Abstract
To identify cold-, drought-, high-salinity-, and/or abscisic acid (ABA)-inducible genes in rice (Oryza sativa), we prepared a rice cDNA microarray including about 1,700 independent cDNAs derived from cDNA libraries prepared from drought-, cold-, and high-salinity-treated rice plants. We confirmed stress-inducible expression of the candidate genes selected by microarray analysis using RNA gel-blot analysis and finally identified a total of 73 genes as stress inducible including 58 novel unreported genes in rice. Among them, 36, 62, 57, and 43 genes were induced by cold, drought, high salinity, and ABA, respectively. We observed a strong association in the expression of stress-responsive genes and found 15 genes that responded to all four treatments. Venn diagram analysis revealed greater cross talk between signaling pathways for drought, ABA, and high-salinity stresses than between signaling pathways for cold and ABA stresses or cold and high-salinity stresses in rice. The rice genome database search enabled us not only to identify possible known cis-acting elements in the promoter regions of several stress-inducible genes but also to expect the existence of novel cis-acting elements involved in stress-responsive gene expression in rice stress-inducible promoters. Comparative analysis of Arabidopsis and rice showed that among the 73 stress-inducible rice genes, 51 already have been reported in Arabidopsis with similar function or gene name. Transcriptome analysis revealed novel stress-inducible genes, suggesting some differences between Arabidopsis and rice in their response to stress.
Drought, high salinity, and low temperature are the most common environmental stress factors that influence plant growth and development and place major limits on plant productivity in cultivated areas worldwide. To overcome these limitations and improve crop yield under stress conditions, it is important to improve stress tolerance in crops. The responses of plants to various abiotic stresses have been important subjects of physiological studies (Levitt, 1980) and, more recently, of molecular and transgenic studies (Bajaj et al., 1999; Hasegawa et al., 2000; Zhang et al., 2000). The identification of novel genes, determination of their expression patterns in response to the stresses, and an improved understanding of their functions in stress adaptation will provide us the basis of effective engineering strategies to improve stress tolerance (Cushman and Bohnert, 2000).
A number of genes have been reported to be induced by drought, high-salinity, and low-temperature stresses, and their products are thought to function in stress tolerance and response (Bray, 1997; Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Many stress-inducible genes are responsive to both water stress and low temperature. Some of these genes are induced only by water stress, and several genes respond only to low temperature. Abscisic acid (ABA) is produced under such environmental stresses and plays an important role in the tolerance of plants to the stresses (Ingram and Bartels, 1996; Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2002). Analyses of the expression of these stress-inducible genes in Arabidopsis have indicated that ABA-dependent and -independent signal pathways function in the induction of the stress-inducible genes. These indicate the existence of complex regulatory mechanisms between perception of abiotic stress signals and gene expression (Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2002).
Plant science has entered a new era after the completion of the entire genomic sequence of Arabidopsis and rice (Oryza sativa), representing model systems for dicot and monocot plants, respectively. Research during this post genomic era is targeted to identify the specific functions of the predicted approximately 20,000 to 40,000 plant genes and their expression profiles. Sequencing projects have been producing not only increasing numbers of genomic sequences for many organisms but also large numbers of expressed sequence tags (ESTs) and full-length cDNA sequences for many plant species. There are many opportunities to use this sequence information to accelerate the progress toward a comprehensive understanding of genetic mechanisms that control plant growth and development and their response to the environments.
Several methods are currently being employed to analyze the profiles of gene expression in plants. DNA microarray technology is one of the most powerful techniques recently developed to bridge the gap between sequence information and functional genomics. Microarray technology allows for the determination of transcript abundance for many or all transcripts in a genome by comparing control and experimental states. The RNAs from different treatments are distinguished by the incorporation of different fluorescent labels (Schuchardt et al., 2000; Deyholos and Galbraith, 2001). With appropriate controls and repeated experiments, significant data are obtained on gene expression profiles under various conditions or in various organs. The microarray data have already been analyzed concerning a number of plant processes, such as seed development (Girke et al., 2000), wounding responses (Reymond et al., 2000), brassinosteroids (Goda et al., 2002), pathogen signaling (Schenk et al., 2000), nutrient-dependent changes in expression profiles (Wang et al., 2000; Thimm et al., 2001), and environmental stress responses (Kawasaki et al., 2001; Seki et al., 2001, 2002a, 2002b; Kreps et al., 2002; Ozturk et al., 2002). However, the DNA microarray data is not sufficient for determining correct expression profiles due to limited accuracy of the obtained data. Different results are sometimes obtained between the microarray and RNA gel-blot analyses as to the same genes. A combination of the microarray and RNA gel-blot analyses may be necessary to obtain high-degree accuracy of transcriptome data.
Rice, one of the most important crops, has now emerged as an ideal model species for the study of crop genomics due to its commercial value, relatively small genome size (approximately 430 Mb), diploid origin (2x = 24), and close relationship to other important cereal crops. Currently, over 75,000 rice EST sequences are available in the dbEST database (Reddy et al., 2002). Although computational analysis of these EST sequences allows annotation of putative gene functions through similarity searches in nucleic acid and protein databases, over two-thirds of the possible rice coding sequences have no matches in current public databases and, hence, remain classified as novel sequences with unknown functions. Even among the annotated sequences, the majority has only a low level of similarity to known genes, indicating that these sequences represent different genes with similar biochemical functions or that the sequences belong to the same gene family. Determination of the biological functions of these genes is among the greatest challenge for post-genomic research. A multidisciplinary approach using studies on structural similarities, expression profiles, and mutant phenotypes is required for assignment of gene functions. Considering the universal constraint to crop productivity represented by drought, low-temperature, and high-salinity conditions, we are conducting studies to improve stress tolerance in plants. We have initiated transcriptional monitoring of stress-inducible rice genes in response to dehydration, high salinity, low temperature, and ABA application. In this study, we report the analysis of transcriptional expression profiles of more than 1,700 rice cDNAs in response to a variety of abiotic stresses using cDNA microarray. We analyzed stress-inducible rice genes, cross talk among these abiotic stress responses, and cis-acting elements in stress-inducible promoters. We also discuss the similarity and difference of stress-inducible genes between rice and Arabidopsis.
