qPCR SuperMix-UDG; Invitrogen, Carlsbad, CA) was used make real-time PCR 25 μl reaction mixtures per the manufacturer's instructions, and reactions were run on an iCycler (Bio-Rad, Hercules, CA). Standard curves for each sequence were constructed from PCR reactions using serial dilutions of known amounts of plasmid DNA containing the MHC sequence of interest as the template. The cycle threshold where DNA product began to accumulate exponentially was plotted as a function of the number of copies of DNA template. Unknown amounts of mRNA from single fibers were determined by using the standard curve to translate cycle threshold into a number of copies of DNA. Each PCR procedure included samples with known amounts of cDNA of the target sequence as a positive control, and cDNA of the alternate sequence as a negative control. Specificity of positive reactions was confirmed by the analysis of product melting point temperatures.
Statistical analyses.
ANOVA was used to compare mean mRNA copy numbers for the IIX and IIB isoforms from single fibers. Total copy numbers from each fiber were log transformed to correct for a lack of homogeneity of variance between samples (38). A two-way factorial ANOVA was used to compare mean mRNA copy numbers for each isoform (IIX or IIB), with one factor being the specific muscle (BR or TA) and the other being muscle fiber type as determined by SDS-PAGE (IIX, IIX/IIB, or IIB). For pairwise comparisons, a Bonferonni correction was used to adjust the experiment-wise error rate to 0.05.
Linear regression was used to examine the correlation of relative IIX and IIB isoforms in hybrid fiber adjacent segments. In addition, the relative difference in isoform proportion between segment pairs was assessed through descriptive statistics. Statview 5.0.1 (SAS Institute, Cary, NC) was used for all statistical analyses. Values reported within the paper are means ± SE.
RESULTS
General features of IIX, IIB, and IIX/IIB fibers.
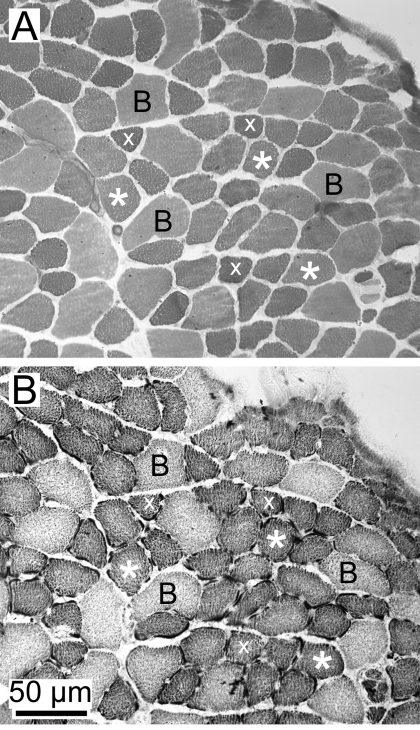
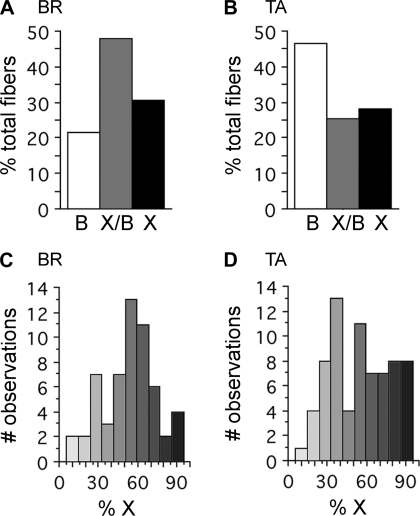
In the present study, we focused on the mouse TA and BR muscles because we had previously identified that these contain high proportions of IIX/IIB hybrid fibers. Using histochemical techniques, we found that the IIB and IIX fiber types existed as a continuum, with many intermediate fibers (Fig. 1). The IIB fibers identified through ATPase histochemistry were larger in diameter and less aerobic, while the IIX fibers were much smaller in diameter and stained intensely with the NADH diaphorase reaction (Fig. 1). However, a large proportion of the fibers identified with these techniques were intermediate to these two extremes and could not be discretely classified as IIB or IIX (Fig. 1, asterisks). We conclude that these intermediate fibers represent the IIX/IIB hybrid fibers. Using single-fiber dissection and SDS-PAGE (Fig. 2), we determined that the TA contained 46.4% IIB, 25.5% IIX/IIB, and 28.1% IIX fibers (n = 392 fibers, n = 22 animals) (Fig. 3A), while the BR contained 21.5% IIB, 47.8% IIX/IIB, and 30.7% IIX fibers (n = 339 fibers, n = 10 animals) (Fig. 3B).
Fig. 1.
Histochemical identification of muscle fiber types in tibialis anterior (TA) muscle. A: ATPase reaction (pH 4.6 preincubation) stains the IIB fibers to an intermediate level (B), while IIX fibers stain a darker hue (x). Many fibers are intermediate in their staining (*) and presumably represent IIX/IIB hybrids. B: NADH-diaphorase reaction stains the IIB fibers lightly (B), while the IIX (x) and IIX/IIB (*) fibers stain more intensely. Type IIB fibers have the largest diameters, IIX/IIB fibers are intermediate, and IIX fibers are the smallest in diameter.
Fig. 2.
Continuum of X/B fiber types (SDS-PAGE). A: using our SDS-PAGE conditions, the four adult mouse myosin heavy chains (MHCs) migrate in the order I > B > A > X. The sample on the left is a homogenate of several soleus fibers, while that on the right is from a single IIX/IIB hybrid from the brachioradialis (BR). B: continuum of fiber types observed in single fibers from the TA muscle. The fiber on the left is a pure IIX fiber, while the one on the far right is pure IIB. Between these two extremes, the IIX/IIB hybrids possess varying proportions of the 2 MHC isoforms.
Fig. 3.
