Abstract
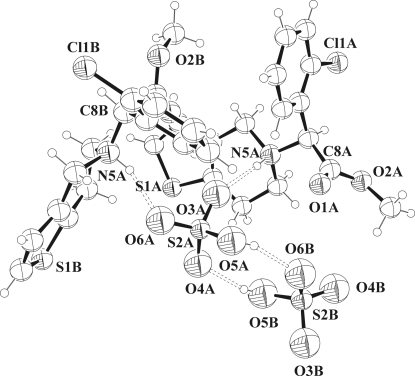
The asymmetric unit of the title compound, C16H17ClNO2S+·HSO4 −, (I) [systematic name: (+)-(S)-5-[(2-chlorophenyl)(methoxycarbonyl)methyl]-4,5,6,7-tetrahydrothieno[3,2-c]pyridin-5-ium hydrogen sulfate], contains two independent cations of clopidogrel and two independent hydrogensulfate anions. The two independent cations are of similar conformation; however, this differs from that observed in orthorhombic form (II) [Bousquet et al. (2003 ▶). US Patent No. 6 504 030]. The H—N—Cchiral—H fragment shows a trans conformation in both independent cations in (I) and a gauche conformation in (II). In (I), classical intermolecular N—H⋯O and O—H⋯O hydrogen bonds link two independent cations and two independent anions into an isolated cluster, in which two cations interact with one anion only via N—H⋯O hydrogen bonds. Weak intermolecular C—H⋯O hydrogen bonds further consolidate the crystal packing.
Related literature
For the characterization of six polymorphic forms of Clopidogrel hydrogensulfate, see: Badorc & Frehel (1989 ▶) (form I); Bousquet et al. (2003 ▶) (orthorhombic form II); Lifshitz-Liron et al. (2006 ▶) (forms III-VI). For recent studies of forms I and II, see: Raijada et al. (2010 ▶); Zupancic et al. (2010 ▶); Srivastava et al. (2010 ▶); Song et al. (2010 ▶). For details of the indexing algorithm, see: Werner et al. (1985 ▶). The methodology of the refinement (including applied restraints and constraints) was described in detail by Chernyshev et al. (2009 ▶).
Experimental
Crystal data
C16H17ClNO2S+·HSO4 −
M r = 419.89
Monoclinic,
a = 10.4315 (12) Å
b = 15.3345 (18) Å
c = 12.6320 (16) Å
β = 113.28 (2)°
V = 1856.1 (5) Å3
Z = 4
Cu Kα1 radiation, λ = 1.54059 Å
μ = 4.23 mm−1
T = 295 K
Flat sheet, 15 × 1 mm
Data collection
Guinier camera G670 diffractometer
Specimen mounting: thin layer in the specimen holder of the camera
Data collection mode: transmission
Scan method: continuous
2θmin = 4.00°, 2θmax = 90.00°, 2θstep = 0.01°
Refinement
R p = 0.019
R wp = 0.025
R exp = 0.015
R Bragg = 0.049
χ2 = 2.982
8601 data points
205 parameters
155 restraints
H-atom parameters not refined
Data collection: G670 Imaging Plate Guinier Camera Software (Huber, 2002 ▶); cell refinement: MRIA (Zlokazov & Chernyshev, 1992 ▶); data reduction: G670 Imaging Plate Guinier Camera Software; method used to solve structure: simulated annealing (Zhukov et al., 2001 ▶); program(s) used to refine structure: MRIA; molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: MRIA and SHELXL97 (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810028783/lh5082sup1.cif
Rietveld powder data: contains datablocks I. DOI: 10.1107/S1600536810028783/lh5082Isup2.rtv
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N5A—H5A⋯O3A | 0.91 | 1.91 | 2.785 (16) | 161 |
| N5B—H5B⋯O6A | 0.91 | 1.94 | 2.795 (19) | 157 |
| O5A—H51⋯O6B | 0.82 | 1.85 | 2.640 (17) | 161 |
| O5B—H52⋯O4A | 0.82 | 1.82 | 2.567 (17) | 152 |
| C4A—H4A1⋯O4Bi | 0.97 | 2.35 | 3.17 (2) | 142 |
| C4A—H4A2⋯O1B | 0.97 | 2.52 | 3.225 (17) | 129 |
| C3B—H3B⋯O4Bii | 0.93 | 2.41 | 3.28 (2) | 154 |
| C6A—H6A2⋯O3Bi | 0.97 | 2.31 | 3.175 (19) | 149 |
| C4B—H4B2⋯O3Biii | 0.97 | 2.23 | 3.13 (2) | 154 |
Symmetry codes: (i) 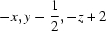 ; (ii)
; (ii) 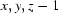 ; (iii)
; (iii) 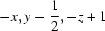 .
