Abstract
The Arabidopsis cell wall-associated kinase (WAK) and WAK-like kinase (WAKL) family of receptor-like kinase genes encodes transmembrane proteins with a cytoplasmic serine/threonine kinase domain and an extracellular region containing epidermal growth factor-like repeats. Previous studies have suggested that some WAK members are involved in plant defense and heavy metal responses, whereas others are required for cell elongation and plant development. The WAK/WAKL gene family consists of 26 members in Arabidopsis and can be divided into four groups. Here, we describe the characterization of group 2 members that are composed of a cluster of seven tandemly arrayed WAKL genes. The predicted WAKL proteins are highly similar in their cytoplasmic region but are more divergent in their predicted extracellular ligand-binding region. WAKL7 encodes a truncated WAKL isoform that is predicted to be secreted from the cytoplasm. Ratios of nonsynonymous to synonymous substitutions suggest that the extracellular region is subject to diversifying selection. Comparison of the WAKL and WAK gene clusters suggests that they arose independently. Protein gel-blot and immunolocalization analyses suggest that WAKL6 is associated with the cell wall. Histochemical analyses of WAKL promoters fused with the β-glucuronidase reporter gene have shown that the expressions of WAKL members are developmentally regulated and tissue specific. Unlike WAK members whose expressions were found predominately in green tissues, WAKL genes are highly expressed in roots and flowers. The expression of WAKL5 and WAKL7 can be induced by wounding stress and by the salicylic acid analog 2,6-dichloroisonicotinic acid in an nonexpressor of pathogenesis-related gene 1-dependent manner, suggesting that they, like some WAK members, are wound inducible and can be defined as pathogenesis-related genes.
The plant cell wall, or extracellular matrix (ECM), is a complex array of carbohydrates, proteins, and proteoglygans surrounding the cell (Carpita and Gibeaut, 1993). The ECM provides structural support, determines cell shape, protects the cell against environmental insults, and mediates cell growth and differentiation (Cosgrove, 1999). The ECM is closely associated with the plasma membrane and functions in mediating communication between neighboring cells and between cells and the environment (Pennell, 1998; Kohorn, 2000). Interactions between ECM components and the plasma membrane are undoubtedly important for the efficient relay of environmental signals to the cytoplasm (Kohorn, 1999). In animal cells, a class of plasma membrane receptors called integrins mediates the attachment of the ECM to the plasma membrane via interactions with adhesive glycoproteins, such as fibronectin, vitronectin, and collagen (Hay, 1981; Hynes, 1981, 1987). Interactions between these molecules and ECM ligands result in the translation of environmental cues into signals that can affect cellular functions (Quaranta and Jones, 1991). In plant cells, proteins immunologically related to vitronectin and fibronectin have been identified in salt-adapted cell walls (Zhu et al., 1993). Associations between the ECM and the plasma membrane have also been observed (Pont-Lezica et al., 1993). The exact nature of these associations and their potential role in mediating communication between the ECM and the cytoplasm is not completely understood. Several plant proteins that are bound to both the ECM and the plasma membrane have been described, including arabinogalactan proteins (Majewska-Sawka and Nothnagel, 2000), cellulose synthases (Pear et al., 1996), an endo-1,3-β-glucanase (Nicol et al., 1998), and cell wall-associated kinase (WAK) proteins (He et al., 1996; Kohorn, 2000; Lally et al., 2001). How or whether these associations function in signaling between the ECM and the cytoplasm is presently unknown.
Other cell surface receptors located in the plasma membrane play a vital role in cell signaling in both plants and animals (Kyriakis, 1999; Schenk and Snaar-Jagalska, 1999; McCarty and Chory, 2000). Many of these are receptor-like protein kinases (RLKs), which typically contain an extracellular region that is thought to function in signal perception, a transmembrane domain, and a cytoplasmic protein kinase domain for transducing the signal (Walker, 1993; Hardie, 1999). RLKs function in a diverse array of signaling processes in plants, including phytohormone responses (Chang et al., 1993; Li and Chory, 1997; Clark et al., 1998; Gamble et al., 1998; Woeste and Keiber, 1998), reproduction (Stein et al., 1991; Mu et al., 1994; Takasaki et al., 2000), developmental regulation (Becraft et al., 1996; Clark et al., 1997; Yokoyama et al., 1998; Jinn et al., 2000), and plant resistance responses (Loh and Martin, 1995; Song et al., 1995). These RLKs can be divided into classes based on sequence relationships between their predicted extracellular regions (Shiu and Bleecker, 2001).
A unique class of RLKs is encoded by a family of genes called the WAKs and WAK-like kinases (WAKLs; He et al., 1996, 1999; Verica and He, 2002). The WAK/WAKL proteins contain a carboxyl-terminal cytoplasmic region with a Ser/Thr kinase active site and an amino-terminal extracellular region containing several motifs, including epidermal growth factor (EGF) repeats, Ca2+-binding EGF domains and EGF2-like domains (He et al., 1996, 1999; Verica and He, 2002). Five WAKL members are predicted to lack the transmembrane domains and may represent truncated, secreted proteins (Verica and He, 2002). The results of topological experiments are consistent with the localization of the WAKs to the plasma membrane. Furthermore, they are tightly associated with the ECM (He et al., 1996). There is evidence to suggest that WAKs are covalently bound to pectin in the cell wall (Wagner and Kohorn, 2001). In addition, WAK1 has been shown to be a component of an approximately 500-kD protein complex with the potential for signaling. This complex includes WAK1, a Gly-rich extracellular protein, AtGRP-3 (Park et al., 2001), and a cytoplasmic type 2C protein phosphatase, KAPP (Anderson et al., 2001), which can associate with a Rho-related protein (Trotochaud et al., 1999).
WAK members are shown to be involved in stress responses and in development. Induction of WAK1 expression is required for plants to survive after infection by the bacterial pathogen Pseudomonas syringae (He et al., 1998). Moreover, the application of salicylic acid (SA) or its analog, 2,6-dichloroisonicotinic acid (INA), induces WAK1 expression. This induction requires nonexpresser of pathogenesis-related (PR) genes (NPR1), demonstrating that WAK1 is a PR gene (He et al., 1998). Transgenic studies have shown that WAKs are also required for cell expansion (Lally et al., 2001; Wagner and Kohorn, 2001). For example, Lally et al. (2001) showed that expression of a WAK4 antisense gene caused a significant decrease in WAK proteins, resulting in inhibition of cell elongation. WAK1 is an aluminum early-responsive gene, and its overexpression renders aluminum tolerance, suggesting that WAK1 is involved in heavy metal responses (Sivaguru et al., 2003). Collectively, these results support a role for WAKs in signal transduction between the ECM and the cytoplasm during both stress responses and cell expansion.
More than 80% of the WAK/WAKL family members exist as tight gene clusters, and 19 of the 26 WAK/WAKL genes are located in chromosome 1 (Verica and He, 2002). The 26 WAK/WAKL gene members can be further placed into four groups according to their sequence and structural similarities (Verica and He, 2002). The group 1 genes consisting of the five tandemly arrayed genes, WAK1 to WAK5, were described before (He et al., 1999). Here, we report the identification and characterization of a new cluster of seven WAKL genes, WAKL1 to WAKL7. The WAKLs, with the exception of WAKL7, belong to the group 2 and are predicted to encode RLKs with a cytoplasmic Ser/Thr protein kinase domain and an extracellular domain containing EGF-like repeats. WAKL7 represents a truncated WAKL family member that is predicted to encode a secreted protein. Our studies have revealed that the WAKL members respond to environmental stresses and are expressed in tissue-specific and developmentally regulated manners.
RESULTS
Genomic Organization and Structural Features of the WAKLs
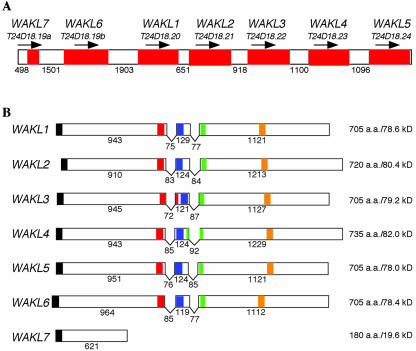
Twenty-six WAK/WAKL genes have been identified in Arabidopsis genome through extensive analyses (Verica and He, 2002). Although many RLKs are found in tandem repeats (Shiu and Bleecker, 2001), the WAK/WAKL family represents a unique case. WAK/WAKL family was found to have more than 80% of its members in tandem repeats that are present on chromosome I within a region spanning less than 12 centiMorgans (Verica and He, 2002). Approximately 4.5 centiMorgans distal to the previously described WAK1 to WAK5 cluster (He et al., 1999), a cluster of seven tandemly arrayed WAKL genes was found to be positioned within a 23-kb region. All seven genes are predicted to be transcribed in the same direction (Fig. 1A). Annotation of the BAC (T24D18) sequences that contained the seven WAKLs includes the predicted intron splice sites for each of the genes (S. Theologis, Plant Gene Expression Center, Albany, CA). cDNAs for all of the WAKL genes were cloned via reverse transcriptase (RT)-PCR. Sequence analysis confirmed the two predicted intron sites for WAKL1 to WAKL5. T24D18.19 was predicted to encode a long open reading frame (ORF) containing eight exons and seven introns; however, our analysis shows that this region contains two separate genes. For example, we found that the first intron, predicted to reside between nucleotides 52,113 and 52,159, was not present. A stop codon was also found in this region at nucleotide 52,118 bp, resulting in an ORF of 621 bp (T24D18.19a, WAKL7). In the second gene, the predicted initiation codon resides at 53,642 bp, resulting in a 2,196-bp ORF divided by two introns (T24D18.19b, WAKL6).