RESULTS AND DISCUSSION
Rice cDNA Microarray
We have constructed three cDNA libraries prepared from rice after exposure to cold, drought, or high-salinity stress (Dubouzet et al., 2003). From these cDNA libraries, we isolated 1,501 independent rice cDNAs, 72% of which showed significant sequence similarity to previously characterized genes from a wide range of organisms. A cDNA microarray was prepared using these 1,501 cDNAs, 185 cDNA clones from the Rice Genome Project, 32 rice cDNAs of previously reported stress-inducible genes, the PCR-amplified fragment from lambda control template DNA fragment (TaKaRa, Kyoto) as an external control, and a mouse (Mus musculus) nicotinic acetylcholine receptor epsilon-subunit (nAChRE) gene, which has no substantial homology to any sequences in the rice database, to assess for nonspecific hybridization as a negative control. A method described previously (Eisen and Brown, 1999) was used to array PCR-amplified cDNA fragments onto glass slides. The resulting microarrays comprised a total of 1,720 transcripts spotted on each glass slide.
Identification of Cold-, Drought-, High-Salinity-, and/or ABA-Inducible Genes by cDNA Microarray
cDNA microarrays were hybridized with Cy3- and Cy5-labeled probe pairs of cold-treated plants plus unstressed plants, drought-stressed plants plus unstressed plants, high-salinity-stressed plants plus unstressed plants, and ABA-treated plants plus untreated plants prepared as described in experimental procedures. Hybridized microarrays were scanned by two separate laser channels for Cy3 and Cy5 emissions from each DNA element. The ratio of the two fluorescent signal intensities of each DNA element was then measured as a relative measure to determine changes in the differential expression of genes represented by cDNA spots on the microarrays. The Cy3/Cy5 signal intensities from different samples were normalized with the help of an exogenously added lambda control template DNA fragment that had been placed in different sections of the microarray slides to compensate for variable background levels.
Rice seedlings were exposed to cold, drought, or high-salinity stress or applied ABA for 5, 10, and 24 h. mRNAs from these stress- or ABA-treated plants and unstressed plants were used for preparation of Cy3- and Cy5-labeled cDNA probes, respectively. These cDNA probes were mixed together and hybridized with the cDNA microarray. To ascertain the reproducibility of the changes in transcripts of a particular sample, at least three separate hybridization experiments were performed with two separately prepared probes from the same RNA samples applied to the same copies of the cDNA microarray. In this study, we regarded genes with an expression ratio (stressed to unstressed) greater than 3-fold that of control genes at least for one time point in each stress treatment as stress-inducible genes.
Data analysis revealed that the genes on the microarray showed differential expression profiles in response to various abiotic stresses. A total of 141 genes were identified as stress-inducible genes. Among them, 64, 75, 48, and 45 genes are candidates for cold-, drought-, high-salinity-, and ABA-inducible genes, respectively. A significant number of transcripts were exclusively up-regulated by any one of the stresses, similar to the previous findings in Arabidopsis (Seki et al., 2002a). For example, 36/64 or 56% of the cold-inducible genes were cold specific. Similarly, 28/75 or 37% of the drought-inducible genes were drought specific and so on. However, northern-blot analysis revealed that these identified genes exhibited overlapping expression patterns, which means the number of transcripts specific to any one of the stresses remained negligible.
Cold-, Drought-, High-Salinity-, and/or ABA-Inducible Genes Identified by cDNA Microarray and RNA Gel-Blot Analyses
We selected 64, 75, 48, and 45 genes as candidates for cold-, drought-, high-salinity-, and ABA-inducible genes, respectively, using the cDNA microarray. Expression of these candidates was further analyzed by RNA gel-blot hybridization (Fig. 1). We confirmed that a total of 73 genes were stress inducible, and among them, 36, 62, 57, and 43 genes were cold-, drought-, high-salinity-, and ABA-inducible genes, respectively (Table I). Seki et al. (2001) found that a similar degree of variability was observed for cold- and drought stress-regulated transcripts using an Arabidopsis full-length cDNA microarray (1,300). They identified around 50% (44) of the genes as real drought-inducible genes after performing RNA gel-blot analysis of 80 putative drought-inducible genes selected by cDNA microarray analysis. The inconsistency between microarray and RNA gel-blot results on several genes may be due to the weak expression of genes, high background, and/or cross-hybridization.
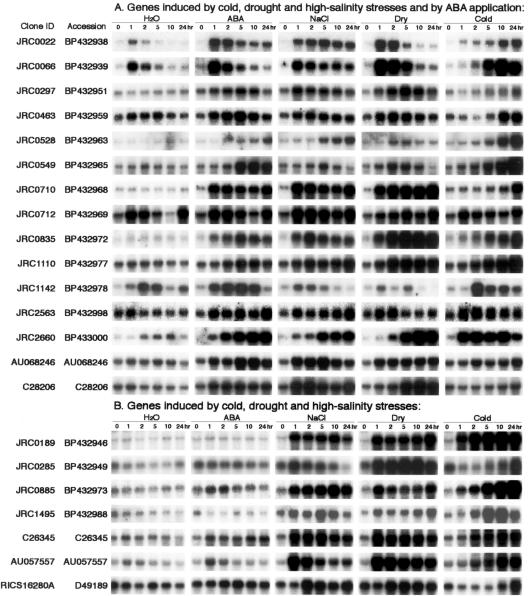
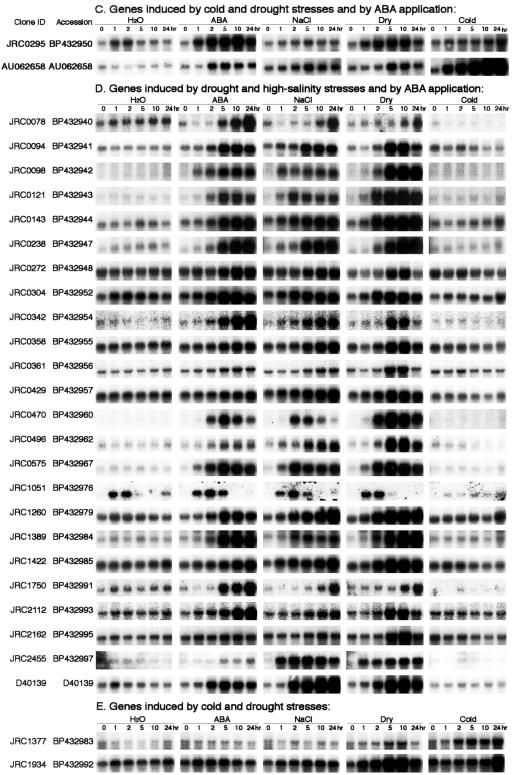
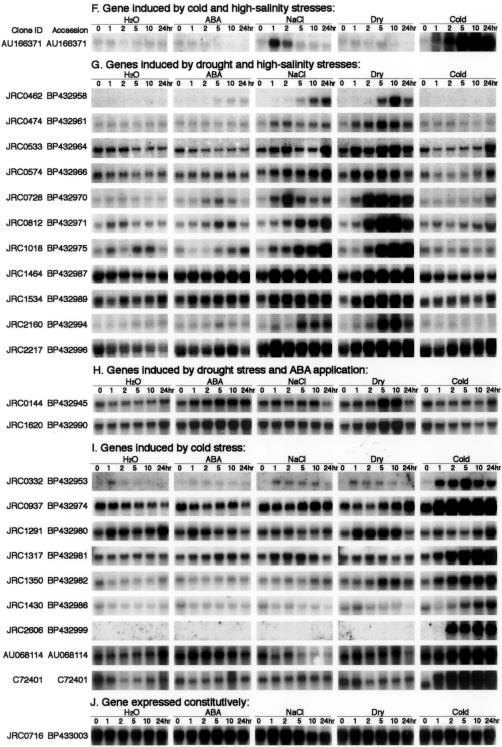
Figure 1.