.
supplementary crystallographic information
Comment
Clopidogrel hydrogensulfate is an antiplatelet drug, which acts by selective and irreversible inhibition of ADP-induced platelet aggregation. The drug is available in the market as oral solid dosage form. Six different polymorphs are known for the drug - I (Badorc & Frehel, 1989), II (Bousquet et al., 2003) and III-VI (Lifshitz-Liron et al., 2006). However, only polymorphs I and II are used in pharmaceutical formulations (Bousquet et al., 2003), and, therefore, they are under intensive studies (Raijada et al., 2010; Zupan˘ci˘c et al., 2010; Srivastava et al., 2010; Song et al., 2010). The crystal structure of orthorhombic polymorph II has been reported by Bousquet et al. (2003). Herewith we report the crystal structure of the monoclinic polymorph I.
The asymmetric unit of I (Fig. 1), contains two independent cations of clopidogrel and two independent hydrogensulfate anions. The two independent cations are of similar conformation, which, however, differs from that observed in II. The H–N—Cchiral–H fragment shows a trans conformation in both independent cations in I and a gauche conformation in II.
The hydrogen-bonding motifs in I and II are essentially different too. In I, the classical intermolecular N—H···O and O—H···O hydrogen bonds (Table 1) link two independent cations and two independent anions into isolated cluster, where two cations interact with one anion only via N—H···O hydrogen bonds (Fig. 1). Weak intermolecular C—H···O hydrogen bonds (Table 1) consolidate further the crystal packing of I. In II, O—H···O hydrogen bonds link anions into linear chains, while N—H···O hydrogen bond attach one cation to one anion. These differences in crystal packings of Forms I and II may explain why II exhibits a lower solubility (and is more stable) than I.
Experimental
The title compound I was synthesized in accordance with the known procedure (Badorc & Frehel, 1989), and obtained as a white polycrystalline powder. Optical rotation [α]D +53.8° (c<ι> 1.9, CH3OH).
Refinement
During the exposure, the specimen was spun in its plane to improve particle statistics. The monoclinic unit-cell dimensions were determined with the indexing program TREOR (Werner et al., 1985), M20=37, using the first 30 peak positions. The same monoclinic unit-cell dimensions were reported in 2003 by Martin Vickers at http://img.chem.ucl.ac.uk/www/reports/clopi/clopi.htm.
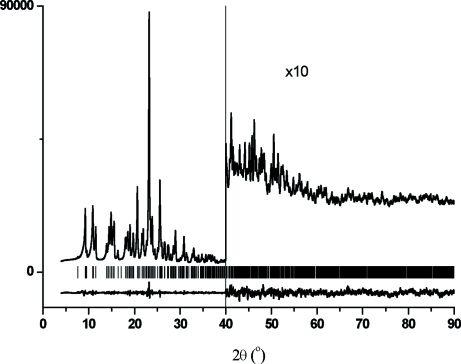
The structure of was solved by simulated annealing procedure (Zhukov et al., 2001) and refined following the methodology described in (Chernyshev et al., 2009). For non-H atoms, ten independent Uiso parameters were refined - six for six independent Cl and S atoms, two common Uiso for two groups of anion' oxygen atoms, and two common Uiso for the rest atoms in independent cations. H atoms were placed in geometrically calculated positions and not refined. The diffraction profiles and the differences between the measured and calculated profiles are shown in Fig. 2.
Figures
Fig. 1.
The content of asymmetric unit of I showing the atomic labeling and 40% probability displacement spheres. Dashed lines denote classical N—H···O and O—H···O hydrogen bonds.
Fig. 2.
The Rietveld plot, showing the observed and difference profiles for I. The reflection positions are shown above the difference profile.
