Abstract
Despite numerous physiological studies addressing the interactions between brassinosteroids (BRs) and auxins, little is known about the underlying molecular mechanisms. We studied the expression of IAA5 and IAA19 in response to treatment with indole acetic acid (IAA) or brassinolide (BL), the most active BR. Exogenous IAA induced these genes quickly and transiently, whereas exogenous BL induced them gradually and continuously. We also found that a fusion of DR5, a synthetic auxin response element, with the GUS (β-glucuronidase) gene was induced with similar kinetics to those of the IAA5 and IAA19 genes in response to both IAA and BL treatment of transgenic plants. These results suggest that the IAA genes are induced by BL, at least in part, via the activation of the auxin response element. Endogenous IAA levels per gram fresh weight did not increase when seedlings of Arabidopsis wild type (WT) or the BR-deficient mutant det2 were treated with BL. Furthermore, the levels of IAA transcripts were lower in the det2 mutant than in the WT, even though endogenous IAA levels per gram fresh weight were higher in the det2 mutant than in the WT. In conclusion, the lack of evidence for auxin-mediated activation of early auxin-inducible genes in response to BL suggests that the BR and auxin signaling pathways independently activate the transcriptional system of the IAA and DR5-GUS genes.
Exogenous application of brassinosteroids (BRs) to plants at nanomolar to micromolar concentrations produces a wide spectrum of physiological effects. These include promotion of cell elongation and division, enhancement of tracheary element differentiation, delaying of abscission, enhancement of gravity-induced bending, promotion of ethylene biosynthesis, and enhancement of stress resistance, as reviewed by Clouse and Sasse (1998) and Sasse (1999). A number of BR-deficient mutants have been identified in Arabidopsis, pea (Pisum sativum), and tomato (Lycopersicon esculentum; for review, see Clouse and Feldmann, 1999; Clouse, 2002; Fujioka and Yokota, 2003). These mutants exhibit dwarfism when grown in either light or dark conditions. Many of these mutants also have dark-green leaves, reduced fertility, a prolonged life span, and abnormal skotomorphogenesis. BR-insensitive mutants have been identified in Arabidopsis, pea, tomato, and rice (Oryza sativa; for review, see Clouse and Feldmann, 1999; Clouse, 2002; Fujioka and Yokota, 2003). The molecular mechanisms of BR action, however, remain unclear.
It has been suggested that the actions of BRs are related to auxin action (Mandava, 1988; Sasse, 1999). Synergistic interactions between BRs and auxins occur in elongating tissues and cells in dicots (Yopp et al., 1981; Katsumi, 1985; Sala and Sala, 1985) and in monocots (Yopp et al., 1981). Such synergism is also found in the bending responses of dicots (Yopp et al., 1981; Cohen and Meudt, 1983; Meudt, 1987) and monocots (Takeno and Pharis, 1982; Fujioka et al., 1998). Several authors have proposed that BR-induced effects might be mediated via auxin, with BR treatment altering the levels of endogenous auxin or enhancing sensitivity to auxin (Mandava, 1988; Sasse, 1999). Although numerous physiological studies have addressed the interactions between BRs and auxins, little is known about the underlying molecular mechanisms. Clouse et al. performed extensive comparisons of the physiological effects of BRs and auxins and molecular analyses of auxin-inducible genes and auxin-insensitive mutants in soybean (Glycine max), tomato, and Arabidopsis. In soybean and tomato, members of the SAUR and GH3 gene families were not rapidly induced during BR-promoted cell expansion but were induced by BR at later time points, even after the beginning of cell elongation, with different kinetics than those induced by auxin treatment (Clouse et al., 1992; Zurek et al., 1994). When free indole acetic acid (IAA) levels in BR-treated tissues were analyzed using mass spectrometry (MS), it was found that free IAA levels decreased in BR-treated soybean epicotyls (Zurek et al., 1994). Therefore, it was concluded that BR does not stimulate SAUR gene transcription via increased IAA levels. The auxin-insensitive tomato mutant dgt (Zurek et al., 1994) and the Arabidopsis mutant axr1-3 (Clouse et al., 1993) are sensitive to BRs. Several studies have concluded that the BR promotion of cell elongation in soybean and tomato and the BR inhibition of root elongation in Arabidopsis are likely not mediated by auxin signaling pathways (Clouse et al., 1992, 1993; Zurek et al., 1994). On the other hand, McKay et al. (1994) reported that IAA levels are reduced in the youngest internodes but not in the apical portion of the pea BR-insensitive mutant lka and the BR-deficient mutant lkb (Nomura et al., 1997) when compared with the WT. Therefore, the possibility cannot be excluded that the endogenous BRs increases the endogenous IAA content.
Recently, we found that a number of early auxin-inducible genes, belonging to the GH3, SAUR (Hagen and Guilfoyle, 2002), and IAA gene families (Abel et al., 1995; Liscum and Reed, 2002), are quickly induced by brassinolide (BL) treatment in Arabidopsis seedlings (Goda et al., 2002). Therefore, it is evident that some auxin-inducible genes respond to BL more quickly than previously reported. The BL induction kinetics of these genes are similar to those of genes involved in cell elongation and cell wall organization (Goda et al., 2002). Among these genes, the SAURAC1 gene (Gil et al., 1994) is induced by BL within 30 min and was found to be one of the earliest responding BL-inducible genes (Goda et al., 2002). Therefore, it is possible that BRs activate auxin biosynthesis or activate a part of the auxin signaling pathway during BR-induced growth promotion in Arabidopsis seedlings or that these genes represent shared signaling components of the auxin and BR signaling pathways. The IAA, GH3, and SAUR genes are classified into at least four groups depending on their sensitivity to IAA and BRs (Goda et al., 2002): those that are sensitive to IAA but insensitive to BRs, those that are sensitive to both IAA and BRs, those that are sensitive to BRs but insensitive to IAA, and those that are repressed by BRs. In contrast to the BL induction of some early auxin-inducible genes, none of the late-auxin-inducible genes are significantly induced by BL within a 3-h treatment period, even though a number of probes for such genes were examined due to their presence on the Arabidopsis Genome Array (Goda et al., 2002).
