Abstract
We recently demonstrated that mRNA levels of three members of the CCAAT/enhancer binding factor (C/EBP) family of transcription factors are increased in skeletal muscle following 12 days of spaceflight. In the present study, we further explored the expression of C/EBP-δ in atrophying fast skeletal muscle by examining its expression in muscle from food-deprived (FD) mice, and investigated its role in regulating the expression of the secreted antigrowth factor myostatin. C/EBP-δ mRNA and protein levels were significantly increased by 2 days of food deprivation in the tibialis anterior (TA) muscle, and expression of both myostatin and C/EBP-δ mRNA during food deprivation was attenuated by injection with the glucocorticoid inhibitor RU486. The increase in myostatin mRNA levels with food deprivation appears to be at least partially transcriptionally driven, since levels of myostatin pre-mRNA were significantly increased in the TA muscle. C/EBP-δ mRNA levels and promoter activity were significantly increased by transfection of C2C12 myotubes with a glucocorticoid receptor construct and 24 h of treatment with the synthetic glucocorticoid dexamethasone. Furthermore, activity of the C/EBP-δ promoter was significantly increased with as little as 1 h of dexamethasone treatment, while activity of the mouse myostatin promoter was only significantly increased with longer treatment periods of 24 h or more. Activity of the myostatin promoter-reporter construct was significantly increased in C2C12 myotubes by cotransfection with expression constructs for C/EBP-α, -β, and -δ, with C/EBP-δ having the greatest effect. The myostatin promoter contains two potential C/EBP binding sequences, a CCAAT box, and a C/EBP binding element (CBE). Mutation of the CCAAT box attenuated basal myostatin promoter activity but potentiated C/EBP-δ-activated myostatin promoter activity in C2C12 myotubes in vitro, while mutation of the CBE abolished glucocorticoid receptor and C/EBP-δ responsiveness. The present results support a model in which glucocorticoid-induced increases in C/EBP-δ expression may contribute to myostatin transcription during atrophic states.
Keywords: fasting, hindlimb suspension, metabolism
the secreted autocrine/paracrine factor myostatin is a major modulator of skeletal muscle growth. Myostatin is expressed and secreted by skeletal muscle and potently inhibits muscle stem cell proliferation and differentiation and muscle fiber protein accretion (17, 23, 26, 37, 40). Inactivating mutations to the myostatin gene are associated with profound muscle growth in several mammalian species (15, 24, 25, 34) and greatly attenuate muscle responsiveness to some atrophic states in the mouse (14, 35). Finally, expression of myostatin has been shown to be increased in a wide variety of atrophy models (3, 7–9, 18, 30). Thus, a wealth of data supports a central, critical role for myostatin in muscle atrophy.
Despite this vast array of data on the expression and function of myostatin during atrophic states, surprisingly little is known regarding the molecular mechanisms regulating myostatin expression during skeletal muscle atrophy. Myostatin expression and activity are likely regulated on many levels, but myostatin transcription may represent a critical node of regulation. Transcriptional activity of the myostatin promoter is upregulated by glucocorticoids (21) and by members of the FoxO transcription factor family (1) in vitro, both of which are implicated in atrophy-associated changes in gene expression. However, at present it is not clear whether other transcription factors may contribute to the regulation of myostatin expression during atrophic states.
One intriguing candidate for regulating myostatin transcription during muscle atrophy is the CCAAT/enhancer binding factor (C/EBP) family of transcription factors. The C/EBP transcription factors are expressed in a wide range of mammalian tissues and play key roles in regulating processes such as inflammation and metabolism (31). We recently demonstrated that expression of three members of the C/EBP family, C/EBP-α, -β, and -δ, was increased along with myostatin in response to 12 days of spaceflight unloading of mouse skeletal muscle, with C/EBP-δ showing the greatest response (3). Moreover, expression and binding activity of C/EBP-β and -δ are also significantly increased in skeletal muscle during sepsis and following glucocorticoid administration (29, 39), while myostatin expression is also induced by glucocorticoids (21, 22). Thus the C/EBP transcription factors may represent another potential transcriptional circuit for regulating the expression of myostatin and/or other atrogenes during skeletal muscle atrophy, particularly in response to glucocorticoid signaling.
The purpose of the present work was to examine the relationship between glucocorticoid signaling, C/EBP-δ expression, and myostatin transcription in skeletal muscle cells and the potential role of C/EBP-δ in regulating transcription of the myostatin gene during food deprivation. We hypothesized that the C/EBP family of transcription factors may contribute to the increased expression of myostatin during food deprivation. We previously demonstrated that myostatin mRNA and protein expression are increased in the fast-twitch tibialis anterior (TA) muscle but not the slow-twitch soleus muscle with food deprivation (5). We demonstrate here that C/EBP-δ mRNA and protein levels were also significantly increased in the TA muscle with 2 days of food deprivation similar to the increase in myostatin mRNA and protein with food deprivation (5). In addition, the increases in both myostatin and C/EBP-δ mRNA levels with food deprivation were attenuated by inhibition of glucocorticoid signaling with RU486. Furthermore, the increase in myostatin mRNA with food deprivation appears to be, in part, transcriptionally driven, since food deprivation also resulted in an increase in the amount of myostatin pre-mRNA in the TA. Finally, we demonstrate that C/EBP-δ transcription is activated by glucocorticoids and, in turn, increases activity of the mouse and human myostatin promoters in C2C12 myotubes in vitro, while mutation of two different potential C/EBP binding sites has differential effects on basal, glucocorticoid receptor (GR)-, and C/EBP-dependent myostatin promoter activity. Our data support the hypothesis that the increase in myostatin expression during periods of atrophy may depend at least in part on the action of glucocorticoid-induced increases in C/EBP-δ expression.
METHODS
Experimental animals.
All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Colorado, Boulder, CO, and complied with the guidelines of the American Physiological Society on the use of laboratory animals. Male wild-type C57/BL6J mice were obtained from our breeding colony in the Department of Integrative Physiology at the University of Colorado, Boulder.
For the food-deprivation studies, 3-mo-old male C57/BL6J mice (n = 7 each) either had ad libitum access to food (Fed), had the food removed from the wire tops of their cages for 48 h [food deprived (FD)], or had the food removed from the wire tops of their cages for 48 h followed by ad libitum access to food for an additional 48 h (FD + Refed) as described (5). All animals had ad libitum access to water throughout the entire experimental period. At the end of the treatment periods, mice were killed and TA muscle was isolated, weighed, frozen in liquid nitrogen, and stored at −80°C until use. We chose to focus on the TA muscle because we previously demonstrated that mass and protein content were decreased and myostatin expression was significantly increased in the TA muscle but not the slow-twitch soleus with food deprivation (5).
For the food deprivation study with RU486 injection, male C57/BL6J mice were divided into four groups (n = 4 each): 1) mice fed ad libitum and injected daily with polyethylene glycol 400 vehicle; 2) mice fed ad libitum and injected daily with the progesterone/glucocorticoid inhibitor RU486; 3) mice food deprived for 2 days and injected with PEG400; 4) mice food deprived for 2 days and injected with RU486. PEG400 vehicle alone or vehicle + RU486 were injected subcutaneously daily for the 2 days of the experiment. RU486 was used at a concentration of 20 mg/kg body wt in 100 μl of PEG400; this concentration has been previously used to inhibit atrophic gene expression in skeletal muscle in response to dexamethasone (DEX) injection (28). At the end of the 2-day food-deprivation period, mice were killed and the TA muscle was isolated, weighed, frozen, and stored as described above.
RT-PCR for pre-mRNA.
