Abstract
The title compound, C6H5NO3, crystallizes in the triclinic system with six independent molecules in the asymmetric unit. In a previous study, the structure of the title compound was determined in the monoclinic P21/n space group at 100 K [Valerga et al. (2009 ▶). Acta Cryst. E65, o1979]. All six independent molecules display an E configuration about the C=C double bond, with the dihedral angles between the planes of the furan rings and the nitroalkenyl groups ranging from 0.61 (7) to 5.03 (7)°. The crystal structure is stabilized by intermolecular C—H⋯O hydrogen-bonding interactions.
Related literature
For the use of nitroalkenes in organic synthesis, see: Ranu et al. (2005 ▶); Ballini & Bosica (2005 ▶); Ono (2005 ▶). For the structure of the monoclinic polymorph, see: Valerga et al. (2009 ▶).
Experimental
Crystal data
C6H5NO3
M r = 139.11
Triclinic,
a = 9.8407 (14) Å
b = 13.4270 (19) Å
c = 15.300 (2) Å
α = 91.105 (1)°
β = 108.603 (2)°
γ = 91.172 (1)°
V = 1914.8 (5) Å3
Z = 12
Mo Kα radiation
μ = 0.12 mm−1
T = 298 K
0.12 × 0.10 × 0.10 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1997 ▶) T min = 0.976, T max = 0.988
13307 measured reflections
8182 independent reflections
4235 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.058
wR(F 2) = 0.149
S = 0.92
8182 reflections
541 parameters
H-atom parameters constrained
Δρmax = 0.15 e Å−3
Δρmin = −0.22 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT-Plus (Bruker, 2001 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053681002492X/rz2466sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681002492X/rz2466Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯O10i | 0.93 | 2.58 | 3.382 (3) | 145 |
| C16—H16⋯O13 | 0.93 | 2.60 | 3.370 (3) | 141 |
| C26—H26⋯O14ii | 0.93 | 2.59 | 3.281 (3) | 131 |
| C28—H28⋯O8iii | 0.93 | 2.59 | 3.405 (3) | 146 |
| C34—H34⋯O4 | 0.93 | 2.59 | 3.383 (3) | 143 |
Symmetry codes: (i) 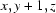 ; (ii)
; (ii) 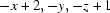 ; (iii)
; (iii) 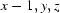 .
.
Acknowledgments
Financial support of this work by the Natural Science Foundation of Hubei Province(2008CDB036) is greatly appreciated.
supplementary crystallographic information
Comment
Nitroalkenes are good substrates for Michael addition reactions because of the stronger electrowithdrawing property of the nitro group (Ranu et al., 2005). At the same time, the nitro group can provide a good nitrogen source for the synthesis of many useful organic molecules (Ballini & Bosica, 2005; Ono, 2005). Our group focus on new organic transformations obtained by nitroalkene as substrates. In this paper, we report the structure of a triclinic polymorph of the title compound. Recently, the structure of the title compound was determined in the monoclinic P21/n space group at 100 K (Valerga et al., 2009).