In our experimental system, BL significantly promoted the growth of Arabidopsis seedlings in both the WT and deetiolated2 (det2) mutant, a BR-deficient mutant blocked early in BR biosynthesis (Fujioka et al., 1997). The respective fresh weight of BL-treated det2 or WT seedlings reached 150% and 115% that of mock-treated seedlings that had undergone a 12-h BL treatment. We focused on members of the IAA gene family to gain insight into interactions between auxin and BRs. The IAA3 gene has been shown to be induced within 10 min after treatment with 20 μm IAA (Abel et al., 1995) and within 1 h after treatment with 10 nm BL (Goda et al., 2002). Therefore, induction of the IAA3 gene seems to be more rapid with IAA treatment than with BL treatment, although its responses to the two hormones have not yet been compared under the same conditions. Very recently, we have used the Affymetrix GeneChip to perform microarray screening for early auxin-inducible genes (Sawa et al., 2002) and early BL-inducible genes (Goda et al., 2002) under the same experimental conditions. Comparison of the two comprehensive studies revealed that both IAA and BL rapidly induce the IAA5 and IAA19 genes. In this manuscript, we analyzed the induction kinetics of these two genes after treatment with various concentrations of IAA or BL. IAA5 has been reported to be quickly induced by IAA (Conner et al., 1990; Abel et al., 1995), although the dose dependence of its induction kinetics has not been studied. We show that both of these IAA genes are induced with different kinetics by auxin and BL. The timing of the maximum expression after auxin induction occurred more quickly than that after BL induction with all concentrations tested. To gain further insight, we also used the DR5-GUS (β-glucuronidase) reporter system, which has been widely used as a marker to study endogenous auxin distribution. Finally, we analyzed endogenous IAA levels using gas chromatography (GC)-MS to discern when BL activates the DR5-GUS, IAA5 or IAA19 genes most efficiently. Based on these studies, the signaling interactions between auxin and BRs are discussed.
RESULTS
Induction of IAA5 and IAA19 Genes by BL and IAA Treatment
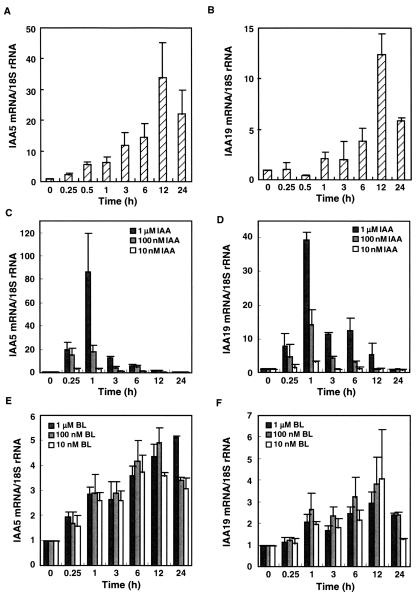
We studied expression of the IAA5 and IAA19 genes in response to treatment with exogenous auxin and BR. Seven-day-old det2 seedlings were treated with 10 nm BL, and the induction kinetics of the two genes were analyzed with real-time quantitative (RTQ) reverse transcriptase (RT)-PCR (Fig. 1, A and B). The IAA5 and IAA19 genes were induced by BL treatment with kinetics similar to the responses of IAA3, SAUR-AC1, and BRU6/GH3-2 to BL, as well as similarly to genes implicated in cell wall organization, as described previously (Goda et al., 2002). IAA5 was induced 34-fold and IAA19 12-fold, and levels of both transcripts peaked at 12 h after BL treatment. The induction of the IAA5 gene was significant 30 min after BL treatment, whereas that of IAA19 was significant 6 h after the treatment (Student's t test, P < 0.05). The IAA5 is one of the earliest responding BL-inducible genes known because no gene was induced more than 2-fold by BL within 15 min in our microarray analysis (Goda et al., 2002).
Figure 1.
The induction kinetics of IAA5 and IAA19 genes after treatment with auxin or BR. A and B, Seven-day-old det2 mutant seedlings treated with 10 nm BL. C and D, Wild-type (WT) seedlings treated with 1 μm, 100 nm, or 10 nm IAA. E and F, WT seedlings treated with 1 μm, 100 nm, or 10 nm BL. The transcript abundance of IAA5 (A, C, and E) and IAA19 (B, D, and F) was analyzed using Taq-Man RTQ RT-PCR. Transcript levels are presented as values relative to those at 0 h, defined as 1, after normalization to 18S ribosomal RNA levels. The data are the means ± se of the results of three independent hormone treatment experiments.
To compare the induction kinetics of the IAA genes by BL treatment with those in response to IAA, WT (Columbia [Col-0]) seedlings were treated with 10 nm, 100 nm, or 1 μm BL or IAA for up to 24 h. Both IAA genes were quickly induced by IAA treatment and showed stronger responses with higher IAA concentrations (Fig. 1, C and D). The IAA5 and IAA19 genes were induced by over 80- and nearly 40-fold, respectively, and transcript levels peaked 1 h after IAA treatment. In contrast, BL induced both IAA genes with much different kinetics than did IAA. The maximum induction of these genes by BL in WT plants ranged from 3- to 5-fold (Fig. 1, E and F). The point of maximum transcript accumulation of these IAA genes after BL treatment occurred later than did the maximum point after auxin induction. The BL induction kinetics in the WT were similar to the BL induction kinetics of the det2 mutant. The timing and the level of peak induction by BL were independent of BL concentration.
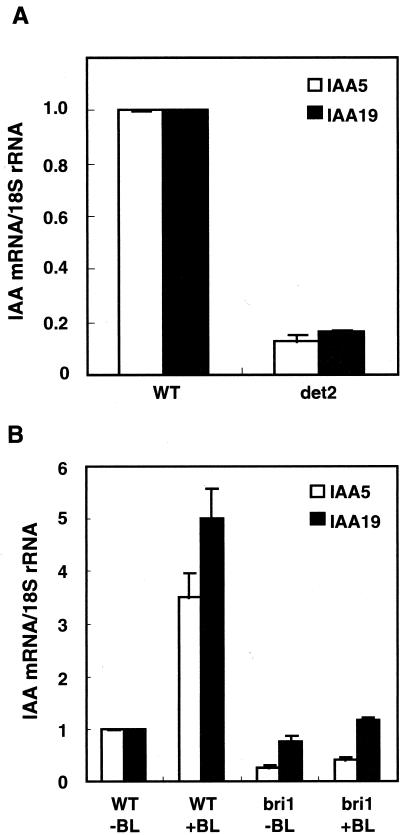
The levels of the IAA5 and IAA19 transcripts were compared between WT plants and det2 mutant plants. The levels were higher in WT plants than in det2 mutant plants (Fig. 2A), indicating that these genes are down-regulated in the det2 mutant, possibly because this mutant has a lower endogenous level of BR (Fujioka et al., 1997). BL induction of these genes was analyzed in bri1 (BR insensitive 1) mutant. Both transcript abundance and BL induction of two genes were impaired in the bri1 mutant (Fig. 2B), indicating that the BL responses of the IAA genes are dependent on the bri1 gene. Therefore, the BL responses presented here are dependent on the BR-specific signaling pathway because the BRI1 gene encodes a critical component of the BR receptor (Wang et al., 2001).