Crystal data
| C16H17ClNO2S+·HSO4− | Dx = 1.503 Mg m−3 |
| Mr = 419.89 | Melting point: 455(3) K |
| Monoclinic, P21 | Cu Kα1 radiation, λ = 1.54059 Å |
| a = 10.4315 (12) Å | µ = 4.23 mm−1 |
| b = 15.3345 (18) Å | T = 295 K |
| c = 12.6320 (16) Å | Particle morphology: plate |
| β = 113.28 (2)° | white |
| V = 1856.1 (5) Å3 | flat sheet, 15 × 1 mm |
| Z = 4 | Specimen preparation: Prepared at 295 K and 101 kPa |
| F(000) = 872 |
Data collection
| Guinier camera G670 diffractometer | Data collection mode: transmission |
| Radiation source: line-focus sealed tube | Scan method: continuous |
| Curved Germanium (111) | 2θmin = 4.00°, 2θmax = 90.00°, 2θstep = 0.01° |
| Specimen mounting: thin layer in the specimen holder of the camera |
Refinement
| Refinement on Inet | Profile function: split-type pseudo-Voigt (Toraya, 1986) |
| Least-squares matrix: full with fixed elements per cycle | 205 parameters |
| Rp = 0.019 | 155 restraints |
| Rwp = 0.025 | 42 constraints |
| Rexp = 0.015 | H-atom parameters not refined |
| RBragg = 0.049 | Weighting scheme based on measured s.u.'s |
| χ2 = 2.982 | (Δ/σ)max = 0.001 |
| 8601 data points | Background function: Chebyshev polynomial up to the 5th order |
| Excluded region(s): none | Preferred orientation correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1A | 0.4095 (4) | 0.6092 (3) | 1.1269 (4) | 0.0731 (15)* | |
| S1A | −0.2971 (4) | 0.6556 (3) | 0.6216 (4) | 0.0642 (15)* | |
| O1A | 0.2960 (8) | 0.9086 (7) | 0.9643 (8) | 0.065 (3)* | |
| O2A | 0.2551 (8) | 0.8678 (6) | 1.1168 (7) | 0.065 (4)* | |
| C2A | −0.2033 (15) | 0.5796 (11) | 0.5834 (13) | 0.066 (6)* | |
| H2A | −0.2406 | 0.5448 | 0.5181 | 0.079* | |
| C3A | −0.0682 (14) | 0.5760 (10) | 0.6594 (12) | 0.065 (6)* | |
| H3A | −0.0030 | 0.5365 | 0.6547 | 0.078* | |
| C4A | 0.1068 (13) | 0.6546 (11) | 0.8382 (12) | 0.065 (5)* | |
| H4A1 | 0.1257 | 0.6125 | 0.8999 | 0.078* | |
| H4A2 | 0.1751 | 0.6463 | 0.8048 | 0.078* | |
| N5A | 0.1194 (10) | 0.7458 (9) | 0.8865 (9) | 0.065 (4)* | |
| H5A | 0.1212 | 0.7831 | 0.8310 | 0.078* | |
| C6A | −0.0077 (15) | 0.7685 (9) | 0.9103 (12) | 0.066 (6)* | |
| H6A1 | 0.0063 | 0.8255 | 0.9464 | 0.079* | |
| H6A2 | −0.0161 | 0.7266 | 0.9647 | 0.079* | |
| C7A | −0.1428 (14) | 0.7696 (10) | 0.8044 (12) | 0.065 (6)* | |
| H7A1 | −0.2214 | 0.7728 | 0.8269 | 0.078* | |
| H7A2 | −0.1455 | 0.8201 | 0.7574 | 0.078* | |
| C8A | 0.2536 (14) | 0.7586 (10) | 0.9906 (12) | 0.065 (6)* | |
| H8A | 0.2491 | 0.7262 | 1.0559 | 0.078* | |