RNA gel-blot analysis of stress-inducible genes. Each lane was loaded with 10 μg of total RNA isolated from 2-week-old rice seedlings that were exposed to water, dehydration, 250 mm NaCl, 100 μm ABA, and 4°C cold treatment for 1, 2, 5, 10, and 24 h. RNA was analyzed by gel-blot hybridization with gene-specific probes of selected stress-inducible clones by rice cDNA microarray. Stress-inducible clones were classified into various groups on the basis of their expression patterns in RNA gel-blot analysis under each stress treatment. Some of the inducible genes were induced by all four stress treatments; some of them were up-regulated by cold, drought, and high salinity; some of them were induced by drought and high salinity; some of them were induced by cold only, etc.
Table I.
No. of clones involved in different functional groups up-regulated by cold, drought, and/or high-salinity stress and/or ABA application
| Functional Category | No. | Description |
|---|---|---|
| Transcriptional factor | 6 | bZIP DNA-binding protein, C2H2-type zinc finger DNA-binding protein, C3HC4-type RING finger protein, Myb type DNA-binding protein, andNAC-type DNA-binding protein |
| Receptor-like protein kinase | 1 | Receptor-like protein kinase |
| Protein phosphatase | 1 | Protein phosphatase 2C |
| Compatible solutes | 6 | LEA protein, dehydrin, lectin |
| Detoxification | 3 | Catalase, O-methyltransferase, aldehyde dehydrogenase |
| Photosynthesis | 1 | Chlorophyll a/b-binding protein |
| Membrane protein | 1 | Chloroplast membrane protein |
| Carbohydrate metabolism | 7 | Glycoside hydrolase, glycosyl transferase, phosphoglycerate kinase, Pyruvate dehydrogenase kinase 1, trehalose-6-phosphate phosphatase, UDP-Glc 4-epimerase, and carboxyphosphonoenolpyruvate mutase |
| Electron transport system | 1 | Thioredoxin |
| Amino acid metabolism | 2 | 4-Hydroxyphenylpyruvate dioxygenase and S-adenosylmethionine decarboxylase |
| Fatty acid metabolism | 3 | Choline kinase, lipase, and lipoxygenase |
| Nucleotide synthesis | 1 | Adenylate kinase |
| Hormone biosynthesis | 1 | Zeaxanthin epoxidase |
| F-box protein | 1 | F-box protein |
| Protease inhibitor | 1 | Protease inhibitor |
| Protease | 1 | Papain Cys protease |
| Dehydrogenase | 3 | 3-Hydroxyacyl-CoA dehydrogenase, dihydroorotate dehydrogenase, and glutamate dehydrogenase |
| Iron homeostasis | 2 | Ferritin, metallothionein-like type 2 |
| Cytoskeleton | 2 | Actin, actin-deporimerizing factor |
| Transporter | 1 | Sugar transporter |
| Unknown protein | 28 | Unknown protein |
The number of cold stress-inducible genes was less than that of either drought- or high-salinity-inducible genes. These results are consistent with earlier findings by Seki et al. (2001, 2002) that the number of cold-inducible genes was less than that of drought- and high-salinity-inducible genes in Arabidopsis. However, Kreps et al. (2002) reported that cold induced nearly twice as many changes as high-salinity stress in Arabidopsis. These inconsistencies may be attributed to the biological differences among plant species used, stress treatment condition, their response to abiotic stresses, and/or detection methodologies. The complete list of identified cold-, drought-, high-salinity-, and/or ABA-inducible genes is available (Supplemental Table available in the online version of this article at http://www.plantphysiol.org).
In total, 73 stress-inducible genes were identified by both cDNA microarray and RNA gel-blot analysis. Fifteen (20%) of these genes are known and have been reported previously as responsive to abiotic stress in rice. These genes included Asr1, dehydrin, calmodulin, catalase, LEA protein, LIP9, LIP19, metallothionein-like protein, Myb transcription factor, RAB-16C, salT, WSI76, and WSI724 (Supplemental Table). Fifty-eight (80%) of our up-regulated genes have not been reported previously as stress-inducible genes in rice. These genes included aldehyde dehydrogenase, lipoxygenase, NAC6 transcription factor, O-methyltransferase, RING finger protein, sugar transporter, zinc finger protein, etc. (Supplemental Table). We also recognized 28 constitutively expressed genes with almost the same expression levels under each stress treatment after RNA gel-blot analysis (Supplemental Table). These constitutively expressed genes may be useful as internal control genes in the cDNA microarray and RNA gel-blot analyses.
Relationship between Each Stress
Stress-inducible genes identified by RNA gel-blot analysis have been classified into groups on the basis of their expression patterns. A comparison of expression profiles from all four stresses revealed 15 genes induced by any of four stresses that are likely to be regulated by the same or overlapping stress signaling pathways (Fig. 1A). Among these genes, we found five well-known rice stress-inducible genes: Asr1, LIP9, OsNAC6, salT, and WSI724 (Aguan et al., 1991; Takahashi et al., 1994; Garcia et al., 1998; Vaidyanathan et al., 1999; Kikuchi et al., 2000). Rice cDNA clones JRC (JIRCAS Rice cDNA clone) 0549, JRC0712, JRC0835, AU068246, and C28206 that encode Glycoside hydrolase; pyruvate dehydrogenase kinase 1; Zeaxanthin epoxidase; OsABA2; Papain Cys protease; and UDP-Gal-4-epimerase, respectively, and five cDNAs (JRC0297, JRC0710, JRC2563, JRC1110, and JRC1142), whose function is unknown, were also included in this group.