Figure 2.
IAA5 and IAA19 transcripts in WT, det2, and bri1 seedlings. A, Transcript abundance of IAA5 and IAA19 in WT (Col) and det2 seedlings. B, Seven-day-old bri1 or WT (Wassilewskija) mutant seedlings were treated with 10 nm BL for 6 h. The transcript abundance was analyzed using Taq-Man RTQ RT-PCR. Transcript levels are presented as values relative to those of the WT, defined as 1, after normalization to 18S ribosomal RNA levels. The data are the means ± se of the results of three independent hormone treatment experiments.
DR5, an Artificial Auxin Response Element (AuxRE), Responds to BL
AuxREs, which consist of a TGTCTC sequence and an adjacent or overlapping coupling element, were defined based on the auxin-responsive promoter of the soybean GH3 gene (Liu et al., 1994; Ulmasov et al., 1995). Gain-of-function experiments with minimal promoter-GUS reporter genes have shown that a single copy of an AuxRE is sufficient to confer auxin responsiveness to reporter genes (Ulmasov et al., 1995). The IAA5 and IAA19 genes have one and three putative AuxREs (TGTCTC elements), respectively, in the 500-bp fragments upstream of their start codons. DR5, an artificial AuxRE containing the TGTCTC element, has increased auxin responsiveness (Ulmasov et al., 1997). The GUS reporter gene fused to a minimal cauliflower mosaic virus 35S promoter, and the DR5 AuxRE has been used widely as a marker to monitor endogenous IAA distribution because the resulting GUS activity coincides with endogenous IAA distribution (Sabatini et al., 1999; Casimiro et al., 2001).
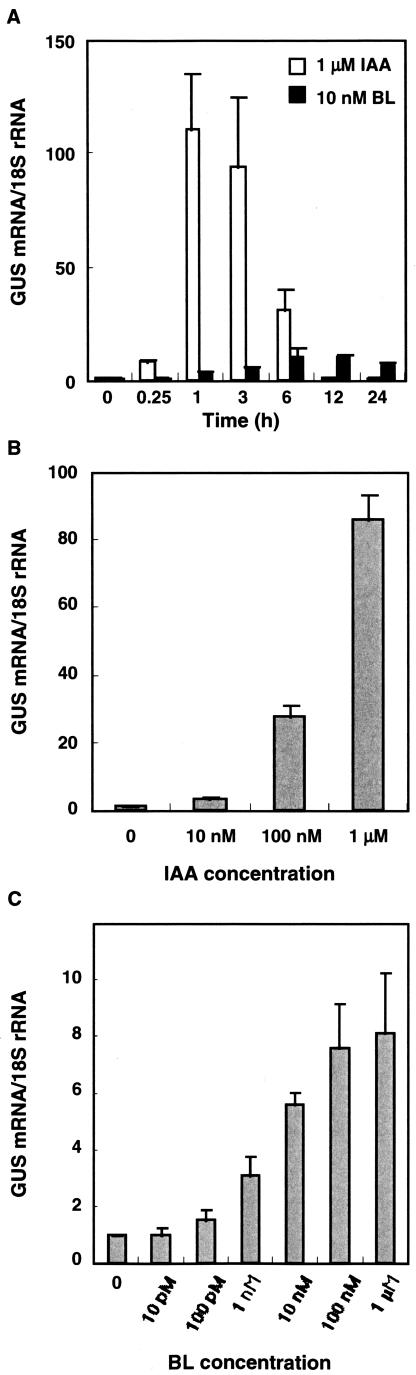
To gain insight into the mechanisms of BL induction of primary auxin-responsive genes, we studied the involvement of the DR5 AuxRE in BL-induced gene expression. Transgenic Arabidopsis seedlings containing the DR5-GUS reporter gene were treated with either 1 μm IAA or 10 nm BL, and the abundance of GUS transcripts was analyzed by RTQ RTPCR (Fig. 3A). IAA quickly induced GUS transcript accumulation, with similar kinetics to the induction of the IAA5 and IAA19 genes by IAA. GUS transcript abundance peaked 1 h after IAA treatment and returned to the basal level by 12 h after treatment, with a maximum induction of about 100-fold. In contrast, BL significantly induced the DR5-GUS gene at 12 h (Student's t test, P < 0.05) with peak induction of approximately 10-fold. These observations indicated that both BL and IAA induce the DR5-GUS reporter gene at the transcriptional level. We also tested the dose dependence of DR5-GUS induction by both IAA and BL at the time of the maximum induction in Figure 3A (1 h for IAA induction and 12 h for BL induction). IAA induced the DR5-GUS gene dose dependently between 10 nm and 1 μm (Fig. 3B). We also tested BL dose response of the DR5-GUS gene between 10 pm and 1 μm (Fig. 3C). At lower BL concentrations, i.e. between 1 pm and 10 nm, BL induction of the DR5-GUS gene was dose dependent. At 10 nm BL, we sometimes observed saturated responses and sometimes did not (individual data not shown). Therefore, 10 nm BL was close to the saturation level. In contrast, the BL induction was less dose dependent between 100 nm and 1 μm (Fig. 3C). Based on these observations, the time- and dose-dependent induction kinetics of the DR5-GUS gene seems to be similar to those of the IAA5 and IAA19 genes in response to IAA or BL, although maximum induction of the DR5-GUS was higher than that of the IAA5 and IAA19 genes. These observations suggest that exogenously applied BL induces primary auxin-responsive genes, at least in part, by activating the same cis-element as auxin, the AuxRE. The above observations suggest that exogenous BL increases endogenous auxin levels, or exogenous BL increases the sensitivity of seedlings for auxin, or the AuxRE is a shared cis-element that functions in both the auxin and BR signaling pathways. The lag period for BL-induced gene expression may be due to the time needed to induce auxin biosynthesis, to activate the auxin signaling pathway, or to directly modulate the transcriptional system of AuxRE without involvement of auxin. It may be difficult to clearly distinguish the latter two cases.
