Abstract
Posttranslational glycosylphosphatidylinositol (GPI) lipid anchoring is common not only for animal and fungal but also for plant proteins. The attachment of the GPI moiety to the carboxyl-terminus after proteolytic cleavage of a C-terminal propeptide is performed by the transamidase complex. Its four known subunits also have obvious full-length orthologs in the Arabidopsis and rice (Oryza sativa) genomes; thus, the mechanism of substrate protein processing appears similar for all eukaryotes. A learning set of plant proteins (substrates for the transamidase complex) has been collected both from the literature and plant sequence databases. We find that the plant GPI lipid anchor motif differs in minor aspects from the animal signal (e.g. the plant hydrophobic tail region can contain a higher fraction of aromatic residues). We have developed the “big-Π plant” program for prediction of compatibility of query protein C-termini with the plant GPI lipid anchor motif requirements. Validation tests show that the sensitivity for transamidase targets is approximately 94%, and the rate of false positive prediction is about 0.1%. Thus, the big-Π predictor can be applied as unsupervised genome annotation and target selection tool. The program is also suited for the design of modified protein constructs to test their GPI lipid anchoring capacity. The big-Π plant predictor Web server and lists of potential plant precursor proteins in Swiss-Prot, SPTrEMBL, Arabidopsis, and rice proteomes are available at http://mendel.imp.univie.ac.at/gpi/plants/gpi_plants.html. Arabidopsis and rice protein hits have been functionally classified. Several GPI lipid-anchored arabinogalactan-related proteins have been identified in rice.
Posttranslational modification with a glycosylphophatitylinositol lipid anchor is an important alternative, widely distributed mechanism for tethering proteins to the lumenal side of the endoplasmic reticulum (ER) membrane and, after vesicular transport, to the extracellular leaflet of the plasma membrane. It has been known for a variety of animals, their viruses, and fungal organisms for about 15 years (for review, see Vai et al., 1993; Udenfriend and Kodukula, 1995a; Eisenhaber et al., 1999; Ferguson, 1999). The modification is executed by the transamidase complex located at the lumenal side of the ER membrane. A glycosylphosphatidylinositol (GPI) moiety (a composite structure including an inositol phospholipid and an oligosaccharide with, at least, one phosphoethanolamine substitution) is attached to the nascent carboxyl terminus (ω-site) of the substrate polypeptide after proteolytic cleavage of a C-terminal propeptide (Ferguson, 1999; Kinoshita and Inoue, 2000). Knowledge about a protein's GPI lipid modification is extremely valuable because this alone determines its cellular localization and limits its range of possible molecular functions.
The first indications for the possible existence of GPI lipid-anchored proteins (Stöhr et al., 1995; Morita et al., 1996; Kuntze et al., 1997; Takos et al., 1997; Sherrier et al., 1999) in plants came relatively recently (for review, see Borner et al., 2002). The first examples of unambiguously verified GPI-anchored proteins and ω-sites were NaAGP1 from Nicotiana alata and PcAGP1 from pear (Pyrus communis; Youl et al., 1998; Oxley and Bacic, 1999). As with other taxa (Eisenhaber et al., 1999), the number of plant protein examples with complete experimental verification of GPI anchoring is low and grows slowly because the required experimental procedures are laborious. Definitive experimental verification means that there are: (a) unambiguous evidence for GPI lipid anchoring and (b) exact determination of the major and possibly alternative (Yan and Ratnam, 1995; Bucht and Hjalmarsson, 1996) ω-sites (cleavage sites) in the proprotein sequence. The issue is further complicated by the less than 100% efficiency of GPI lipid anchor attachment for a number of precursor sequences (Bucht and Hjalmarsson, 1996; Coussen et al., 2001) and the fact that even close sequence homology does not guarantee GPI lipid anchoring throughout a protein family (for review of the folate receptor case, see Eisenhaber et al., 1999). The most direct approach involves protease digestion of the modified protein, the separation of the GPI-anchored peptide, and its sequencing with tandem mass spectrometry. Other methods for the physicochemical characterization of the protein and the potentially GPI-modified amino acid residue are more indirect and leave room for interpretation. These techniques include radioactive labeling with GPI lipid anchor components, site-directed mutagenesis, protein solubilization with phospholipases C or D, amino acid composition analysis, NMR, mass spectrometry fingerprinting, and the like (Killeen et al., 1988; Clayton and Mowatt, 1989; Misumi et al., 1990; Stahl et al., 1990; Moran et al., 1991; Nuoffer et al., 1991; Sugita et al., 1993; Udenfriend and Kodukula, 1995a). Additional significance is achieved by the combination of various indirect techniques.
Because large-scale experimental screening for GPI lipid anchored plant proteins with currently available methods is difficult, theoretically derived criteria for the preselection of potential candidates from their amino acid sequence are desirable. Characteristic properties of the recognition signal encoded in protein sequences that are substrates for the transamidase complex can be derived: (a) from the sequence variability of known substrates, and (b) from knowledge of the enzymes and auxiliary proteins involved in catalysis.
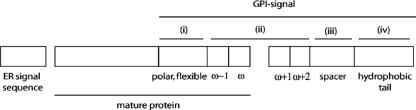
The transamidase complex consists of four components in human cells: PIG-K, a member of the C13/C14 clade of proteases that are specific for polypeptide regions with small residues; PIG-T, the auxiliary protein most tightly bound to PIG-K and, apparently, gating the access to PIG-K's active site; and two other proteins, PIG-S and GPAA1, with as yet unknown molecular function (Ohishi et al., 2001; Eisenhaber et al., 2003). In all sequenced eukarya including Arabidopsis and rice (Oryza sativa), there are obvious full-length orthologs for the four components of the transamidase complex (see Eisenhaber et al., 2001, 2003; http://mendel.imp.univie.ac.at/SEQUENCES/gpi-biosynthesis/). Therefore, it is justified to suggest that the general scheme of the recognition signal in substrate sequences discovered for metazoa and protozoa (Eisenhaber et al., 1998) is also valid for fungi and plants. Both an export signal for translocation into the ER (for example, an N-terminal signal peptide) and the recognition signal for interaction with the transamidase complex are required. The classical sequence motif for the latter (Fig. 1) is located in the approximately 30 to 40 C-terminal residues of the substrate proprotein and consists of four necessary elements (Eisenhaber et al., 1998): (i) a polar and flexible linker region of about 11 residues (ω - 11... ω - 1) without intrinsic secondary structural preference, (ii) a region of small residues (ω - 1.... ω + 2) with the ω-site, (iii) a spacer region (ω + 3... ω + 9) of moderately polar residues with sufficient backbone flexibility, and (iv) a tail beginning with ω + 9 or ω + 10 up to the C-terminal end with a long, sufficiently hydrophobic stretch.
Figure 1.