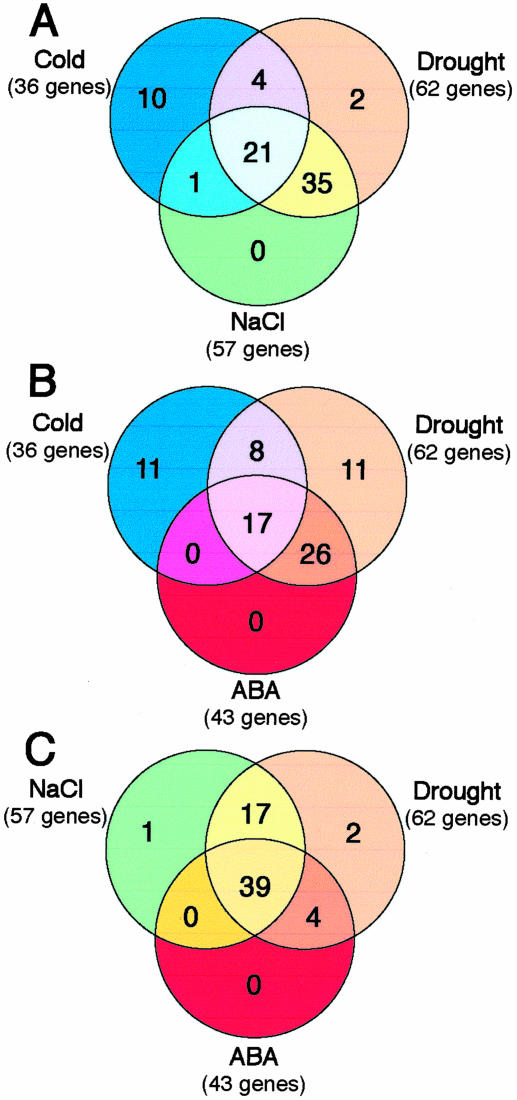
Based on Venn diagram analysis, we analyzed differences and cross talk of gene expression among cold-, drought-, and high-salinity stress responses and ABA response in rice. As shown in Figure 2, we identified 36, 62, 57, and 43 genes as cold-, drought-, high-salinity-, and ABA-inducible genes by RNA gel-blot analysis, respectively. Fifty-six genes were induced by both drought and high salinity, 25 genes were induced by both drought and cold stress, and 22 genes were induced by cold and high-salinity stresses. Similarly, 43 genes were up-regulated by both drought and ABA application, whereas only 17 genes were identified as cold- and ABA-inducible genes. More than 98% of the high-salinity- and 100% of ABA-inducible genes were also induced by drought stress, which indicates a strong relationship not only between drought and high-salinity responses but also between drought and ABA responses. These results indicate the existence of a substantial common regulatory system or a greater cross talk between drought and high-salinity stress and between drought and the ABA signaling process than that between cold and high-salinity stress or between cold and the ABA signaling process. Our results in rice are consistent with previous observations on the overlap of drought- and high-salinity-responsive gene expression in Arabidopsis (Shinozaki and Yamaguchi-Shinozaki, 1999, 2000; Seki et al., 2002a, 2002b). However, our results were contradictory to the observations made by Kreps et al. (2002) in cold- and high-salinity-stressed Arabidopsis and to those made by Ozturk et al. (2002) in drought- and high-salinity-stressed barley (Hordeum vulgare). In their studies, the majority of stress-regulated changes appeared to be stress specific and not part of a general stress response common to cold, drought, and high-salinity stress. These discrepancies may be attributable to the difference in plant species used, stress treatment, plant growth condition, array strategy, or detection methodologies.
Figure 2.
Venn diagrams showing the classification of genes inducible by cold, drought, and high-salinity stresses and by ABA application identified on the basis of microarray and RNA gel-blot analyses: In total, 36 cold-inducible, 62 drought-inducible, 57 high-salinity-inducible, and 43 ABA-inducible genes were identified by cDNA microarray and confirmed by RNA gel-blot analysis. The identified genes were classified into various groups, such as cold-stress-inducible and drought-stress-inducible, genes that were up-regulated by cold, drought, and high-salinity stresses; genes that were induced by cold or drought stress and ABA application; genes that were up-regulated by cold and drought stresses; and genes that were induced by drought and high-salinity stresses. A, Intersection of genes that were up-regulated by cold stress with those that were either up-regulated by drought stress or high-salinity stress. B, Intersection of genes that were up-regulated by cold stress with those that were either up-regulated by drought stress or ABA application. C, Intersection of genes that were up-regulated by high-salinity stress with those that were either up-regulated by drought stress or ABA application.
Characterization of Drought-, Cold-, High-Salinity-, and/or ABA-Inducible Genes
We identified 36 cold stress-inducible, 62 drought stress-inducible, 57 high-salinity stress-inducible, and 43 ABA-inducible genes in our study (Fig. 2) The list for these drought-, cold-, high-salinity- and ABA-inducible genes identified are available (Supplemental Table). Their gene products were classified into two groups. The first group consists of functional proteins or proteins that probably function in stress tolerance. They were late embryogenesis-abundant (LEA) proteins, water stress-inducible (WSI76 and WSI724) proteins, dehydration-inducible (RD and ERD) proteins, cold-acclimation (COR) proteins, osmoprotectant biosynthesis-related proteins, carbohydrate metabolism-related proteins, sugar transporters, detoxification enzymes, protease inhibitor, catalase, ferritin, salT proteins, etc. (Table I; Supplemental Table). LEA proteins are thought to play a role as desiccation protectants and have been shown to be involved in protecting macromolecules such as enzymes and lipids (Moons et al., 1997; Shinozaki and Yamaguchi-Shinozaki, 1999). Several COR and RD genes encode hydrophilic polypeptides, which hypothetically play a role in protecting cells from low temperature and water deficit such as drought and high-salinity stress conditions (Artus et al., 1996; Shinozaki and Yamaguchi-Shinozaki, 1999). Sugar transporters are thought to function in the transport of sugars through plasma membranes and tonoplast to adjust the osmotic pressure under stress conditions. The protease inhibitor may perform a defensive role against the proteases. Ferritin may play a role to protect cells from oxidative damage caused by various stresses by sequestering intracellular iron involved in the generation of various reactive hydroxyl radicals through a Fenton reaction (Bajaj et al., 1999). Fatty acid metabolism-related genes may participate in the repair of stress-induced damage in membranes, perhaps to regulate permeability to toxic ions and the fluidity of the membrane (Torres-Schumann et al., 1992; Holmberg and Bülow, 1998). Genes encoding plant catalases are expected to play an important role in the antioxidant defense in response to environmental and physiological oxidative stress (Scandalios, 1990; Iwamoto et al., 1998).