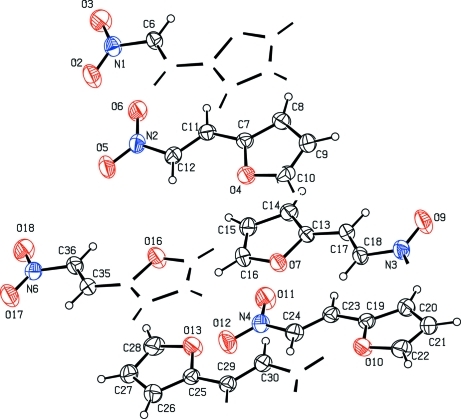
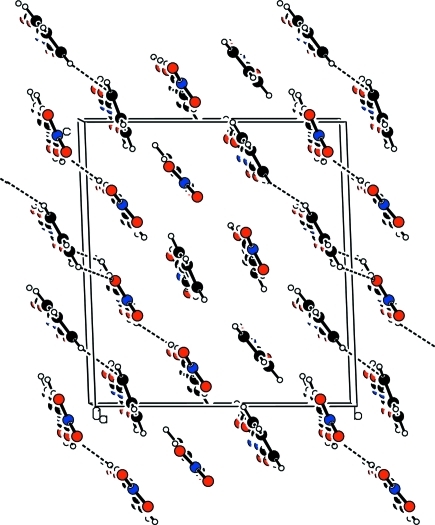
In the asymmetric unit of the title compound, there are six independent molecules (Fig.1). All molecules display an E configuration about the C═C double bond. Bond lengths and angles are in normal ranges and are comparable with those found in the monoclinic polymorph. The dihedral angles between the planes of the furan rings and the nitroalkenyl groups range from 0.61 (7) to 5.03 (7)°. The crystal structure (Fig. 2) is stabilized by intermolecular C—H···O hydrogen bonding interactions (Table 1).
Experimental
Furfural (0.1 mol) and nitromethane (0.1 mol) were dissolved in 30 ml CH3OH with stirring under ice bath, and a few ml of a NaOH in CH3OH solution was added dropwise. After stirring for 1 h, ice water was added and the solution neutralized with a diluted hydrochloric acid solution. The yellowish-brown solid precipitate was filtered and recrystallized from C2H5OH (yield 92%). Crystals suitable for X-ray analysis were obtained by slow evaporation of a dichloromethane/hexane (1:1 v/v) solution at 283 K.
Refinement
All hydrogen atoms were placed in geometrically idealized positions with C–H = 0.93 Å, and constrained to ride on their parent atoms with Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The asymmetric unit of the title compound showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Crystal packing for the title compound, with C—H···O interactions drawn as dashed lines.
Crystal data
| C6H5NO3 | Z = 12 |
| Mr = 139.11 | F(000) = 864 |
| Triclinic, P1 | Dx = 1.448 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.8407 (14) Å | Cell parameters from 2345 reflections |
| b = 13.4270 (19) Å | θ = 1.2–25.9° |
| c = 15.300 (2) Å | µ = 0.12 mm−1 |
| α = 91.105 (1)° | T = 298 K |
| β = 108.603 (2)° | Block, colourless |
| γ = 91.172 (1)° | 0.12 × 0.10 × 0.10 mm |
| V = 1914.8 (5) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 8182 independent reflections |
| Radiation source: fine-focus sealed tube | 4235 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| φ and ω scans | θmax = 27.0°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1997) | h = −12→12 |
| Tmin = 0.976, Tmax = 0.988 | k = −16→17 |
| 13307 measured reflections | l = −14→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.149 | H-atom parameters constrained |
| S = 0.92 | w = 1/[σ2(Fo2) + (0.0705P)2] where P = (Fo2 + 2Fc2)/3 |
| 8182 reflections | (Δ/σ)max = 0.002 |
| 541 parameters | Δρmax = 0.15 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.9416 (2) | 0.92135 (16) | 0.24460 (16) | 0.0650 (6) | |