Relative proportions of IIX/IIB isoforms in TA and BR muscles (A and B), relative proportions of IIX, IIX/IIB, and IIB fiber types. A: brachioradialis muscle had 47.8% of its fibers as IIX/IIB hybrids (n = 339 fibers). B: TA muscle had 25.5% IIX/IIB hybrids (n = 392 fibers). The remaining fibers in both muscles were either pure IIB or IIX fibers. C and D: relative proportions of IIX and IIB isoforms within single IIX/IIB hybrid fibers. IIX/IIB hybrid fibers within the BR muscle exhibit a range of MHC isoform proportions that are fairly normally distributed ∼50:50 mix of IIX/IIB (n = 57 fibers) (C). Hybrids within the TA muscle display a similar range of IIX/IIB isoform proportions (n = 71 fibers) (D). Shading corresponds to the relative levels of IIB (white) or IIX (black) MHC present in hybrid fibers.
Relative abundance of IIX and IIB MHCs in single muscle fibers.
Using single-fiber SDS-PAGE combined with densitometry, we determined the relative proportions of the IIX and IIB isoforms within single IIX/IIB fibers (Figs. 3, C and D). In both the TA and BR muscles, a continuum of IIX/IIB fiber types existed from those predominantly expressing the IIX MHC, to those mostly expressing IIB MHC. The relative abundance of these fiber types was highest in those fibers expressing a roughly equal mixture of the two isoforms in both of the muscles (Figs. 3, C and D) (n = 71 for TA and 57 for BR).
Expression levels of IIX and IIB isoforms in single fibers determined by real-time PCR.
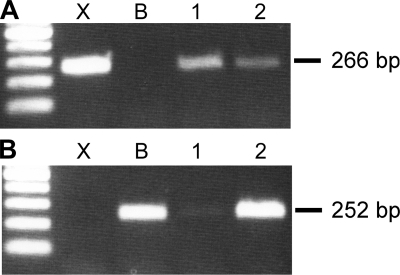
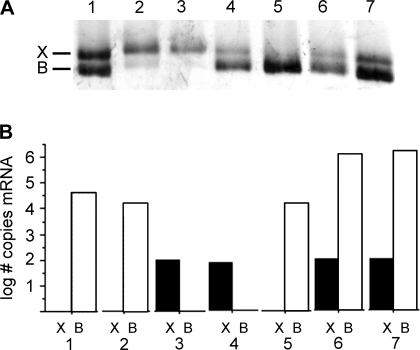
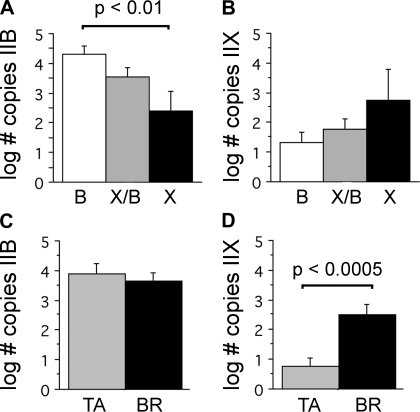
We measured the abundance of IIX and IIB mRNA levels within single fibers using real-time PCR n = 50 fibers: 24 TA, 26 BR. We initially used standard end-point PCR to verify that specific mRNA MHC sequences could be amplified from single fibers and to demonstrate primer specificity for the IIB and IIX sequences (Fig. 4). For real-time measurements, adjacent fiber segments from the same fiber were also used to determine the MHC protein fiber type in parallel with these measurements (Fig. 5). Collectively, the IIB mRNA isoform was expressed at significantly higher levels than the IIX mRNA isoform (P < 0.0001, BR and TA fibers, direct comparison not shown). When fibers were segregated by fiber type at the protein level, we determined that the IIB mRNA expression was significantly greater in fibers classified as IIB than in fibers classified as IIX (P < 0.01, BR and TA fibers, Fig. 6A). Fibers classified as IIX/IIB hybrids expressed intermediate levels. MHC IIX mRNA levels exhibited an opposite trend, although the differences between fiber types did not reach statistical significance (P > 0.11; BR and TA fibers Fig. 6B). When we compared expression levels between the TA and BR muscles, we found that while IIB mRNA expression was not significantly different (P > 0.54; Fig. 6C), the IIX mRNA isoform was expressed to a greater extent in the BR fibers (P < 0.0005; Fig. 6D). In fibers of each type (IIX, IIX/IIB, and IIB), we observed coexpression of both the IIB and IIX isoforms. In IIX fibers, the IIX mRNA expression levels were not significantly different from IIB mRNA levels (P > 0.74; BR and TA fibers direct comparison not shown).
Fig. 4.
RT-PCR of IIX and IIB transcripts from single TA fibers. A: PCR products amplified with IIX primers. cDNA encoding the IIX isoform (X) was used as a positive control and the cDNA encoding the IIB isoform (B) was used as a negative control. Two single fibers (1, 2) were used to isolate RNA as template for the RT reaction. B: PCR products from the same samples, but using IIB primers. Fiber 1 is predominantly expressing IIX, while fiber 2 expresses IIB>IIX.
Fig. 5.
Example of the correspondence between protein and mRNA content in single BR fibers. A: SDS-PAGE gel of fiber segments to identify IIX and IIB protein content. One fiber is pure IIX (3), one is pure IIB (5), while most have varying proportions of each isoform. B: expression levels of IIX (black) and IIB (white) mRNAs measured with real-time PCR from segments of the same fibers. Overall, there is an imprecise match between the relative IIX and IIB content at the protein and mRNA levels.
Fig. 6.