Figure 3.
Induction kinetics of the DR5-GUS gene after auxin and BL treatment. A, Time course of the DR5-GUS gene response to 10 nm BL or 1 μm IAA treatment. Seven-day-old transgenic Arabidopsis seedlings containing the DR5-GUS gene were treated with BL or IAA for up to 24 h. B, Dose-dependent induction of the DR5-GUS gene in response to IAA treatment. Seven-day-old transgenic Arabidopsis seedlings containing the DR5-GUS gene were treated with 10 nm, 100 nm, or 1 μm IAA for 1 h. C, Dose-dependent induction of the DR5-GUS gene in response to BL. Seven-day-old transgenic Arabidopsis seedlings containing the DR5-GUS gene were treated with BL for 12 h. Levels of GUS gene transcripts were analyzed using Taq-Man RTQ RT-PCR. Transcript levels are presented as values relative to those at 0 h (A) or mock treatment (B and C), both defined as 1, after normalization to 18S ribosomal RNA levels. The data are the means ± se of the results of three independent hormone treatment experiments. The experiments in each panel were performed independently; therefore, the results differ, even under the same conditions.
Endogenous IAA Levels after BL Treatment
The above findings prompted us to determine the endogenous free IAA levels in response to treatment with exogenous BL. We used det2 mutant seedlings and WT seedlings not only because det2 shows clearer IAA gene induction in response to BL treatment (Fig. 1) but also because effects of endogenous BRs can be studied. Seven-day-old det2 and WT seedlings were treated with 10 nm BL for 12 h and then analyzed for fresh weight and endogenous free IAA levels (Table I). The fresh weight per seedling of BL-treated det2 or BL-treated WT seedlings was 150% or 115%, respectively, that of mock-treated seedlings, indicating that exogenous BL strongly promotes seedling growth. The amount of IAA per plant increased as the fresh weight increased. However, IAA levels per gram fresh weight did not change in response to BL treatment of either det2 or WT seedlings. This result indicates that exogenous BL induces the DR5-GUS reporter gene without detectable increases in IAA levels per gram fresh weight. It also indicates that lower endogenous BR levels in the det2 mutant do not result in reduced IAA levels but instead result in higher IAA levels.
Table I.
Endogenous IAA levels in the det2 and WT seedlings in response to BL
| Treatments | Fresh Wt | IAA | IAA |
|---|---|---|---|
| mg plant−1 | pg plant−1 | ng g fresh wt−1 | |
| det2 (80)a | |||
| Mock | 1.02 ± 0.01 | 12.27 ± 1.94 | 11.98 ± 1.86 |
| 10 nm BL | 1.54 ± 0.08 | 17.30 ± 2.60 | 11.09 ± 1.18 |
| WT (60)a | |||
| Mock | 1.92 ± 0.02 | 13.13 ± 0.54 | 6.83 ± 0.32 |
| 10 nm BL | 2.27 ± 0.04 | 17.30 ± 2.02 | 7.60 ± 0.79 |
No. of plants used for the experiment. The data are the means ± se from three independent hormone treatment experiments.
Specific Induction of DR5-GUS by Both IAA and BL
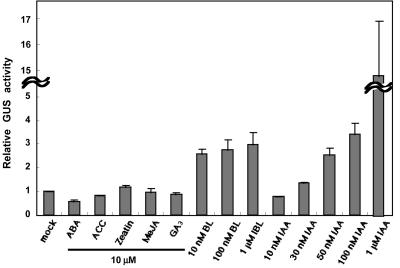
Natural AuxRE D1-4 is stimulated specifically by auxin and not by other plant hormones (cytokinin, GA3, ethylene, abscisic acid [ABA], and jasmonate) in carrot (Daucus carota) protoplasts (Ulmasov et al., 1995), but its stimulation by BR has not been studied yet. Because the DR5 AuxRE has a 2-bp substitution outside of the TGTCTC element of the natural AuxRE D1-4 and the previous report with D1-4 used carrot protoplasts, we examined the response of DR5 AuxRE to various plant hormones in Arabidopsis seedlings. We first confirmed that in response to a 50 μm 1-naphthalene acetic acid (NAA) treatment, GUS activity was more than 100 times that in mock-treated plants. This result was consistent with the induction in carrot protoplasts in the previous report (Ulmasov et al., 1997). Next, the DR5-GUS transgenic seedlings were treated with either 10 nm to 1 μm BL, 10 nm to 1 μm IAA, or 10 μm 1-aminocyclopropane-1-carboxylate, ABA, zeatin, methyl jasmonate, or GA. BL was the most effective hormone, behind auxins, to stimulate the DR5-reporter gene construct (Fig. 4). BL induced GUS activity dose independently between 10 nm and 1 μm (the induction was significant at 10 nm BL based on Student's t test, P < 0.05). GUS activities in BL-treated plants were about 3-fold higher than those in the mock-treated plants after 24 h of treatment (Fig. 4). In contrast, IAA induced GUS activity dose dependently between 10 nm and 1 μm. The GUS activation of about 3-fold triggered by the 10 nm BL treatment was equivalent to the GUS activation with 50 nm IAA treatment (Fig. 4). GUS activities were measured at 24 h, a relatively later time point, to reflect how much GUS protein was synthesized by each hormone treatment because each hormone may induce the DR5 with different kinetics (e.g. IAA induction is transient, whereas BL induction is continuous).
Figure 4.
Induction of GUS activity in DR5-GUS transgenic seedlings in response to treatment with various plant hormones. Seven-day-old transgenic Arabidopsis seedlings harboring the DR5-GUS gene were treated or mock treated with plant hormones for 24 h; 10 μm solutions of the hormones ABA, 1-aminocyclopropane-1-carboxylate (ACC), zeatin, methyl jasmonate (MeJA), and GA3 were used. BL and IAA were used at concentrations of 10 nm to 1 μm. GUS activities were determined and presented as values relative to mock-treated plants after normalization to the protein content. The data are the means ± se of the results of three independent hormone treatment experiments, except for IAA treatment at 1 μm (six experiments).