The second group consists of regulatory proteins, that is, protein factors involved in further regulation of signal transduction and gene expression that probably function in stress responses. They are various kinds of transcription factors such as RING finger, zinc finger, MYB, NAC, and basic region/Leu zipper motif (bZIP) family transcription factors; protein kinases; protein phosphatases; and enzymes involved in phospholipid metabolism (Table I; Supplemental Table). It appears that the RING finger transcriptional factors are involved in mediating protein-protein interactions and, in some cases, multiprotein complexes, which might depend on the presence of other proteins and/or domains (Saurin et al., 1996). The MYB family is one of the largest families of transcriptional factors characterized in plants. MYB-related transcriptional activators are believed to be involved in the regulation of secondary metabolism, control of cellular morphogenesis, and regulation of meristem formation and the cell cycle (Jin and Martin, 1999). bZIP transcription factors regulate diverse biological processes in plants such as pathogen defense, light and stress signaling, seed maturation, and flower development (Martinez-Garcia et al., 1998; Jakoby et al., 2002). Zinc finger proteins have been shown to act as the DNA-binding domain of transcription factors (Margolin et al., 1994; Cook et al., 1999). NACs are a family of genes specific to plants and are found to play a role in a diverse set of developmental processes including formation and maintenance of shoot apical meristem and floral morphogenesis (Kikuchi et al., 2000; Xie et al., 2000). Among protein kinase genes, we found the receptor-like protein kinase gene. These regulatory proteins are thought to function in further regulating various functional genes under stress conditions.
A gene involved in biosynthesis of ABA have also been identified as a stress-inducible gene in rice. Aldehyde dehydrogenase gene, genes related to secondary metabolism, genes involved in various cellular metabolic processes, genes encoding membrane protein, and genes related to O-methyltransferase and metallothionein-like protein were also identified as abiotic stress-inducible genes (Table I; Supplemental Table). The functions of most of these genes are not fully understood at present. We also found many stress-inducible genes whose functions are unknown.
Various Expression Profiles of Stress-Inducible Genes during Stress Treatment
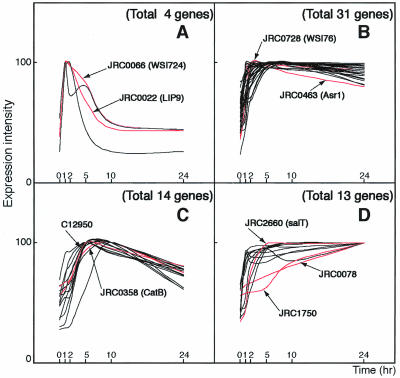
Expression profiles of cold-, drought-, high-salinity-, and/or ABA-inducible genes were classified into several gene groups based on RNA gel-blot analysis. Analysis of gene expression profiles during drought treatment showed the existence of four groups with different expression profiles (Fig. 3). In one group containing four genes, gene induction was rapid and transient in response to drought treatment, reached a maximum at 1 to 2 h, and then decreased (Fig. 3A). Among the four genes, we found a cDNA (JRC0022) showing sequence identity with low temperature-inducible protein (LIP9), a cDNA (JRC0066) having identity with water stress-inducible protein (WSI724), and a cDNA (JRC1142) whose function is unknown. In the second group consisting of 31 genes, their expression increased after drought treatment within 1 to 2 h, and the level was kept relatively constant (Fig. 3B). This group contained a cDNA (JRC0728) having sequence similarity to a water stress-inducible protein (WSI76), a cDNA (JRC0463) related to a rice ABA- and stress-inducible protein (Asr1), and a number of cDNAs with unknown functions. In the third group, consisting of 14 genes, gene expression was induced after drought stress, reached a maximum at 5 or 10 h, and then decreased (Fig. 3C). The genes in this group contained a cDNA (JRC0358) showing identity with rice CatB gene for catalase, a cDNA (C12952) having sequence homology to a putative protein with similarity to rice phi-1, etc. In the fourth group consisting of 13 genes, their expression increased slowly and gradually after drought treatment and reached a maximum at 24 h of stress (Fig. 3D). This group included a cDNA (JRC0078) having sequence similarity to water stress-inducible protein, dehydrin, a cDNA (JRC1750) related to rice LEA protein, a cDNA (JRC2660) showing sequence identity to salt-induced gene (salT), and many other genes with unknown functions.
Figure 3.
Classification of drought-inducible genes on the basis of expression patterns: Stress-inducible genes were divided into four groups on the basis of their expression patterns under drought stress. A, In the first group consisting of LIP9 and WSI724, expression was induced rapidly after drought stress, reached a maximum at 1 to 2 h after stress, and then decreased. B, In the second group consisting of Asr1 and WSI76, expression was induced within 1 to 2 h of drought stress, and the level was kept relatively constant. C, In the third group consisting of CatB, expression was induced after drought stress, reached a maximum at 5 or 10 h, and then declined. D, In the fourth group consisting of salT, expression was induced after drought stress and reached a maximum at 24 h.
Analysis of expression profiles of genes during cold stress treatment also exhibited the existence of four groups showing different expression profiles (Supplemental Fig. 1, available in the online version of this article at http://www.plantphysiol.org). In one group containing a cDNA JRC1142 encoding unknown protein, gene induction was rapid and transient after cold stress treatment, reached a maximum at 1 to 2 h, and then decreased. In the second group, containing five genes, expression increased after cold treatment within 1 to 2 h, and the level was kept relatively constant. In this group, we found two cDNAs (JRC2606 and JRC2660) showing sequence similarity with Glu dehydrogenase 2 and salT, respectively. In the third group, consisting of four genes, gene expression was induced after cold treatment, reached a maximum at 5 or 10 h, and then decreased. The genes in this group were WSI724, zinc finger transcription factor, MYB family transcription factor, and a cDNA (JRC0463) showing sequence similarity with an ABA- and stress-inducible protein, Asr1. In the fourth group, consisting of 26 genes, expression increased slowly and gradually after cold stress and reached a maximum at 24 h of stress treatment. This group consisted of well-known stress-related genes in rice such as LIP9, LIP19, NAC6, and a number of genes with unknown functions. These genes may function as regulatory protein factors involved in the regulation of signal transduction and gene expression functioning in stress responses. Like cold- and drought-inducible genes, the analysis of expression patterns of high salt- and ABA-inducible genes during stress treatments also exhibited the existence of four groups showing different expression profiles (Supplemental Figs. 2 and 3).