IIX and IIB MHC mRNA levels measured by real-time PCR. Levels of mRNA segregated by muscle fiber type (IIB, IIX/IIB, or IIX) as determined by SDS-PAGE (A and B). A: IIB MHC mRNA levels were significantly higher in IIB fibers than in IIX fibers (P < 0.01), while IIX/IIB hybrids were intermediate in these levels. B: IIX MHC mRNA showed the opposite trend, with expression levels increasing toward the pure IIX fiber types. However, these differences were not statistically significant (P > 0.11). Levels of mRNA segregated by muscle (TA or BR) (C and D). C: IIB mRNA levels were not different between TA and BR fibers (P > 0.54). D: 26 BR fibers expressed significantly greater levels of the IIX isoform than 24 TA fibers (P < 0.0005).
Relative abundance of IIX and IIB isoforms in adjacent fiber segments.
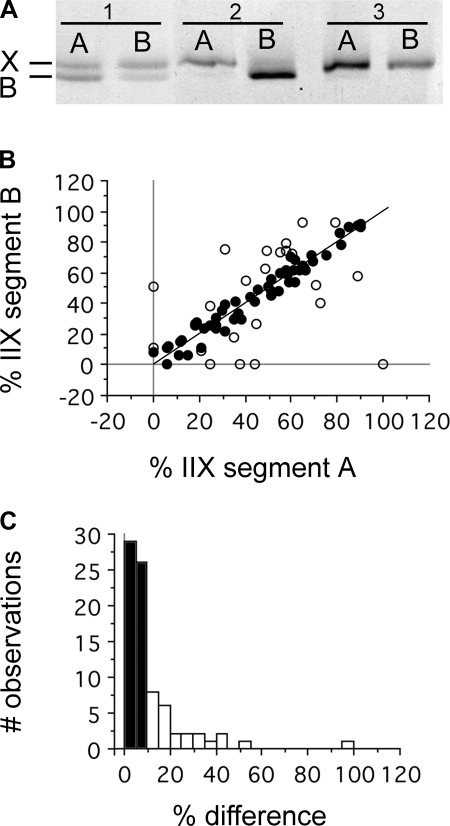
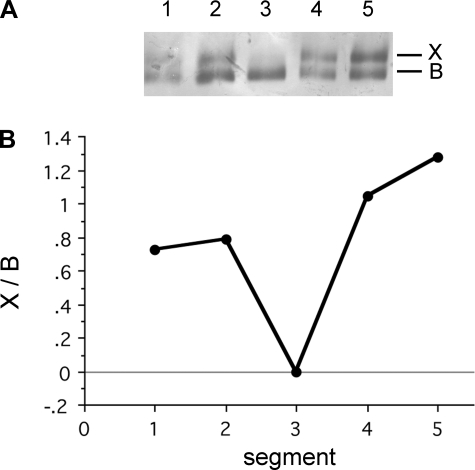
To examine the distribution of IIX and IIB MHC isoforms within single fibers, we divided single fibers into ∼0.5- to 2-mm segments and determined their protein content using SDS-PAGE (Fig. 7A). In most cases, fibers were divided into two segments, but for ∼25 fibers, we were able to determine relative isoform composition for 3 to 5 continuous segments (Fig. 8). Our analyses revealed that although the majority of IIX/IIB hybrids exhibited uniform MHC isoform proportions between adjacent segments, in ∼30% of the fiber pairs examined (n = 80 pairs: 32 TA, 48 BR), we detected significant asymmetries in MHC content. Plotting the relative MHC content of adjacent segment pairs from IIX/IIB hybrids showed that the MHC proportions were significantly correlated (% in B = 8.44 + 0.81*% in A; P < 0.0001), but with a relatively weak correlation (r2 = 0.55) (Fig. 7B). When only those segment pairs differing in MHC composition by ≥ 10% were used in the analysis (Fig. 7B, white circles, n = 25), there was no significant correlation between MHC content in adjacent fiber segments (P > 0.13; r2 = 0.10). Plotting the difference in MHC isoform proportion between segment pairs as a frequency histogram showed that the degree of difference conformed to something like a probability distribution, with 31% of pairs having differences of ≥ 10% (Fig. 7C). Control analyses of MHC content in replicates of samples containing two MHC isoforms to assess measurement error indicated that the error averaged 5.4 ± 0.8%. In some instances where single fibers were divided into three to five segments, MHC isoform content differed significantly along the length of the fiber (Fig. 8).
Fig. 7.
Asymmetries in IIX and IIB isoform proportions within single fibers. A: SDS-PAGE showing the relative proportions of IIX and IIB MHC isoforms in adjacent fiber segments. Fiber 1 is a IIX/IIB hybrid expressing different relative proportions of the IIX and IIB isoforms. Fiber 2 is a IIX/IIB hybrid expressing completely different isoforms within each segment. Segment 2A is IIX in phenotype, while segment 2B is IIB. Fiber 3 is a pure IIX fiber in both segments. B: correlation between isoform proportions within adjacent segments of single fibers. The line represents the line of unity, where the proportion of MHC is precisely the same for each segment. Of the segment pairs, 69% differed in isoform proportion by < 10% (●), while 31% differed by ≥10% (○) C: % differences between adjacent segments presented as a histogram demonstrates the distribution of pairs differing by < 10% (black bars), and the 31% of the segments exhibiting differences ≥10% (white bars). This distribution suggests an overall likelihood of adjacent segments of the same fiber containing roughly equal proportions of the IIX and IIB MHC isoforms, but with some proportion of normal fibers exhibiting significant differences (n = 80 segment pairs 32 TA, 48 BR).
Fig. 8.
Example of a single TA fiber exhibiting major shifts in fiber type along its length. A: 5 fragments were obtained from the fiber, and the MHC isoform content was determined using SDS-PAGE. B: relative proportion of the IIX and IIB isoforms was determined using densitometry and demonstrates significant changes along the length of the fiber.