The C-terminal GPI lipid anchor signal. The scheme illustrates the two signals that are necessary for GPI lipid anchoring: the N-terminal ER export signal and the C-terminal transamidase recognition signal. The latter consists of four regions (i–iv, see text). The mature protein that remains after N- and C-terminal processing is also indicated.
Despite the presence of these four common elements (Fig. 1), the substrate sequence-specific catalytic efficiency of the transamidase complex varies among taxa, although there is some overlap. For example, parasitic protozoa and human systems have similar but not identical sequence requirements (Moran and Caras, 1994). Similarly, in plants, both exemplary fungal and plant GPI sequence signals were successfully processed in tobacco (Nicotiana tabacum) cells, whereas a mammalian version was a poor target for anchor addition (Takos et al., 2000). It is, therefore, not surprising that the predictor for the C-terminal GPI anchor signal in metazoan sequences (Eisenhaber et al., 2000) does not predict a number of known plant targets. For example, the C-terminal signals of PcAGP1 from pear (Youl et al., 1998), LeAGP1 from tomato (Lycopersicon esculentum; Takos et al., 2000), and AtAGP10 (Schultz et al., 2000) and SKU5 (Sedbrook et al., 2002) from Arabidopsis are not recognized. In contrast, the putative GPI lipid anchored COBRA in Arabidopsis (Schindelman et al., 2001) is predicted as a transamidase substrate with a significant score, and NaAGP1 from N. alata (Youl et al., 1998) is recognized as twilight zone/borderline hit.
In this work: (a) We analyze the C-terminal sequence pattern in known GPI lipid anchored plant proteins, (b) we describe an algorithm that recognizes the GPI lipid anchor signal in C-termini of plant protein sequences, (c) we describe the performance of its computer implementation in large-scale tests, and (d) we describe its application on the proteomes of Arabidopsis (Arabidopsis Genome Initiative, 2000) and rice (Yuan et al., 2003), the genomes of which have recently been sequenced, and the classification of the identified proteins into families based on predicted function. To distinguish between suitable and nonpermissive C-termini in the query proteins, a mathematical decision criterion is used that rates the concordance with the physical property pattern for all four signal elements as derived from a learning set of plant sequences.
The big-Π plant predictor is offered in the form of a Web server (http://mendel.imp.univie.ac.at/gpi/plants/gpi_plants.html and links therein) and provides a new service for the plant research community. It will allow researchers to identify possible C-terminal GPI anchor signals in their sequences of interest and to analyze the influence of amino acid substitutions when designing mutation experiments. Increased confidence that a protein is putatively GPI lipid anchored should encourage more researchers to experimentally verify this modification, which in turn will allow the construction of a predictor with even higher accuracy in the future.
RESULTS
The results of this work are: (a) a verified learning set of substrate proteins, (b) the plant-specific C-terminal motif properties, (c) an online-accessible prediction tool big-Π for cleavage and attachment sites of plant proproteins, (d) prediction accuracy estimates for this version of big-Π, and (e) the results of applying big-Π onto plant proteomes.
Collection of a Learning Set of Example Substrates
We collected a learning set of protein sequences that are known to be associated with the desired biological property (here, GPI lipid anchoring) with the goal: (a) to characterize plant-specific features of the C-terminal recognition signal, and (b) to extrapolate conserved sequence properties in the recognition signal to uncharacterized query sequences (via construction of a score function for knowledge-based prediction of potential GPI lipid anchoring from sequence alone).
The assembly of a reliable learning set is an important result in itself because the data are scattered in the literature, and the reported verification status for many plant sequences needs updating. Swiss-Prot (Bairoch and Apweiler, 2000) is generally the prime source of annotated protein sequences (Eisenhaber et al., 1998; Nielsen et al., 1999; Maurer-Stroh et al., 2002b), but, at the time when the learning set was completed, none of the plant proteins in Swiss-Prot were annotated as being GPI lipid anchored. Thus, we turned to the original scientific literature and to collaborators.
The final learning set contains a total of 219 entries: 40 are supported with at least some experimental data, and 179 are based on theoretical consideration (for procedures, see “Materials and Methods”; for sequence lists and verification status as of October 2002, see http://mendel.imp.univie.ac.at/gpi/plants/l_set/plants.learn.html). Except for three classical arabinogalactan (AG) proteins (AGPs) and five AG peptides with experimentally analyzed ω-site (see above), a putative ω-site was assigned with theoretical considerations (with rules from Eisenhaber et al., 1998, 1999).
Although our learning set represents the current state of the field, it is certainly not an ideal starting point for predictor construction. We would prefer a learning set composed of proproteins with sufficiently diverse C-terminal sequences that are GPI lipid anchor modified in vivo and that are experimentally verified with the same stringent procedure (also including ω-site determination). The inclusion of sequences based on compatibility with the animal signal criterion and the large number of AGPs may favor the prediction of certain classes of anchored proteins. However, the procedures used in assembling the learning set ensure that there are few, if any, incorrectly included proteins.
Plant-Specific Aspects of the C-Terminal GPI Lipid Anchor Motif
A physical model of substrate protein binding by the transamidase complex can be derived from the learning set of substrate proteins by observing naturally occurring sequence variation. This model can be expressed as a pattern of position-specific physicochemical requirements to amino acid residues characteristic for good substrates. Such an analysis has been carried out with animal sequence sets (Eisenhaber et al., 1998, 1999) and was repeated with the plant learning set in this work. This involved the analysis of correlation of amino acid property scales with amino acid type occurrences at positions of the gapless C-terminal alignment of substrate sequences (see the Web site for detailed data on amino acid scale correlations). In this alignment, the sequences were centered on the ω-sites assigned to each sequence in the data set.
This analysis showed that the amino acid type occurrences in region ω - 11... ω - 1 are similar to those found in turns, loops, and isolated extended (poly-Pro-II like; Adzhubei and Sternberg, 1993) stretches known from globular structures. The predominant amino acid type in this segment is Pro (approximately 15% regardless of alignment position). In contrast, animal sequences do not have a clear single preferential amino acid type in this region. Possibly, this effect is due to many AGPs in the plant learning set. It should be noted that many Pro residues in AGPs are modified to Hyp, and this may occur before the AGPs are GPI lipid anchored. To conclude, the result is in agreement with the interpretation of a sufficiently polar, but not highly charged, conformationally extended linker region without intrinsic preference for α-helical or β-sheet structures.