| C9A | 0.3733 (14) | 0.7242 (10) | 0.9634 (12) | 0.065 (6)* | |
| C10A | 0.4519 (12) | 0.6534 (11) | 1.0198 (12) | 0.065 (6)* | |
| C11A | 0.5690 (13) | 0.6242 (10) | 1.0020 (12) | 0.065 (6)* | |
| H11A | 0.6223 | 0.5766 | 1.0406 | 0.078* | |
| C12A | 0.5986 (14) | 0.6727 (11) | 0.9213 (12) | 0.065 (6)* | |
| H12A | 0.6703 | 0.6529 | 0.9013 | 0.078* | |
| C13A | 0.5311 (13) | 0.7470 (11) | 0.8691 (12) | 0.065 (5)* | |
| H13A | 0.5617 | 0.7792 | 0.8213 | 0.078* | |
| C14A | 0.4160 (14) | 0.7729 (9) | 0.8893 (12) | 0.065 (6)* | |
| H14A | 0.3669 | 0.8226 | 0.8536 | 0.078* | |
| C15A | 0.2739 (15) | 0.8538 (10) | 1.0203 (12) | 0.065 (5)* | |
| C16A | 0.2910 (14) | 0.9531 (10) | 1.1684 (13) | 0.066 (6)* | |
| H16A | 0.2731 | 0.9553 | 1.2373 | 0.098* | |
| H16B | 0.2357 | 0.9964 | 1.1151 | 0.098* | |
| H16C | 0.3881 | 0.9642 | 1.1873 | 0.098* | |
| C17A | −0.1513 (14) | 0.6881 (10) | 0.7374 (12) | 0.065 (6)* | |
| C18A | −0.0391 (13) | 0.6396 (10) | 0.7466 (12) | 0.065 (6)* | |
| Cl1B | 0.4361 (4) | 0.7397 (3) | 0.3437 (4) | 0.0692 (14)* | |
| S1B | −0.3489 (4) | 0.8614 (3) | 0.1741 (4) | 0.0670 (16)* | |
| O1B | 0.1492 (9) | 0.6692 (7) | 0.5994 (8) | 0.072 (4)* | |
| O2B | 0.3318 (9) | 0.6156 (6) | 0.5793 (8) | 0.072 (4)* | |
| C2B | −0.2713 (14) | 0.9258 (11) | 0.1059 (12) | 0.072 (6)* | |
| H2B | −0.3189 | 0.9664 | 0.0493 | 0.086* | |
| C3B | −0.1318 (13) | 0.9106 (12) | 0.1440 (12) | 0.072 (6)* | |
| H3B | −0.0732 | 0.9378 | 0.1146 | 0.087* | |
| C4B | 0.0592 (15) | 0.8170 (11) | 0.2903 (13) | 0.072 (6)* | |
| H4B1 | 0.1222 | 0.8665 | 0.3124 | 0.086* | |
| H4B2 | 0.0822 | 0.7819 | 0.2363 | 0.086* | |
| N5B | 0.0783 (11) | 0.7634 (8) | 0.3955 (10) | 0.072 (5)* | |
| H5B | 0.0669 | 0.8006 | 0.4472 | 0.086* | |
| C6B | −0.0383 (15) | 0.6974 (10) | 0.3650 (13) | 0.072 (6)* | |
| H6B1 | −0.0191 | 0.6578 | 0.4293 | 0.086* | |
| H6B2 | −0.0411 | 0.6634 | 0.2993 | 0.086* | |
| C7B | −0.1796 (14) | 0.7400 (12) | 0.3363 (12) | 0.072 (6)* | |
| H7B1 | −0.2535 | 0.6985 | 0.2974 | 0.086* | |
| H7B2 | −0.1883 | 0.7587 | 0.4065 | 0.086* | |
| C8B | 0.2241 (14) | 0.7255 (10) | 0.4537 (13) | 0.072 (6)* | |
| H8B | 0.2438 | 0.6882 | 0.3989 | 0.086* | |
| C9B | 0.3321 (15) | 0.7989 (10) | 0.4937 (13) | 0.071 (6)* | |
| C10B | 0.4343 (15) | 0.8091 (11) | 0.4515 (13) | 0.072 (6)* | |
| C11B | 0.5344 (14) | 0.8730 (10) | 0.4922 (12) | 0.072 (6)* | |
| H11B | 0.6013 | 0.8796 | 0.4613 | 0.087* | |
| C12B | 0.5345 (15) | 0.9267 (11) | 0.5787 (12) | 0.072 (6)* | |