Promoter Analysis of Stress-Inducible Genes
Cis- and trans-acting elements involved in dehydration-induced gene expression have been analyzed extensively (Ingram and Bartels, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1999). The dehydration-responsive element (DRE) with the core sequence A/GCCGAC was identified as a cis-acting promoter element in regulating gene expression in response to drought, high-salinity, and cold stresses in Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 1994). A similar motif was identified as CRT (C-repeat) and LTRE (low-temperature-responsive element) in cold-inducible genes (Baker et al., 1994; Jiang et al., 1996). Many stress-inducible genes are also induced by exogenous application of ABA treatment. These genes contain potential ABA-responsive elements (ABREs) in their promoter regions (Ingram and Bartels, 1996; Shinozaki and Yamaguchi-Shinozaki, 2000). In this study, 73 genes were identified as cold-, drought-, ABA-, and/or high-salinity-inducible genes. We compared the 5′ sequence data of identified stress-inducible genes with those of genomic sequences of rice using GenBank and the Rice Genome Project databases. Among them, 69 stress-inducible genes were identified having complete genomic sequences including promoter regions in the databases. Table II summarizes ABRE and DRE sequences observed in the 27 drought-, cold-, ABA-, and/or high-salinity-inducible genes identified by RNA gel-blot analyses. Twenty-two genes contained DRE (A/GCCGAC) in their promoter regions, whereas eight genes contained ABRE (PyACGTGG/TC) in their promoters, suggesting that they were ABA inducible. The rest of the genes did not contain any ABRE and/or DRE motifs within the 1-kb region of their promoter sequences.
Table II.
ABRE and DRE sequences observed in the promoter regions of stress-inducible genes identified by cDNA microarray and RNA gd-blot analyses
| Clone | Putative Gene Identification | ABRE (PyACGTG(G/T)C) | DRE (A/GCCGACNN) |
|---|---|---|---|
| JRC0022 | Dehydrin | - | tGCCGACac (−310 to −302) |
| cGCCGACct (−384 to −376) | |||
| JRC0066 | Dehydrin | - | tGCCGACgc (−913 to −905) |
| tGCCGACta (−959 to −951) | |||
| JRC0098 | Unknown protein | cACGTGGa (−201 to −194) | gGCCGACca (−170 to −162) |
| cACGTGTc (−243 to −236) | |||
| JRC0121 | Protein phosphatase 2C | - | gaGTCGGTg (−808 to −816) |
| tGCCGACga (−975 to −967) | |||
| JRC0144 | Unknown protein | cACGTGTc (−533 to −526) | tcGTCGGCt (−160 to −168) |
| JRC0189 | Unknown protein | - | caGTCGGCc (−323 to −331) |
| cGCCGACcc (−794 to −786) | |||
| cGCCGACca (−881 to −873) | |||
| JRC0272 | Unknown protein | - | aACCGACaa (−280 to −272) |
| tGCCGACtc (−376 to −368) | |||
| JRC0304 | Unknown protein | cACGTGTc (−232 to −225) | acGTCGGCt (−639 to −647) |
| cACGTGGc (−659 to −652) | |||
| JRC0332 | C2H2-type zinc finger DNA-binding protein | - | aACCGACga (−658 to −650) |
| JRC0361 | Unknown protein | cACGTGGc (−304 to −297) | - |
| JRC0462 | O-methyltransferase | - | tACCGACca (−688 to −680) |
| JRC0463 | Unknown protein | - | tgGTCGGTg (−651 to −659) |
| tGCCGACac (−715 to −707) | |||
| JRC0528 | NAC-type DNA-binding protein | cACGTGTc (−166 to −159) | - |
| cACGTGTc (−258 to −251) | |||
| cACGTGTc (−444 to −437) | |||
| JRC0533 | Lipoxygenase | - | cGCCGACcg (−726 to −718) |
| JRC0710 | Unknown protein | cACGTGGc (−839 to −832) | - |
| JRC0812 | Lipase | - | aACCGACgg (−149 to −141) |
| JRC0885 | Unknown protein | - | atGTCGGCt (−239 to −247) |
| tcGTCGGCa (−397 to −405) | |||
| JRC1260 | 4-hydroxyphenulpyruvate dioxygenase | - | cGCCGACgc (−370 to −362) |
| cGCCGACct (−454 to −446) | |||
| gGCCGACgc (−508 to −500) | |||
| cGCCGACgc (−517 to −509) | |||
| gcGTCGGCg (−621 to −629) | |||
| cGCCGACgc (−790 to −782) | |||
| JRC1389 | Dehydrin | - | gcGTCGGCt (−864 to −872) |
| JRC1430 | Unknown protein | - | tGCCGACca (−868 to −860) |
| JRC1750 | LEA protein | tACGTGTc (−340 to −333) | - |
| tACGTGGa (−371 to −364) | |||
| cACGTGTc (−556 to −549) | |||
| JRC2160 | Unknown protein | - | aACCGACtt (−106 to −98) |
| JRC2455 | Sugar transporter | - | aGCCGACta (−784 to −776) |
| JRC2563 | Unknown protein | tACGTGGc (−217 to −210) | - |
| AU068246 | Papain cysteine protease | - | cGCCGACcc (−163 to −155) |
| C12952 | Unknown protein | - | cGCCGACcg (−623 to −615) |
| D49189 | Ferritin | - | gACCGACtc (−254 to −246) |
We identified 36 cold-inducible, 62 drought-inducible, 57 high-salinity-inducible, and 43 ABA-inducible genes in our study, and obtained the promoter sequence for 33, 58, 55, and 40 genes, respectively. Among them, several genes including JRC0078, JRC0358, JRC0575, JRC0728, JRC1018, JRC1317, JRC1495, and JRC2660 did not contain any ABRE and/or DRE or DRE-related CCGAC core motif in their promoters. These results suggest the existence of novel cis-acting elements involved in stress-inducible gene expression in their promoters. The studies are underway not only to analyze the cis-acting elements involved in drought-, cold-, high-salinity-, or ABA-responsive expression in the promoter regions of these selected genes but also to identify early responsive and strong stress-inducible promoters in rice crop.
Comparison of Identified Stress-Inducible Genes in Rice with Those of Arabidopsis
Rice and Arabidopsis are model plants for monocot and dicot crops, respectively. Comparative analysis of two species is quite useful not only for understanding the genomic similarities across the monocot and dicot species but also for discovering important genes for genetic engineering for various kinds of crops. We compared the identified stress-inducible genes of rice with those of Arabidopsis, which have been reported as connected to abiotic stress responses. These Arabidopsis genes are thought to be involved in plant responses or tolerance to environmental stresses. Our comparison of rice and Arabidopsis stress-inducible genes revealed a considerable level of similarities in stress responses between the two genomes at a molecular level. Among 73 identified stress-inducible genes in rice, 51 (70%) with similar functions or gene names have already been reported in Arabidopsis (Supplemental Table). Our analysis has revealed that rice has a lot of stress-inducible genes in common with Arabidopsis, even though these two plants have evolved separately for a million years. Common stress-inducible genes include dehydration-inducible genes (RD and ERD), cold acclimation proteins (COR and KIN), dehydrins, LEA proteins, aldehyde dehydrogenase, MYB transcription factor, NAC transcription factor, zinc finger transcription factor, lipoxygenase, protein phosphatase 2C, receptor-like protein kinase 4, sugar transporter, and metallothionein-like proteins. All these genes have been found to be up-regulated in response to at least one of the abiotic stresses in rice and reported as stress-inducible genes in Arabidopsis. Previous studies have also shown some collinearity or synteny between rice and Arabidopsis genomes at both the genetic and physical map levels (Liu et al., 2001).