Levels of myostatin pre-mRNA were evaluated using the technique developed by Elferink and Reiners (11) and adapted for skeletal muscle by Huey et al. (16). Total RNA was isolated from TA muscle with Trizol reagent (Invitrogen) using standard techniques (1–4). Then 30 μg of total RNA was incubated with 5 units RQ1 RNase-free DNase (Promega) for 30 min at 37°C to eliminate contaminating DNA. RNA was then reextracted with 150 μl RNase-free H2O, 750 μl Trizol, and 250 μl chloroform. RNA was pelleted by the addition of 800 μl of 100% isopropanol and microfuged at maximum speed for 10 min. The pellet was washed twice with 75% ethanol, air dried for 15 min, frozen overnight at −40°C, and then resuspended in 20 μl of RNAse-free H2O. RNA concentration was determined spectrophotometrically at A260 and brought to a concentration of 50 ng/μl. Approximately 200 ng of RNA was then used for each reaction in a 25-μl reaction volume. RT-PCR was carried out using the One-Step RT-PCR kit from Qiagen according to the manufacturer's instructions. The primer sequences consisted of an exonic forward primer (5′-TAGGGCCATGAAAGGAAAAATGAAGTCTA-3′) and an intronic reverse primer (5′-TCTCCGGGACCTCTTGGGTGTG-3′); as a control, primers for GAPDH were used (forward: 5′-ATCACCATCTTCCAGGAGCGA-3′; reverse: 5′-AGTCTTCTGGGTGGCAGTGAT-3′). The reverse transcription reaction was carried out using the 5′ primer for 30 min at 50°C; after this, the 3′ primer was added and was followed by the initial PCR activation step of 95°C for 25 min, 36 cycles of denaturation (1 min, 94°C), annealing (45 s, 56°C), and extension (45 s, 72°C) was followed by 10 min of final extension at 72°C. Then 5 μl of 6× loading buffer was added to each sample, and the entire reaction was run on a 1.5% agarose gel containing 10 μg/ml ethidium bromide and visualized and photographed using a gel documentation system. Gel photos were scanned, and the bands for myostatin pre-mRNA and GAPDH were quantified by densitometry using National Institutes of Health Image software.
Quantitative real-time RT-PCR.
RNA was isolated from skeletal muscle samples as described above. The RT reaction was carried out using 0.5 μg of RNA using the cDNA Archive kit (Applied Biosystems) according to the manufacturer's protocol. Primer and probe sets for myostatin, C/EBP-δ, and β-actin were obtained from Applied Biosystems. All real-time PCR procedures were run in triplicate to correct for variances in loading. In addition, a standard curve ranging from 10- to 0.001-μg dilutions of mouse TA muscle cDNA was run in duplicate for every assay to produce a standard curve for quantification. All values are expressed as the mean of the triplicate measure for the experimental (myostatin) divided by the mean of the triplicate measure of β-actin for each sample.
Western blot analysis.
Western blot analysis was carried out on TA muscle homogenates from Fed and FD mice. Briefly, muscles were homogenized in RIPA buffer (Pierce) containing protease inhibitors using a handheld Tissuemiser homogenizer (Fisher Scientific). Following centrifugation at 1,000 g for 10 min, the supernatant was collected and protein concentration was determined using the Bradford method. Approximately 50 μg of total protein was loaded per lane on a 12% polyacrylamide gel and electrophoresed for ∼2 h at 150 V. Proteins were then transferred to polyvinylidene difluoride membrane by electrophoresis, and membranes were dried and then rewetted with 100% methanol and blocked in Tris-buffered saline (TBS) containing 0.1% Tween-20 and 5% BSA. Blots were incubated in rabbit anti-mouse C/EBP-δ antibody (Cell Signaling Technology) at 1/1,000 dilution in blocking solution for 1 h at room temperature. After several rinses in TBS, blots were incubated in a goat anti-rabbit peroxidase conjugated secondary at 1/1,000 dilution for 1 h at room temperature. Blots were rinsed several times in TBS and then visualized using the SuperSignal West Femto visualization kit (Thermo Scientific) and scanned on a computer densitometric imaging and analysis system. After being scanned, blots were stripped in Restore Plus stripping solution (Pierce) and reprobed with an antibody to β-actin (Abcam), restained as described above, and scanned.
Cloning and mutagenesis.
The mouse and human myostatin upstream promoter regions were cloned from mouse and human genomic DNA, respectively, using PCR and cloned into the MluI and XhoI sites of pGL3 basic as described previously (1, 4). Mutagenesis to alter the CCAAT and C/EBP binding element (CBE) sites within the mouse myostatin promoter region were carried out using standard techniques. For the CCAAT site, inverse PCR was used to substitute an AvrII (underlined) site in the CCAAT core motif using the primers 5′-TATTGGCCTAGGCATAGATCCTGACGACACTTG-3′ and 5′-TAGTGGCCTAGGTCCTGCTCCACAATGAATCTC-3′. After ligation and transformation, positive colonies were screened by cutting with AvrII. For the CBE site (underlined), the primer 5′-CGGTACATGCACTAATATTTCACTTGGCGtTcACTCAAAAGCAAAAAGAAG-3′ (altered nucleotides are in lower case), and its complement were used to amplify plasmid DNA using PCR, and the resulting PCR products were subjected to DpnI digestion. DpnI-resistant colonies were then selected and screened by sequencing at the University of Colorado Sequencing Facility, Boulder, CO.
The C/EBP-α expression construct was kindly provided by Dr. Alan Friedman of Johns Hopkins University, Baltimore, MD. The C/EBP-β expression construct, originally created by Dr. Shizuo Akira, Osaka University, Osaka, Japan, was kindly provided by Dr. Dov Zipori of the Weizmann Institute of Science, Rehovot, Israel. The C/EBP-δ expression construct was kindly provided by Dr. Peter Rotwein of the Oregon Health Sciences University, Portland, OR. The GR expression construct was kindly provided by Dr. Stoney Symons of the National Institutes of Health. The 2.1-kb mouse C/EBP-δ promoter driving luciferase expression in pGL2 was kindly provided by Dr. Jim Dewille of Ohio State University, Columbus, OH.
Cell culture and transfection.
C2C12 myoblasts were plated on 0.75% gelatin-coated six-well plates in proliferation medium consisting of DMEM supplemented with 20% FBS and 1% penicillin/streptomycin (pen/strep). FBS that had been charcoal/dextran treated to remove endogenous glucocorticoids (Hyclone) was used for all studies. Cells were split after reaching 90% confluence onto four to six gelatin-coated 24-well plates and were transfected with Lipofectamine 2000 as previously described (1, 4). Briefly, for each well, 1.5 μl of Lipofectamine 2000 and 1.0 μg of DNA was mixed in 100 μl of FBS-free and pen/strep-free DMEM and allowed to complex for 30 min. The transfection mix was added to wells containing proliferation medium and allowed to remain on cells for 1–2 days until they reached confluence, at which time the medium was removed and replaced with differentiation medium consisting of DMEM plus 1% horse serum for 2 days to induce differentiation into myotubes, at which time > 90% of cells had differentiated into myotubes. Cells were harvested by washing in PBS and then adding 100 μl of passive lysis buffer (Promega) to each well. Luciferase activity was determined for all cell culture experiments using a luminometer and firefly luciferase reagent (Promega) as previously described (1, 4).
For the DEX-treatment studies, after 2 days of differentiation, myotubes were incubated in charcoal dextran-scrubbed FBS to remove endogenous glucocorticoids for 6 h, and they were then treated with either ethanol or DMSO vehicle or with 1 μm DEX for 1, 6, or 24 h. At the end of the treatment period, myotubes were washed once with PBS and then lysed in passive lysis buffer. Luciferase activity was determined for all cell culture experiments as described above.