| H12B | 0.6045 | 0.9683 | 0.6083 | 0.086* | |
| C13B | 0.4337 (13) | 0.9206 (11) | 0.6231 (13) | 0.072 (6)* | |
| H13B | 0.4310 | 0.9599 | 0.6783 | 0.086* | |
| C14B | 0.3367 (14) | 0.8543 (11) | 0.5828 (13) | 0.072 (6)* | |
| H14B | 0.2724 | 0.8464 | 0.6162 | 0.087* | |
| C15B | 0.2303 (14) | 0.6716 (12) | 0.5555 (13) | 0.072 (6)* | |
| C16B | 0.3275 (14) | 0.5400 (11) | 0.6458 (12) | 0.072 (6)* | |
| H16D | 0.4071 | 0.5038 | 0.6579 | 0.108* | |
| H16E | 0.2436 | 0.5076 | 0.6049 | 0.108* | |
| H16F | 0.3289 | 0.5583 | 0.7190 | 0.108* | |
| C17B | −0.1919 (15) | 0.8163 (11) | 0.2605 (13) | 0.072 (6)* | |
| C18B | −0.0874 (14) | 0.8485 (11) | 0.2336 (12) | 0.072 (6)* | |
| S2A | 0.0232 (4) | 0.9234 (3) | 0.6166 (4) | 0.0572 (13)* | |
| O3A | 0.0642 (12) | 0.8444 (8) | 0.6881 (9) | 0.115 (5)* | |
| O4A | −0.1156 (11) | 0.9512 (8) | 0.6065 (10) | 0.115 (5)* | |
| O5A | 0.1279 (11) | 0.9964 (8) | 0.6804 (10) | 0.115 (5)* | |
| H51 | 0.1260 | 1.0045 | 0.7439 | 0.173* | |
| O6A | 0.0282 (11) | 0.9101 (10) | 0.5049 (10) | 0.115 (4)* | |
| S2B | −0.0311 (5) | 1.0685 (3) | 0.8780 (4) | 0.0692 (16)* | |
| O3B | −0.0818 (11) | 1.1589 (9) | 0.8583 (11) | 0.132 (5)* | |
| O4B | −0.0199 (13) | 1.0398 (9) | 0.9921 (11) | 0.133 (5)* | |
| O5B | −0.1420 (12) | 1.0105 (9) | 0.7869 (10) | 0.133 (5)* | |
| H52 | −0.1264 | 1.0087 | 0.7283 | 0.200* | |
| O6B | 0.1001 (12) | 1.0588 (9) | 0.8650 (10) | 0.133 (5)* |
Geometric parameters (Å, °)
| Cl1A—C10A | 1.721 (17) | O2B—C16B | 1.44 (2) |
| S1A—C2A | 1.709 (18) | C2B—C3B | 1.359 (19) |
| S1A—C17A | 1.716 (13) | C2B—H2B | 0.9313 |
| O1A—C15A | 1.18 (2) | C3B—C18B | 1.41 (2) |
| O2A—C15A | 1.33 (2) | C3B—H3B | 0.9305 |
| O2A—C16A | 1.443 (18) | C4B—C18B | 1.49 (2) |
| C2A—C3A | 1.357 (17) | C4B—N5B | 1.51 (2) |
| C2A—H2A | 0.9313 | C4B—H4B1 | 0.9717 |
| C3A—C18A | 1.41 (2) | C4B—H4B2 | 0.9710 |
| C3A—H3A | 0.9304 | N5B—C6B | 1.511 (19) |
| C4A—N5A | 1.51 (2) | N5B—C8B | 1.519 (17) |
| C4A—C18A | 1.522 (16) | N5B—H5B | 0.9092 |
| C4A—H4A1 | 0.9699 | C6B—C7B | 1.52 (2) |
| C4A—H4A2 | 0.9674 | C6B—H6B1 | 0.9690 |
| N5A—C8A | 1.508 (15) | C6B—H6B2 | 0.9711 |
| N5A—C6A | 1.51 (2) | C7B—C17B | 1.49 (2) |
| N5A—H5A | 0.9102 | C7B—H7B1 | 0.9698 |
| C6A—C7A | 1.512 (17) | C7B—H7B2 | 0.9703 |
| C6A—H6A1 | 0.9690 | C8B—C15B | 1.51 (2) |
| C6A—H6A2 | 0.9712 | C8B—C9B | 1.53 (2) |
| C7A—C17A | 1.49 (2) | C8B—H8B | 0.9795 |
| C7A—H7A1 | 0.9694 | C9B—C10B | 1.38 (3) |
| C7A—H7A2 | 0.9697 | C9B—C14B | 1.40 (2) |