Organ-Specific Analysis of BL and IAA Inductions
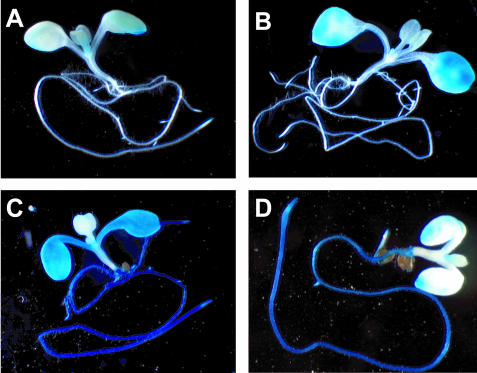
To analyze the organ specificity of the DR5-GUS reporter activity induced by BL or IAA treatment, 7-d-old DR5-GUS transgenic seedlings were treated with either of these hormones for 12 h and then stained for GUS activity (Fig. 5). In the control plants, GUS activity was detectable only at the edges of cotyledons and root tips (Fig. 5A) under our staining conditions. When seedlings were treated with 1 μm IAA, both shoots and roots stained (Fig. 5C), similar to the staining after 50 μm NAA treatment observed by Ulmasov et al. (1997). When seedlings were treated with a lower concentration of IAA (50 nm), roots stained more strongly than shoots (Fig. 5D), and the staining pattern was similar to the pattern seen with shorter staining after 50 μm NAA treatment by Ulmasov et al. (1997). These observations, together with those of Ulmasov et al. (1997), indicate that the DR5-reporter fusion gene is more sensitive to auxin in roots than in shoots. In contrast, treatment with 10 nm BL induced GUS activity mainly in cotyledons, whereas GUS activity was not detectable in roots (Fig. 5B). A higher concentration (100 nm) or longer treatment (3 d) of BL did not change the staining pattern of the DR5-GUS reporter fusion (data not shown). These observations indicate that the DR5-GUS gene responds to BL mainly in cotyledons.
Figure 5.
Histochemical staining of GUS activity in DR5-GUS transgenic Arabidopsis seedlings in response to IAA or BL treatment. Seven-day-old DR5-GUS transgenic seedlings were mock treated (A), treated with 10 nm BL for 12 h (B), treated with 1 μm IAA (C), or treated with 50 nm IAA (D) for 12 h. Seedlings were stained for 24 h for GUS activity in staining buffer containing 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide.
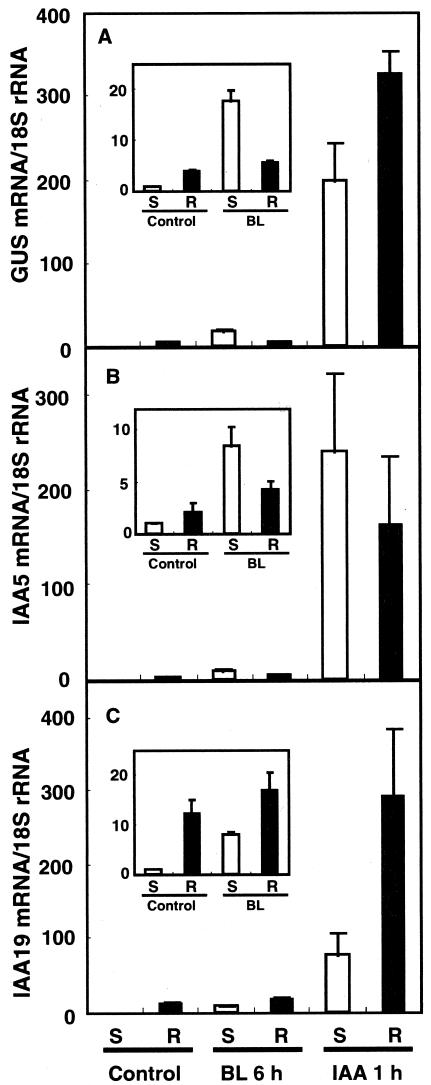
The organ-specific induction of the IAA and DR5-GUS genes was analyzed by RTQ RT-PCR. Seven-day-old DR5-GUS transgenic seedlings were treated with either 10 nm BL for 6 h or 1 μm IAA for 1 h; then, the seedlings were divided into shoots and roots. All three genes were expressed more strongly in roots than in shoots (Fig. 6, A-C, insets). The induction of these three genes in response to BL treatment was stronger in shoots, whereas their induction was limited in roots (Fig. 6, A-C, insets). This result was consistent with the histochemical staining of the DR5-GUS reporter (compare Fig. 5, B with A). Conversely, IAA induced these genes in both shoots and roots, although the degree of the induction in shoots and roots varied with each gene (Fig. 6). The maximum induction shown in Figure 6 is higher than in the previous figures. This may be because the transcript levels shown in Figure 6 were normalized to those in shoots of control seedlings, in which the maximum level of DR5-GUS transcripts was lower than in roots.
Figure 6.
Organ-specific expression of the DR5-GUS and IAA genes. Seven-day-old WT seedlings transgenic for a DR5-GUS gene were dissected into shoots (S) and roots (R) before (control) and after treatment with 10 nm BL for 6 h or 1 μm IAA for 1 h. The abundance of GUS (A), IAA5 (B), and IAA19 (C) transcripts was analyzed using Taq-Man RTQ RT-PCR. Transcript levels are presented as values relative to those for control shoots, defined as 1, after normalization to the 18S ribosomal RNA levels. The data are the means ± se of the results of three independent hormone treatment experiments.