Statistical analysis.
All in vivo studies represent a group of n = 4–7 animals per condition, and data from these studies are reported as means ± SE. All in vitro studies represent two to three independent experiments consisting of 4–8 wells for each condition per experiment, and data from these studies are reported as means ± SE. Statistically significant differences between Fed, FD, and FD+Refeed groups, and RU486-treated body mass, tissue mass, or mRNA expression were determined by ANOVA and Fisher's post hoc test, with an alpha level of 0.05 taken as significant. Statistically significant differences in myostatin or C/EBP-δ promoter activity between the green fluorescent protein (GFP) vehicle, GFP DEX, GR vehicle, and GR DEX and between GFP and C/EBP-α, -β, and -δ transfected myotubes were determined by ANOVA and Fisher's post hoc test, with an alpha level of 0.05 taken as significant. Differences between wild-type and CCAAT and CBE-mutated myostatin promoter activity were determined by ANOVA and Fisher's post hoc test, with an alpha level of 0.05 taken as significant.
RESULTS
C/EBP-δ levels increase in the TA muscle with food deprivation.
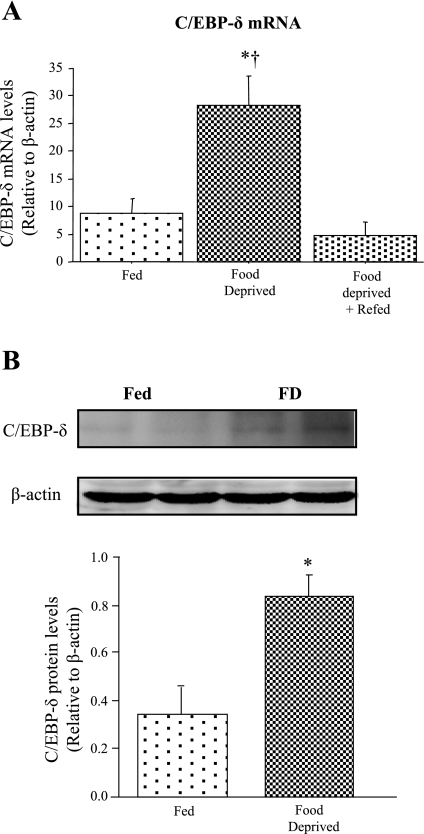
We recently demonstrated that 2 days of food deprivation significantly decreased TA muscle mass and protein content by ∼20–25% and significantly increased myostatin mRNA levels approximately threefold and myostatin protein levels by approximately twofold in the mouse (5). We therefore examined here the effects of 2 days of food deprivation on C/EBP-δ mRNA and protein expression. Two days of food deprivation significantly increased C/EBP-δ mRNA levels approximately threefold relative to the Fed control and 2 days of refeeding returned these values to baseline in the mouse TA muscle (Fig. 1A). In addition, as shown in Fig. 1B, we observed an increase in the intensity of a band migrating at ∼29 kDa, which is the reported size of C/EBP-δ, in TA muscle homogenates from FD mice (Fig. 1, B, top). Levels of β-actin protein did not appear to change with food deprivation (Fig. 1, B, bottom). Quantification of the C/EBP-δ band relative to β-actin revealed a significant increase in the C/EBP-δ protein band following food deprivation (Fig. 1B).
Fig. 1.
CCAAT/enhancer binding factor-δ (C/EBP-δ) expression is increased by food deprivation. A: C/EBP-δ mRNA expression in tibialis anterior (TA) muscle from Fed, food-deprived (FD), and FD + Refed mice. Bars represent means ± SE for n = 4 animals/group *Significantly different from Fed, P < 0.05. †Significantly different from FD + Refed, P < 0.05. B: representative Western blots showing 2 homogenate samples each from Fed TA muscle and FD TA muscle using antibodies to C/EBP-δ and β-actin. All samples for both Fed and FD mice were run on the same gel so as to avoid problems with quantification across gels. Bar graph shows the difference in C/EBP-δ protein expression between Fed and FD TA muscle. Bars represent means ± SE for 3 animals/group.*Significantly different from Fed, P < 0.05.
Myostatin pre-mRNA levels increase in response to food deprivation.
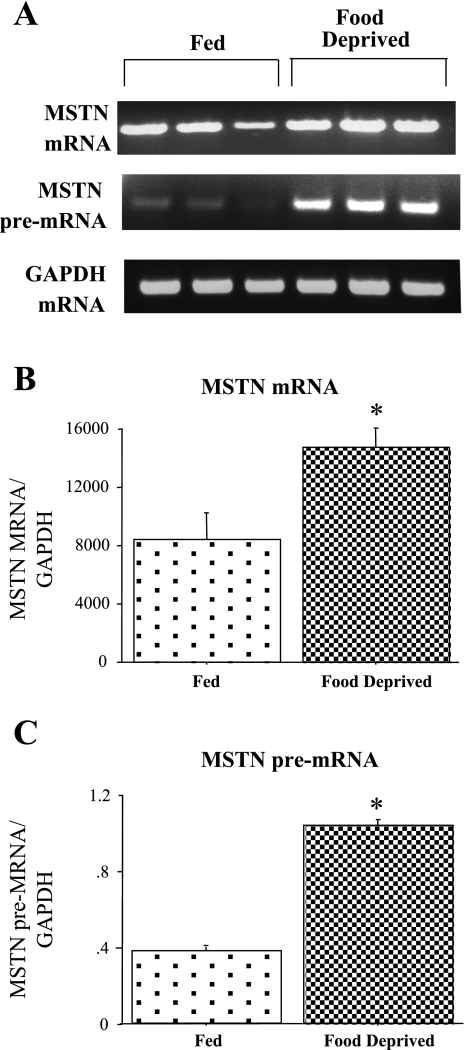
Similar to our previous results (5), myostatin mRNA levels significantly increased with 2 days of food deprivation (Fig. 2, A and B). Similarly, 2 days of food deprivation significantly increased levels of myostatin pre-mRNA in the TA, while GAPDH mRNA levels were unaffected (Fig. 2, A and C). The two- to threefold increase in myostatin pre-mRNA is identical in magnitude to the two- to threefold increase in mature myostatin mRNA reported in the present study and from these same samples using QRT-PCR in another study (5). Thus the increase in myostatin mRNA with food deprivation appears to be at least partly due to increased transcription and/or processing of this gene product.
Fig. 2.
Myostatin (MSTN) pre-mRNA in Fed and FD TA muscle. A: gel shows MSTN pre-mRNA and GAPDH mRNA in TA muscle from Fed and FD mice. Top, gel results from RT-PCR using a exonic primer to amplify MSTN mature mRNA for 3 Fed and 3 FD TA muscle samples; middle, gel results from RT-PCR intronic-exonic primer pair for MSTN to amplify MSTN pre-mRNA for 3 Fed and 3 FD samples; and bottom, exonic-exonic GAPDH primers to amplify processed GAPDH as a control for RNA loading for 3 Fed and 3 FD samples. B: bar graph shows quantification of densitometrically scanned results for MSTN mRNA. C: bar graph shows quantification of densitometrically scanned results MSTN pre-mRNA. In both cases, the values were normalized to GAPDH mRNA. Bars in B and C represent means ± SE for 3 animals/group. *Statistically significantly different from Fed, P < 0.05.
Glucocorticoid inhibition attenuates the increase in myostatin and C/EBP-δ mRNA with food deprivation.