| C8A—C15A | 1.50 (2) | C10B—C11B | 1.37 (2) |
| C8A—C9A | 1.52 (2) | C11B—C12B | 1.37 (2) |
| C8A—H8A | 0.9803 | C11B—H11B | 0.9312 |
| C9A—C10A | 1.38 (2) | C12B—C13B | 1.38 (3) |
| C9A—C14A | 1.40 (2) | C12B—H12B | 0.9299 |
| C10A—C11A | 1.40 (2) | C13B—C14B | 1.38 (2) |
| C11A—C12A | 1.39 (2) | C13B—H13B | 0.9301 |
| C11A—H11A | 0.9303 | C14B—H14B | 0.9306 |
| C12A—C13A | 1.36 (2) | C16B—H16D | 0.9596 |
| C12A—H12A | 0.9298 | C16B—H16E | 0.9613 |
| C13A—C14A | 1.38 (2) | C16B—H16F | 0.9609 |
| C13A—H13A | 0.9307 | C17B—C18B | 1.36 (2) |
| C14A—H14A | 0.9297 | S2A—O6A | 1.447 (14) |
| C16A—H16A | 0.9589 | S2A—O4A | 1.466 (13) |
| C16A—H16B | 0.9591 | S2A—O3A | 1.470 (13) |
| C16A—H16C | 0.9607 | S2A—O5A | 1.549 (12) |
| C17A—C18A | 1.35 (2) | O5A—H51 | 0.8200 |
| Cl1B—C10B | 1.733 (18) | S2B—O6B | 1.449 (15) |
| S1B—C2B | 1.710 (18) | S2B—O4B | 1.468 (15) |
| S1B—C17B | 1.714 (14) | S2B—O3B | 1.469 (15) |
| O1B—C15B | 1.18 (2) | S2B—O5B | 1.549 (12) |
| O2B—C15B | 1.302 (19) | O5B—H52 | 0.8200 |
| C2A—S1A—C17A | 91.5 (7) | C18B—C3B—H3B | 124.2 |
| C15A—O2A—C16A | 117.1 (12) | C18B—C4B—N5B | 110.7 (14) |
| C3A—C2A—S1A | 112.3 (12) | C18B—C4B—H4B1 | 109.4 |
| C3A—C2A—H2A | 123.8 | N5B—C4B—H4B1 | 109.6 |
| S1A—C2A—H2A | 123.9 | C18B—C4B—H4B2 | 109.5 |
| C2A—C3A—C18A | 111.5 (14) | N5B—C4B—H4B2 | 109.7 |
| C2A—C3A—H3A | 124.3 | H4B1—C4B—H4B2 | 107.9 |
| C18A—C3A—H3A | 124.2 | C4B—N5B—C6B | 109.1 (10) |
| N5A—C4A—C18A | 110.4 (11) | C4B—N5B—C8B | 113.2 (13) |
| N5A—C4A—H4A1 | 109.4 | C6B—N5B—C8B | 114.7 (11) |
| C18A—C4A—H4A1 | 109.5 | C4B—N5B—H5B | 106.5 |
| N5A—C4A—H4A2 | 109.6 | C6B—N5B—H5B | 106.4 |
| C18A—C4A—H4A2 | 109.7 | C8B—N5B—H5B | 106.4 |
| H4A1—C4A—H4A2 | 108.3 | N5B—C6B—C7B | 112.3 (13) |
| C4A—N5A—C8A | 112.0 (10) | N5B—C6B—H6B1 | 109.2 |
| C4A—N5A—C6A | 110.4 (11) | C7B—C6B—H6B1 | 109.2 |
| C8A—N5A—C6A | 112.4 (11) | N5B—C6B—H6B2 | 109.0 |
| C4A—N5A—H5A | 107.1 | C7B—C6B—H6B2 | 109.1 |
| C8A—N5A—H5A | 107.3 | H6B1—C6B—H6B2 | 107.9 |
| C6A—N5A—H5A | 107.2 | C17B—C7B—C6B | 108.7 (14) |
| N5A—C6A—C7A | 114.3 (13) | C17B—C7B—H7B1 | 109.9 |
| N5A—C6A—H6A1 | 108.7 | C6B—C7B—H7B1 | 110.0 |
| C7A—C6A—H6A1 | 108.7 | C17B—C7B—H7B2 | 109.9 |
| N5A—C6A—H6A2 | 108.6 | C6B—C7B—H7B2 | 109.9 |
| C7A—C6A—H6A2 | 108.7 | H7B1—C7B—H7B2 | 108.3 |
| H6A1—C6A—H6A2 | 107.6 | C15B—C8B—N5B | 108.6 (13) |
| C17A—C7A—C6A | 108.6 (12) | C15B—C8B—C9B | 110.1 (12) |
| C17A—C7A—H7A1 | 109.9 | N5B—C8B—C9B | 110.1 (12) |
| C6A—C7A—H7A1 | 110.1 | C15B—C8B—H8B | 109.3 |