Figure 1.
Genomic organization of the WAKL gene cluster. A, Cartoon of the region of chromosome 1 containing the WAKL cluster. The identities of each gene (red shaded boxes) and the corresponding BAC gene from which it was derived are given above. The arrow above each gene corresponds to the direction in which it is transcribed. The unshaded areas correspond to the intergenic regions. The lengths of each intergenic region are given in the number of nucleotide base pairs between the stop codon of the preceding gene and the predicted start codon of the gene that follows. B, Cartoon of each of the WAKL genes. The identity of each gene is indicated at the left. Exons are shown as boxes. The “V” between each exon indicates the introns. Regions of each sequence displaying similarity with predicted functional domains are indicated with shaded boxes as follows: N-terminal signal sequence (black), EGF2-like domain (red), calcium-bind EGF domain (blue), transmembrane domain (green), and Ser/Thr protein kinase active site (orange). The numbers under each exon and intron correspond to their respective lengths in nucleotide base pairs. The size and molecular mass (excluding the signal peptide) for each of the predicted proteins are shown at the right.
Each of the WAKL genes, with the exception of WAKL7, contains two introns whose positions are highly conserved (Fig. 1B). The intron sizes range from 72 to 85 bp for the first intron and 77 to 92 bp for the second intron. This is in contrast to the WAKs, which showed no variation in the sizes of each intron, 80 and 76 bp, respectively (He et al., 1999). The WAKLs have shorter intergenic regions (0.50–1.90 kb) than the WAKs (1.95–3.24 kb). It is of interest to note that the intergenic regions preceding WAKL2 and WAKL7 are 0.65 and 0.50 kb, respectively. Accounting for the 5′-untranslated region of each gene and the 3′-noncoding region of the gene preceding each, the promoters for these genes may be less than 0.4 to 0.5 kb in length.
We analyzed the intergenic regions for potential transacting factor-binding sites in an effort to gain insight into how expression of the various WAKL genes may be regulated. All of the WAKL genes contained putative TATA and CAAT boxes in a region spanning 33 to 92 bp (data not shown). The putative TATA box of each gene was located in a region between -91 to -239 bp, relative to the predicted start codon. A variety of other regulatory elements were also found to be in common with several of the WAKL genes. For example, WAKL1, WAKL2, WAKL4, and WAKL5 all contained the AS-1 element (TGACG). This element was previously shown to be required for the SA-induced expression of the PR gene, PR-1 (Lebel et al., 1998). In addition, all of the WAKL promoters, except WAKL4, contained multiple copies of the W-box (TTGAC), an element similar to the AS-1 element, which was shown to be required for the SA induction of the tobacco (Nicotiana tabacum) class I chitinase gene (Yang et al., 1999). These findings, along with our previous observation that WAK1 expression is induced by SA and its analog INA, suggest that some WAKL genes may also be INA inducible (see below).
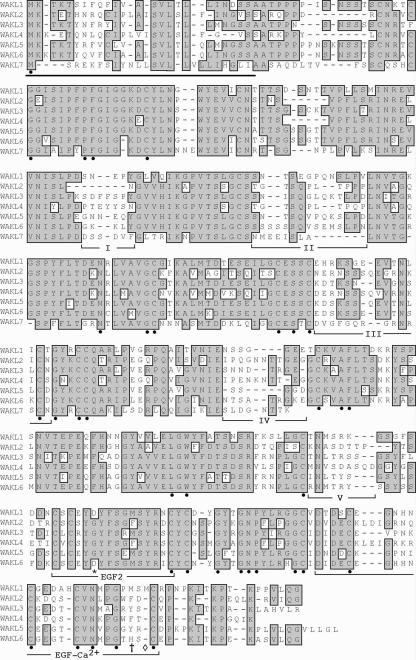
Comparison of the predicted WAKL protein sequences shows that they are highly similar to each other (Fig. 2). They are 65% to 88% identical in the cytoplasmic region (74%–91% similar). They are less similar in the extracellular region, showing 51% to 77% identity (62%–82% similarity). Pair wise comparison of the WAKLs and WAKs shows that they are 40% to 45% identical (60%–64% similar) to each other in the cytoplasmic region but only 18% to 22% identical (30%–36% similar) in their extracellular regions. Thus, the proteins that comprise each group are more similar to each other than they are to those of the other group.
Figure 2.
Multiple sequence alignment of the predicted extracellular region of the WAKL proteins. Amino acids that are identical among the WAKLs are shaded. Amino acids that are identical among the WAKs and WAKLs are indicated by the black dots. The predicted N-terminal signal sequences are underlined. The five variable domains analyzed for nonsynonymous to synonymous substitution ratios are underlined and indicated by roman numerals (I–V). The EGF2-like domain (EGF2) and calcium-binding EGF-like domain (EGF-Ca2+) are underlined. The amino acid substitutions in the EGF2-like domain are indicated by the asterisk. The cross indicates the amino acid substitutions in the EGF-Ca2+-like domain. The additional amino acid inserted before the last conserved Cys in the WAKL1 calcium-binding EGF-like domain is indicated with a diamond.
Overall, there are 144 amino acids that are identical between both the WAKLs and the WAKs (excluding the missing portion of WAKL7). Of the identical amino acids, 111 are in the cytoplasmic region and one is in the transmembrane domain. The predicted extracellular region contains 35 conserved residues, 23 of which are Cys or Gly (Fig. 2). The conserved Cys residues could facilitate the formation of disulfide bonds, particularly in the EGF-like domains (see below) where such roles have been shown in animals (Appella et al., 1987; Rebay et al., 1991; Stenflo et al., 2000). These results further suggest that the various WAK and WAKL isoforms have similar structural conformations. The divergence between the two groups in their cytoplasmic regions is most apparent in the area between the Ser/Thr kinase domain and the C terminus. This suggests that the WAKLs and WAKs may be able to interact with different cytoplasmic proteins.
The WAKLs, with the exception of WAKL7, encode proteins that are similar in size (705–735 amino acids) and molecular mass (78.0–82.0 kD; Fig. 1B). Analysis of their predicted protein products shows many structural features in common with the WAKs (He et al., 1999). They contain a short hydrophobic segment at their N termini that could serve to target the proteins to the endoplasmic reticulum, a transmembrane region characteristic of plasma membrane-spanning proteins, and the signature domain of Ser/Thr protein kinases (Prosite: PS00108) in their predicted cytoplasmic tail. Each also contains an EGF2-like domain (CxCx(2)[GP][FYW]x(4,8) C; Prosite: PS01186) and a calcium-binding EGF-like domain ([DEQN]x[DEQN](2) Cx(3,14) Cx(3,7) Cx[DN]x(4)[FY]xC; Prosite: PS01187; Fig. 2); however, their order within the sequence is reversed relative to the WAK proteins. WAKL7 encodes a truncated WAKL protein of 180 amino acids (19.6 kD) that lacks many of these structural features and is predicted to be secreted from the cell. The EGF-like regions in some of the WAKLs are slightly degenerate. For example, WAKL1 and WAKL6 have all of the consensus residues for the EGF2-like domain, except for a Gly to Asp substitution at the sixth position. Similarly, their calcium-binding EGF-like domains each have a Met or His residue in place of a Phe/Tyr residue. In addition, WAKL1 has one additional amino acid inserted before the last conserved Cys (Fig. 2). Because EGF-like domains have not been well characterized in plants, it is not clear whether the domains present in WAKL1 and WAKL6 function as EGF-like domains or whether such functions have been lost.
WAKLs Are Subject to Diversifying Selection
Although the EGF-like domains are good candidates for mediating interactions with potential ligands, similar roles for other regions of the WAKLs cannot be ruled out. Comparison of the WAKL protein sequences shows that there are several stretches of amino acids that appear to be highly variable, particularly in the central part of the extracellular region (Fig. 2). These variable regions contain a number of hydrophilic amino acids that may comprise a solvent-exposed ligand-binding domain. Because binding regions encoded by members of gene families involved in recognizing diverse ligands have been shown to have higher ratios of nonsynonymous to synonymous substitutions (dn/ds; Hughes and Yeager, 1998; McDowell et al., 1998; Meyers et al., 1998b; Wang et al., 1998; Ellis et al., 1999), we examined the central variable regions and the EGF-like domains of the WAKLs for nonsynonymous and synonymous substitution ratios (dn/ds). As a control, a region overlapping the Ser/Thr kinase site was analyzed. The truncated WAKL7 is missing many of these regions and was therefore not included in this analysis. Dn/ds ratios significantly less than one indicate a higher rate of synonymous substitutions, and thus conservation of the sequence. Ratios significantly greater than one indicate a higher rate of nonsynonymous substitutions, and thus diversification of the sequence.