DISCUSSION
The respective fresh weight per seedling of the BL-treated det2 mutant and the WT reached 150% and 115% that of mock-treated seedlings after a 12-h BL treatment (Table I). During the growth promotion induced by BL treatment, the IAA5 gene was induced as a primary BL-responsive gene, similar to the SAUR-AC1 gene. Because the IAA5 and IAA19 genes are induced by BL as quickly as the genes involved in cell elongation and cell wall organization (Goda et al., 2002), it is possible that these IAA genes are involved in growth acceleration in response to BL. A precise dose-dependent kinetics study of the IAA5 and IAA19 genes after IAA treatment (Fig. 1) revealed that induction of the IAA genes was as rapid as that observed in previous studies (Conner et al., 1990; Abel et al., 1995). In contrast, BL induced IAA genes more moderately and more continually than IAA treatment in both det2 and the WT (Fig. 1). The timing of maximum transcript accumulation after BL induction was independent of BL concentration between 10 nm and 1 μm and later than that after IAA induction. The magnitude of maximum transcript accumulation was also independent of BL concentration at these concentrations. From these observations, it is apparent that IAA and BL induce the two IAA genes with different kinetics and different dose dependency.
Xu et al. (1995) reported that the Arabidopsis TCH4 gene, which encodes a xyloglucan endotransglycosylase, was induced both by BL and IAA with similar kinetics as the IAA genes presented here. Very recently, they found that transgenic plants harboring -147 to -45 TCH4::LUC respond to BR but not to IAA (Iliev et al., 2002). In our study, both the BL and IAA induction kinetics of the DR5-GUS gene seem to be similar to those of the IAA5 and IAA19 genes in the timing of the peaks, the relative peak heights, and the dose dependency. The IAA5 and IAA19 genes both have putative AuxREs containing a TGTCTC sequence. Therefore, we speculate that BL activates these IAA genes at the transcriptional level, at least in part through the AuxRE. The DR5-GUS gene has been used to monitor endogenous IAA levels because the resulting GUS activity coincides with endogenous IAA distribution as determined by MS measurement (Sabatini et al., 1999; Casimiro et al., 2001). We found that more DR5-GUS transcripts accumulated in roots than in shoots (Fig. 6). This is consistent with the findings that the endogenous free IAA levels per gram fresh weight are higher in roots than in shoots in 7-d-old Arabidopsis seedlings (T. Koshiba, unpublished data) and are higher in roots than in cotyledons in 10-d-old Arabidopsis seedlings (Kowalczyk and Sandberg, 2001). Conversely, DR5-GUS reporter activity appears to be higher in cotyledons than in roots in 6- to 7-d-old Arabidopsis seedlings (Fig. 3 in Ulmasov et al., 1997). In 20-d-old Arabidopsis seedlings, the downstream BRs, 6-deoxocastasterone and castasterone, accumulate to higher levels in shoots than in roots (Bancos et al., 2002; Shimada et al., 2003). Therefore, the distribution of endogenous BR appears consistent with the organ-specific DR5-GUS reporter activity. The inconsistency between the DR5-GUS reporter activity and the DR5-GUS transcript accumulation in roots and shoots might be due to highly localized expression of the DR5-GUS gene in root tips.
In this study, the IAA5 and IAA19 genes (Fig. 1) and the DR5-GUS reporter gene (Figs. 2 and 3) were induced within 30 min and remained activated up to 24 h posttreatment. A 12-h BL treatment accelerated seedling growth (Table I). However, the endogenous free IAA levels per gram fresh weight did not change in either det2 or WT seedlings (Table I). It is possible that endogenous IAA was increased in response to BL treatment, but the increase was below the limits of detection. For example, it was increased mainly in cotyledons as a result of transport from other organs. Alternatively, endogenous IAA might not increase in response to BL treatment. If this is the case, the AuxRE is not specific to auxin but responds to both auxins and BRs. Of note, endogenous IAA levels were higher in the BR-deficient mutant det2 than in the WT (Table I). This indicates that the lower endogenous BRs in the det2 mutant do not result in lower endogenous IAA levels. Moreover, the abundance of IAA5 and IAA19 transcripts was lower in det2 seedlings than in WT seedlings (Fig. 2), suggesting that decreased BRs down-regulate IAA5 and IAA19 gene expression in the det2 mutant not via decreased endogenous IAA levels but with increased IAA levels. Müssig et al. (2002) also reported that IAA2, IAA3, IAA13, and IAA22 transcript levels are lower in BR-deficient plants than in the WT. These findings support the possibility that BL regulates IAA genes and the transcriptional activity of AuxRE without altering the levels of auxin molecules. The increased IAA levels in det2 seedlings may be independent of their BR content because IAA levels did not change in response to exogenous BL treatment. One possible explanation is that the higher IAA levels in det2 are due to the larger seedling number per gram fresh weight in det2 as compared with the WT, given that det2 seedlings are smaller than WT seedlings. Another explanation is that the lower level of endogenous BRs in the det2 mutant results in decreased auxin sensitivity, which in turn causes increased IAA levels.
Although both IAA and BL activate the IAA5, IAA19, and DR5-GUS genes, a number of characteristics distinguish BL and IAA induction. IAA induction is steep and transient, consistent with the proposed hypothesis that IAA proteins function as self-repressors (Ulmasov et al., 1997; Gray and Estelle, 2000; Hagen and Guilfoyle, 2002). In contrast, BL induction is gradual and continuous. There is less evidence for a self-repressing system in BL induction. BL might induce these genes with a different mechanism from that of auxins. Meanwhile, IAA induction is dose dependent and is highest at higher IAA concentrations (1 μm, Figs. 1 and 3), whereas BL induction is dose independent between 10 nm and 1 μm (Figs. 1 and 3). This is probably because the BL response is saturated at BL treatments exceeding 10 nm. We found that BL induction of the DR5-GUS gene starts at about 100 pm and saturates between 10 and 100 nm (Fig. 3C). Interestingly, the endogenous free BL level is <400 pm in light-grown plants (Choe et al., 2001) and is less in light-grown seedlings (Fujioka et al., 2002) if one calculates this level on the assumption that free BL is distributed uniformly in plants. Similarly, IAA induction starts at between 30 to approximately 50 nm (Fig. 4), which is equivalent to the endogenous free IAA level in seedlings (Table I) calculated using the same assumption. Therefore, each hormone induces the DR5-GUS gene roughly at its physiological concentration, and BL induction starts at a lower concentration than IAA induction dose.