| C17A—C7A—H7A2 | 109.9 | N5B—C8B—H8B | 109.3 |
| C6A—C7A—H7A2 | 110.0 | C9B—C8B—H8B | 109.4 |
| H7A1—C7A—H7A2 | 108.4 | C10B—C9B—C14B | 117.3 (14) |
| C15A—C8A—N5A | 109.6 (11) | C10B—C9B—C8B | 122.5 (15) |
| C15A—C8A—C9A | 110.4 (13) | C14B—C9B—C8B | 120.0 (16) |
| N5A—C8A—C9A | 108.7 (12) | C11B—C10B—C9B | 121.7 (16) |
| C15A—C8A—H8A | 109.3 | C11B—C10B—Cl1B | 119.3 (14) |
| N5A—C8A—H8A | 109.5 | C9B—C10B—Cl1B | 119.0 (11) |
| C9A—C8A—H8A | 109.4 | C12B—C11B—C10B | 119.3 (16) |
| C10A—C9A—C14A | 119.0 (15) | C12B—C11B—H11B | 120.4 |
| C10A—C9A—C8A | 122.2 (15) | C10B—C11B—H11B | 120.3 |
| C14A—C9A—C8A | 118.3 (13) | C11B—C12B—C13B | 121.7 (14) |
| C9A—C10A—C11A | 123.2 (16) | C11B—C12B—H12B | 119.3 |
| C9A—C10A—Cl1A | 115.6 (12) | C13B—C12B—H12B | 119.0 |
| C11A—C10A—Cl1A | 120.8 (11) | C12B—C13B—C14B | 117.8 (16) |
| C12A—C11A—C10A | 114.0 (13) | C12B—C13B—H13B | 121.1 |
| C12A—C11A—H11A | 122.8 | C14B—C13B—H13B | 121.1 |
| C10A—C11A—H11A | 123.2 | C13B—C14B—C9B | 122.1 (16) |
| C13A—C12A—C11A | 125.4 (16) | C13B—C14B—H14B | 118.9 |
| C13A—C12A—H12A | 117.3 | C9B—C14B—H14B | 119.1 |
| C11A—C12A—H12A | 117.3 | O1B—C15B—O2B | 122.7 (16) |
| C12A—C13A—C14A | 118.2 (16) | O1B—C15B—C8B | 128.3 (14) |
| C12A—C13A—H13A | 121.0 | O2B—C15B—C8B | 108.3 (14) |
| C14A—C13A—H13A | 120.8 | O2B—C16B—H16D | 109.6 |
| C13A—C14A—C9A | 119.9 (13) | O2B—C16B—H16E | 109.5 |
| C13A—C14A—H14A | 120.0 | H16D—C16B—H16E | 109.4 |
| C9A—C14A—H14A | 120.1 | O2B—C16B—H16F | 109.6 |
| O1A—C15A—O2A | 124.9 (15) | H16D—C16B—H16F | 109.4 |
| O1A—C15A—C8A | 125.7 (15) | H16E—C16B—H16F | 109.3 |
| O2A—C15A—C8A | 109.3 (14) | C18B—C17B—C7B | 125.1 (13) |
| O2A—C16A—H16A | 109.5 | C18B—C17B—S1B | 110.7 (12) |
| O2A—C16A—H16B | 109.4 | C7B—C17B—S1B | 123.0 (12) |
| H16A—C16A—H16B | 109.6 | C17B—C18B—C3B | 113.9 (13) |
| O2A—C16A—H16C | 109.3 | C17B—C18B—C4B | 122.3 (14) |
| H16A—C16A—H16C | 109.5 | C3B—C18B—C4B | 123.7 (15) |
| H16B—C16A—H16C | 109.5 | O6A—S2A—O4A | 111.8 (7) |
| C18A—C17A—C7A | 123.8 (12) | O6A—S2A—O3A | 111.7 (8) |
| C18A—C17A—S1A | 111.0 (12) | O4A—S2A—O3A | 109.3 (8) |
| C7A—C17A—S1A | 124.1 (11) | O6A—S2A—O5A | 108.6 (8) |
| C17A—C18A—C3A | 113.7 (11) | O4A—S2A—O5A | 107.7 (7) |
| C17A—C18A—C4A | 123.6 (14) | O3A—S2A—O5A | 107.6 (6) |
| C3A—C18A—C4A | 122.7 (13) | S2A—O5A—H51 | 109.5 |
| C2B—S1B—C17B | 91.7 (8) | O6B—S2B—O4B | 111.6 (7) |
| C15B—O2B—C16B | 117.0 (13) | O6B—S2B—O3B | 111.8 (8) |
| C3B—C2B—S1B | 112.1 (12) | O4B—S2B—O3B | 109.5 (8) |