DISCUSSION
This is the first study focusing on mouse single fibers coexpressing the MHC IIX and IIB isoforms as a model of hybrid muscle fibers. The IIX/IIB hybrids are by far the most abundant hybrid fiber type in both rat and mouse muscles, representing from ∼50 to 75% of the total fibers in certain muscles (16, 25). We had previously identified the mouse TA muscle as containing a significant proportion of the IIX/IIB hybrids (∼25%) (25), and here we report that the BR muscle in the mouse has an even higher proportion of these hybrids (∼50%). While these two muscles are representatives of mouse muscles with a high IIX/IIB content, there are likely many others with similar fiber composition that have not yet been identified. These IIX/IIB hybrids appear to form a phenotypic continuum, as determined by their morphological and histochemical characteristics (Fig. 1), and in terms of the relative amounts of the IIX and IIB isoforms present within single fibers (Figs. 2 and 3). Type IIB fibers are larger in diameter and less aerobic than the IIX fibers, while IIX/IIB hybrid fibers possess a range of intermediate characteristics between the two (Fig. 1). The kind of continuum in fiber type reported here has also been identified in the muscles of other species (32–34, 57–59) and fits within the phenotypic continuum in single-fiber types proposed by Pette and Staron (44–46). Although certain experimental conditions seem to produce mismatched hybrids that do not follow this continuum (5a, 15, 19, 64), our results support previous observations that the proportion of these mismatched fibers in normal muscles is low (16, 25).
Using real-time PCR and SDS-PAGE, we were able to quantify isoform-specific mRNA levels and protein isoform composition from single fibers. These analyses revealed that although a general correlation exists between the mRNA isoforms expressed and the protein isoform(s) present within single fibers, there is not always a close match between these two indicators of muscle fiber type (Figs. 5 and 6). This pattern is consistent with previous observations indicating that MHC mRNA and protein distribution patterns in single fibers are often poorly correlated (6, 34, 35, 48). In rat muscle, the IIX and IIB MHC genes are cooperatively regulated by a bidirectional promoter that produces an antisense IIX RNA, while activating the IIB MHC gene (49). This coordinate regulation of the IIX and IIB genes illustrates the complexity in the control of muscle fiber types and may help explain the weak correlation between mRNA and protein content in single fibers. Finally, we measured the relative proportions of the IIX and IIB isoforms present along the length of single hybrid fibers and discovered that a substantial proportion of these fibers possessed significant asymmetries in the distribution of the proteins (Fig. 7). In certain cases, muscle fiber type differed dramatically from one region to another within a single fiber (Figs. 7 and 8).
Hybrid single muscle fibers have been recognized for decades (3, 55, 57–59, 67), but their significance remains a matter of debate (16, 60). Compared with limb muscles, mammalian laryngeal and extraocular fibers exhibit striking complexities in MHC combinations, which may relate to their distinct developmental origins and complex innervation patterns (l0a, 27a, 52a, 52d). Many studies focusing on hybrid limb muscle fibers have dealt with their role in phenotypic transitions, since a fiber switching from one type to another must contain multiple MHC isoforms during the period of transition. A variety of conditions known to influence muscle fiber type clearly do lead to changes in the proportion of hybrid fibers. These include exercise (25, 27, 29, 68), neural input (2, 41, 63–65), hormonal environment (2, 13, 15, 17), and development (19). Although hybrid fibers definitely play a role in muscle fiber type transitions, a number of studies have now shown that hybrid fibers also form a common component of normal muscles (1, 16, 25, 60). Many of these hybrid fibers represent very stable phenotypes, even in the face of different types of exercise (25, 28, 40). The relative proportion of hybrid fibers varies among different muscles (1, 16, 25) and even among anatomical regions within the same muscle (16, 25). It seems likely that hybrid fibers may simply represent an intermediate phenotype along the continuum of fiber types. Since the alpha motor neurons that control these fibers possess a range of physiological properties (21), it may be that motor neurons with intermediate properties are the ones that innervate hybrid muscle fibers.
Asymmetric distribution of MHC isoforms.
This is the first quantitative analysis of single fibers in normal mammalian muscles that possess significant MHC asymmetries along their length (Figs. 7 and 8). A number of studies in other species have reported similar patterns in MHC isoform distribution, but in some cases these patterns were thought to represent the consequence of experimental manipulation (53, 56, 71) or as a result of aging (5). Our results indicate that as many as one third of the hybrid fibers in these normal mouse muscles possess significant MHC differences along their length. We observed multiple examples of fibers that would be classified as completely different fiber types, depending on which region of the fiber was examined (Figs. 7 and 8). Peuker and Pette (48) assessed MHC mRNA isoforms in single rabbit-fiber fragments and found that one-third of the fibers possessed unambiguous variations in isoform expression along their lengths. Similar differences in MHC isoform content along the length of single muscle fibers have been reported in electrically stimulated mammalian limb muscles (56), mammalian extraocular muscles (27b, 52a, 52b), bird flight muscles (10, 52), and limb muscles of frogs (32, 33). However, it is unclear how common or extensive these asymmetries are generally in skeletal muscles, or particularly within the limb muscles of mammals. In some cases, alternate myofibrillar isoforms are asymmetrically distributed within the same thick filament (66), or segregated among myofibrils within single fibers (24, 36, 37). Our approach in the present studies relied on MHC analyses of relatively long fiber segments (∼0.5–2 mm) so that we could only detect large-scale shifts in fiber types along the longitudinal axis of the fibers. Fine-scale differences along the length of the fibers, or asymmetries across fiber diameters, would be missed with this approach. It is also possible that longitudinal asymmetries were missed because both adjacent segments were divided in a way that produced two asymmetric segments, each with the same relative proportions of MHC isoforms (e.g., see IIX/IIB hybrid in Fig. 8).
Potential role of myonuclear domains in MHC asymmetries.