Because IAA induction of the IAA5, IAA19, and DR5-GUS genes did not show saturation, the expression of these genes might be related to a response to higher IAA levels. The most remarkable physiological response to treatment with higher IAA concentrations is growth retardation, which is most significant in roots. This appears consistent with the tissue-specific induction of the DR5-GUS reporter; IAA induces DR5-GUS more strongly in roots than in shoots (Fig. 5). In contrast, the application of approximately 1 μm BL promotes shoot growth (e.g. Li et al., 1996; Seto et al., 2002) and retards root growth (e.g. Clouse et al., 1993; Li et al., 2001). Because BL induces DR5 mainly in cotyledons, activation of the DR5 element by BL might be related to the promotion of growth in cotyledons.
Conclusions
The early auxin-inducible genes, IAA5, IAA19, and DR5-GUS, are induced by BL in Arabidopsis seedlings, without increases in IAA levels per gram fresh weight. Endogenous IAA levels were higher in the det2 mutant than in the WT, whereas the levels of IAA5 and IAA19 transcripts were lower in the det2 mutant than in the WT. The lack of evidence for auxin-mediated activation of early auxin-inducible genes in response to BL suggests that the BR and auxin signaling pathways activate the transcriptional system of the IAA and DR5-GUS genes independently. Very recently, we demonstrated that AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis (Nakamura et al., 2003). Functional characterization of IAA genes and insights into ubiquitin-mediated system will further extend understanding signal interaction between BR and auxin.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis ecotype Col-0 was used as the WT in this study unless otherwise noted. The Arabidopsis mutant det2-1 (Chory et al., 1991) was used as the BR-deficient mutant, and bri1-5 (Noguchi et al., 1999) was used as the BR-insensitive mutant. The DR5-GUS transgenic plant has been described by Ulmasov et al. (1997). Seedlings were grown for 7 d in one-half-strength Murashige and Skoog (Murashige and Skoog, 1962) liquid medium (Life Technologies/Gibco-BRL, Cleveland) supplemented with 1.5% (w/v) Suc, with shaking at 120 rpm at 22°C under continuous fluorescent light of 100 μE m-2 s-1. The seedlings were then treated with hormones or mock treated with 0.1% (v/v) dimethyl sulfoxide in the medium.
Isolation of RNAs and RTQ RT-PCR
Isolation of RNAs and RTQ RT-PCR were performed essentially as described previously (Shimada et al., 2001, 2003). In brief, total RNAs were extracted from Arabidopsis seedlings using the guanidine-hydrochloride method. The RNAs were then treated with DNase I and converted to cDNAs with random primers using the Super Script First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Quantitative RT-PCR was performed using real-time-monitoring Taq-Man technology (Holland et al., 1991) with a model 7700 sequence detector and a Taq-Man Universal PCR Master Mix (Perkin-Elmer Applied Biosystems, Foster City, CA). The gene-specific Taq-Man probes and PCR primers were designed to avoid homology to other members of the IAA gene family, using the program Primer Express version 1.5 (Perkin-Elmer Applied Biosystems) and the BLAST program (Altschul et al., 1990). The IAA5 gene was analyzed using the forward primer (5′-AAGAGTCAAGTTGTGGGTTGGC-3′), the reverse primer (5′-AATGCAGCTCCATCTACACTCACT-3′), and the Taq-Man probe (5′-FAM-CGAACTTTTGGTCCGTTCGAGACTGTTC-TAMRA-3′; complement). IAA19 was analyzed with the forward primer (5′-GAGCATGGATGGTGTGCCTTAT-3′), the reverse primer (5′-TTCGCAGTTGTCACCATCTTTC-3′), and the Taq-Man probe (5′-FAMATAAGCTCTTCGGTTTCCGTGGCATCG-TAMRA-3′) or with the forward primer (5′-TGAATATGACGTCGTCGGGTAGTA-3′), the reverse primer (5′-CCGGCGAATCATTAACCTTCT-3′), and the Taq-Man probe (5′-FAM-CCAGATGAAACGACGCCGCTTTCA-TAMRA-3′; complement). Because the sequences of the IAA5 and IAA19 genes are the two most closely related in the IAA gene family (Liscum and Reed, 2002), we tested the risk of cross amplification. Solutions of IAA5 or IAA19 cDNAs were used as templates for RTQ-PCR with the probe and primers specific for the other gene. We found that neither probe-primer set recognizes the cDNA template of the other gene (data not shown). The GUS gene was analyzed with the forward primer (5′-GGCAGGCCAGCGTATCG-3′), the reverse primer (5′-TTGACCCACACTTTGCCGTA-3′), and the Taq-Man probe (5′-FAM-CTGCGTTTCGATGCGGTCACTCA-TAMRA-3′). Primers and a probe for 18S ribosomal RNA were described previously (Goda et al., 2002). There was no or little signal in RTQ-PCR when the RT reactions were omitted.
Histochemical Localization of GUS Activity
The DR5-GUS construct and transgenic plants were described by Ulmasov et al. (1997). Histochemical GUS staining was performed by incubating whole seedlings in GUS staining buffer containing 50 mm sodium phosphate (pH 7.0), 0.5 mm potassium ferrocyanide, 10 mm EDTA, 0.1% (v/v) Triton X-100, 2% (v/v) dimethyl sulfoxide, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C for 24 h (Jefferson, 1987).
GUS Activity Assay
GUS activities were measured by using the GUS activity kit (Sigma, St. Louis) according to the manufacturer's instructions. Plants were ground in liquid nitrogen, then homogenized in 500 μL of extraction buffer containing 50 mm sodium phosphate (pH 7.0), 10 mm EDTA, 10 mm β-mercaptoethanol, 0.1% (w/v) sodium n-lauroylsarcosine, and 0.1% (v/v) Triton X-100. To assay GUS activity, 25 μL of the extracts was mixed with 75 μL of 0.7 mm 4-methylumbelliferyl β-d-glucuronide in extraction buffer prewarmed to 37°C. The mixture was incubated at 37°C for 90 min, and 10 μL of the mixture was mixed with 200 μL of stop solution (0.2 m sodium carbonate). The resulting fluorescence was measured with a multilabel counter, the Wallac 1420 ARVOsx2 (Perkin Elmer Life Science, Boston) at 355 (excitation) and 460 (emission) nm. Protein content was determined according to the Bradford method (Bradford, 1976) to normalize GUS activity.