The ω-site region (ω - 1... ω + 2) is expected to enter the catalytic cleft in the protease structure of PIG-K. Not surprisingly, this region is generally occupied by small amino acids. There is even a preference for tiny amino acids in the region (ω... ω + 2). Ser (at ω - 1 and ω) and Ala (at ω + 1 and ω + 2) are specifically favored. At the ω-site, we observed Ser (55%), Gly, Ala, Asn, and Asp (but no Cys). The plant sequences show a similar volume compensation effect for the region (ω - 1... ω + 2) that was first described by Eisenhaber et al. (1998; see Fisher criterion with Eq. 3 therein). It affects the combinations of all four positions (ω - 1... ω+2), pairs [(ω - 1, ω + 1) and (ω - 1, ω+2)], and the triple (ω - 1, ω + 1, ω + 2). To our astonishment, the amino acid type occurrences in the spacer region (ω + 3... ω + 9) correlate also with the property “tiny” (correlation coefficient in the range 0.70... 0.88 except for positions ω + 5 and ω + 9, which tend to be more hydrophobic). Thus, the spacer region is conformationally flexible, restricted in volume, and polar but not highly charged.
The C-terminal tail (starting with ω + 9) has a length between nine and 24 residues and is occupied preferentially by hydrophobic residues. It remains unclear whether this hydrophobic segment interacts with transmembrane (TM) regions of the transamidase complex or with the ER membrane compartment. In the case of very long tails, we find that a hydrophobic stretch in the first two-thirds is always present, but some polar or even a few charged residues may occur in the most terminal one-third of the tail. Although Leu was the dominant amino acid in metazoan sequences (with up to 50% of the amino acid type occurrence at the respective alignment position), plant sequences contain much less Leu, which is substituted by Ala and aromatics (mostly Phe). In general, the aromatic residues are not sequentially clustered. The lower Leu content (and higher content of aromatic amino acids) in the tail of many plant sequences is one of the major reasons why many potential plant precursor sequences do not pass the big-Π predictor for animal sequences.
The Big-Π Predictor for Plant Sequences
The motif properties derived from the learning set were used to design a composite score function for distinction between true transamidase substrate C-termini and non-substrate C-termini (see “Materials and Methods”). Amino acid type preferences (evaluated in a profile term Sprofile) have been shown to be insufficient for the characterization of the GPI lipid anchor sequence motif (Eisenhaber et al., 1999). The GPI anchor signal sequence is more completely described using additional terms of physical properties, sometimes involving interactions of several sequence positions (evaluated in the physical property term Sppt). Following a previously described approach (Eisenhaber et al., 1999; Maurer-Stroh et al., 2002a), simple analytical functions are used to formulate conditions for the query sequence that correspond to the transamidase-binding model derived in the substrate protein motif analysis in the previous chapter. Parameters of the prediction function were calculated from the plant learning set with proper reduction of redundancy caused by homologous protein groups. Finally, the score S (calculated as sum of Sprofile and Sppt) is translated into a probability estimate of false positive prediction (see “Materials and Methods”).
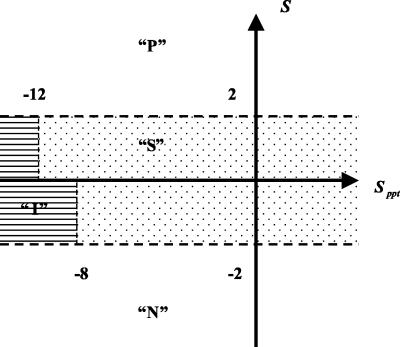
For each query sequence, the big-Π plant predictor returns the score of the best putative ω-site and its probability of false positive prediction. If a secondary site is available, the same information is generated for it. A reliably predicted substrate protein has amino acid-type preferences that are similar to the learning set consensus (positive Sprofile) and no strong deviation from the physical property pattern (zero or only slightly negative Sppt; for illustration, see Fig. 2). We consider predictions with score S ≥ 2 as predicted transamidase substrates (quality “P”). With 2 > S ≥ 0 and Sppt < -12 or with S < 0 and Sppt < -8, we consider a query as not predicted for GPI lipid anchoring due to serious disagreement with general physical properties of the motif region (quality “I”). Otherwise, hits with 2 > S ≥ -2 and Sppt ≥ -12 are considered twilight zone predictions (quality “S”; these examples appear to be of special interest for experimental testing). Queries evaluated with S < -2 are clearly rejected as targets (quality “N”). For large-scale tests, we consider all hits with quality “P” or “S” (with positive total score) as predicted and all others as not predicted.
Figure 2.
Prediction quality classes and decision scores. The different prediction quality classes are shown in the coordinate system spanned by the score component Sppt and the total score S. It should be noted that Sppt is never positive for real sequences due to the construction of the mathematical expression involved (Eisenhaber et al., 1999). “P” is considered predicted; “S” denotes twilight zone predictions. With scores in the “I” and “N” zones, sequences are not predicted to have the C-terminal GPI lipid anchor motif. For more detail, see text.
Prediction Accuracy of the Plant Big-Π Predictor Version
Self-Consistency Test
We have executed standardized statistical tests (see Table I and sequence-specific results on the Web site) to assess the accuracy of the new prediction function (Eisenhaber et al., 1999; Maurer-Stroh et al., 2002a). In the self-consistency test, the complete learning set is used for deriving score function parameters. Of the 219 sequences, 215 are confidently predicted (98.2%). Three other examples pass the test only with quality “S,” that is, with a positive total score but low Sppt.
Table I.
Prediction performance of the score function S in statistical test
| Prediction Quality | Self-Consistency Test (219 Sequences) | Jackknife Test I (Sprofile and Sppt) (219 Sequences) | Jackknife Test II (Only Sppt) (179 Sequences) |
|---|---|---|---|
| “P” (S ≥ 2) | 215 (98.2%) | 202 (92.2%) | 175 (97.8%) |
| “S” (2 > S ≥ —2, Sppt ≥ —12) | 3 (1.4%) | 7 (3.2%) | 3 (1.7%) |
| “S” with positive score | 3 (1.4%) | 4 (1.8%) | 3 (1.7%) |
| “S” with negative score | 0 | 3 (1.4%) | 0 |
| “I” (S < 2, Sppt < —12) | 1 (0.4%) | 7 (3.2%) | 1 (0.5%) |
| “N” (S < —2) | 0 | 3 (1.4%) | 0 |
| Predicted | 218 (99.6%) | 206 (94.0%) | 178 (99.5%) |
| Not predicted | 1 (0.4%) | 13 (6%) | 1 (0.5%) |
Only the Arabidopsis sequence CAA74765.1 (At4g16120) is clearly rejected by the predictor due to strongly negative side chain volume terms in the ω-site region. Apparently, the large residues in the sequence... MRSSQH... flanking the potential attachment site are much too voluminous compared with the sequence consensus of the remaining learning set. It should be noted that this sequence contradicts the rules of Udenfriend and Kodukula (1995a, 1995b) of having small residues at both the ω and ω + 2 sites. This COBRA-like sequence also differs dramatically at the Arg and at the Gln positions from the COBRA family alignment (see Web site). It should be noted that phosphatidylinositol-phospholipase C (PIPLC) sensitivity (Borner et al., 2003) points to possible anchor attachment for this protein. There may be a whole class of as yet undiscovered transamidase substrate proteins with larger than normal accompanying residues for which the predictor is simply not trained due to the absence of examples. Other possibilities include: (a) slight sequence errors in the CAA74765.1 C terminus (an unlikely possibility because several expressed sequence tags cover this region), (b) lower efficiency of precursor processing, or (c) the occurrence of alternative splicing forms or RNA editing.