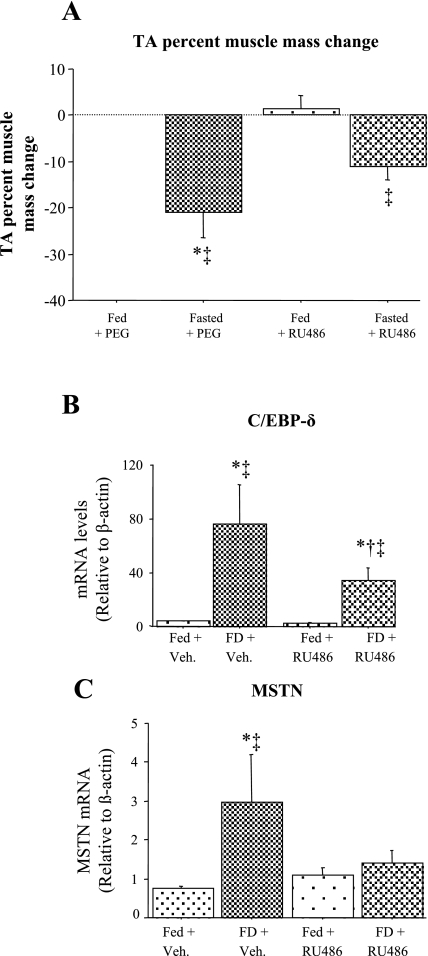
One signal that is believed to regulate the muscle atrophic response to food deprivation is increased systemic glucocorticoid levels (38). We therefore investigated the role of glucocorticoids on expression of C/EBP-δ and myostatin in the TA muscle in response to food deprivation. RU486 treatment resulted in a significant attenuation of the atrophy response to food deprivation, suggesting that it inhibited glucocorticoid activity (Fig. 3A). RU486 treatment of Fed mice had no significant effect on basal C/EBP-δ and myostatin mRNA levels (Fig. 3, B and C). C/EBP-δ mRNA levels were again significantly increased with food deprivation in the vehicle-injected mice, and RU486 injection significantly attenuated the increase in C/EBP-δ mRNA levels in response to food deprivation such that they were significantly different from the FD + vehicle treated group (Fig. 3B). Similarly, food deprivation was again associated with a significant increase in myostatin mRNA levels in vehicle-injected mice (Fig. 3C), but injection with RU486 almost completely abolished the food deprivation-induced increase in myostatin mRNA levels, such that they were significantly different from FD + vehicle but not significantly different from the Fed + RU486 mice (Fig. 3C).
Fig. 3.
Effects of glucocorticoid inhibition on muscle mass (A) C/EBP-δ (B) and MSTN (C) mRNA levels in TA muscle. A: effects of RU486 treatment with and without food deprivation on TA muscle mass. RU486 significantly attenuated the decrease in TA muscle mass with food deprivation. B: effects of RU486 treatment with and without food deprivation on C/EBP-δ mRNA levels in the TA muscle. RU486 treatment had no effect on C/EBP-δ mRNA levels in the Fed TA muscle but significantly attenuated the increase in C/EBP-δ mRNA levels with food deprivation. C: effects of RU486 treatment with and without food deprivation on MSTN mRNA levels in the TA muscle. RU486 treatment had no effect on MSTN mRNA levels in the Fed TA muscle but abolished the increase in MSTN mRNA levels with food deprivation. Bars represent means ± SE for 4 animals/group. *Significantly different from Fed vehicle (Veh)-injected mice, P < 0.05. †Significantly different from FD vehicle injected, P < 0.05. ‡Significantly different from Fed RU486-injected, P < 0.05.
Glucocorticoids acutely activate the C/EBP-δ but not the myostatin promoter in vitro.
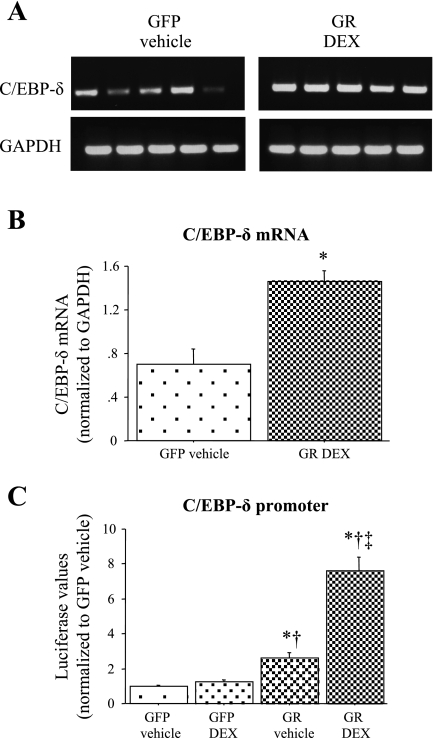
To begin to resolve the relationship between glucocorticoids and C/EBP-δ and myostatin expression, we switched to an in vitro approach. Previous work has established that glucocorticoids increase skeletal muscle C/EBP-δ mRNA, protein levels, and DNA binding activity in L6 myotubes (39). To confirm these results in C2C12 myotubes, we quantified C/EBP-δ mRNA levels and promoter activity from untreated and GR- and DEX-treated cells. Twenty four hours of GR + DEX treatment significantly increased C/EBP-δ mRNA levels approximately two- to threefold (Fig. 4A and B). Similarly, GR transfection without and with DEX cotreatment significantly increased C/EBP-δ promoter activity, with GR + DEX having the greatest effect (Fig. 4C).
Fig. 4.
Effects of glucocorticoids on C/EBP-δ mRNA levels and promoter activity. A: gel showing the effects of 24 h of vehicle + green fluorescent protein (GFP (left) and glucocorticoid receptor (GR) + 1 μM dexamethasone (DEX) treatment (right) on C/EBP-δ (top) and GAPDH (bottom) mRNA levels. B: quantification of C/EBP-δ mRNA levels. Twenty four hours of GR + DEX treatment significantly increased C/EBP-δ mRNA levels relative to GFP + vehicle controls. Bars represent means ± SE for 5 independent transfections. *Significantly different from GFP vehicle-treated control, P < 0.05. C: mouse C/EBP-δ promoter activity after 24 h of treatment with GFP cotransfection and vehicle alone (GFP vehicle), GFP cotransfection and 1 μM DEX, GR cotransfection and vehicle alone (GR vehicle), or glucocorticoid receptor cotransfection with 1 μM DEX (GR DEX). GR cotransfection with and without DEX cotreatment significantly increased activity of the mouse C/EBP-δ promoter at 24 h effect on mouse MSTN promoter activity with or without DEX cotreatment at either time point. Bars represent means ± SE for 3 independent transfections with 6 wells/transfection. *Significantly different from GFP vehicle-treated control, P < 0.05. †Significantly different from GFP DEX treated, P < 0.05. ‡Significantly different from GR vehicle treated, P < 0.05.