Our analysis shows that the region overlapping the Ser/Thr kinase site was highly conserved, with dn/ds ranging from 0.03 to 0.15 (Table I), suggesting that this region is subject to purifying selection (Hughes and Yeager, 1998; Meyers et al., 1998b). Conservation of this region is consistent with its proposed effector function. In the EGF-like domains, the dn/ds ratios were also less than one (0.10–0.85). Thus, if the EGF domains are involved in protein-protein interactions, they are more likely to be involved in binding similar sequences. In the five areas in the central part of the extracellular region, dn/ds was more variable, depending on which two WAKL sequences were compared. In addition, there did not appear to be any correlation between dn/ds values and the overall distance relationships between the WAKL sequences. In three pair wise comparisons, dn/ds was less than or close to 1.0 (WAKL1 to WAKL2, WAKL4 to WAKL5, and WAKL4 to WAKL6). In the remaining pair wise comparisons, dn/ds was greater than 1.0, ranging from 1.24 to 3.18, highly suggesting that diversifying selection is acting on these regions.
Table I.
Nonsynonymous to synonymous substitution ratios (dn/ds) for WAKL1-WAKL6 STK, Codon-aligned sequences of a 159-base region overlapping the Ser/Thr kinase active site. EGF, Codon aligned sequences of the regions corresponding to the EGF-like domains. Extracellular, Codon-aligned sequences corresponding to the five variable areas in the central part of the WAKL extracellular region (see Fig. 2). nd, Not determined. In some cases, the proportion of observed synonymous substitutions has reached mutational saturation. As such, dn/ds can not accurately be estimated (Nei and Gojobori, 1986).
| Gene Pair | STK | EGF | Extracellular |
|---|---|---|---|
| WAKL1-WAKL2 | 0.06 | nd | 1.03 |
| WAKL1-WAKL3 | 0.15 | 0.10 | 1.82 |
| WAKL1-WAKL4 | 0.09 | 0.24 | 1.26 |
| WAKL1-WAKL5 | 0.09 | 0.39 | 1.32 |
| WAKL1-WAKL6 | 0.10 | 0.20 | 2.08 |
| WAKL2-WAKL3 | 0.07 | 0.65 | 1.49 |
| WAKL2-WAKL4 | 0.03 | 0.46 | 1.24 |
| WAKL2-WAKL5 | 0.03 | 0.38 | 1.49 |
| WAKL2-WAKL6 | 0.03 | 0.36 | 1.24 |
| WAKL3-WAKL4 | 0.11 | 0.41 | 1.36 |
| WAKL3-WAKL5 | 0.11 | 0.85 | 3.18 |
| WAKL3-WAKL6 | 0.12 | 0.25 | 2.42 |
| WAKL4-WAKL5 | 0.06 | 0.34 | 1.08 |
| WAKL4-WAKL6 | 0.06 | 0.60 | 0.96 |
| WAKL5-WAKL6 | 0.08 | 0.32 | 1.67 |
WAKL6 Is Associated with the Cell Wall
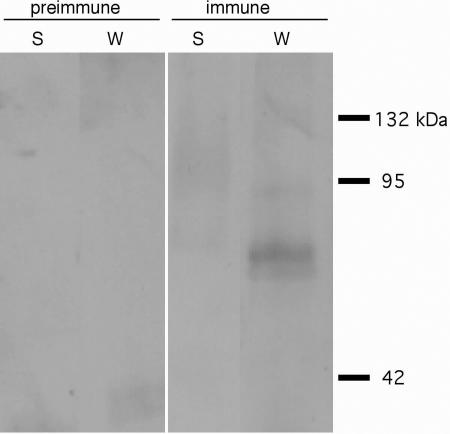
Previous findings demonstrated that the various WAK proteins are physically associated with the cell wall (He et al., 1996; Kohorn, 2000; Park et al., 2001). These findings, along with the sequence similarity between the WAK and WAKL extracellular domains, suggest that the WAKLs may also be tightly associated with the cell wall. To test this possibility, we generated polyclonal antibodies against a peptide corresponding to a unique region of the WAKL6 extracellular domain. WAKL6 was chosen because multiple WAKL6 ESTs were found in the database and our analyses suggest WAKL6 was consistently expressed in all organs tested. Protein gel-blot analysis shows that WAKL6 protein, like the WAKs (He et al., 1996), was present only in the insoluble protein fraction, and not in the soluble protein fraction (Fig. 3). The molecular mass of the detected major band is about 80 kD, consistent with the predicted size of WAKL6 protein. A slightly lower faint band can also be detected (Fig. 3). This size shift could be due to a different phosphorylation status of WAKL6. Extraction of WAKL6 from the insoluble fraction, like that of WAK1 (He et al., 1996), required boiling in 50 mm dithiothreitol and 4% (w/v) SDS. Control blots using preimmune serum showed no cross-reacting proteins (Fig. 3).
Figure 3.
Cell wall-association of WAKL6. Protein gel blots were performed on soluble (S) and wall-associated (W) leaf proteins. Protein gel blots were performed using either the preimmune serum or immune serum containing polyclonal antibodies directed against a unique region of the WLK6 extracellular domain. Lines on the right indicate molecular mass markers.
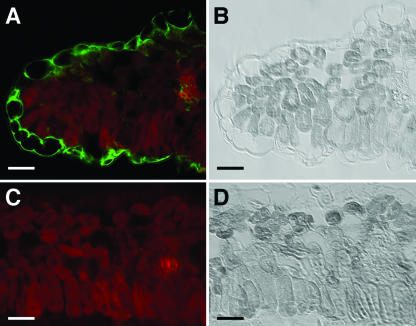
The WAKL6 antibody was further used to immunolocalize WAKL6 in leaf tissue sections from 14-d-old plants. Figure 4 shows that the WAKL6 antibody, as indicated in green fluorescence, bound to the cell surface in a region overlapping the cell wall and plasma membrane. The green fluorescence can be best seen in epidermal cells where there is little auto fluorescence from chlorophylls. Only faint green fluorescence can be observed in mesophyll cells due to strong chlorophyll autofluorescence in these cells (Fig. 4A). These results, taken together with previous findings for WAK proteins (He et al., 1996; Lally et al., 2001; Park et al., 2001), are consistent with the localization of WAKL6 to the cell wall. The similarities between their extracellular domains of other WAKL proteins suggest that they are also likely to be cell wall-associated.
Figure 4.
Immunolocalization of WAKL6. A and C, Sections through 14-d-old leaves were probed with WAKL6 antiserum (A) or preimmune serum (C). Antibody binding was detected with a fluorescein-conjugated secondary antibody. Green fluorescence indicating WAKL6 protein and autofluorescence of chloroplasts (red) in the mesophyll cells are superimposed. B and D, Bright-field images of the sections in A and C, respectively. Bars = 25 μm.
Expression of the WAKL Genes
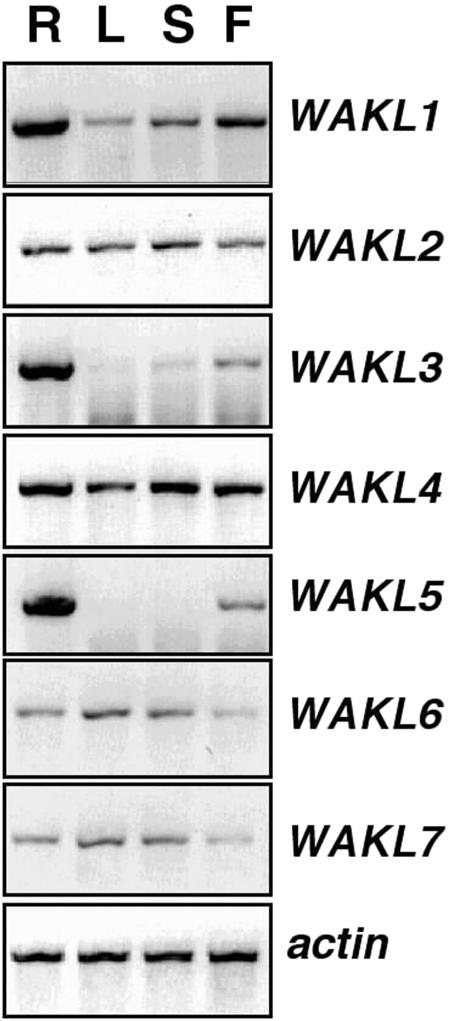
As a first step in functional analysis, expression patterns of the WAKL genes were characterized. Gene-specific primers were used in RT-PCR reactions on RNA isolated from individual organs (roots, leaves, stems, and flowers). Pairs of primers used for the RT-PCR were designed in a unique region to assure their specificity and were confirmed by sequencing of the RT-PCR products. Six of the seven WAKL genes were expressed in all organs examined; however, their relative levels in each organ varied (Fig. 5). The lowest steady-state level of WAKL1 expression was observed in leaves, with slightly higher levels in the stems and flowers. The highest level of WAKL1 expression was observed in roots. WAKL3 and WAKL5 both had similar expression profiles in roots and flowers. Both were expressed highly in roots and to a lesser degree in flowers. In leaves and stems, WAKL3 was observed at trace levels, and WAKL5 was not detected. The remaining WAKLs (WAKL2, WAKL4, WAKL6, and WAKL7) were all expressed to similar levels in all organs examined, with only slight differences being detected (Fig. 5).