Given that single fibers exhibit regional differences in MHC distribution, an important question is how different regions of the same cell acquire distinct myofibrillar proteins. There is good reason to speculate that the asymmetrical distribution of MHC isoforms stems from the unique characteristic of muscle fibers being multinucleate, possessing hundreds or thousands of individual nuclei within the same cell (4). These nuclei arise from distinct cellular populations and may possess a predisposition to express a particular MHC isoform. During development, single myoblasts fuse to form a single myotube, and these founding cells possess differences in terms of the expression of specific MHC isoforms (21). In adult muscle fibers, new nuclei are dynamically incorporated into existing fibers as satellite cells fuse with the fiber during processes of fiber repair or hypertrophy (8, 21, 72). A fundamental question in skeletal muscle biology is how well coordinated multiple myonuclei within the same cell are in their expression of specific myofibrillar genes (4). Cultured fibers explanted from intact and regenerating mouse muscles exhibit stochastic patterns of expression within different regions of the same fiber (39). Experimentally implanting slow fibers into the fast breast muscle in the chicken results in hybrid single fibers that coexpress both fast and slow tropomyosins (71), indicating that even after fusing with a new fiber type, myonuclei continue to preferentially express distinct myofibrillar isoforms. Subsequent studies of natural hybrid fibers demonstrated abrupt changes in fiber type along the length of fibers and that the protein composition can even differ within the same myofibril (37). Myofibrillar mRNA and proteins are restricted in their movement away from the transcriptionally active nucleus, meaning that regional differences in myofibrillar protein content within single fibers likely reflect differences in myonuclear expression (37, 42, 52c, 71). Overall, these patterns suggest that although the myonuclei within a single fiber experience similar intracellular signals, the transcriptional responses of individual nuclei to these signals may remain fairly distinct. It is noteworthy that when neural input to a muscle is removed, its fibers switch from their normal MHC expression patterns and become hybrids, coexpressing multiple MHC isoforms (18, 41, 53, 61–65). This shift toward hybrid phenotypes suggests that neural activity provides a coordinating or synchronizing influence over the multiple nuclei within a fiber, and removing this influence serves to unmask the native potential of the myonuclei to express different myofibrillar programs in an uncoordinated fashion. The present findings of regional MHC asymmetries are consistent with the idea that the regional myonuclear domains within a single fiber may routinely express distinct MHC isoforms. Similar patterns reported for single fibers from several different species (10, 32, 33, 52, 56) suggests that this nonuniform expression pattern may be a general characteristic of skeletal muscles.
Functional implications of MHC asymmetries.
From a functional perspective, it is unclear how regional differences in MHC content might influence muscle mechanics. The conventional perspective is that each fiber within the motor unit is of the same phenotype and that when the motor neuron sends an action potential, each of the fibers contracts simultaneously. The existence of regional MHC differences within single fibers raises the question of how MHC motors with different kinetic properties influences contraction within single fibers. Mechanical studies of single fibers indicate that hybrid fibers possess contractile properties intermediate to those of pure muscle fiber types (9, 14, 31). However, other studies of single fiber segments clearly indicate that different regions of the same fiber can possess distinct contractile properties (22, 23, 33). If multiple isoforms are assembled into the same thick filaments, then myosin crossbridges would be expected to cycle at different rates within the same sarcomere. When isoforms are segregated among different myofibrils within the same fiber (24, 36, 37), then tension development and shortening across the fiber diameter should vary. If MHC isoforms change along the length of a fiber, then contractile properties at one part of a single fiber may be distinct from other regions. This type of MHC isoform asymmetry can occur along the length of whole muscles (5, 10, 22, 23, 32, 33, 52, 53, 56) (Figs. 7 and 8), but the prevalence and functional significance of these MHC distribution patterns are not well understood. The results from the present study indicate that significant regional asymmetries in the distribution of MHC isoforms within hybrid fibers may be common, occurring in ∼30% of hybrid IIX/IIB fibers.
Perspectives and Significance
Although hybrid skeletal muscle fibers have been recognized for years, their existence continues to challenge our understanding of fundamental issues of skeletal muscle organization and plasticity (16, 60). Hybrid fibers represent a common component of many muscles from a variety of species, and in many cases these appear to represent normal stable phenotypes. In the end, these fibers may simply represent functional intermediates along the continuum of muscle fiber types, paired with motor neurons possessing intermediate physiological properties. Whatever the precise reason for their occurrence, hybrid fibers present a unique and useful model for understanding the factors that impact muscle fiber type, and the cellular and molecular mechanisms that control these processes. One area in particular that can benefit from the study of hybrid fibers is the coordination of gene expression among the multiple myonuclei within single fibers. Although some fibers clearly exhibit differences in MHC isoform content along their length, the prevalence and significance of these patterns deserve further study.