Jackknife Tests I and II
The jackknife cross-validation tests check whether the parameters of the prediction function are robustly determined by the learning set of sequences (see Table I); i.e. it is a check for “over-training.” In this procedure, the sequence to be predicted is removed from the learning set for the computation of a specified set of score function parameters. In the jackknife test I, the whole score function is completely recalculated from the reduced learning set after removal of the query sequence. We find that, nevertheless, 94% of the learning set still remain predicted (92.2% still have confidence level “P”). Therefore, 94% can be considered as estimate of sensitivity (defined as the ability to recognize true targets) for the big-Π plant predictor. As we have discussed previously (Eisenhaber et al., 1999; Maurer-Stroh et al., 2002a), the number of sequences in the learning set is too small for reliable parametrization of the profile term Sprofile. Therefore, we carried out the jackknife test II where the query sequence was removed only for the computation of the Sppt parameters. Because they are computed only from the largest subset of proteins with sequentially non-related C-termini in the learning set (here, 179 of 219 proteins), the jackknife II test produces only 179 predictions. As expected, the results are very similar to the self-consistency test; 99.5% of the examples are predicted, and only the same Arabidopsis protein Atg416120 does not fit the predictor requirements.
For two AGP-related proteins in tobacco and pear (Youl et al., 1998), five in Arabidopsis (C.J. Schultz, K.L. Ferguson, and A. Bacic, personal communication) and one in tomato (Takos et al., 2000), the ω-sites have been experimentally determined. When these sequences were subjected to the three statistical tests described, the predicted primary (and in one case, the predicted secondary) sites as analyzed by big-Π were identical with the experimentally found ones. For the remaining sequences, the putative ω-site was assigned by theoretical considerations. In each test, at least 96% of the a priori-defined sites were recovered as primary or, at least, as secondary ω-site by the prediction algorithm.
Rate of False Positive Prediction
The sensitivity of a prediction algorithm defined as the ability to recognize true examples can be estimated relatively easily from its application over the learning set. In contrast, i.e. the estimation of the selectivity, the fraction of false positive predictions relative to the total number of query sequences is more difficult. Obviously, a GPI lipid anchor attachment signal is secondary to several other sequence signals and can become functional only after export into the ER because the lumenal side of the ER membrane is the only known cellular localization with a GPI lipid anchor biosynthesis and attachment machinery (Eisenhaber et al., 2003). Therefore, the GPI lipid anchor signal may be contained in a non-substrate; for example, cytoplasmic or nuclear protein without harming its function because only the combination with an ER export signal leads to anchor attachment. Ideally, such proteins should be excluded from the test set, but this is difficult in practice due to the absence of verified sets of non-ER proteins. It might even be of interest to test the capacity of such C-termini for anchoring in synthetic constructs. In “Materials and Methods,” we show how the extrapolation from the distributions of negative scores in the Swiss-Prot database leads to the assessment of the false positive prediction rate of the order of one of 1,000 analyzed query sequences. An upper limit of this rate can be calculated independently from sequence sets with known cellular localizations. Among plant proteins with annotated nuclear localization in Swiss-Prot (380 entries), we found no hits, neither with quality “P” nor with quality “S,” in agreement with the above estimate (see also the functional analysis of the Arabidopsis and rice proteome hits below).
Application on Protein Databases and the Proteomes of Arabidopsis and Rice
Due to its sensitivity and selectivity, the big-Π plant predictor can be reasonably applied for scanning large sequence datasets for potentially GPI lipid-anchored proteins (for detailed lists of data, see Web site). We emphasize that the big-Π plant predictor tests only the existence of the C-terminal signal for anchor attachment. In addition to our new tool, we also applied the SIGNALP predictor version 1.2 (Nielsen et al., 1997, 1999) to check for signal leader peptides. It should be noted that the sensitivity of SIGNALP is only in the range of 70% for nonredundant sequence sets and that alternative export mechanisms do exist (for example, with N-terminal myristoylation and palmitoylation; Denny et al., 2000).
In Swiss-Prot (release 40), we find 22 hits. The 13 examples with a signal peptide have functions that are compatible with GPI lipid anchoring. Among them are early nodulins, blue copper proteins, and an indole-3-acetic acid-Ala resistance factor. In the plant section of SP-TrEMBL, we detected 403 proteins (as of July 2002) with potential C-terminal GPI lipid anchor signal. The number of confidently predicted hits (quality “P”) is 344; 236 of those have a predicted N-terminal ER signal leader.
The estimation of the fraction of lipid-modified proteins in the proteome of completely sequenced plant species is of great interest (Table II). In the case of Arabidopsis, 245 proteins are predicted to have a potential C-terminal anchor attachment signal with quality “P” or “S” (with a positive score) or 0.94% (180 or 0.69% with predicted signal peptide) of the total proteome (as of June 2002). For rice, 294 of such proteins (198 with signal peptide) have been found (0.75% and 0.51%, respectively, of the total proteome as of June 2002). Such numbers have to be considered preliminary because some protein sequences in the proteome lists may be incomplete or incorrectly annotated (especially at the termini), not all splicing versions might be present, and conceptual translations of pseudogenes might be included. Nevertheless, the fractions are higher than the typical value of 0.5% (independent of the ER export signal status) in other eukarya (Eisenhaber et al., 2000, 2001). One possibility is that lipid modifications have a greater role in the plant physiology than in other taxa. Previously, the fraction of N-terminally N-myristoylated proteins in plant genomes was reported to be significantly higher than in animal genomes (Maurer-Stroh et al., 2002a).
Table II.
Prediction rates in complete plant genomes
| Categories of Proteins | Arabidopsis | Rice |
|---|---|---|
| Proteins in the proteome | 26,184 | 39,389 |
| Proteins submitted to predictiona | 26,063 | 38,490 |
| Predicted GPI anchor signalsb | ||
| Total | 245 (0.94%) | 294 (0.75%) |
| with predicted signal peptide | 180 (0.69%) | 198 (0.51%) |
| “P” | 219 | 251 |
| “S” | 42 | 71 |
| “S” with positive score | 26 | 43 |
| “S” with negative score | 16 | 28 |
Not all proteins in proteome can be submitted to the big-∏ plant predictor. At least the C-terminal 55 residues must be assigned to individual amino acids (and not B, X, etc.). Finally, a minimum total sequence length is required (55 residues)
The fraction of proteins with predicted GPI anchor signal is calculated relative to the total no. of proteins with minimal length (26,180 and 39,343, respectively)
We have also functionally classified the proteins with both predicted N-terminal signal leader peptide and C-terminal GPI lipid anchoring motif for both completely sequenced plant species, Arabidopsis and rice. We used available database annotations, sequence comparisons with BLASTP searches in the nonredundant protein database (Altschul et al., 1994) and comparisons with the PFAM domain (Bateman et al., 2000) library (for total nos., see Table III; for accession no. listings, see Web site). Sequence comparison methods are unsuitable for identifying many AGP-related proteins in rice due to their long sequence regions with compositional bias toward polar residues and Pro. Therefore, we applied the methodology described by Schultz et al. (2002).