Figure 5.
Organ-specific expression of the WAKL genes. Gene-specific RT-PCR analyses were performed in the following organs: roots (R), leaves (L), stems (S), and flowers (F). Actin was analyzed as a control.
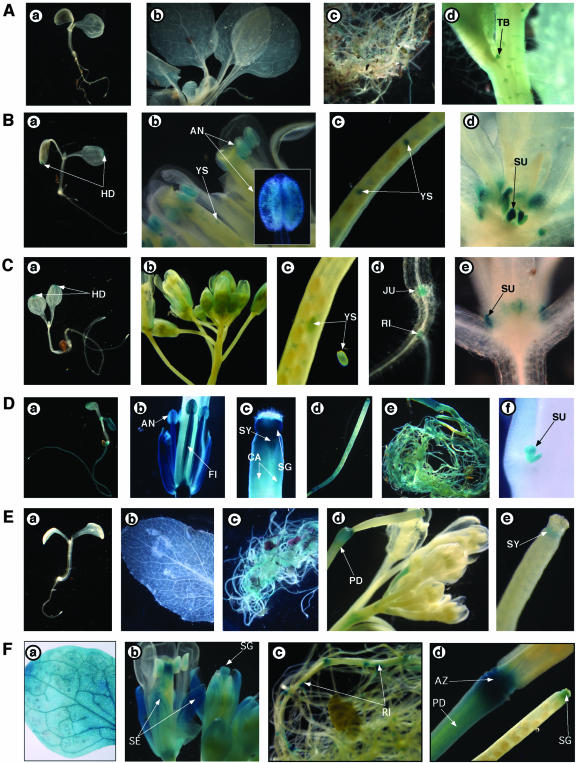
To further investigate the expression profile for the WAKL members, promoters for WAKL1 to WAKL7 genes were fused to the β-glucuronidase (GUS) reporter gene, and the resulting constructs were used to transform wild-type Arabidopsis (Col-0). More than 10 independent transgenic lines have been generated for each WAKL promoter-GUS construct. Histochemical analysis of these transgenic plants indicates that GUS expression occurs in a developmentally regulated and tissue-specific manner (Fig. 6). In general, the GUS expression patterns in different organs were consistent with what were suggested by the RT-PCR results. WAKL1 promoter appeared to be slightly active in leaves and moderately active in roots of mature plants (Fig. 6A), and WAKL3 promoter showed weak GUS activities in most of the organs examined (data not shown). The rest of WAKL members were strongly expressed and showed both overlapping and distinct tissue-specific expression patterns (Fig. 6). GUS activity driven from the WAKL2 promoter was associated with all of the organs tested, including anthers, young embryos/seeds, and rosette stipules (Fig. 6B). In young seedlings, WAKL2::GUS was predominantly active in the cotyledon hydathodes. This expression was observable soon after germination and did not appear to increase relative to the size of the growing cotyledon. Although very little GUS staining was detected in mature WAKL2::GUS rosette leaves, heavy GUS staining could be found in rosette stipules (Fig. 6B, d). The detection of WAKL2 transcripts in leaves by RTPCR was likely due to WAKL2 expression in the rosette stipules that were harvested together when total RNA was prepared for the analysis (Fig. 5). Like WAKL2 promoter in young seedlings, WAKL4 promoter was also solely expressed in hydathodes (Fig. 6C, e). In mature plants, apart from young embryos/seeds and rosette stipules, WAKL4::GUS activities were specifically found in flower petals, hypocotyl-root junctions, and lateral root initiation sites (Fig. 6C, b and d).
Figure 6.
Developmental and tissue-specific expressions of WAKL1, WAKL2, WAKL4, WAKL5, WAKL6, and WAKL7. Histochemical analyses of GUS reporter expression from WAKL promoters. A, WAKL1::GUS activity in transgenic plants. 5-Bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc) staining for 8-d-old seedling (a), mature leaves (b), roots (c), and trichomes (d) is shown. B, WAKL2::GUS activity in transgenic plants. X-Gluc staining for 8-d-old seedling (a), flower (a), silique (c), and leaf/stipule (c) is shown. C, WAKL4::GUS activity in transgenic plants. X-Gluc staining for 8-d-old seedling (a), flower (b), silique/young seeds (c), shoot-root junction/lateral root (d), and leaf/stipule (e) is shown. D, WAKL5::GUS activity in transgenic plants. X-Gluc staining for 8-d-old seedling (a), flower (b), silique (c and d), root (e), and stipule (f) is shown. E, WAKL6::GUS activity in transgenic plants. X-Gluc staining for 8-d-old seedling (a), leaf (b), roots (c), flower (d), and silique (e) is shown. F, WAKL7::GUS activity in transgenic plants. X-Gluc staining for mature leaf (a), flower (b), root (c), and silique (d) is shown. AN, Anther; CA, carpel; FI, filament; HD, hydathode; JU, hypocotyl-root junction; PD, pedicel; RI, lateral root initiation; SG, stigmatic tissue; ST, style; SU, stipule; SY, style; TB, trichome base; YS, young seeds.
High levels of WAKL5::GUS activities were consistently observed in young seedlings, roots, cauline leaf stipules, flowers, and at the abscission zone of developing seed pods (Fig. 6D). Particularly, strong WAKL5::GUS activities were seen in roots of both young seedlings and mature plants (Fig. 6D, a and e) but not in mature leaves and stems (data not shown), consistent with the RT-PCR results (Fig. 5). Heavy GUS staining was also found in many parts of the flower including anthers, stamen filaments, styles, and stigmatic tissues (Fig. 6D, b and c). WAKL6::GUS expression appears to be more developmentally regulated (Fig. 6E). GUS staining for WAKL6 promoter could be detected in all organs of the mature plants but not in young seedlings (Fig. 6E). Close examination revealed that WAKL6::GUS activities were specifically present at the distal ends of the styles, just below the stigmatic tissues (Fig. 6E, e) and at the distal end of the pedicel (Fig. 6E, d).
WAKL7 is one the five Arabidopsis abbreviated WAKL genes predicated to encode truncated receptor-like proteins (RLPs) that could be secreted into the ECM. Whether or how these abbreviated WAKL genes are expressed were totally unknown (Verica and He, 2002). Our RT-PCR analyses suggested that WAKL7 transcripts were present in roots, leaves, stems, and flowers (Fig. 5). As shown in Figure 6F, histochemical analyses confirmed that the WAKL7 promoter activity could be seen in these organs. Specifically, WAKL7 promoter was highly active in sepals, stigmatic tissues, lateral root branching sites, pedicels, and at the distal end of the pedicel (Fig. 6F). The heavy GUS staining in the tip of the stigma and in flower sepals but not petals was unique to WAKL7. In roots, WAKL7 promoter was particularly active at the lateral root branching points. Although WAKL4 promoter also showed activities at the lateral root branching points (Fig. 6C, d), WAKL7 promoter demonstrated a much more prominent GUS staining at these locations (Fig. 6F, c).