GRANTS
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases grant RAR-053666A (to S. Medler). We sincerely appreciate the insightful comments from three anonymous reviewers of this manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1.Acevedo LM, Rivero JL. New insights into skeletal muscle fibre types in the dog with particular focus towards hybrid myosin phenotypes. Cell Tissue Res 323: 283–303, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Adams GR, McCue SA, Zeng M, Baldwin KM. Time course of myosin heavy chain transitions in neonatal rats: importance of innervation and thyroid state. Am J Physiol Regul Integr Comp Physiol 276: R954–R961, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Aigner S, Gohlsch B, Hamalainen N, Staron RS, Uber A, Wehrle U, Pette D. Fast myosin heavy-chain diversity in skeletal-muscles of the rabbit–heavy chain-IID, not chain-IIB predominates. Eur J Biochem 211: 367–372, 1993 [DOI] [PubMed] [Google Scholar]
- 4.Allen DL, Roy RR, Edgerton VR. Myonuclear domains in muscle adaptation and disease. Muscle Nerve 22: 1350–1360, 1999 [DOI] [PubMed] [Google Scholar]
- 5.Andersen JL. Muscle fibre type adaptation in the elderly human muscle. Scand J Med Sci Sports 13: 40–47, 2003 [DOI] [PubMed] [Google Scholar]
- 5a.Anderson JL, Gruschy-Knudsen T, Sandri C, Larsson L, Schiaffino S. Bed rest increases the amount of mismatched fibers in human skeletal muscle. J Appl Physiol 86: 455–460, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Andersen JL, Schiaffino S. Mismatch between myosin heavy chain mRNA and protein distribution in human skeletal muscle fibers. Am J Physiol Cell Physiol 272: C1881–C1889, 1997 [DOI] [PubMed] [Google Scholar]
- 7.Andersen JL, Terzis G, Kryger A. Increase in the degree of coexpression of myosin heavy chain isoforms in skeletal muscle fibers of the very old. Muscle Nerve 22: 449–454, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Anderson JE. The satellite cell as a companion in skeletal muscle plasticity: currency, conveyance, clue, connector and colander. J Exp Biol 209: 2276–2292, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Andruchov O, Andruchova O, Wang Y, Galler S. Kinetic properties of myosin heavy chain isoforms in mouse skeletal muscle: comparison with rat, rabbit, and human correlation with amino acid sequence. Am J Physiol Cell Physiol 287: C1725–C1732, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Bartnik BL, Waldbillig DM, Bandman E, Rosser BWC. Persistent expression of developmental myosin heavy chain isoforms in the tapered ends of adult pigeon pectoralis muscle fibres. Histochem J 31: 321–329, 1999 [DOI] [PubMed] [Google Scholar]
- 10a.Bicer S, Reiser PJ. Myosin isoform expression in dog rectus muscles: patterns in global and orbital layers and among single fibers. Invest Ophthalmol Vis Sci 50: 157–167, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Blough E, Rennie ER, Zhang F, Reiser PJ. Enhanced electrophoretic separation and resolution of myosin heavy chains in mammalian and avian skeletal muscles. Anal Biochem 233: 31–35, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Bolster DR, Kimball SR, Jefferson LS. Translational control mechanisms modulate skeletal muscle gene expression during hypertrophy. Exerc Sport Sci Rev 31: 111–116, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Caiozzo V, Haddad R, Baker M, McCue S, Baldwin KM. MHC polymorphism in rodent plantaris muscle: effects of mechanical overload and hypothyroidism. Am J Physiol Cell Physiol 278: C709–C717, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Caiozzo VJ. Plasticity of skeletal muscle phenotype: mechanical consequences. Muscle Nerve 26: 740–768, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Caiozzo VJ, Baker M, Baldwin K. Novel transitions in MHC isoforms: separate and combined effects of thyroid hormone and mechanical unloading. J Appl Physiol 85: 2237–2248, 1998 [DOI] [PubMed] [Google Scholar]
- 16.Caiozzo VJ, Baker MJ, Huang K, Chou H, Wu Y, Baldwin K. Single-fiber myosin heavy chain polymorphism: how many patterns and what proportions? Am J Physiol Regul Integr Comp Physiol 285: R570–R580, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Caiozzo VJ, Baker MJ, Baldwin KM. Novel transitions in MHC isoforms: separate and combined effects of thyroid hormone and mechanical unloading. J Appl Physiol 85: 2237–2248, 1998 [DOI] [PubMed] [Google Scholar]
- 18.Cormery B, Pons F, Marini JF, Gardiner PF. Myosin heavy chains in fibers of TTX-paralyzed rat soleus and medial gastrocnemius muscles. J Appl Physiol 88: 66–76, 2000 [DOI] [PubMed] [Google Scholar]
- 19.Di Maso NA, Caiozzo VJ, Baldwin KM. Single-fiber myosin heavy chain polymorphism during postnatal development: modulation by hypothyroidism. Am J Physiol Regul Integr Comp Physiol 278: R1099–R1106, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Dubowitz V, Sewry CA. Muscle Biopsy: A Practical Approach. New York: Elsevier, 2007 [Google Scholar]
- 21.Edgerton VR, Bodine-Fowler S, Roy RR, Ishihara A, Hodgson JA. Neuromuscular adaptation. In: Handbook of Physiology Section 12: Exercise: Regulation and Integration of Multiple Systems, edited by Rowell LB, Shepherd JT. New York: Oxford University Press, 1996 [Google Scholar]