Table III.
Functional grouping of proteins in Arabidopsis and rice with predicted N-terminal ER export and C-terminal GPI lipid anchor attachment signal The functional grouping of proteins is similar to Borner et al. (2002). Where available, the PFAM entry characterizing the functional group is included. The second and third columns list the no. of entries in the respective genomes. In this table, we considered all proteins predicted by big-Π with classes either “P” or “S” that additionally pass the SIGNALP (Nielsen et al., 1999) predictor (version 1.2). In parentheses, we indicate if proteins of the same group with C-terminal signal but without predicted signal peptide have been found. The complete lists with accession nos. are available at the associated Web site http://mendel.imp.univie.ac.at/gpi/plants/gpi_plants.html.
| Functional group | Arabidopsis | Rice |
|---|---|---|
| AGP related and fasciclin-like (also including AGPs, AG peptides, and Lys rich)a | 38b | 20 (+2)b |
| Extensin-like | 2 | 2 |
| Phytocyanin-like (stellacyanin-like, uclacyanin-like, early nodulin-like, PF02298) | 25 | 36 (+3) |
| Phytochelatin synthethase-like (COBRA, COBRA-like, PF04833) | 6 (+3) | 5 |
| Glycerophosphodiesterase-like (PF03009) | 5 | 3 |
| Glycohydrolases (families 1, 5, 8, or 17; PF00232, PF00150, PF01501, and PF00332) | 18 (+1) | 20 (+2) |
| Pectin metabolism related | 3 | 4 (+1) |
| Proteases | ||
| Aspartyl proteases (PF00026) | 8 (+1) | 7 |
| Metalloproteinases (PF00413, PF03933) | 3 | 1 |
| Cys proteinases (PF00112) | 1 | 1 (+1) |
| Putative carboxy-terminal proteinase (PF03080) | None | 1 (+2) |
| Lipid transfer protein-like/seed storage/protease inhibitor (PF00234) | 18 | 16 |
| Kazal-type Ser protease inhibitors (PF00050) | 2 | None |
| Multicopper oxidase/BP10-like (PF00394) | 2 | 3 |
| Glc/sorbosone dehydrogenase/hedgehog interacting-like | 1 | 2 |
| Signaling receptor-like | 7 (+2) | 9 (+2) |
| Thaumatin-like (PF00314) | 2 | 5 |
| Cation transporters | 2 (+3) | 1 (+2) |
| Other functions | 10 | 18 |
| Hypothetical/unknown function | 34 | 48 |
| Total | 187 | 202 |
Following AGP-related protein definition of Schultz et al. (2002)
Additional AGP-related entries might be hidden among the hypothetical proteins with sequential bias
Not surprisingly, most of the proteins detected belong to functional families for cell wall and extracellular matrix synthesis and remodeling and to signaling protein groups. The majority of the families were already identified by Borner et al. (2002). For both plant species, we find AGP-related proteins (classical AGPs, AG peptides, and Lys-rich and fasciclin-like proteins; Schultz et al., 2002), extensin-like proteins (Schultz et al., 2002), phytocyanins/plastocyanins and related proteins, the COBRA-like group, glycerophosphodiesterases, glycohydrolases of family 17 (in both species) and of families 1, 5, and 8 (in rice only), proteins of pectin metabolism, extracellular proteases (aspartyl, Cys and metalloproteinases), lipid transfer protein-like examples, multicopper oxidases/BP10/SKU5-like proteins, signaling receptors, and sugar dehydrogenases/hedgehog-interacting-like proteins. New classes include, for example, kazal-type protease inhibitors and cation transporters (Table III).
Numerous hypothetical proteins with unknown function were detected (20%–25% of all hits with both signal peptide and predicted C-terminal GPI lipid anchor signal for both studied plant proteomes). The group classified as “other functions” includes proteins characterized only phenotypically and doubtful protein hits with ER or Golgi localization (for example, one cytochrome P450 in Arabidopsis). At least 10 rice hits have to be classified as potentially false positives due to their predicted cellular localization (see Web site). If we assume the estimated rate of about one false positive prediction per 1,000 queries, we expect approximately 30 false hits for Arabidopsis (total proteome of 26,184 proteins) and approximately 40 mispredictions for rice (total of 39,389 proteins) that are expected mainly among the “other” and “hypothetical” groups; thus, the estimate of false positive predictions is within the expected magnitude.
DISCUSSION
The big-Π predictor provides the first online tool, to our knowledge, for the prediction of C-terminal proprotein cleavage and GPI lipid anchor attachment sites in plant sequences. This study has shown that there are minor but significant differences between animal and plant GPI lipid anchor signals, although the overall four-region structure of the recognition signal is conserved (Fig. 1). The estimated accuracy of the big-Π plant predictor is sufficiently high (sensitivity approximately 95% for known targets) with high selectivity against non-related sequences (only approximately one false positive prediction among 1,000 query sequences).
It should be noted that specialized tools such as the big-Π predictor are necessary because traditional sequence analytic methods relying on the homology concept, and the conservation of protein domain folds and secondary structural elements are not applicable due to large numbers of false positive predictions. Many motifs for localization targeting or posttranslational modifications (including the GPI lipid anchor signal) are located in non-globular regions of proteins. In addition, these sequence signals are relatively short (a dozen or a few dozens of residues, approximately 40 residues in the GPI anchor signal case) compared with a typical globular domain (100–150 residues). Finally, a single residue mutation can render an otherwise unchanged GPI lipid anchor signal nonfunctional; also, homologous forms of one and the same protein may behave differently with respect to anchoring (for review, see Eisenhaber et al., 1999).