Responses of WAKL5 and WAKL7 to Environmental Stresses
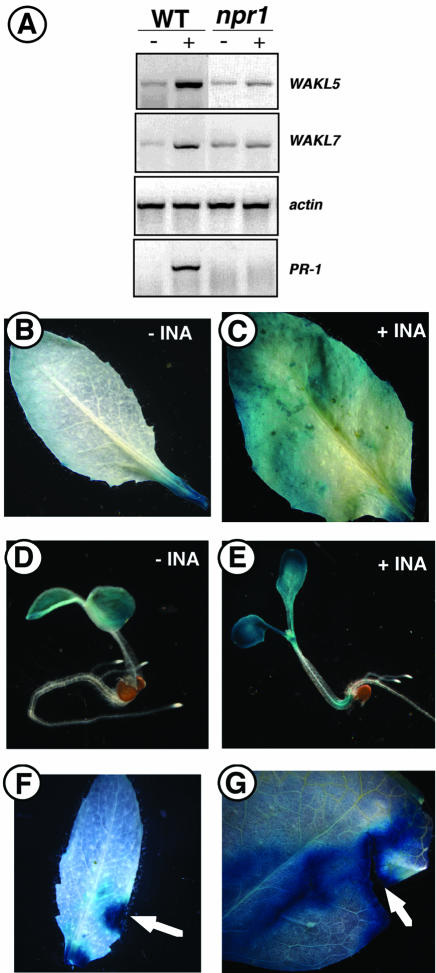
Induction of WAK1 expression by SA was previously shown to be required for Arabidopsis plants to survive infection by the bacterial pathogen P. syringae. In addition, SA and its analog, INA, induce WAK1 expression in an NPR1-dependent manner (He et al., 1998). SA is a signaling molecule that accumulates in plants in response to pathogen attack and is required for the establishment of systemic acquired resistance (SAR; Gaffney et al., 1993). SAR involves the activation of a number of genes collectively called PR genes. NPR1, which functions downstream of the SA signal, is required for the activation of the PR genes (Cao et al., 1997; Dong, 1998). Analysis of the WAKL promoter regions showed that some of the genes contained an AS-1 element and/or a W element, suggesting that they may be SA/INA inducible. As such, we analyzed their steady-state expression levels before and after treatment with INA. With the exception of WAKL3, the WAKLs were all induced by INA (data not shown). WAKL5 and WAKL7 were the members that were most noticeably induced (Fig. 7A). The enhanced expressions for the two genes were further confirmed by examining their promoter::GUS activities in responding to INA treatment (Fig. 7, B–E). In the presence of INA, GUS staining was increased in mature leaves of WAKL5::GUS transgenic plants and WAKL7::GUS seedlings when compared with their controls.
Figure 7.
Expression of the WAKL5 and WAKL7 genes is regulated by environmental stimuli. A, RT-PCR analyses of WAKL5 and WKAL7 in responding to INA. RNA from whole plants was assayed for WAKL5 and WKAL7 expression before (-) and after (+) treatment with INA in both wild-type (WT) and npr1-2 (npr1) mutant plants. The expression profile of PR-1 was included for the INA induction control, and actin was analyzed as a control. B through E, Activities of WAKL5::GUS and WAKL7::GUS in responding to INA. GUS staining for transgenic WAKL5::GUS leaves treated without (B) or with (C) INA and for transgenic WAKL7::GUS seedlings grown on media containing no INA (D) or 100 μm INA (E) is shown. F and G, Activities of WAKL5::GUS and WAKL7::GUS are responding to wounding. WAKL5 expression is induced by wounding. Mature leaves from transgenic WAKL5::GUS plants (F) and WAKL7::GUS plants (G) were wounded before being immediately stained for GUS activities.
The induced expression of the WAKL genes by INA suggests that they may be PR genes. We further tested this possibility by examining their expression levels in the npr1-2 mutant background (Cao et al., 1997). As a control, we also examined the expression of PR-1, a gene that has previously been shown to require NPR1 for induction by INA (Bowling et al., 1994). As shown in Figure 7A, the expression of PR-1 was eliminated in npr1-2 mutant. The INA-induced expression of both WAKL5 and WAKL7 was also eliminated in the npr1-2 background. Collectively, these results indicate that both WAKL5 and WAKL7 are PR genes.
SA is one of the chemical signals that are also involved in wounding response. Promoters for both WAK1 and WAK2 were shown to be responding to wounding (Wagner and Kohorn, 2001). To test whether any WAKL genes were also wound inducible, GUS staining was performed to analyze for WAKL promoter activities in responding to wounding stress. GUS activities driven by WAKL5 and WAKL7 promoters are highly enhanced in wounded leaf tissues (Fig. 7, F and G), indicating that, like WAK1 and WAK2, WAKL5 and WAKL7 are also involved in wounding stress responses.
DISCUSSION
WAK/WAKL proteins are good candidates to physically link the cell wall to the intracellular compartment and to transmit signals across the plasma membrane. Our characterization of the WAKL1 to WAKL7 gene cluster offers further evidence suggesting that various WAK/WAKL members possess differential tissue-specific and developmentally regulated expression patterns and may provide Arabidopsis with the potential to respond to a broad range of ligands. Our analyses suggested the WAK1 to WAK5 and WAKL1 to WAKL7 gene clusters arose independently and the extracellular regions of the WAKL members were subject to diversifying selection. WAKL6 was shown to be associated with the cell wall, implying that WAKL6 and likely other nonabbreviated WAKLs share similar topology with that of the WAKs. The abbreviated WAKL7 gene is predicted to encode a small secreted protein. Our expression analyses have shown that both WAKL7 and WAKL5 are PR genes that are also responding to wound stresses. The different expression profiles for WAKL1 to WAKL7 genes suggest their possible diverse functions in plant stress tolerance and in plant development and serve as a good basis in further defining their specific functional roles.
Origin of the WAKL Gene Cluster
The structural similarities between the WAKL and WAK genes suggest that they may have a common evolutionary origin. Members from each group make up a cluster, a feature common to plant R genes from a variety of species, including the members of the Arabidopsis RPP1 and RPP5 loci (Botella et al., 1998; Noel et al., 1999), and the tomato (Lycopersicon esculentum) Pto and compare with disease resistance genes (Martin et al., 1993; Parniske et al., 1997). The WAK and WAKL clusters both contain tandemly arrayed genes that are transcribed in the same direction. In addition, each of the genes (excluding the truncated WAKL7) contains two introns, which are located in conserved positions. There are several possibilities to explain how these clusters were generated. For example, misalignment of chromosomes during meiosis followed by unequal crossing-over could result in a duplication, giving rise to two clusters. Similar scenarios have been proposed for duplications in clusters of plant R genes, such as the Xa21 resistance cluster in rice (Oryza sativa; Song et al., 1997), the major resistance gene cluster (Dm) in lettuce (Lactuca sativa; Meyers et al., 1998a), and the Rp1 complex in maize (Zea mays; Hulbert, 1997). Alternatively, a single progenitor WAK gene could have been duplicated. Each of these genes could then serve as a progenitor for an independent gene cluster through a series of duplications (Nei et al., 1997; Michelmore and Meyers, 1998). Although we cannot rule out either of these possibilities, the latter is more consistent with the observations that the genes within each cluster are more similar to each other than they are to those of the other cluster and that they are tandemly arranged (Ronald, 1998). In addition, none of the genes flanking the WAKL cluster was found to be similar to those flanking the WAK cluster (Verica and He, 2002).
EGF Repeats in WAKL Extracellular Domains
The unique structural feature for the WAK/WAKL RLK subfamily is their extracellular EGF repeats. All of the WAKs have an Asp/Asn hydroxylation site (Cx[DN]x(4)[FY]xCxC; Prosite: PS00010) overlapping their calcium-binding EGF domain. This domain is absent in all of the WAKLs. Many animal coagulation proteases with calcium-binding EGF domains also have an associated Asp/Asn hydroxylation site (Selander-Sunnerhagen et al., 1992). It was initially thought that hydroxylation of the conserved Asp/Asn residue was required for calcium binding; however, both hydroxylated and nonhydroxylated forms of coagulation proteases have equal affinities for calcium at physiological concentrations (Sunnerhagen et al., 1993). As such, the role of this posttranslational modification is presently unclear. It is speculated that the hydroxyl group may be involved in hydrogen bonding in protein-protein interactions mediated by the EGF-like domain (Stenflo et al., 2000). If this holds true for the WAKLs and WAKs, it is possible that they may bind different ligands or, alternatively, may have different binding affinities for a common ligand.
Analysis of other EGF-containing proteins from plants and animals suggests several possible roles for the EGF-like domains in the WAKs/WAKLs. In the plant vacuolar sorting protein BP-80, the EGF domains were shown to alter the structural conformation of the ligand-binding domains and thereby increased their affinity for ligand binding (Cao et al., 2000). EGF-like domains in some proteins from animals directly participate in protein-protein interactions (Appella et al., 1987; Rebay et al., 1991; Kuroda and Tanizawa, 1999; Stenberg et al., 1999). For example, the fruitfly (Drosophila melanogaster) transmembrane protein Notch contains 36 tandem EGF-like repeats. Notch interacts with other EGF-containing transmembrane proteins, including Delta and Serrate. Two of the Notch EGF repeats have been shown to be both necessary and sufficient for mediating these interactions (Rebay et al., 1991). The formation of both homodimeric and heterodimeric receptor complexes has been proposed for a large number of receptor kinases (Heldin, 1995), including members of the EGF family of receptor kinases in animals (Yarden and Schlessinger, 1987; Sliwkowski et al., 1994) and the CLV1 receptor complex in plants (Clark et al., 1997; Trotochaud et al., 1999). It remains to be determined whether the EGF-like domains in WAK/WAKL proteins are involved in protein-protein interactions.