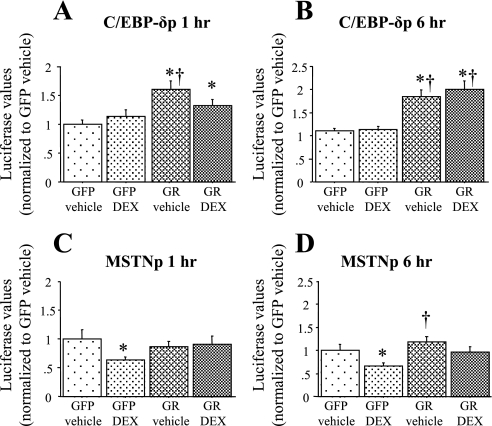
We therefore wished to compare the effects of acute glucocorticoid stimulation on the C/EBP-δ and the myostatin promoters so as to establish the temporal pattern of expression of these two genes. We chose 1 and 6 h of treatment because these would be less likely to include secondary effects of glucocorticoids on other downstream genes that might, in turn, affect expression of either C/EBP-δ or myostatin. As can be seen in Fig. 5, A and B, cotransfection of C2C12 myotubes with a GR expression construct alone or with cotreatment with DEX significantly increased activity of the mouse C/EBP-δ promoter in C2C12 myotubes, and this effect was obvious as early as 1 h posttreatment (Fig. 5, A and B). In contrast, GR cotransfection with or without DEX cotreatment did not significantly increase activity of the myostatin promoter at either of these early time points (Fig. 5, C and D). By 24 h of treatment, the C/EBP-δ promoter was induced 2.6- and 7.6-fold in the GR + vehicle (Fig. 4C) and GR + DEX groups, respectively, while the myostatin promoter was modestly but significantly induced 1.5-fold by GR + DEX at this later time point (Fig. 6C). Thus, glucocorticoids appear to activate transcription of the C/EBP-δ promoter at earlier time points than the myostatin promoter, which is temporally consistent with a glucocorticoid-mediated increase in C/EBP-δ expression contributing to the transcriptional regulation of the myostatin gene.
Fig. 5.
Effects of glucocorticoids on activity of the mouse C/EBP-δ and MSTN promoters in C2C12 myotubes in vitro. Mouse C/EBP-δ promoter activity after 1 h (A) or 6 h (B) of treatment. GR cotransfection with and without DEX cotreatment significantly increased activity of the mouse C/EBP-δ promoter at both 1 and 6 h. Mouse MSTN promoter activity after 1 h (C) or 6 h (D) of treatment with GFP vehicle, GFP DEX, GR vehicle, or GR DEX (see Fig. 4 legend for explanation of groups). GR cotransfection had no significant effect on mouse MSTN promoter activity with or without DEX cotreatment at either time point. Bars represent means ± SE for 2–3 independent transfections with 6 wells/transfection. *Significantly different from GFP vehicle-treated control, P < 0.05. †Significantly different from GFP DEX treated, P < 0.05.
Fig. 6.
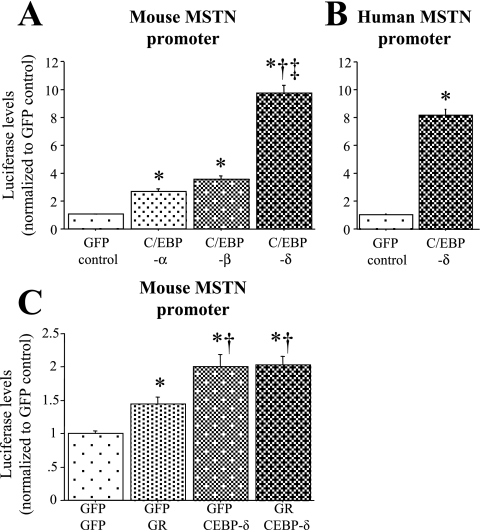
Effects of C/EBP cotransfection on MSTN promoter activity in C2C12 myutubes. A: effects of C/EBP-α, -β, and -δ cotransfection on activity of the ∼1,200 bp mouse MSTN promoter in C2C12 myutubes . All C/EBP expression constructs significantly increased mouse MSTN promoter activity relative to cotransfection with the GFP control, but C/EBP-δ had the greatest effect. B: effects of C/EBP-δ cotransfection on activity of the ∼1,000 bp human MSTN promoter in C2C12 myutubes. C/EBP-δ cotransfection also significantly increased activity of the human MSTN promoter. Bars for A and B represent means ± SE for n = 3 independent transfections with 4–6 wells/transfection. *Significantly different from CMV-GFP cotransfected control, P < 0.05. †Significantly different from C/EBP-α cotransfected, P < 0.05. ‡Significantly different from C/EBP-β cotransfected, P < 0.05. C: effects of GR and C/EBP-δ cotransfection on mouse MSTN promoter activity in C2C12 myutubes. Myotubes were transfected with the mouse MSTN promoter and either GFP alone (GFP GFP), GR (GFP GR), C/EBP-δ (GFP C/EBP-δ), or GR and C/EBP-δ (GR C/EBP-δ). Bars represent means ± SE for n = 3 independent transfections with 4–6 wells/transfection. *Significantly different from GFP GFP cotransfected control, P < 0.05. †Significantly different from GFP GR transfected, P < 0.05.
C/EBP-δ activates the mouse myostatin promoter in C2C12 myotubes in vitro.
We next tested whether myostatin transcription could be activated by C/EBP family members in C2C12 myotubes in vitro in the absence of increased glucocorticoid signaling. Cotransfection with C/EBP-α, -β, or -δ expression constructs significantly increased activity of the ∼1,200 bp mouse myostatin promoter construct relative to cotransfection with a cytomegalovirus (CMV)-GFP control (Fig. 6A). Cotransfection with the C/EBP-δ expression construct had the greatest effect on mouse myostatin promoter activity, increasing it a significantly greater amount (nearly 10-fold) than cotransfection with either the C/EBP-α or -β expression constructs (Fig. 6A). A similarly dramatic effect was observed when a reporter plasmid containing ∼1,000 bp of the human myostatin promoter was cotransfected with the C/EBP-δ expression construct (Fig. 6B). Thus increased C/EBP-δ expression alone is sufficient to increase myostatin transcription in C2C12 myotubes in vitro.
To further explore the relationship among glucocorticoid signaling, C/EBP-δ, and myostatin transcription, we cotransfected the myostatin promoter with the GR expression construct, the C/EBP-δ expression construct, or both. Cotransfection with the GR expression construct produced a modest but significant increase in myostatin promoter activity compared with cotransfection with a GFP control plasmid (Fig. 6C). Cotransfection with the C/EBP-δ construct resulted in an approximately twofold increase in myostatin promoter activity in this assay, which used less cotransfected C/EBP-δ expression construct DNA than the assays above, and cotransfection with both the GR and C/EBP-δ expression constructs together did not further increase this beyond the increase observed for C/EBP-δ alone (Fig. 6C). Thus overexpression of C/EBP-δ alone is sufficient to activate the myostatin promoter and GR cotransfection does not further increase activity of the myostatin promoter, suggesting that these two events are serial events in the same pathway.
Identification of potential C/EBP binding sites in the myostatin promoter region.
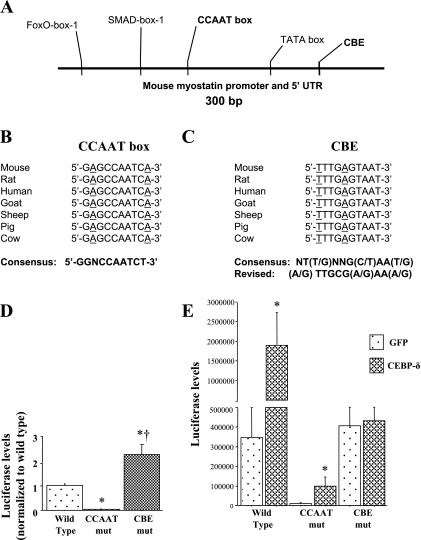
Bioinformatic analysis of the myostatin promoter region revealed two potential C/EBP binding sites, both of which show high degrees of conservation across species (Fig. 7, A–C). First, there is a near-match to the CCAAT box consensus (GGNCCAATCT) (27) found ∼100 bp upstream of the TATA box in all mammalian species examined (Fig. 7A); this sequence differs from the consensus by just two nucleotide substitutions in the flanking arms of this element, while the core CCAAT sequence is 100% identical to the consensus for this site (Fig. 7B). In addition, a sequence exactly matching the consensus for C/EBP binding [NT(T/G)NNG(C/T)AA(T/G)] (32), which we have named CBE to distinguish it from the CCAAT box, is found on the (−) strand ∼50 bp downstream of the TATA box but upstream of the transcription start site (Fig. 7A). This sequence differs at two nucleotides from that of the revised consensus [(A/G)TTGCG(A/G)AA(A/G)] for all C/EBP family members reported by Osada et al. (28); however, these authors reported that C/EBP-δ had a lower overall affinity that relaxed its requirements for binding to this sequence and, in particular, showed greater binding promiscuity than C/EBP-α or -β with respect to changes to these two nucleotides (28). The myostatin CBE sequence is 100% conserved across the species examined (Fig. 7C).