Comparison with Predictions of Borner et al. (2002, 2003)
In a previous work, Borner et al. (2002) predicted that 208 GPI-anchored proteins are encoded in the Arabidopsis genome (originally 210, but two sequence pairs are identical after sequence correction). An updated analysis using more recent database annotation (November 2002) suggested 248 GPI-anchored proteins (Borner et al., 2003). Among them, 30 examples were supported by PI-PLC sensitivity data. The screen of the protein database involved only evaluating C-terminal hydrophobicity and compliance with the rules of Udenfriend and Kodukula (1995a, 1995b) for the ω-site. This much simpler description of the GPI lipid anchor modification signal provided a list of candidate proteins, which were then selected if they possessed an ER signal peptide and had no internal TM domains. In the 2002 dataset, some examples also have been included by homology considerations. Borner et al. applied great care to correct gene annotations and especially to extend protein termini if possible (10 cases in the 2003 sets). In their original list (Borner et al., 2002), of 208 plant proteins (see Web site), 141 (68%) are also predicted by the big-Π plant predictor (among them, 126 with quality “P” and 15 with quality “S”). The updated list of Borner et al. (2003) with 248 proteins (see Web site) contains 192 examples that pass the big-Π plant predictor test (77% of 248, 171 among the 192 are with quality “P”). The majority of the proteins not predicted by big-Π deviate from the physical consensus pattern of the learning set, mainly in the ω-site region or tail properties but sometimes also in the other two motif regions (for details, see Web site).
Among the 187 big-Π hits with either quality “P” or “S” and with predicted N-terminal signal (in the Arabidopsis proteome as of June 2002), 132 are identical with the 2002 set and 165 reoccur in the 2003 set of Borner et al. (2002, 2003; see Web site). The overlap between the big-Π predictions and the Borner et al. sets is further enlarged if sequences corrected at the termini but not the older database versions are analyzed by big-Π and if the same database version is used.
Generally, a potential substrate predicted by big-Π having a signal peptide also passes the Borner et al. selection criteria (except for those with internal TM regions). At the same time, the big-Π tool predicts generally fewer hits due to a more detailed C-terminal motif description (but on the contrary, proteins with internal TM regions are not excluded). The big-Π predictor is based on a more complete model of substrate protein interaction with the transamidase complex because we have included sequence properties in all four motif regions. The disagreement between the big-Π predictions and the Borner et al. reflects that their prediction sets contain both sequences that strictly adhere to the GPI lipid anchoring motif (as derived in this work) and other sequences that miss some of the sequence properties. Thus, the Borner et al. criteria are more of a necessary nature but, in the light of this work, appear not sufficient for positive prediction. For example, the effect of mutations close to the ω-site region cannot be evaluated with the Borner et al. criteria but with big-Π (Eisenhaber et al., 1999). At the same time, big-Π does extrapolate more conservatively to other sequences. Thus, positively predicted proteins are very likely to be true substrates. This does not exclude that there might be substrate sequences with very deviant C-terminal signals, but they do not comply with the transamidase binding model as assumed in this version of big-Π.
To conclude, to which extent the criteria of Borner et al. for the GPI lipid anchor attachment pattern are not stringent enough or whether the learning set for the big-Π predictor is overly restricted and conservative will become clearer as more experimental evidence (especially mutational series) is gathered. Most importantly, the functional protein families with numerous GPI lipid-anchored members are mostly the same in the studies of Borner et al. (2002; for Arabidopsis) and in this work (for Arabidopsis and rice). This provides support to the view that many proteins of these families are likely to become GPI lipid anchored and that these are the major families of GPI lipid-anchored proteins in plants.
The Big-Π Predictor. Outlook
We expect that the first version of the big-Π plant predictor as described here will assist the experimental plant research community in deciding whether a target sequence has a potential GPI lipid anchor signal. The algorithm has implemented the physical pattern of the C-terminal GPI anchor signal as generalized consensus of the learning set. As with all prediction tools, users will have to keep in mind the following possible limitations. First, the big-Π plant predictor analyzes only the C terminus for its compatibility with the GPI lipid anchor motif. A positive answer does not necessarily mean that the query protein is definitely GPI lipid anchored because it must also contain a sequence signal for an ER export mechanism. Second, any knowledge-based predictor cannot be better than the learning set it relies on. In our case, we have a bias toward Arabidopsis and AGP proteins. The number of sequences is still small, most of them are not sufficiently studied experimentally, and the ω-site assignment largely relies on theoretical considerations. As a result, the parametrization is preliminary especially for the profile component. Proteins with close to zero Sppt can be considered reasonably as probable targets even if their profile score is low.
The big-Π program can be easily updated: (a) when improved learning sets become available, and (b) when new requirements to protein substrates become clear from a better understanding of the binding site and the catalytic mechanism of the transamidase complex. With respect to the prediction of GPI lipid-anchored plant proteins, increasing information from other taxa is helpful from the viewpoint of general motif properties only. Because the sequence specificity of plant transamidases is distinct, although overlapping with that of animal enzyme counterparts, dedicated experimental and theoretical studies in plants are necessary.
The plant community is now invited to submit their query sequences to the Web server. Prediction results will be returned instantly. In the event that sequences are not predicted as GPI lipid anchored, the output describes the sequence features that are responsible for rejection of a query as potential target (terms with large negative scores). Further, the Web server can used to test different site-directed mutations for their ability to abolish potential GPI lipid anchoring capacity.
MATERIALS AND METHODS
The Construction of the Learning Set
40 Sequences with Experimental Support
We found descriptions of a total of seven examples of GPI lipid-modified proteins where there is at least some indirect experimental confirmation and an established amino acid sequence. Six of these seven proteins were included in the learning set. These include: case 1, NaAGP1 from Nicotiana alata; and case 2, PcAGP1 from pear (Pyrus communis; Youl et al., 1998), still the best analyzed targets with confirmed ω-site. Four other examples are: case 3, LeAGP1 from tomato (Lycopersicon esculentum; Takos et al., 2000); case 4, AtSKU5 (Sedbrook et al., 2002); case 5, AtAGP10 (Schultz et al., 2000); and case 6, COBRA (Schindelman et al., 2001) from Arabidopsis. The PI-PLC test was used to support GPI lipid anchoring for cases 3 and 4, cellular localization was checked for proteins (cases 4 and 6), mutations at the putative ω-site were tested (for case 3), and mass spectrometric fingerprinting was carried out for case 5. For all six sequences, the canonical four-element motif for GPI lipid signal recognition is present, and a putative ω-site can be unambiguously assigned. The same type of C-terminal motif is visible also in a number of homologs in other species.
The seventh suggested protein in the literature, an alkaline phosphatase from the aquatic plant Spirodela oligorrhiza (Morita et al., 1996; Nakazato et al., 1998), was not included in the learning set because there is contradictory evidence. The GPI lipid anchoring was concluded from incorporation of a radioactively marked GPI anchor component into a specific electrophoretic band on a protein gel. However, these data are inconsistent with the resistance of the membrane-bound phosphatase fraction to PI-PLC. The possibly incomplete protein sequence was published subsequently (Nishikoori et al., 2001) and lacks the established canonical motif, especially the long hydrophobic tail. None of the characterized homologous alkaline phosphatases in plants, such as At2g16430 in Arabidopsis, have the canonical C-terminal motif. For many other early reports of GPI-modified plant proteins, the sequences are still not available, and their posttranslational modification status needs further confirmation.