Diversifying Selection of WAKL Extracellular Regions
Our analyses suggested that diversifying selection is acting on specific regions in the ECM domains of the WAKLs. Previous experiments have suggested the WAK/WAKL members play important roles in a variety of cellular events, including cell elongation, pathogen response, and aluminum response (He et al., 1998; Lally et al., 2001; Sivaguru et al., 2003). The WAK/WAKL members may serve as signaling molecules in both developmental processes and environmental responses. The presence of 26 WAK/WAKL members and their distinct spatial and temporal expression patterns are consistent with their diverse functional roles. The diversifying selection on the extracellular domains, predicted to be signal-sensing domains, perhaps serve to generate variety in recognition specificities for various ligands (Parniske et al., 1997; Michelmore and Meyers, 1998; Noel et al., 1999). If these central regions are involved in ligand recognition, there may also be some redundancy or closely related binding specificities among WAKL isoforms with dn/ds ratios less than or close to 1.0. Further biochemical and genetic analyses of the specific functions for the WAKL members will provide some insights to this question.
Spatial and Temporal Regulation of WAKL Expressions
Previous analysis of the WAKs showed that they are mainly expressed in green organs, namely the leaves and stems; however, low levels of WAK2 expression are observed in flowers and roots (He et al., 1999). All of the WAKLs were expressed in roots and flowers, and the steady-state levels of some were low in green organs. The different expression profiles of the various WAK/WAKL genes suggest that they have evolved to provide similar but distinct functions in different regions of the plant. The distinct functions may reflect differences in the organs in which the various WAKs/WAKLs are expressed or diversity of the signals to which the organs are exposed.
Induction of WAK1 expression was previously shown to be required for Arabidopsis plants to survive infection by the bacterial pathogen P. syringae. In addition, SA and its analog, INA, induce WAK1 expression in an NPR1-dependent manner (He et al., 1998). SA is a signaling molecule that accumulates in plants in response to pathogen attack and is required for the establishment of SAR (Gaffney et al., 1993). SAR involves the activation of a number of genes collectively called PR genes. NPR1, which functions downstream of the SA signal, is required for the activation of the PR genes (Cao et al., 1997; Dong, 1998). Analysis of the WAKL promoter regions showed that some of the genes contained an AS-1 element and/or a W element, suggesting that they may be INA inducible. Our analyses suggest that, with the exception of WAKL3, the WAKLs were all induced by INA. The induced expression of the WAKL genes by INA suggests that they are PR genes. Our further examination for WAKL5 and WAKL7 on their expression levels in responding to INA in the npr1-2 mutant background confirmed they are PR genes.
The WAK proteins have been shown to be required for cell elongation and for plants to survive P. syringae infection (He et al., 1996; Lally et al., 2001; Wagner and Kohorn, 2001). The data presented here strongly suggest that the WAKL proteins are likely to function in a similar manner. For example, WAKs and WAKLs share many structural and developmental features. They both encode RLKs with extracellular domains containing EGF repeats, are induced by the SA analog, INA, in an NPR1-dependent manner, and encode proteins that are tightly associated with the cell wall. The challenge for future research is to determine precisely how these proteins function in two pathways (cell elongation and pathogenesis) that, on the surface, are seemingly unrelated.
A common feature of these processes is that both involve alteration of the cell wall. For example, successful pathogen attack must begin with the breaching of the cell wall, and elongation involves structural modification of the cell wall (Cosgrove, 1997; Grant and Mansfield, 1998). Responses to both of these events require the synthesis and secretion of new wall material to either seal off pathogen-damaged cells or to maintain the structural integrity of the recently expanded wall (McQueen-Mason, 1997; Scheel, 1998). One possibility suggested by 'these commonalities is that the tight association of WAKs and WAKLs to the ECM could allow them to function by responding to environmental changes in the wall that are similar in both cell elongation and pathogenesis. If this model is correct, it would be expected that the WAKs and WAKLs might be able to mediate a response to a variety of insults that alter the integrity of the ECM, including biotic as well as abiotic factors. This possibility is further supported by the presence of putative cis-elements in the WAKL promoters that are involved in responses to environmental stresses. We are currently testing these ideas.
Our GUS reporter gene experiments have suggested that whereas some WAKL members showed overlapping tissue-specific expression patterns, others demonstrated distinct profiles. These tissue-specific expression patterns may indicate possible overlapping as well as distinct functions for individual WAKL members. In young seedlings, both WAKL2 and WAKL4 showed consistent expressions in the hydathodes, a region that coincides with the end point of the main vascular tract of the cotyledon. Little GUS activities were detected in the mature leaves for WAKL2::GUS and WAKL4::GUS plants, suggesting that the two WAKL members may play a role in early seedling development. Promoters for WAKL2, WAKL4, and WAKL5 were active in Arabidopsis stipules, small masses of cells that appear on either side of emerging leaf primordia in both rosette and cauline leaf development. In particular, WAKL4 was active in rosette leaf stipules but not cauline leaf stipules (Fig. 6I). In contrast, WAKL5 was expressed in cauline leaf stipules and not in rosette leaf stipules (Fig. 6O). The WAKL2 promoter appeared to be active in both tissues types (Fig. 6D). The function of stipules in Arabidopsis is currently unknown. Further biochemical and genetic experiments will help to understand the relationship between these WAKL members and the stipule functions.
Our expression analyses suggested the WAKL promoters appeared to be developmentally regulated. Both WAKL2 and WAKL4 promoters are active in young developing seeds but not in mature seeds. WAKL5 showed good activities in young leaves, but not in mature leaves. Conversely, WAKL6 had little expression in young leaves, but a detectable level of expression in mature leaves. The expressions of WAK1 to WAK5 genes were found predominately in green tissues (He et al., 1999). In contrast, in this study, we have shown that the WAKL1 to WAKL7 promoters are mostly active in roots and flowers and only modestly active in green tissues. The differences in their spatial expression patterns may reflect distinct functions for the WAKs and WAKLs.
Abbreviated WAKL Members and RLPs
In Arabidopsis, there are five abbreviated WAKLs genes that are predicted to encode small secreted proteins resembling the extracellular domains of the nonabbreviated WAKs/WAKLs (Verica and He, 2002). Genes predicted to encode truncated RLPs that lack the cytoplasmic kinase domains exist in many subfamilies of the RLK super gene family (Jeong et al., 1999). Some RLPs have a transmembrane domain and may be positioned to physically interact with related RLKs. Others do not have a transmembrane domain and may serve as a mobile ligand. The Arabidopsis CLAVATA3 (CLV3) is a secreted transmembrane RLP that contains Leu-rich repeats (LRRs). CLV3 activates the CLV1/CLV2 receptor complex in underlying cells, and this interaction is critical in restricting the size of the stem cell population in shoot and floral meristems (Jeong et al., 1999). Both CLV1 and CLV2 are LRR-containing LRRs. Another LRR-containing transmembrane RLP, Too Many Mouths, is involved in stomatal patterning (Nadeau and Sack, 2002). The S locus glycoprotein is very similar to the extracellular portion of its receptor counterpart, S locus receptor kinase, and molecular and genetic analyses have shown the interactions between the two play an important role in self-incompatibility in Brassica sp. (Cui et al., 2000). Our studies have suggested that WAKL7 is highly expressed in most tissues. It is interesting to note that WAKL7 expression is also regulated by INA induction and wound stresses. How and if WAKL7 functions with other WAK/WAKL RLK counterparts is currently not clear. Because it lacks the EGF repeats and a transmembrane domain, WAKL7 is not expected to be directly associated with the plasma membrane and could work as a mobile ligand. The combination of WAK/WAKL RLP members with the nonabbreviated WAK/WAKL members may provide a high capacity for Arabidopsis to respond to various environmental and developmental signals. Our results provide the first evidence, to our knowledge, that a WAKL RLP was highly expressed in various tissues. Our findings establish a foundation from which to pursue the mechanisms of how WAK/WAKL members function during plant development and environmental responses.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis ecotype Columbia seeds or npr1-2 seeds (Columbia) were cold treated at 4°C for 48 h and grown in pots on Metro-Mix 220 (Grace Sierra Horticultural Products, Milpitas, CA) in a growth chamber maintained at 22°C. Light (100 μmol m-2 s-1) was provided under an 16-h/8-h light/dark cycle.
Sequence Analysis
The WAKL1 to WAKL7 cluster was identified using the WAK1 cDNA sequence as a query in a BLAST (Altschul et al., 1990) search against the nonredundant nucleotide databases available at GenBank. Sequence translation, multiple sequence comparisons, and distance analyses were done using the MacVector software package (Genetics Computer Group, Madison, WI). Analysis of the intergenic regions for putative transfactor binding sites was done using the PlantCare (Rombauts et al., 1999) and PLACE (Higo et al., 1999) databases. Ratios of nonsynonymous to synonymous substitutions (dn/ds) were done using the SNAP method (Nei and Gojobori, 1986; Ota and Nei, 1994).
RNA Extraction and RT-PCR Analysis
Verification of the predicted intron splice sites was determined by sequencing cDNAs of each of the WAKL genes. Expression profiles of the WAKL genes were determined by RT-PCR. For expression profiles, the indicated organs were harvested and immediately frozen in liquid nitrogen. For INA treatment, wild-type and npr1-2 plants (just before bolting) were sprayed with a 1.5 mm INA solution and returned to the growth chamber. Whole plants were harvested 72 h later as above.