Fig. 7.
Sequence analysis of the MSTN promoter showing the 2 potential C/EBP binding sites. A: schematic of ∼300 bp of the upstream promoter region of the mouse MSTN gene. The TATA box, SMAD-box-1, and FoxO-box-1 previously identified by us (1) are shown along with the CCAAT box and the CBE (both in bold) identified by sequence analysis in the present study. Sequence alignment of the CCAAT box (B) and CBE (C) showing the conservation of these sites. The consensus CCAAT box sequence reported by Nussinov (27) is shown at the bottom in bold. On the right, the consensus reported by Ryden and Beemon (32) and the revised consensus reported by Osada et al. (28) are shown. Underlined nucleotides for the CCAAT box are ones that do not match the consensus sequence reported by Nussinov (27); underlined nucleotides for the CBE are ones that do not match the consensus sequence reported by Osada et al. (28). D: effects of mutation of the CCAAT box or CBE on basal MSTN promoter activity. Bars represent means ± SE for 3 independent transfections with 4–6 wells/transfection. *Significantly different from wild-type control, P < 0.05. †Significantly different from CCAAT box mutated, P < 0.05. E: effects of mutation of the CCAAT box or CBE on C/EBP-δ-dependent MSTN promoter activity. Bars represent means ± SE for 3 independent transfections with 4–6 wells/transfection. *Significantly different from GFP transfected, P < 0.05. †Significantly different from wild-type CMV-GFP cotransfected, P < 0.05. ‡Significantly different from CCAAT mutation (mut) construct C/EBP-β cotransfected, P < 0.05.
We therefore used mutagenesis to alter these elements within the context of the 1,200 bp mouse myostatin promoter to determine their effect on basal and C/EBP-δ-dependent promoter activity. Interestingly, these two elements showed complementary effects on basal and C/EBP-δ-dependent promoter activity. Mutation of the CCAAT box almost completely abolished basal activity of the mouse myostatin promoter relative to the wild-type construct, while mutation of the CBE surprisingly increased basal myostatin promoter activity approximately twofold relative to wild type (Fig. 7D). In contrast, though it attenuated the absolute value compared with that of the wild-type construct, mutation of the CCAAT box did not alter responsiveness to C/EBP-δ cotransfection, as activity of the myostatin CCAAT mutation construct was still induced severalfold by C/EBP-δ cotransfection similar to the wild-type myostatin construct (Fig. 7E). However, mutation of the CBE almost completely abolished responsiveness of the myostatin promoter construct to C/EBP-δ cotransfection (Fig. 7E) and to GR cotransfection (data not shown).
DISCUSSION
Food deprivation is associated with dramatic changes in skeletal muscle mass and gene expression, and we previously demonstrated that myostatin expression is increased with food deprivation and that the atrophy associated with 2 days of food deprivation is at least partially myostatin-dependent (5). We show here that both C/EBP-δ mRNA and protein levels are similarly elevated by food deprivation in the TA. Taken with our previously published data on C/EBP-δ mRNA increases following 12 days of spaceflight (3), these data suggest that C/EBP-δ expression is increased in both loading-dependent and loading-independent modes of muscle atrophy.
Pre-mRNA levels have been used as a readout of transcription (11); this approach is particularly useful to explore transcriptional mechanisms in skeletal muscle (16), from which nuclear isolation for nuclear run-on assays can be quite difficult. Despite a wealth of published data on changes in myostatin mature mRNA levels in a variety of species and in response to a variety of physiological manipulations, and despite considerable published analysis of the myostatin promoter, particularly in vitro (1, 2, 4, 21), we are unaware of any other published data implicating transcriptional mechanisms in the regulation of myostatin expression through examination of pre-mRNA levels, through inhibition of transcription with actinomycin D, or through nuclear run-on assays. The present data thus provide evidence that the increase in myostatin mRNA with food deprivation may be transcriptionally regulated, although this does not preclude contributions from posttranscriptional mechanisms to myostatin expression in this or other models of physiological adaptation.
An alternative explanation for the increase in myostatin pre-mRNA is that the rate of splicing/processing of myostatin is altered by food deprivation. However, we feel that this is unlikely for two reasons. First, while the pattern of splicing can obviously be regulated (i.e., alternative splicing), the rate of splicing has not been well established as a regulatory step between transcription and translation. Second, in their paper outlining the use of pre-mRNA as a readout of transcriptional activity, Elferink and Reiners (11) state that if the magnitude of the change between pre-mRNA and mature mRNA is similar and/or identical, it strongly suggests that the processing rate was not affected, and in the present study both pre- and mature myostatin mRNA were increased approximately two to threefold by food deprivation, suggesting that processing was not altered. Thus, while it remains possible that a decrease in myostatin mRNA processing may have contributed to some or all of the increase in myostatin pre-mRNA observed in the present study, we feel that the more likely explanation for this increase is an increase in transcription of the myostatin gene in response to food deprivation.
We then chose to examine the role of glucocorticoids in food deprivation-mediated increases in myostatin and C/EBP-δ expression for two reasons. First, increased glucocorticoid secretion is a hallmark of the food-deprivation state. Second, expression of myostatin (21, 22) and C/EBP-δ (6, 29, 39) is known to be regulated by glucocorticoids. Treatment with the glucocorticoid inhibitor RU486, which binds to and inhibits glucocorticoids (29), significantly attenuated the food deprivation-induced increase in both myostatin and C/EBP-δ mRNA levels (Fig. 3). Our results are thus consistent with the interpretation that expression of both myostatin and C/EBP-δ is glucocorticoid dependent in the food-deprived TA.
To better understand the relationship between glucocorticoids, C/EBP-δ, and myostatin expression, we therefore moved to an in vitro model to explore how glucocorticoids might regulate expression of C/EBP-δ and/or myostatin. We first examined whether glucocorticoid signaling affects transcription of C/EBP-δ, myostatin, or both genes. Our hypothesis was that glucocorticoids actually influence myostatin transcription indirectly by first increasing expression of C/EBP-δ, and our data support this hypothesis. Activity of the C/EBP-δ promoter was induced much earlier in response to glucocorticoid signaling than the myostatin promoter, increasing significantly with as little as 1 h of DEX treatment. The finding is consistent with the rapid induction of C/EBP-δ transcription in response to DEX in rat intestinal epithelial cells, in which DEX treatment increased C/EBP-δ transcription as assayed by nuclear run-on with as little as 30 min of treatment (6). Given its great responsiveness at this very early time point, the most likely explanation is that C/EBP-δ transcription is directly activated by GR, although this still needs to be determined empirically through binding assays and mutagenesis of the C/EBP-δ promoter, and that C/EBP-δ, in turn, contributes to the transcriptional activation of the myostatin promoter by glucocorticoids. The fact that cotransfection with both GR and C/EBP-δ had no additional effect on myostatin promoter activity than C/EBP-δ cotransfection alone is also consistent with the interpretation that glucocorticoid signaling acts via increasing C/EBP-δ expression and that overexpressing C/EBP-δ obviates this effect.