| C3B—C2B—H2B | 123.8 | O6B—S2B—O5B | 108.6 (8) |
| S1B—C2B—H2B | 124.0 | O4B—S2B—O5B | 107.6 (8) |
| C2B—C3B—C18B | 111.5 (15) | O3B—S2B—O5B | 107.6 (7) |
| C2B—C3B—H3B | 124.3 | S2B—O5B—H52 | 109.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5A—H5A···O3A | 0.91 | 1.91 | 2.785 (16) | 161 |
| N5B—H5B···O6A | 0.91 | 1.94 | 2.795 (19) | 157 |
| O5A—H51···O6B | 0.82 | 1.85 | 2.640 (17) | 161 |
| O5B—H52···O4A | 0.82 | 1.82 | 2.567 (17) | 152 |
| C4A—H4A1···O4Bi | 0.97 | 2.35 | 3.17 (2) | 142 |
| C4A—H4A2···O1B | 0.97 | 2.52 | 3.225 (17) | 129 |
| C3B—H3B···O4Bii | 0.93 | 2.41 | 3.28 (2) | 154 |
| C6A—H6A2···O3Bi | 0.97 | 2.31 | 3.175 (19) | 149 |
| C4B—H4B2···O3Biii | 0.97 | 2.23 | 3.13 (2) | 154 |
Symmetry codes: (i) −x, y−1/2, −z+2; (ii) x, y, z−1; (iii) −x, y−1/2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5082).
References
- Badorc, A. & Frehel, D. (1989). US Patent No. 4 847 265.
- Bousquet, A., Castro, B. & Germain, J. S. (2003). US Patent No. 6 504 030.
- Chernyshev, V. V., Machula, A. A., Kukushkin, S. Y. & Velikodny, Y. A. (2009). Acta Cryst. E65, o2020–o2021. [DOI] [PMC free article] [PubMed]
- Huber (2002). G670 Imaging Plate Guinier Camera Software Huber Diffraktionstechnik GmbH, Rimsting, Germany.
- Lifshitz-Liron, R., Kovalevski-Ishai, E., Wizel, S., Avhar-Maydan, S. & Lidor-Hadas, R. (2006). US Patent No. 7 074 928.
- Raijada, D. K., Prasad, B., Paudel, A., Shah, R. P. & Singh, S. (2010). J. Pharm. Biomed. Anal.52, 332–344. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, L., Li, M. & Gong, J. (2010). J. Chem. Eng. Data DOI:10.1021/je100022w.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Srivastava, A., Mishra, S., Tandon, P., Patel, S., Ayala, A. P., Bansal, A. K. & Siesler, H. W. (2010). J. Mol. Struct.964, 88–96.
- Toraya, H. (1986). J. Appl. Cryst.19, 440–447.
- Werner, P.-E., Eriksson, L. & Westdahl, M. (1985). J. Appl. Cryst.18, 367–370.
- Zhukov, S. G., Chernyshev, V. V., Babaev, E. V., Sonneveld, E. J. & Schenk, H. (2001). Z. Kristallogr.216, 5–9.
- Zlokazov, V. B. & Chernyshev, V. V. (1992). J. Appl. Cryst.25, 447–451.
- Zupancic, V., Kotar-Jordan, B., Plevnik, M., Smrkolj, M. & Vrecer, F. (2010). Pharmazie, 65, 388–389. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810028783/lh5082sup1.cif
Rietveld powder data: contains datablocks I. DOI: 10.1107/S1600536810028783/lh5082Isup2.rtv
Additional supplementary materials: crystallographic information; 3D view; checkCIF report