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA) according to the manufacturer's instructions. RT-PCR analysis was performed on 0.1 to 1.0 μg total RNA using the OneStep RT-PCR Kit (Qiagen USA). Reverse transcription was carried out at 50° for 30 min. Reaction mixtures were heated at 95° for 15 min to activate HotStar TaqDNA Polymerase. PCR was performed for 25 to 30 cycles under the following conditions: 94° for 1 min, 60° to 68° for 1 min (primer dependent), and 72° for 1 min, followed by a final extension at 72° for 10 min. Samples were visualized on 3% (w/v) agarose gels using a UV transilluminator. To eliminate the possibility of DNA contamination, RNA samples were treated with RNase-free DNase I (Panvera, Madison, WI). In addition, we performed control PCR experiments using RNA samples in which no RT was added. Under these conditions, no PCR bands were observed.
The following primers were employed to verify the predicted intron splice sites: WAKL1, WAKL1F (5′-TCATCAGCAGCAACACCTCCACCTCCAAT-3′) and WAKL1–1120R (5′-GTTTAAATAGCCAGAATAGCCCAAC-3′); WAKL2, WAKL2F (5′-AGCCAAGAAGGAAGAAACAAAATCTGCAA-3′) and WAKL2-1300R (5′-ACAGTGCCTTGACCGCCTTGCCCAAG-3′); WAKL3, WAKL3-540F (5′-GTAATGAAGTAGGTAACTCGCTTTGTAAC-3′) and WAKL3-1160R (5′-CTCCATTACGTTTTGAAGAACTTCA-3′); WAKL4, WAKL4F (5′-CTTATCTCTGTTTGGCGTCTCATCAGCAA-3′) and WAKL4-1300R; WAKL5, WAKL5-640F (5′-ATAGAGAGCTCAGGAGGAGATGGATGCAA-3′) and WAKL5-1100R (5′-TCCCAACAACTAAGAACAATAGTCCCATT-3′); WAKL6, WAKL6-840F (5′-ACATGACGCGGTACAGTTCTTACAG-3′) and W11-1910R (5′-ATCCAGTTGCCAATCCTCTTCCTTGACAGT-3′); and WAKL7, WAKL7-70F (5′-CATCTGCTCAAGATCTCACTGTCTT-3′), WAKL7-650R (5′-ATTTAAAGGAGAATATGCCTCGTCTGT-3′).
The reverse primers used in the intron verification were designed to anneal to DNA encoding the cytoplasmic protein kinase domain. Because of the high degree of sequence identity in these regions, these primers typically were not specific to a single WAKL gene. As such, we designed addition reverse primers that were specific to each WAKL gene. These primers were employed to establish WAKL expression profiles: WAKL1, WAKL1F, WAKL1R (5′-GTATGGTTCGTTACTGTCTGGGAGAGAGA); WAKL2, WAKL2F, WAKL2R (5′-CATTTAAACTTGAATACTTGTCGCTCGTC-3′); WAKL3, WAKL3F (5′-TTCCAAAATCTGACTTCTTCAGCCCTTAC-3′) and WAKL3R (5′-GTTACAAAGCGAGTTACCTACTTCATTAC-3′); WAKL4, WAKL4F, WAKL4R (5′-AGAGTAGTACTCAGTAGGATCTGGAAGAT-3′); WAKL5, WAKL5F (5′-CATCTCTCTTCCAGAAGGTAATAATGAAC-3′) and WAKL5R (5′-TTGCATCCATCTCCTCCTGAGCTCTCTAT-3′); WAKL6, WAKL6-660F (5′-AACACTAGTGCCACAAGAGGGAAAG-3′) and WAKL6-840R (5′-CTGTAAGAACTGTACCGCGTCATGT-3′); and WAKL7, WAKL7-70F (5′-CATCTGCTCAAGATCTCACTGTCTT-3′) and WAKL7-280R (5′-GCCCAAAGACATCGCTTGAGTCGTC-3′).
The following control primers were employed: PR-1, PR1F (5′-ACAACCAGGCACGAGGAGCGGTA-3′) and PR1R (5′-TCTCGTTCACATAATTCCCACGAGGATCA-3′); ACTIN, ACTF (5′-TATTGAATTCTTTTTGTGTGTTTGCAGC-3′) and ACTR (5′-TGGTGGATCCCAACCATGACACCATG-3′).
Protein Gel-Blot Analysis and Immunolocalization
WAKL6 polyclonal antibodies were raised in rabbits against a synthetic peptide corresponding to a unique region of the WAKL6 extracellular domain (GCESSCEDSKSSEE; Genemed Synthesis, South San Francisco, CA). Protein fractionation and protein gel blotting were performed as previously described (He et al., 1996; Lally et al., 2001). Nitrocellulose membranes with blotted proteins were blocked for 2 h in 1× casein solution (Vector Laboratories, Burlingame, CA), followed by overnight incubation in 1:500 dilutions of either preimmune or immune serum. Membranes were washed three times for 5 min each in 1× casein solution and developed using the Vectastain chemiluminescent detection systems (Vector Laboratories).
Promoter Fusions and GUS Activity
Promoter regions were defined as the intergenic sequences between the start site of each WAKL gene sequence and the end site of the preceding gene. The following primer sets were used to clone promoters of the noted lengths from genomic DNA using PCR (He et al., 1999). The introduced restriction sites used for cloning are indicated at the end of the primer sequences. WAKL1 (1,969 bp): 5′-CTCGAGTATCTCCATCCAACTCTCACTGTTG-3′ (XhoI), 5′-TCTAGACTTCTCCCTTTCCCTCTCTTTATTTTTCT-3′ (XbaI); WAKL2 (650 bp): 5′-GTCGACTCTTGAACCGCGTTCATCAGATTTTCTGA-3′ (SalI), 5′-GGATCCATCTTGTTCTTGTTCTCTCAACTTGCCA-3′ (BamH1); WAKL3 (917 bp): 5′-GTCGACTGATCAACTCAAAACACTGCATATGTATA-3′ (SalI), 5′-GGATCCATGAGCAGAGTTAGGACAGACGCTACAA-3′ (BamH1); WAKL4 (1,159 bp): 5′-GTCGACCGTCGTAAATCAGACTTTGCTTTAATA-3′ (SalI), 5′-AGATCTTGTTTCTCAACTCGTCAAGTCGGTCTT-3′ (BglII); WAKL5 (1,035 bp): 5′-GTCGACCCTCATCTGATGAAAAAATGTTATGT-3′ (SalI), 5′-GGATCCAATTCTTGTATTTTTACTTTGGTCCTTGA-3′ (BamH1); WAKL6 (1,501 bp): 5′-GTCGACTGGGTGCAGAGTTGCTTTCTTGACA-3′ (SalI), 5′-AGATCTTCTGCATTTTTTTAATCTGATTGATCT-3′ (BglII); and WAKL7 (511 bp): 5′-GTCGACTAAAGTGATATAGAGCCAGTCTTTCTAAA-3′ (SalI), 5′-GGATCCTTATGCTCTGTTTTTTTTTCTTTGTAGA-3′ (BamH1). The PCR products were inserted into the promoter-less GUS transformation vector pBI101.2. The resulting constructs were sequenced for confirmation and introduced into Arabidopsis wild-type plants (Col-0) via Agrobacterium sp.-mediated transformation as described before (Lally et al., 2001). More than 10 independent lines were generated for each construct, and at least three to six T2 or T3 homozygous lines were used for each promoter. To assay GUS activities, dissected samples were incubated with X-Gluc solution as described by Campisi et al. (1999). Samples were vacuum infiltrated in the X-Gluc solution for 10 min at room temperature and then incubated at 37°C for 14 h. The treated samples were transferred to 70% (v/v) ethanol. Photomicrographs were taken as described before (Lally et al., 2001).
Acknowledgments
We thank Dr. Maureen Whalen (San Francisco State University) and other members of the He Lab (David Lally, Boni Cruz, Emanuel Kwahk, and Judy Zhu) for helpful discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.028530.