Given the evidence that C/EBP-δ expression appears to be activated by glucocorticoids, we next examined whether C/EBP-δ cotransfection alone was sufficient to increase myostatin promoter activity independent of glucocorticoid signaling. Cotransfection with C/EBP-α, -β, or -δ significantly increased activity of the mouse myostatin promoter, with C/EBP-δ showing the greatest effect (Fig. 6A). While the most conservative interpretation of this data is that members of the C/EBP family bind to C/EBP recognition sequences in the myostatin promoter region and activate its transcription, it is also possible that C/EBPs increase myostatin promoter activity through an indirect effect, either by partnering with another transcription factor and binding to its site or by activating expression of another transcription factor that then binds to its recognition sequence in the myostatin promoter to activate myostatin transcription.
To begin to distinguish between these possibilities, we mutated two potential C/EBP binding sites within the myostatin promoter: a CCAAT box and a sequence matching another C/EBP recognition sequence that we refer to as a CBE, and determined the effects of these mutations on both basal and C/EBP-dependent myostatin promoter activity. Mutation of the CCAAT box almost completely abolished basal myostatin promoter activity, while mutation of the CBE abolished C/EBP-δ responsiveness. The decrease in basal myostatin promoter activity due to the CCAAT mutation may reflect the fact that CCAAT box elements along with TATA boxes typically act as staging areas for assemblage of the transcriptional machinery (12), while the decrease in C/EBP-δ responsiveness of the CBE mutation strongly suggests that C/EBP-δ binds to this element to produce its effect on myostatin transcription, although this still needs to be confirmed with binding assays. The reason for the potentiating effects of the two mutants on basal and C/EBP-δ-induced myostatin promoter activity are not clear at present but may reflect the disruption of binding of other transcription factors to these sites by these mutations.
We also demonstrate that mutation of the CBE element also abolishes all responsiveness to GR signaling. The fact that GR signaling had such a modest effect on wild-type myostatin promoter activity, increasing it by ∼50% with 24 h of DEX treatment, was somewhat surprising given that others have demonstrated that both the sheep (10) and human (21) myostatin promoters are glucocorticoid responsive. However, in both of these studies myostatin promoter responsiveness was also rather small, increasing 20–60% in the work of Du et al. (10) and approximately twofold in the work by Ma et al. (21), which is within the range of the results of the present study. Also, both Du et al. and Ma et al. used incubation durations much longer (40 h for Du et al., 96 h for Ma et al.) than the 24-h incubation used in the present study. Thus the increase in myostatin promoter activity in response to 24 h of DEX treatment observed in the present study is comparable with these previous findings. Moreover, our data is consistent with the hypothesis that mutation of the CBE abolishes transcriptional responsiveness of the mouse myostatin promoter to GR as well as C/EBP-δ, which strongly suggests that glucocorticoids signal through activation of C/EBP-δ expression and binding of C/EBP-δ to the CBE element and not through a canonical glucocorticoid response element as has been suggested (10). However, future studies will need to definitively identify the protein(s) binding to the CBE for this hypothesis to be fully confirmed.
The results of the present study complement several previous studies on glucocorticoids, C/EBPs, and myostatin, and provide a more complete picture of the relationship between these three factors. For example, previous work has established that glucocorticoids increase expression and DNA binding ability of C/EBP family members and C/EBP-δ in particular (39), and sepsis induces an increase in C/EBP-δ expression and DNA binding activity that is glucocorticoid dependent as it can be attenuated by treatment with RU486 (29). In the present study, we show that C/EBP-δ mRNA and protein levels were also increased in response to food deprivation, and that the increase in C/EBP-δ mRNA in response to food deprivation is also glucocorticoid dependent. Moreover, we demonstrate that the effects of glucocorticoids on C/EBP-δ expression in muscle cells in vitro appear to be transcriptional, as cotransfection with a GR expression construct significantly increased mouse C/EBP-δ promoter activity in C2C12 myotubes. The results of the present study therefore confirm and extend previous findings on the relationship between glucocorticoid signaling and C/EBP-δ expression in skeletal muscle. Similarly, previous work has established that glucocorticoids also regulate myostatin expression both in vitro and in vivo (21, 22) and that transcriptional mechanisms may contribute to this effect since myostatin promoter activity is also glucocorticoid responsive (21). Here we provide several lines of evidence to suggest that glucocorticoids may act at least in part through C/EBP-δ to achieve this effect, which ties in with the data on glucocorticoid regulation of C/EBP-δ expression above. Finally, inactivation of the myostatin gene abolishes muscle atrophy in response to glucocorticoids in vivo (14), a finding further supported by the mechanistic connection between glucocorticoid signaling and myostatin expression elaborated in the present study.
However, several caveats need to be mentioned. First, while our in vitro data is consistent with the hypothesis that glucocorticoids regulate myostatin transcription through increased expression of C/EBP-δ, and, as mentioned above, previous studies have shown that glucocorticoid treatment in vivo increases muscle C/EBP-δ expression and DNA binding activity (39), it is not yet certain whether the effect of glucocorticoids in vivo is a direct one or whether it is a secondary consequence of effects of glucocorticoids on some other tissue that then affect C/EBP-δ expression in muscle.
Second, while the present work posits a role for C/EBP family members in glucocorticoid-mediated myostatin expression, it does not exclude contributions from other signaling pathways in general or other glucocorticoid-mediated signaling pathways in particular. For example, expression of the FoxO1/FKHR transcription factor is increased by food deprivation (13) and glucocorticoids (33), and we have shown that FoxO1 increases myostatin mRNA levels and promoter activity (1). Moreover, insulin antagonizes the effects of glucocorticoids on atrophic gene expression through activation of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathway (20), which results in increased phosphorylation and nuclear export of FoxO1 (19, 36), and during food deprivation insulin levels decrease dramatically. It is likely that the increased expression of myostatin during food deprivation represents the collective action of these and other as-yet undescribed pathways.
Third, the present work suggests that glucocorticoids have their effect on myostatin expression primarily via increased expression (and in particular transcription) of C/EBP-δ, consistent with the work of others (29), but this does not preclude additional mechanisms, including direct interactions between the GR and C/EBP-δ proteins, which may foster transcription of the myostatin gene. We are aware of no data to suggest that the GR and C/EBP-δ interact, but an interaction between the GR and C/EBP-α has been described that is necessary for transactivation of the 11β-hydroxysteroid dehydrogenase type 1 gene in human amnion fibroblasts (41). Finally, the present work does not exclude a contribution of posttranscriptional mechanisms in regulating expression of myostatin in response to either glucoroticoids or food deprivation. A glucocorticoid-dependent increase in C/EBP-δ expression may therefore reflect just one mechanism by which glucocorticoids mediate expression of the myostatin gene.
Perspectives and Significance
Our data support a model in which food deprivation-induced atrophy is associated with an glucocorticoid-mediated increase in C/EBP-δ expression, which in turn may directly activate transcription of the myostatin gene via the CBE. The participation of C/EBP-δ in myostatin transcription and/or muscle atrophy is a novel finding of this work, as this transcription factor has not been previously established to regulate expression of atrophy-associated genes. It will be of interest to determine whether C/EBP family members target other genes regulating muscle growth. Nevertheless, C/EBP-δ represents a new circuit by which myostatin expression may be regulated during atrophic states.
GRANTS
J. M. Reed and S. F. Lindsay were supported by the University of Colorado Undergraduate Research Opportunity Program. This work was supported in part by an Innovative Seed Grant from the University of Colorado, Boulder. D. L. Allen was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases K01-AR-050505-01.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
D. L. Allen thanks the Department of Integrative Physiology for providing funds to support this research and for purchasing the quantitative real-time PCR system that made much of the data collection possible in the present study, and the following from the University of Colorado, Boulder: Dr. Douglas Seals for providing the use of his cell culture hood for the cell culture studies, and Drs. Gary McCall, Fadia Haddad, and Kenneth Baldwin for their invaluable assistance in designing the myostatin pre-mRNA primers and running the pre-mRNA RT-PCR assays.