- 22.Edman KAP, Reggiani C, Schiaffino S, te Kronnie G. Maximum velocity of shortening related to myosin isoform composition in frog skeletal-muscle fibers. J Physiol 395: 679–694, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edman KAP, Reggiani C, te Kronnie G. Differences in maximum velocity of shortening along single muscle-fibers of the frog. J Physiol 365: 147–163, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauthier GF. Differential distribution of myosin isoforms among the myofibrils of individual developing muscle-fibers. J Cell Biol 110: 693–701, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser B, You G, Zhang M, Medler S. Relative proportions of hybrid fibers are unaffected by 6 weeks of running exercise in mouse skeletal muscles. Exp Physiol 95: 211–221, 2010 [DOI] [PubMed] [Google Scholar]
- 26.Hamalainen N, Pette D. The histochemical profiles of fast fiber types IIB, IID, and IIA in skeletal muscles of mouse, rat, and rabbit. J Histochem Cytochem 41: 733–743, 1993 [DOI] [PubMed] [Google Scholar]
- 27.Harber MP, Gallagher PM, Trautmann J, Trappe SW. Myosin heavy chain composition of single muscle fibers in male distance runners. Int J Sports Med 23: 484–488, 2002 [DOI] [PubMed] [Google Scholar]
- 27a.Hoh JFY. Laryngeal muscle fibre types. Acta Physiol Scand 183: 133–149, 2005 [DOI] [PubMed] [Google Scholar]
- 27b.Jacoby J, Ko K, Weiss C, Rushbrook JI. Systematic variation in myosin expression along extraocular muscle fibres of the adult rat. J Muscle Res Cell Motil 11: 25–40, 1989 [DOI] [PubMed] [Google Scholar]
- 28.Kesidis N, Metaxas TI, Vrabas IS, Stefanidis P, Vamvakoudis E, Christoulas K, Mandroukas A, Balasas D, Mandroukas K. Myosin heavy chain isoform distribution in single fibres of bodybuilders. Eur J Appl Physiol 103: 579–583, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Kohn TA, Essen-Gustavsson B, Myburgh KH. Exercise pattern influences skeletal muscle hybrid fibers of runners and nonrunners. Med Sci Sports Exerc 39: 1977–1984, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Kucera J, Walro JM. Nonuniform expression of myosin heavy-chain isoforms along the length of cat intrafusal muscle-fibers. Histochemistry 92: 291–299, 1989 [DOI] [PubMed] [Google Scholar]
- 31.Larsson L, Moss RL. Maximum velocity of shortening in relation to myosin isoform composition in single fibers from human skeletal muscles. J Physiol 472: 595–614, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz GJ, Bremner SN, Bade MJ, Lieber RL. Identification of myosin light chains in Rana pipiens skeletal muscle and their expression patterns along single fibres. J Exp Biol 204: 4237–4248, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Lutz GJ, Sirsi SR, Shapard-Palmer SA, Bremner SN, Lieber RL. Influence of myosin isoforms on contractile properties of intact muscle fibers from Rana pipiens. Am J Physiol Cell Physiol 282: C835–C844, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Medler S, Lilley T, Mykles DL. Fiber polymorphism in skeletal muscles of the American lobster, Homarus americanus: continuum between slow-twitch (S-1) and slow-tonic (S-2) fibers. J Exp Biol 207: 2755–2767, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Medler S, Mykles DL. Analysis of myofibrillar proteins and transcripts in adult skeletal muscles of the American lobster Homarus americanus: variable expression of myosins, actin and troponins in fast, slow-twitch and slow-tonic fibres. J Exp Biol 206: 3557–3567, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Nakada K, Kimura F, Hirabayashi T, Miyazaki JI. Immunohistochemical studies on regulation of alternative splicing of fast skeletal muscle troponin T: non-uniform distribution of the exon X 3 epitope in a single muscle fiber. Cell Tissue Res 299: 263–271, 2000 [DOI] [PubMed] [Google Scholar]
- 37.Nakada K, Miyazaki JI, Saba R, Hirabayashi T. Natural occurrence of fast- and fast/slow-muscle chimeric fibers in the expression of troponin T isoforms. Exp Cell Res 235: 93–99, 1997. [DOI] [PubMed] [Google Scholar]
- 38.Neter J, Wasserman W, Kutner MH. Applied Linear Statistical Models: Regression, Analysis of Variance, and Experimental Designs. Boston: Irwin, 1990 [Google Scholar]
- 39.Newlands S, Levitt LK, Robinson CS, Karpf ABC, Hodgson VRM, Wade RP, Hardeman EC. Transcription occurs in pulses in muscle fibers. Genes Dev 12: 2748–2758, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parcell AC, Sawyer RD, Drummond MJ, O'Neil B, Miller N, Woolstenhulme MT. Single-fiber MHC polymorphic expression is unaffected by sprint cycle training. Med Sci Sports Exerc 37: 1133–1137, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Patterson MF, Stephenson GMM, Stephenson DG. Denervation produces different single fiber phenotypes in fast- and slow-twitch hindlimb muscles of the rat. Am J Physiol Cell Physiol 291: C518–C528, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Pavlath GK, Rich K, Webster SG, Blau HM. Localization of muscle gene-products in nuclear domains. Nature 337: 570–573, 1989 [DOI] [PubMed] [Google Scholar]
- 43.Perry MJ, Tait J, Hu J, White SC, Medler S. Skeletal muscle fiber types in the ghost crab, Ocypode quadrata: implications for running performance. J Exp Biol 212: 673–683, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Pette D, Staron RS. Cellular and molecular diversities of mammalian skeletal muscle fibers. Rev Physiol Biochem Pharmacol 116: 1–76, 1990 [DOI] [PubMed] [Google Scholar]
- 45.Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microsc Res Tech 50: 500–509, 2000 [DOI] [PubMed] [Google Scholar]
- 46.Pette D, Staron RS. The molecular diversity of mammalian muscle-fibers. News Physiol Sci 8: 153–157, 1993 [Google Scholar]
- 47.Pette D, Staron RS. Transitions of muscle fiber phenotypic profiles. Histochem Cell Biol 115: 359–372, 2001 [DOI] [PubMed] [Google Scholar]
- 48.Peuker H, Pette D. Quantitative analyses of myosin heavy-chain mRNA and protein isoforms in single fibers reveal a pronounced fiber heterogeneity in normal rabbit muscles. Eur J Biochem 247: 30–36, 1997 [DOI] [PubMed] [Google Scholar]