The analysis of our transcriptomal data has revealed some differences between two plant species regarding many genes responsive to environmental stress. There are a number of rice genes that have been reported with a similar function or gene name in Arabidopsis but not documented as genes responsive to abiotic stress in Arabidopsis. These consisted of pyruvate dehydrogenase kinase 1, carboxyphosphonoenolpyruvate mutase, S-adenosyl-Met decarboxylase 2, adenylate kinase, chloroplast membrane protein, hydrolase, and a number of proteins with unknown functions.
CONCLUSIONS
In this study, we identified many abiotic stress-inducible genes in rice by microarray and RNA gel-blot analyses. However, the functions of a number of these genes remain unknown. It is important to analyze the function of stress-inducible genes, not only for further understanding of molecular mechanisms of stress tolerance and responses of higher plants but also for improving the stress tolerance of crops by gene manipulation. cDNA microarray in combination with RNA gel-blot analysis confirmed the stress-responsive expression of a number of previously reported stress-inducible genes in rice like Asr1, dehydrin, LIP9, LIP19, NAC6, LEA protein, salT, WSI76, WSI724, etc. Several genes such as homologs of Arabidopsis ERD15, barley ABA-responsive protein, Lotus (Loutus japonicus) RING-finger protein, pea (Pisum sativum) actin, wheat (Triticum aestivum) WCOR719, etc. also have been identified as stress-inducible genes in rice. Our results indicate greater cross talk between the signaling processes for drought stress and high-salinity stress or for drought stress and ABA application than between the signaling processes for cold stress and high-salinity stress or for cold stress and ABA application, which are consistent with a previous observation on the overlap of stress-responsive gene expression in Arabidopsis (Seki et al., 2002a, 2002b). We compared the identified stress-inducible genes of rice with those of Arabidopsis to find that rice has many stress-inducible genes in common with those of Arabidopsis. These results indicate that there are similar molecular mechanisms of stress tolerance and responses between dicots and monocots. Analysis of our data also enabled us to identify a number of promoters and possible cis-acting elements of several stress-inducible genes responsive to a variety of environmental stresses. Further analysis of these stress-inducible genes using transgenic plants will provide more information about not only the functions of the stress-inducible genes involved in stress tolerance but also novel cis-acting promoter elements involved in cold-, drought-, high-salinity-, or ABA-responsive gene expression in the promoter regions of these stress-inducible rice genes.
MATERIALS AND METHODS
Plant Materials and Stress Treatments
Seeds of rice (Oryza sativa) var. Nipponbare were grown under controlled conditions having 28°C day/25°C night temperatures, 12-h-light/12-h-dark cycle, and 83% relative humidity. After 2 weeks of germination, seedlings were exposed to cold (4°C), drought, or high-salinity stress (250 mm), or treated with 100 μm ABA. Root and leaf tissues were harvested after 5, 10, and 24 h of stress treatment, frozen in liquid nitrogen, and stored at -80°C for further analysis. Control plants were harvested at the same time as the stressed plants.
cDNA Clones
In the microarray analysis, we used rice cDNA clones isolated from three cDNA libraries constructed from cold-, drought-, and high-salinity-stressed plants (Dubouzet et al., 2003) The nucleotide sequences of individual cDNA clones were determined on both strands, and their homologies were examined by comparison with those in the GenBank/EMBL/DDBJ database using a BLAST search program.
Preparation of cDNA Microarrays
A total of 1,718 rice clones were used in the cDNA microarray analysis. To monitor the detection sensitivity limit, the PCR-amplified fragment from lambda control template DNA fragment (TX803, TaKaRa) was used as an external control, whereas DNA derived from the mouse (Mus musculus) nAChRE (nicotinic acetylcholine receptor epsilon-subunit) gene was used as a negative control. Inserts of cDNA clones were amplified by PCR using primer pairs as described before (Seki et al., 2002a). The yield and amplification quality of PCR products was confirmed by separating one aliquot of each finished reaction on 1% (w/v) agarose gel as described before. PCR fragments were arrayed from 384-well microtiter plates onto a poly-l-Lys-coated micro slide glass using a microarray stamping machine (model SPBIO2000, Hitachi Software Engineering Co. Ltd., Tokyo) as described before (Seki et al., 2002a). Slides were post-processed according to the manufacturer's protocols (Telechem International Inc., Sunnyvale, CA) and using the detailed procedure previously described by Seki et al. (2002a).
Microarray Analysis
Total RNA was isolated using TRIZOL Reagent (Life Technologies, Rockville, MD). Poly(A+) RNA was isolated using an mRNA isolation kit (Miltenyi Biotec, Auburn, CA) as described by Seki et al. (2002a). For the cDNA microarray analysis, each mRNA sample was reverse transcribed in the presence of Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia, Piscataway, NJ). Hybridization was carried out as described before (Seki et al., 2002a).
Scanning, Data Analysis, and Quantification
Slides were scanned using a ScanArray 4000 (GSI Lumonics, Oxnard, CA) and analyzed by ImaGene III Software (BioDiscovery, Los Angeles). For microarray data analysis, image analysis and signal quantification were performed with QuantArray version 2.0 (GSI Lumonics). Background fluorescence was calculated on the basis of the fluorescence signal of the negative control gene, the mouse nAChRE gene. Lambda control template DNA fragment (TX803, TaKaRa) was used as an external control to equalize hybridization signals generated from different samples. Transcript regulation was expressed as the ratio of intensities between stress and control plants. Changes in signal intensity between stress and control experiments exceeding a 3-fold or higher difference in repeated experiments were considered significant.