For an additional 34 proteins, recent experimental data support GPI lipid anchoring. GPI lipid anchoring and the ω-sites for five AG peptides from Arabidopsis have been shown with mass spectrometric evidence (C.J. Schultz, K.L. Ferguson, and A. Bacic, personal communication). Borner et al. (2003) contributed a further 30 PI-PLC-sensitive proteins (without ω-site determination) from the same species. The latter 30 examples include also SKU5 (also published by Sedbrook et al. (2002).
179 Sequences Included Based on Theoretical Considerations
The learning set was enriched with 21 predicted sequences that have the canonical C-terminal signal structure, an N-terminal ER export peptide, and are similar to experimentally studied proteins. We relied on lists of totally 29 AG-like proteins from different species (Schultz et al., 1998; Schultz et al., 2000, 2002), among which 17 proteins are new and have been included. Among the 12 members of the COBRA family collected in a PSI-BLAST search (started with COBRA=NP_568930.1, inclusion E value = 0.001), 11 have nonidentical C-termini. Five of these, including COBRA, have both terminal signals as tested with SIGNALP (Nielsen et al., 1997) and big-Π (for metazoan sequences; Eisenhaber et al., 2001). The four COBRA-like proteins have also been included in the learning set.
Finally, we scanned the plant sections of Swiss-Prot and SP-TrEMBL and the Arabidopsis (Arabidopsis Genome Initiative, 2000) and rice (Oryza sativa; Yuan et al., 2003) proteomes for highly probable targets for GPI lipid anchoring. We required the existence of a signal leader peptide (as predicted with SIGNALP; Nielsen et al., 1997) and a score larger than one calculated with the metazoan GPI lipid anchor signal prediction function (Eisenhaber et al., 2001). After removal of sequences with identical C-termini from the four search hit lists, we found 185 entries, among which 158 were not contained in any of the lists discussed above. As a control, we checked the function descriptions in databases and in the original literature for all 158 entries. If the entry was a hypothetical translation, we searched for homolog sequences and hits of PFAM domains (Bateman et al., 2000) and analyzed their annotation. If available, the described function was always consistent with possible GPI lipid anchoring.
Description of the Score Function
To develop the plant big-Π predictor, we adapted the score function used for metazoan proteins (Eisenhaber et al., 1999). The score function S consists of two parts: S = Sprofile + Sppt.
The profile-dependent section Sprofile evaluates the concordance with the weak amino acid type preferences in the learning set at single alignment positions. The functional form of Sprofile remains unchanged (see Eisenhaber et al., 1999), but its parameters are calculated from the plant learning set. Redundancy originating from groups of homologous sequences in the learning set was eliminated with the PSIC algorithm during profile calculation (Sunyaev et al., 1999).
As a consequence of our analysis of the plant-specific C-terminal pattern in learning set sequences, we found out that, in addition to the previously described terms in the Sppt component (physical property terms; see Eisenhaber et al., 1999), seven new terms for plant-specific features can be introduced and are described below. It should be noted that, typically, each individual term in Sppt produces an almost zero score for a learning set sequence, but it yields a negative value for a query sequence that deviates from the physical property pattern. Thus, the Sppt components provide a mechanism only for rejecting potentially non-substrate proteins.
The new terms are: (a) The Asp and Glu contents and their clustering in the N-terminal part (ω - 11... ω - 1) are penalized with ρ= -4 (less than 50% of all residues, and, if this threshold is exceeded, maximally two successive ones). (b) An increased average hydrophobicity of the same region is penalized with a term according to Equation 14 in (Eisenhaber et al., 1999). (c) The previous term T2 imposes a residue volume penalty for the position pair (ω - 1, ω + 1) in the metazoan function. Here, an additional penalty for the pair (ω - 1, ω+2) in accordance with Equation 10 in (Eisenhaber et al., 1999) is introduced. Both terms enter the score with weight = 0.5. (d) The number of charged residues (DEKRH) in the region (ω + 3... ω + 11) is limited to three; otherwise, a penalty of ρ = -4 is added to the score. (e) The PVI content of the spacer region (ω + 3... ω + 8) is evaluated with a functional form identical to that of the metazoan T4 term. (f) In the C-terminal tail (from ω + 9 on), the aromatic residue content is restricted to 40%, and, if this value is exceeded, the occurrence of aromatic clusters is penalized. (g) With a metazoan T10-like function, the occurrence of windows with many small residues (volume threshold) in the C-terminal hydrophobic domain is penalized. Finally, the original terms T12 and T14 are applied only on the first two-thirds of the C-terminal tail if its length exceeds eight residues. The LVI contents threshold in the former term T13 is reduced to 15%.
In total, the Sppt component has less than 40 parameters, of which about 20 are calculated as means or dispersions of physical properties from the learning set of plant sequences. To remove redundancy from the learning set for the computation of Sppt parameters, a largest subset of nonredundant sequences was obtained from the learning set (for detail, see Eisenhaber et al., 1999). Further detail on the function parametrization is available on the Web site.
The score is translated into the probability of false positive prediction with a generalized extreme-value distribution (see Eqs. 2 and 4 in Eisenhaber et al., 2001). For its parametrization, the set of putatively non-GPI lipid anchored proteins; the plant sequences from Swiss-Prot with score below zero (release 40, approximately 8,000 sequences) were taken. Thus, score S = 0 for a query sequence corresponds to a false prediction probability of about 0.0005.
This work was supported by the Boehringer Ingelheim (continuous support to B.E., M.W., and F.E.), in part by the Fonds zur Förderung der Wissenschaftlichen Forschung Österreichs (grant no. FWF P15037), by the Austrian National Bank (Österreichische Nationalbank), by the Austrian Ministry of Economic Affairs (BMWA), by the Austrian Gen-AU Program (bioinformatics integration network grant), by the Australian Research Council (Discovery grant no. DP0343454 to C.S.), by the University of Adelaide (partial support for a visit to Vienna for this project), by the Biotechnology and Biological Sciences Research Council (research studentship to G.H.H.B.), and by the Studienstiftung des Deutschen Volkes (scholarship to G.H.H.B.).