REFERENCES
- 1.Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol 292: C188–C199, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Allen DL, Cleary AS, Speaker KJ, Lindsay SF, Uyenishi J, Reed JM, Madden MC, Mehan RS. Myostatin, activin receptor IIb, and follistatin-like-3 gene expression are altered in adipose tissue and skeletal muscle of obese mice. Am J Physiol Endocrinol Metab 294: E918–E927, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Allen DL, Bandstra ER, Harrison BC, Thorng S, Stodieck LS, Kostenuik PJ, Morony S, Lacey DL, Hammond TG, Leinwand LL, Argraves WS, Bateman TA, Barth JL. Effects of spaceflight on murine skeletal muscle gene expression. J Appl Physiol 106: 582–595, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen DL, Du M. Comparative functional analysis of the cow and mouse myostatin genes reveals novel regulatory elements in their upstream promoter regions. Comp Biochem Physiol B Biochem Mol Biol 150: 432–439, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Allen DL, Cleary AS, Lindsay SF, Loh A, Reed JM. Myostatin expression is increased by food deprivation in a muscle-specific manner and contributes to muscle atrophy during prolonged food deprivation in mice. J Appl Physiol 109: 692–701, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Boudreau F, Blais S, Asselin C. Regulation of CCAAT/enhancer binding protein isoforms by serum and glucocorticoids in the rat intestinal epithelial crypt cell line IEC-6. Exp Cell Res 222: 1–9, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Carlson CJ, Booth FW, Gordon SE. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am J Physiol Regul Integr Comp Physiol 277: R601–R606, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Cao L, Ye J, Zhu D. Upregulation of myostatin gene expression in streptozotocin-induced type 1 diabetes mice is attenuated by insulin. Biochem Biophys Res Commun 388: 112–116, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Dasarathy S, Muc S, Hisamuddin K, Edmison JM, Dodig M, McCullough AJ, Kalhan SC. Altered expression of genes regulating skeletal muscle mass in the portacaval anastomosis rat. Am J Physiol Gastrointest Liver Physiol 292: G1105–G1113, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Du R, An XR, Chen YF, Qin J. Some motifs were important for myostatin transcriptional regulation in sheep (Ovis aries). J Biochem Mol Biol 40: 547–553, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Elferink CJ, Reiners JJ., Jr Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. Biotechniques 20: 470–477, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Etienne J, Brault D, Firmin S. “Cis” and “trans” regulator elements of transcription. Ann Biol Clin (Paris) 48: 681–694, 1990 [PubMed] [Google Scholar]
- 13.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J 375: 365–371, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilson H, Schakman O, Combaret L, Lause P, Grobet L, Attaix D, Ketelslegers JM, Thissen JP. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology 148: 452–460, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74, 1997 [DOI] [PubMed] [Google Scholar]
- 16.Huey KA, Haddad F, Qin AX, Baldwin KM. Transcriptional regulation of the type I myosin heavy chain gene in denervated rat soleus. Am J Physiol Cell Physiol 284: C738–C748, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Ji M, Zhang Q, Ye J, Wang X, Yang W, Zhu D. Myostatin induces p300 degradation to silence cyclin D1 expression through the PI3K/PTEN/Akt pathway. Cell Signal 20: 1452–1458, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Lang CH, Silvis C, Nystrom G, Frost RA. Regulation of myostatin by glucocorticoids after thermal injury. FASEB J 15: 1807–1809, 2001 [DOI] [PubMed] [Google Scholar]
- 19.Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem 280: 2737–2744, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Li BG, Hasselgren PO, Fang CH. Insulin-like growth factor-I inhibits dexamethasone-induced proteolysis in cultured L6 myotubes through PI3K/Akt/GSK-3β and PI3K/Akt/mTOR-dependent mechanisms. Int J Biochem Cell Biol 37: 2207–2216, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S. Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol Endocrinol Metab 281: E1128–E1136, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Ma K, Mallidis C, Bhasin S, Mahabadi V, Artaza J, Gonzalez-Cadavid N, Arias J, Salehian B. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metab 285: E363–E371, 2003 [DOI] [PubMed] [Google Scholar]
- 23.McFarlane C, Plummer E, Thomas M, Hennebry A, Ashby M, Ling N, Smith H, Sharma M, Kambadur R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-κB-independent, FoxO1-dependent mechanism. J Cell Physiol 209: 501–514, 2006 [DOI] [PubMed] [Google Scholar]
- 24.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94: 12457–12461, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387: 83–90, 1997 [DOI] [PubMed] [Google Scholar]
- 26.Morissette MR, Cook SA, Buranasombati C, Rosenberg MA, Rosenzweig A. Myostatin inhibits IGF-I-induced myotube hypertrophy through Akt. Am J Physiol Cell Physiol 297: C1124–C1132, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nussinov R. Sequence signals in eukaryotic upstream regions. Crit Rev Biochem Mol Biol 25: 185–224, 1990 [DOI] [PubMed] [Google Scholar]
- 28.Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem 271: 3891–3896, 1996 [DOI] [PubMed] [Google Scholar]
- 29.Penner G, Gang G, Sun X, Wray C, Hasselgren PO. C/EBP DNA-binding activity is upregulated by a glucocorticoid-dependent mechanism in septic muscle. Am J Physiol Regul Integr Comp Physiol 282: R439–R444, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Plant PJ, Brooks D, Faughnan M, Bayley T, Bain J, Singer L, Correa J, Pearce D, Binnie M, Batt J. Cellular markers of muscle atrophy in chronic obstructive pulmonary disease (COPD). Am J Respir Cell Mol Biol 42: 461–471, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J 365: 561–575, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryden TA, Beemon K. Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein. Mol Cell Biol 9: 1155–1164, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Siriett V, Platt L, Salerno MS, Ling N, Kambadur R, Sharma M. Prolonged absence of myostatin reduces sarcopenia. J Cell Physiol 209: 866–873, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 4: 395–403, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol 296: C1258–C1270, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Wing SS, Goldberg AL. Glucocorticoids activate the ATP-ubiquitin-dependent proteolytic system in skeletal muscle during fasting. Am J Physiol Endocrinol Metab 264: E668–E676, 1993 [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Mammen J, Wei W, Menconi M, Evenson A, Fareed M, Petkova V, Hasselgren PO. Expression and activity of C/EBPβ and -δ are upregulated by dexamethasone in skeletal muscle. J Cell Physiol 204: 219–226, 2005 [DOI] [PubMed] [Google Scholar]
- 40.Yang W, Zhang Y, Li Y, Wu Z, Zhu D. Myostatin induces cyclin D1 degradation to cause cell cycle arrest through a phosphatidylinositol 3-kinase/AKT/GSK-3β pathway and is antagonized by insulin-like growth factor 1. J Biol Chem 282: 3799–3808, 2007 [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Guo C, Zhu P, Li W, Myatt L, Sun K. Role of glucocorticoid receptor and CCAAT/enhancer-binding proteinα in the feed-forward induction of 11β-hydroxysteroid dehydrogenase type 1 expression by cortisol in human amnion fibroblasts. J Endocrinol 195: 241–253, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Zerath E, Holy X, Andre C, Renault S. Effects of space food bar feeding on bone mass and metabolism in normal and unloaded rats. Nutr Res 22: 1309–1318, 2002 [DOI] [PubMed] [Google Scholar]