RNA Gel-Blot Analysis
Total RNA was isolated from 2-week-old rice seedlings, exposed to cold, drought, or salt stress, or applied ABA for 1, 2, 5, 10, or 24 h. RNA gel-blot hybridization was performed as described before (Liu et al., 1998). The PCR-amplified fragments prepared from the rice cDNAs were used as probes for RNA gel-blot hybridization. The probes were labeled with a 32P-dCTP using a BcaBEST DNA labeling kit (TaKaRa) according to the manufacturer's protocol. The expression intensity of each gene was quantified by densitometry as an absolute value from the RNA gel blot and plotted over each time course using a computer program NIH Image 1.62f (National Institutes of Health, Bethesda, MD). Initially, the absolute value in each stress was converted into relative expression value by taking the maximum level of the transcript as 100. Relative values for remaining time courses were calculated as: the absolute value of the remaining time course/absolute value of the time course with maximum expression × 100.
Promoter Analysis
Sequence data of identified stress-inducible clones showing increased expression by microarray and RNA gel-blot analysis were compared with those of genomic sequences of rice using GenBank and MAFF databases. Among them, a number of stress-inducible genes were identified that had complete genomic sequences including promoter regions and their possible cis-acting elements (ABRE/DRE).
Supplementary Material
Acknowledgments
We are grateful for the excellent technical support provided by Fumie Saito, Chitose Kato, and Atsuko Iuchi (JIRCAS). The 185 EST clones of rice were obtained from the Rice Genome Project (Ministry of Agriculture, Forestry and Fisheries, Japan).
This work was supported in part by the Program for the Promotion of Basic Research Activities for Innovative Biosciences; in part by the Ministry of Agriculture, Forestry, and Fisheries, Japan (project grant); and by JIRCAS and Japan Society for the Promotion of Science (visiting research fellowship to M.A.R.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025742.
References
- Aguan K, Sugawara K, Suzuki N, Kusano T (1991) Isolation of genes for low-temperature-induced proteins in rice by a simple subtractive method. Plant Cell Physiol 32: 1285-1289 [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93: 13404-13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj S, Targolli J, Liu LF, Ho THD, Wu R (1999) Transgenic approaches to increase dehydration-stress tolerance in plants. Mol Breed 5: 493-503 [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that control cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701-713 [DOI] [PubMed] [Google Scholar]
- Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2: 48-54 [Google Scholar]
- Cook T, Gebelein B, Belal M, Mesa K, Urrutia R (1999) Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem 274: 29500-29504 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3: 117-124 [DOI] [PubMed] [Google Scholar]
- Deyholos M, Galbraith DW (2001) High-density microarrays for gene expression analysis. Cytometry 43: 229-238 [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression Plant J 33: 751-763 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Brown PO (1999) DNA arrays for analysis of gene expression. Methods Enzymol 303: 179-205 [DOI] [PubMed] [Google Scholar]
- Garcia AB, Engler JA, Claes B, Villarroel R, Montagu MV, Gerats T, Caplan A (1998) The expression of the salt-responsive gene salT from rice is regulated by hormonal and developmental cues. Planta 207: 172-180 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570-1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463-499 [DOI] [PubMed] [Google Scholar]
- Holmberg N, Bülow L (1998) Improving stress tolerance in plants by gene transfer. Trends Plant Sci 3: 61-66 [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377-403 [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Maekawa M, Saito A, Higo H, Higo K (1998) Evolutionary relationship of plant catalase genes inferred from exon-intron structures: isozyme divergence after the separation of monocots and dicots. Theor Appl Genet 97: 9-19 [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106-111 [DOI] [PubMed] [Google Scholar]
- Jiang C, Lu B, Singh J (1996) Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol 30: 679-684 [DOI] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41: 577-585 [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889-905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY (2000) Molecular analysis of the NAC gene family in rice. Mol Gen Genet 262: 1047-1051 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129-2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J (1980) Responses of Plants to Environmental Stress, Ed 2. Academic Press, New York
- Liu H, Sachidanandam R, Stein L (2001) Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res 11: 2020-2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) The transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391-1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJR (1994) Kruppel-associated boxes are potent transcriptional activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 91: 4509-4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Moyano E, Alcocer MJC, Martin C (1998) Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant J 13: 489-505 [DOI] [PubMed] [Google Scholar]
- Moons A, De Keyser A, Van Montagu M (1997) A group 3 LEA cDNA of rice, responsive to abscisic acid, but not to jasmonic acid, shows variety-specific differences in salt stress response. Gene 191: 197-204 [DOI] [PubMed] [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48: 551-573 [DOI] [PubMed] [Google Scholar]
- Reddy AR, Ramakrishna A, Sekhar AC, Ithal N, Babu PR, Bonaldo MF, Soares MB, Bennetzen JL (2002) Novel genes are enriched in normalized cDNA libraries from drought-stressed seedlings of rice (Oryza sativa L. subsp. indica cv Nagina 22). Genome 45: 204-211 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707-719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Borden KL, Boddy MN, Freemont PS (1996) Does this have a familiar RING? Trends Biochem Sci 21: 208-214 [PubMed] [Google Scholar]
- Scandalios JG (1990) Responses of plant antioxidant defense genes to environmental stresses. Adv Genet 28: 1-41 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655-11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchardt J, Beule D, Malik A, Wolski E, Eickhoff H, Lehrach H, Herzel H (2000) Normalization strategies for cDNA microarrays. Nucleic Acids Res 28: E47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses using full-length cDNA microarray. Plant Cell 13: 61-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T et al. (2002b) Monitoring the expression pattern of around 7, 000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282-291 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al. (2002a) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279-292 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (1999) Molecular responses to drought stress. In K Shinozaki, K Yamaguchi-Shinozaki, eds, Molecular Responses to Cold, Drought, Heat and Salt Stress in Higher Plants. R.G. Landes, Austin, TX, pp 11-28
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217-223 [PubMed] [Google Scholar]
- Takahashi R, Joshee N, Kitagawa Y (1994) Induction of chilling resistance by water stress, and cDNA sequence analysis and expression of water stress-regulated genes in rice. Plant Mol Biol 26: 339-352 [DOI] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol 127: 1030-1043 [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571-599 [DOI] [PubMed] [Google Scholar]
- Torres-Schumann S, Godoy JA, Pintor-Toro JA (1992) A probable lipid transfer protein gene is induced by NaCl in stems of tomato plants. Plant Mol Biol 18: 749-757 [DOI] [PubMed] [Google Scholar]
- Vaidyanathan R, Kuruvilla S, Thomas G (1999) Characterization and expression pattern of an abscisic acid and osmotic stress responsive gene from rice. Plant Sci 140: 21-30 [Google Scholar]
- Wang RC, Guegler K, LaBrie ST, Crawford NM, Wang RC (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024-3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JX, Klueva NY, Wang Z, Wu R, Ho TH, Nguyen HT, Ho THD (2000) Genetic engineering for abiotic stress resistance in crop plants. In Vitro Cell Dev Biol Plant 36: 108-114 [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.