References
- Adzhubei AA, Sternberg MJE (1993) Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol 229: 472-493 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Boguski MS, Gish W, Wootton JC (1994) Issues in searching molecular sequence databases. Nat Genet 6: 119-129 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res 28: 45-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL (2000) The Pfam protein families database. Nucleic Acids Res 28: 263-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucht G, Hjalmarsson K (1996) Residues in Torpedo californica acetylcholinesterase necessary for processing to a glycosyl phophatidylinositol-anchored form. Biochim Biophys Acta 1292: 223-232 [DOI] [PubMed] [Google Scholar]
- Clayton CE, Mowatt MR (1989) The procyclic acidic repetitive proteins of Trypanosoma brucei. J Biol Chem 264: 15088-15093 [PubMed] [Google Scholar]
- Coussen F, Ayon A, Le Goff A, Leroy J, Massoulie J, Bon S (2001) Addition of a glycophosphatidylinositol to acetylcholinesterase: processing, degradation, and secretion. J Biol Chem 276: 27881-27892 [DOI] [PubMed] [Google Scholar]
- Denny PW, Gokool S, Russell DG, Field MC, Smith DF (2000) Acylation-dependent protein export in Leishmania. J Biol Chem 275: 11017-11025 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F (1998) Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng 11: 1155-1161 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F (1999) Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292: 741-758 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F (2001) Post-translational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes. Protein Eng 14: 17-25 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Yuan Y, Loffler G, Eisenhaber F (2000) Automated annotation of GPI anchor sites: case study C. elegans. Trends Biochem Sci 25: 340-341 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Maurer-Stroh S, Novatchkova M, Schneider G, Eisenhaber F (2003) Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and posttranslational transfer to proteins. Bioessays 25: 367-385 [DOI] [PubMed] [Google Scholar]
- Ferguson MA (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci 112: 2799-2809 [DOI] [PubMed] [Google Scholar]
- Killeen N, Moessner R, Arvieux J, Willis A, Williams AF (1988) The MRC OX-45 antigen of rat leukocytes and endothelium is in a subset of the immunoglobulin superfamily with CD2, LF-3 and carcinoembryonic antigens. EMBO J 7: 3087-3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Inoue N (2000) Dissecting and manipulating the pathway for glycosylphos-phatidylinositol-anchor biosynthesis. Curr Opin Chem Biol 4: 632-638 [DOI] [PubMed] [Google Scholar]
- Kuntze M, Riedel J, Lange U, Hurwitz R, Tischner R (1997) Evidence for the presence of GPI-anchored PM-NR in leaves of Beta vulgaris and for PN-NR in barley leaves. Plant Physiol Biochem 35: 507-512 [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F (2002a) N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol 317: 541-557 [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F (2002b) N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol 317: 523-540 [DOI] [PubMed] [Google Scholar]
- Misumi Y, Ogata S, Ohkubo K, Hirose S, Ikehara Y (1990) Primary structure of human placental 5′-nucleotidase and identification of the glycolipid anchor in the mature form. FEBS Lett 191: 563-569 [DOI] [PubMed] [Google Scholar]
- Moran P, Caras IW (1994) Requirements for glycosylphosphatidylinositol attachment are similar but not identical in mammalian cells and parasitic Protozoa. J Cell Biol 125: 333-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P, Raab H, Kohr WJ, Caras IW (1991) Glycophospholipid membrane anchor attachment. J Biol Chem 266: 1250-1257 [PubMed] [Google Scholar]
- Morita N, Nakazato H, Okuyama H, Kim Y, Thompson GA Jr (1996) Evidence for a glycosylinositolphospholipid-anchored alkaline phosphatase in the aquatic plant Spirodela oligorrhiza. Biochim Biophys Acta 1290: 53-62 [DOI] [PubMed] [Google Scholar]
- Nakazato H, Okamoto T, Nishikoori M, Washio K, Morita N, Haraguchi K, Thompson GA Jr, Okuyama H (1998) The glycosylphosphatidylinositol-anchored phosphatase from Spirodela oligorrhiza is a purple acid phosphatase. Plant Physiol 118: 1015-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Brunak S, von Heijne G (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng 12: 3-9 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8: 581-599 [DOI] [PubMed] [Google Scholar]
- Nishikoori M, Washio K, Hase A, Morita N, Okuyama H (2001) Cloning and characterization of cDNA of the GPI-anchored purple acid phosphatase and its root tissue distribution in Spirodela oligorrhiza. Physiol Plant 113: 241-248 [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Jenö P, Conzelmann A, Riezmann H (1991) Determinants for glycophospholipid anchoring of the Saccharomyces cerevisae GAS1 protein to the plasma membrane. Mol Cell Biol 11: 27-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Inoue N, Kinoshita T (2001) PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J 20: 4088-4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley D, Bacic A (1999) Structure of the glycosylphosphatidy, inositol anchor of an arabinogalactan protein from Pyrus communis suspension cultured cells. Proc Natl Acad Sci USA 96: 14246-14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Gilson P, Oxley D, Youl J, Bacic A (1998) The GPI-anchors on arabinogalactan-proteins: implication for signalling in plants. Trends Plant Sci 3: 426-431 [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1751-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A (2002) Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiol 129: 1448-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635-1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P (1999) Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20: 2027-2035 [DOI] [PubMed] [Google Scholar]
- Stahl N, Baldwin MA, Burlingame AL, Prusiner SB (1990) Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry 29: 8879-8884 [DOI] [PubMed] [Google Scholar]
- Stöhr C, Schuler F, Tischner R (1995) Glycosyl-phosphatidylinositol-anchored proteins exist in the plasma membrane of Chlorella saccharophila (Krüger) Nadson: plasma-membrane-bound nitrate reductase as an example. Planta 196: 284-287 [Google Scholar]
- Sugita Y, Nakano Y, Oda E, Noda K, Tobe T, Miura NH, Tomita M (1993) Determination of carboxy-terminal residue and disulphide bonds of MACIF (CD59), a glycosyl-phosphatidylinositol-anchored membrane protein. J Biochem 114: 473-477 [DOI] [PubMed] [Google Scholar]
- Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN (1999) PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng 12: 387-394 [DOI] [PubMed] [Google Scholar]
- Takos AM, Dry IB, Soole KL (1997) Detection of glycosylphosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett 405: 1-4 [DOI] [PubMed] [Google Scholar]
- Takos AM, Dry IB, Soole KL (2000) Glycosyl-phosphatidylinositol-anchor addition signals are processed in Nicotiana tabacum. Plant J 21: 43-52 [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K (1995a) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem 64: 563-591 [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K (1995b) Prediction of omega site in nascent precursor of glycosylphosphatidylinositol protein. Methods Enzymol 250: 571-582 [DOI] [PubMed] [Google Scholar]
- Vai M, Lacanà E, Gatti E, Breviario D, Popolo L, Alberghina L (1993) Evolutionary conservation of genomic sequences related to the GGP1 gene encoding a yeast GPI-anchored glycoprotein. Curr Genet 23: 19-21 [DOI] [PubMed] [Google Scholar]
- Yan W, Ratnam M (1995) Preferred sites of glycosylphosphatidylinositol modification in folate receptors and constraints in the primary structure of the hydrophobic portion of the signal. Biochemistry 34: 14594-14600 [DOI] [PubMed] [Google Scholar]
- Youl JJ, Bacic A, Oxley D (1998) Arabinogalactan-proteins from Nicotiana alata und Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc Natl Acad Sci USA 95: 7921-7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quacken-bush J, Buell CR (2003) The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res 31: 229-233 [DOI] [PMC free article] [PubMed] [Google Scholar]