This work was supported by the National Science Foundation (grant no. MCB 9985135). Additional support for this work was provided by the National Institutes of Health (grant nos. MBRS SCORE 5 SO6 GM52533 and RIMI P20 RR11805).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410 [DOI] [PubMed] [Google Scholar]
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD (2001) WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol 47: 197-206 [PubMed] [Google Scholar]
- Appella E, Robinson EA, Ullrich SJ, Stoppelli MP, Corti A, Cassani G, Blasi F (1987) The receptor-binding sequence of urokinase: a biological function for the growth-factor module of proteases. J Biol Chem 262: 4437-4440 [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR (1996) CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406-1409 [DOI] [PubMed] [Google Scholar]
- Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JD (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10: 1847-1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T (1999) Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J 17: 699-707 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57-63 [DOI] [PubMed] [Google Scholar]
- Cao X, Rogers SW, Butler J, Beevers L, Rogers JC (2000) Structural requirements for ligand binding by a probable plant vacuolar sorting receptor. Plant Cell 12: 493-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539-544 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401-5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575-585 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13: 171-201 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1999) Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol 50: 391-417 [DOI] [PubMed] [Google Scholar]
- Cui Y, Bi YM, Brugiere N, Arnoldo M, Rothstein SJ (2000) The S locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc Natl Acad Sci USA 97: 3713-3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1: 316-323 [DOI] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11: 495-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754-756 [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA 95: 7825-7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Mansfield J (1998) Early events in host-pathogen interactions. Curr Opin Plant Biol 2: 312-319 [DOI] [PubMed] [Google Scholar]
- Hardie DG (1999) Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50: 97-131 [DOI] [PubMed] [Google Scholar]
- Hay ED (1981) Collagen and embryonic development. In ED Hay, ed, Cell Biology of the Extracellular Matrix. Plenum Press, New York, pp 379-409
- He ZH, Cheeseman I, He D, Kohorn BD (1999) A cluster of five cell wall-associated receptor kinase genes, WAK1–5, are expressed in specific organs of Arabidopsis. Plant Mol Biol 39: 1189-1196 [DOI] [PubMed] [Google Scholar]
- He ZH, Fujiki M, Kohorn BD (1996) A cell wall-associated, receptor-like protein kinase. J Biol Chem 271: 19789-19793 [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14: 55-63 [DOI] [PubMed] [Google Scholar]
- Heldin CH (1995) Dimerization of cell surface receptors in signal transduction. Cell 80: 213-223 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Yeager M (1998) Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32: 415-435 [DOI] [PubMed] [Google Scholar]
- Hulbert SH (1997) Structure and evolution of the Rp1 complex conferring rust resistance in maize. Annu Rev Phytopathol 35: 293-310 [DOI] [PubMed] [Google Scholar]
- Hynes RO (1981) Fibronectin and its relation to cellular structure and behavior. In ED Hay, ed, Cell Biology of the Extracellular Matrix. Plenum Press, New York, pp 295-334
- Hynes RO (1987) Integrins: A family of cell surface receptors. Cell 48: 549-554 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925-1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108-117 [PMC free article] [PubMed] [Google Scholar]
- Kohorn B (1999) Shuffling the deck: plant signalling plays a club. Trends Cell Biol 9: 381-383 [DOI] [PubMed] [Google Scholar]
- Kohorn B (2000) Plasma membrane-cell wall contacts. Plant Physiol 124: 31-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Tanizawa K (1999) Involvement of epidermal growth factor-like domain of NELL proteins in the novel protein-protein interaction with protein kinase C. Biochem Biophys Res Commun 265: 752-757 [DOI] [PubMed] [Google Scholar]
- Kyriakis JM (1999) Making the connection: coupling of stress-activated ERK/MAPK (extracellular-signal-regulated kinase/mitogen-activated protein kinase) core signaling modules to extracellular stimuli and biological responses. Biochem Soc Symp 64: 29-48 [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH (2001) Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13: 1317-1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16: 223-233 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929-938 [DOI] [PubMed] [Google Scholar]
- Loh YT, Martin GB (1995) The Pto bacterial resistance gene and the Fen insecticide sensitivity gene encode functional protein kinases with serine/threonine specificity. Plant Physiol 108: 1735-1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122: 3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, de Vincente MC, Tanksley SD (1993) High resolution linkage analysis and physical characterization of the Pto bacterial resistance locus in tomato. Mol Plant-Microbe Interact 6: 26-34 [Google Scholar]
- McCarty DR, Chory J (2000) Conservation and innovation in plant signaling pathways. Cell 103: 201-209 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861-1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S (1997) Plant cell walls and the control of growth. Biochem Soc Trans 25: 204-214 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998a) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10: 1817-1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998b) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 11: 1833-1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8: 1113-1130 [DOI] [PubMed] [Google Scholar]
- Mu JH, Lee HS, Kao TH (1994) Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell 6: 709-721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697-1700 [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418-426 [DOI] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T (1997) Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA 94: 7799-7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H (1998) A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563-5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JDG (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099-2112 [PMC free article] [PubMed] [Google Scholar]
- Ota T, Nei M (1994) Variance and covariances of the numbers of synonymous and nonsynonymous substitutions per site. Mol Biol Evol 11: 613-619 [DOI] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK (2001) Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein AtGRP-3. J Biol Chem 276: 26688-26693 [DOI] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kossack KE, Goldstein C, Thomas CM, Jones DA, Harrison K, Wulff BBH, Jones JDG (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821-832 [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637-12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell R (1998) Cell walls: structures and signals. Curr Opin Plant Biol 1: 504-510 [DOI] [PubMed] [Google Scholar]
- Pont-Lezica RF, McNally JG, Pickard BG (1993) Wall-to-membrane linkers in onion epidermis: some hypotheses. Plant Cell Environ 16: 111-123 [Google Scholar]
- Quaranta V, Jones JCR (1991) The internal affairs of an integrin. Trends Cell Biol 1: 2-3 [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67: 687-699 [DOI] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27: 295-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald PC (1998) Resistance gene evolution. Curr Opin Plant Biol 1: 294-298 [DOI] [PubMed] [Google Scholar]
- Scheel D (1998) Resistance response physiology and signal transduction. Curr Opin Plant Biol 1: 305-310 [DOI] [PubMed] [Google Scholar]
- Schenk PW, Snaar-Jagalska BE (1999) Signal perception and transduction: the role of protein kinases. Biochim Biophys Acta 1449: 1-24 [DOI] [PubMed] [Google Scholar]
- Selander-Sunnerhagen M, Ullner M, Persson E, Teleman O, Stenflo J, Drakenberg T (1992) How an epidermal growth factor (EGF)-like domain binds calcium: high resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X. J Biol Chem 267: 19642-19649 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763-10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M, Ezaki B, He Z-H, Tong H, Osawa H, Baluska F, Volkmann D, Matsumoto H (2003) Aluminum induced gene-expression and protein localization of a cell wall-associated receptor protein kinase in Arabidopsis thaliana. Plant Physiol 132: 2256-2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwkowski MX, Schaefer G, Akita RW, Lofgren JA, Fitzpatrick VD, Nuijens A, Fendly BM, Cerione RA, Vandlen RL, Carraway KL (1994) Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J Biol Chem 269: 14661-14665 [PubMed] [Google Scholar]
- Song WY, Pi LY, Wang GL, Gardner J, Holsten T, Ronald P (1997) Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9: 1279-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhe LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21 Science 270: 1804-1806 [DOI] [PubMed] [Google Scholar]
- Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB (1991) Molecular cloning of a putative receptor protein kinase encoded at the self incompatibility locus of Brassica oleracea. Proc Natl Acad Sci USA 88: 8816-8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg Y, Muranyi A, Steen C, Thulin E, Drakenberg T, Stenflo J (1999) EGF-like module pair 3–4 in vitamin K-dependent protein S: modulation of calcium affinity of module 4 by module 3, and interaction with factor X. J Mol Biol 293: 653-665 [DOI] [PubMed] [Google Scholar]
- Stenflo J, Stenberg Y, Muranyi A (2000) Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim Biophys Acta 1477: 51-63 [DOI] [PubMed] [Google Scholar]
- Sunnerhagen MS, Persson E, Dahlqvist I, Drakenberg T, Stenflo J, Mayhew M, Robin M, Handford P, Tilley JW, Campbell ID (1993) The effect of aspartate hydroxylation on calcium binding to epidermal growth factor-like modules in coagulation factors IX and X. J Biol Chem 268: 23339-23344 [PubMed] [Google Scholar]
- Takasaki T, Katsunori H, Suzuki G, Watanabe M, Isogai A, Hinata K (2000) The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913-916 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE (1999) The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11: 393-406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, He ZH (2002) The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol 129: 455-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Kohorn B (2001) Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13: 303-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC (1993) Receptor-like protein kinase genes of Arabidopsis thaliana. Plant J 3: 451-456 [DOI] [PubMed] [Google Scholar]
- Wang GL, Ruan DL, Song WY, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS et al. (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10: 765-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste K, Keiber J (1998) The molecular basis of ethylene signaling in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 29: 1431-1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Chen C, Wang Z, Fan B, Chen Z (1999) A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J 18: 141-149 [Google Scholar]
- Yarden Y, Schlessinger J (1987) Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry 26: 1443-1451 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Katom A, Toriim KU, Komeda Y (1998) The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J 15: 301-310 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Shi J, Singh U, Wyatt SE, Bressan RA, Hasegawa PM, Carpita NC (1993) Enrichment of vitronectin- and fibronectin-like proteins in NaCl-adapted plant cells and evidence for their involvement in plasma membrane-cell wall adhesion. Plant J 3: 637-648 [PubMed] [Google Scholar]