- 49.Rinaldi C, Haddad F, Bodell PW, Qin AX, Jiang WH, Baldwin KM. Intergenic bidirectional promoter and cooperative regulation of the IIx and IIb MHC genes in fast skeletal muscle. Am J Physiol Regul Integr Comp Physiol 295: R208–R218, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rome LC, Funke RP, Alexander RM, Lutz G, Aldridge H, Scott F, Freadman M. Why animals have different muscle fibre types. Nature 335: 824–827, 1988. [DOI] [PubMed] [Google Scholar]
- 51.Rome LC, Lindstedt SL. Mechanical and metabolic design of the muscular system in vertebrates. In: Handbook of Physiology. Comparative Physiology. Bethesda, MD: Am. Physiol. Soc., 1997, sect. 13, vol. II, chapt. 23, p. 1587–1651 [Google Scholar]
- 52.Rosser BWC, Waldbillig DM, Lovo SD, Armstrong JD, Bandman E. Myosin heavy-chain expression within the tapered ends of skeletal-muscle fibers. Anat Rec 242: 462–470, 1995 [DOI] [PubMed] [Google Scholar]
- 52a.Rubinstein NA, Hoh JFY. The distribution of myosin heavy chain isoforms among rat extraocular muscle fiber types. Invest Ophthalmol Vis Sci 41: 3391–3398, 2000 [PubMed] [Google Scholar]
- 52b.Rubinstein NA, Porter JD, Hoh JFY. The development of longitudinal variation of myosin isoforms in the orbital fibers of extraocular muscles of rats. Invest Ophthalmol Vis Sci 45: 3067–3072, 2004 [DOI] [PubMed] [Google Scholar]
- 52c.Russell B, Dix DJ. Mechanisms for intracellular distribution of mRNA: in situ hybridization studies in muscle. Am Physiol Cell Physiol 262: CI–C8, 1992 [DOI] [PubMed] [Google Scholar]
- 52d.Schachat F, Briggs MM. Phylogenetic implications of the superfast myosin in extraocular muscles. J Exp Biol 205: 2189–2201, 2002 [DOI] [PubMed] [Google Scholar]
- 53.Schiaffino S, Gorza L, Pitton G, Saggin L, Ausoni S, Sartore S, Lomo T. Embryonic and neonatal myosin heavy-chain in denervated and paralyzed rat skeletal-muscle. Dev Biol 127: 1–11, 1988 [DOI] [PubMed] [Google Scholar]
- 54.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 76: 371–423, 1996 [DOI] [PubMed] [Google Scholar]
- 55.Staron RS, Pette D. Correlation between myofibrillar ATPase activity and myosin heavy chain composition in rabbit muscle fibers. Histochemistry 86: 19–23, 1986 [DOI] [PubMed] [Google Scholar]
- 56.Staron RS, Pette D. Nonuniform myosin expression along single fibers of chronically stimulated and contralateral rabbit tibialis anterior muscles. Pflügers Arch 409: 67–73, 1987 [DOI] [PubMed] [Google Scholar]
- 57.Staron RS, Pette D. The continuum of pure and hybrid myosin heavy chain-based fiber types in rat skeletal-muscle. Histochemistry 100: 149–153, 1993 [DOI] [PubMed] [Google Scholar]
- 58.Staron RS, Pette D. The multiplicity of combinations of myosin light-chains and heavy-chains in histochemically typed single fibers–rabbit soleus muscle. Biochem J 243: 687–693, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staron RS, Pette D. The multiplicity of combinations of myosin light-chains and heavy-chains in histochemically typed single fibers–rabbit tibialis anterior muscle. Biochem J 243: 695–699, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephenson GMM. Hybrid skeletal muscle fibres: a rare or common phenomenon? Clin Exp Pharmacol Physiol 28: 692–702, 2001 [DOI] [PubMed] [Google Scholar]
- 61.Talmadge RJ. Myosin heavy chain isoform expression following reduced neuromuscular activity: potential regulatory mechanisms. Muscle Nerve 23: 661–679, 2000 [DOI] [PubMed] [Google Scholar]
- 62.Talmadge RJ, Castro MJ, Apple DF, Dudley GA. Phenotypic adaptations in human muscle fibers 6 and 24 wk after spinal cord injury. J Appl Physiol 92: 147–154, 2002 [DOI] [PubMed] [Google Scholar]
- 63.Talmadge RJ, Garcia ND, Roy RR, Edgerton VR. Myosin heavy chain isoform mRNA and protein levels after long-term paralysis. Biochem Biophys Res Commun 325: 296–301, 2004 [DOI] [PubMed] [Google Scholar]
- 64.Talmadge RJ, Roy RR, Edgerton VR. Persistence of hybrid fibers in rat soleus after spinal cord transection. Anat Rec 255: 188–201, 1999 [DOI] [PubMed] [Google Scholar]
- 65.Talmadge RJ, Roy RR, Edgerton VR. Prominence of myosin heavy-chain hybrid fibers in soleus muscle of spinal cord-transected rats. J Appl Physiol 78: 1256–1265, 1995 [DOI] [PubMed] [Google Scholar]
- 66.Taylor LD, Bandman E. Distribution of fast myosin heavy chain isoforms in thick filaments of developing chicken pectoral muscle. J Cell Biol 108: 533–542, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Termin A, Staron RS, Pette D. Myosin heavy-chain isoforms in histochemically defined fiber types of rat muscle. Histochemistry 92: 453–457, 1989 [DOI] [PubMed] [Google Scholar]
- 68.Williamson DL, Gallagher PM, Carroll CC, Raue U, Trappe SW. Reduction in hybrid single muscle fiber proportions with resistance training in humans. J Appl Physiol 91: 1955–1961, 2001 [DOI] [PubMed] [Google Scholar]
- 69.Williamson DL, Godard MP, Porter DA, Costill DL, Trappe SW. Progressive resistance training reduces myosin heavy chain coexpression in single muscle fibers from older men. J Appl Physiol 88: 627–633, 2000 [DOI] [PubMed] [Google Scholar]
- 70.Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem 118: 197–203, 1981 [DOI] [PubMed] [Google Scholar]
- 71.Yao Y, Miyazaki JI, Hirabayashi T. Coexistence of fast-muscle-type and slow-muscle-type troponin-t isoforms in single chimeric muscle-fibers induced by muscle transplantation. Exp Cell Res 214: 400–407, 1994 [DOI] [PubMed] [Google Scholar]
- 72.Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem 54: 1177–1191, 2006 [DOI] [PubMed] [Google Scholar]