Quantification of Endogenous IAA
For GC single-ion monitoring MS analyses of free IAA, fresh plant material was carefully weighed, frozen in liquid nitrogen, and stored at -80°C. The material was then ground in liquid nitrogen using a mortar and pestle. After addition of [13C6]IAA at a rate of 30 pg/1 mg fresh weight (Cambridge Isotope Lab, Andover, MA) as an internal standard, the material was extracted in 80% (v/v) acetone with 0.1 mg mL-1 2,6-di-tert-butyl-4-methylphenol (BHT) for 60 min. After centrifugation, the supernatant was collected. The pellet was re-extracted for 90 min, and the supernatant was brought to a water phase in a rotary evaporator. After adjustment of the pH to 2.0, the aqueous phase was then partitioned twice against ether containing 0.01 mg mL-1 BHT. The combined ether phase, which contained ether-soluble acidic and neutral substances, was concentrated to about 0.6 mL under a nitrogen stream. After adjustment of the pH to 10.0 with 2% (w/v) NaHCO3, vortexing for 1 min, and centrifugation, the organic phase was discarded. The pH was adjusted to 2.0, an equal volume of ether containing 0.01 g mL-1 BHT was added, and the material was dried under a nitrogen stream and dissolved in methanol. IAA was purified by HPLC using a Nucleosil N(CH3)2 column (Senshu, Tokyo) and a mobile phase of methanol with 0.03% (w/v) acetic acid. The purified IAA fraction was dried under a nitrogen stream and trimethylsilylated with N-methyl-N-trimethylsilyltrifluoroacetamide at 60°C for 15 min. Splitless injections were made into a GC-single-ion monitoring-MS system (QP5050A, Shimadzu, Kyoto) equipped with a capillary column (DB-1, 0.25-mm × 30 m i.d., 0.25-μm film thickness; J&W Scientific, Folsom, CA). A linear temperature gradient was applied from 80°C to 280°C with an increase of 20°C min-1. The injection temperature of the GC was 250°C, the ion source temperature of the MS was 250°C, a helium flow of 1.2 mL min-1 was applied, the ionization potential was 70 eV, and the scan time was 0.2 s. The percentages of IAA molecules labeled with 13C were calculated from the relative intensities of m/z 202 to 208 and 319 to 325 ions after subtraction of background.
Acknowledgments
We thank Dr. Tom J. Guilfoyle for providing the DR5-GUS system and Mr. Narumasa Miyauchi for technical assistance with molecular techniques.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.030031.
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533-549 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410 [DOI] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843-852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573-582 [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD (2002) Brassinosteroids. In The Arabidopsis Book. American Society of Plant Biologists. http://www.aspb.org/publications/arabidopsis/toc.cfm DOI/10.1199/tab.0009
- Clouse SD, Feldmann KA (1999) Molecular genetics of brassinosteroid action. In A Sakurai, T Yokota, S Clouse, eds, Brassinosteroids: Steroidal Plant Hormones. Springer-Verlag, Tokyo, pp 163-190
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker ME (1993) Physiological and molecular effects of brassinosteroids on Arabidopsis thaliana. J Plant Growth Regul 12: 61-66 [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427-451 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Zurek DM, McMorris TC, Baker ME (1992) Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol 100: 1377-1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Meudt WJ (1983) Investigation on the mechanism of the brassinosteroid response. Plant Physiol 72: 691-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner TW, Goekjian VH, LaFayette PR, Key JL (1990) Structure and expression of two auxin-inducible genes from Arabidopsis. Plant Mol Biol 15: 623-632 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J et al. (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951-1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Takatsuto S, Yoshida S (1998) Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49: 1841-1848 [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137-164 [DOI] [PubMed] [Google Scholar]
- Gil P, Liu Y, Orbovic V, Verkamp E, Poff KL, Green PJ (1994) Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol 104: 777-784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Estelle M (2000) Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci 25: 133-138 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373-385 [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction product by utilizing the 5′->3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 88: 7276-7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev EA, Xu W, Polisensky DH, Oh MH, Torisky RS, Clouse SD, Braam J (2002) Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli: roles of cis regions and brassinosteroids. Plant Physiol 130: 770-783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387-405 [Google Scholar]
- Katsumi M (1985) Interaction of a brassinosteroid with IAA and GA3 in the elongation of Cucumber hypocotyl sections. Plant Cell Physiol 26: 615-625 [Google Scholar]
- Kowalczyk M, Sandberg G (2001) Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol 127: 1845-1853 [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398-401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387-400 [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645-657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava NB (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39: 23-52 [Google Scholar]
- McKay MJ, Ross JJ, Lawrence NL, Cramp RE, Beveridge CA, Reid JB (1994) Control of internode length in Pisum sativum. Plant Physiol 106: 1521-1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meudt WJ (1987) Investigations on the mechanism of the brassinosteroid response: VI. Effect of brassinolide on gravitropism of bean hypocotyls. Plant Physiol 83: 195-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473-498 [Google Scholar]
- Müssig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241-1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Shimada Y, Goda H, Fujiwara MT, Asami T, Yoshida S (2003) AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis. FEBS Lett 553: 28-32 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T (1997) Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol 113: 31-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463-472 [DOI] [PubMed] [Google Scholar]
- Sala C, Sala F (1985) Effect of brassinosteroid on cell division and enlargement in cultured carrot (Daucus carota L.) cells. Plant Cell Rep 4: 144-147 [DOI] [PubMed] [Google Scholar]
- Sasse J (1999) Physiological actions of brassinosteroids. In A Sakurai, T Yokota, SD Clouse, eds, Brassinosteroids: Steroidal Plant Hormones. Springer-Verlag, Tokyo, pp 137-161
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T (2002) The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32: 1011-1022 [DOI] [PubMed] [Google Scholar]
- Seto H, Hiranuma S, Fujioka S, Koshino H, Suenaga T, Yoshida S (2002) Preparation, conformational analysis and biological evaluation of 6a-carbabrassinolide and related compounds. Tetrahedron 58: 9741-9749 [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126: 770-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeno K, Pharis RP (1982) Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: an auxin-mediated phenomenon. Plant Cell Physiol 23: 1275-1281 [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7: 1611-1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963-1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380-383 [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555-1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yopp JH, Mandava NB, Sasse JM (1981) Brassinolide, a growth-promoting steroidal lactone: I. Activity in selected auxin bioassays. Physiol Plant 53: 445-452 [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD (1994) Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol 104: 505-513 [DOI] [PMC free article] [PubMed] [Google Scholar]