| H1 | 0.9831 | 0.9532 | 0.2056 | 0.078* | |
| C2 | 1.0123 (2) | 0.86624 (16) | 0.31429 (16) | 0.0644 (6) | |
| H2 | 1.1096 | 0.8528 | 0.3329 | 0.077* | |
| C3 | 0.9112 (2) | 0.83226 (15) | 0.35424 (15) | 0.0599 (6) | |
| H3 | 0.9288 | 0.7912 | 0.4047 | 0.072* | |
| C4 | 0.7841 (2) | 0.86944 (14) | 0.30671 (14) | 0.0479 (5) | |
| C5 | 0.6460 (2) | 0.85930 (14) | 0.31761 (14) | 0.0517 (5) | |
| H5 | 0.6387 | 0.8211 | 0.3661 | 0.062* | |
| C6 | 0.5279 (2) | 0.89934 (15) | 0.26493 (14) | 0.0539 (5) | |
| H6 | 0.5308 | 0.9382 | 0.2159 | 0.065* | |
| C7 | 0.7237 (2) | 0.63452 (14) | 0.18843 (14) | 0.0491 (5) | |
| C8 | 0.8029 (2) | 0.67212 (15) | 0.13981 (16) | 0.0638 (6) | |
| H8 | 0.7714 | 0.7146 | 0.0905 | 0.077* | |
| C9 | 0.9416 (2) | 0.63636 (16) | 0.17613 (17) | 0.0669 (7) | |
| H9 | 1.0196 | 0.6501 | 0.1560 | 0.080* | |
| C10 | 0.9392 (2) | 0.57880 (16) | 0.24518 (17) | 0.0657 (6) | |
| H10 | 1.0178 | 0.5452 | 0.2819 | 0.079* | |
| C11 | 0.5770 (2) | 0.64540 (14) | 0.18134 (14) | 0.0515 (5) | |
| H11 | 0.5239 | 0.6880 | 0.1368 | 0.062* | |
| C12 | 0.5094 (2) | 0.60051 (15) | 0.23223 (15) | 0.0567 (6) | |
| H12 | 0.5587 | 0.5573 | 0.2776 | 0.068* | |
| C13 | 0.8059 (2) | 0.37791 (14) | 0.10512 (14) | 0.0484 (5) | |
| C14 | 0.6775 (2) | 0.40843 (14) | 0.05235 (15) | 0.0570 (6) | |
| H14 | 0.6596 | 0.4424 | −0.0024 | 0.068* | |
| C15 | 0.5752 (2) | 0.38043 (15) | 0.09389 (16) | 0.0599 (6) | |
| H15 | 0.4772 | 0.3915 | 0.0726 | 0.072* | |
| C16 | 0.6481 (2) | 0.33457 (16) | 0.17040 (16) | 0.0643 (6) | |
| H16 | 0.6070 | 0.3081 | 0.2121 | 0.077* | |
| C17 | 0.9454 (2) | 0.38754 (14) | 0.09616 (14) | 0.0498 (5) | |
| H17 | 0.9525 | 0.4201 | 0.0446 | 0.060* | |
| C18 | 1.0657 (2) | 0.35470 (14) | 0.15413 (14) | 0.0525 (5) | |
| H18 | 1.0631 | 0.3202 | 0.2057 | 0.063* | |
| C19 | 0.8813 (2) | 0.13701 (14) | 0.01911 (15) | 0.0495 (5) | |
| C20 | 0.9605 (2) | 0.15719 (15) | −0.03563 (16) | 0.0607 (6) | |
| H20 | 0.9267 | 0.1808 | −0.0953 | 0.073* | |
| C21 | 1.1038 (2) | 0.13626 (15) | 0.01342 (18) | 0.0660 (6) | |
| H21 | 1.1826 | 0.1428 | −0.0073 | 0.079* | |
| C22 | 1.1038 (2) | 0.10540 (16) | 0.09513 (18) | 0.0697 (7) | |
| H22 | 1.1852 | 0.0868 | 0.1418 | 0.084* | |
| C23 | 0.7319 (2) | 0.14092 (14) | 0.00502 (15) | 0.0512 (5) | |
| H23 | 0.6750 | 0.1662 | −0.0504 | 0.061* | |
| C24 | 0.6664 (2) | 0.11183 (15) | 0.06361 (15) | 0.0565 (6) | |
| H24 | 0.7199 | 0.0866 | 0.1200 | 0.068* | |
| C25 | 0.7166 (2) | 0.11542 (14) | 0.39786 (15) | 0.0505 (5) | |
| C26 | 0.6371 (2) | 0.08594 (15) | 0.45004 (16) | 0.0640 (6) | |
| H26 | 0.6703 | 0.0533 | 0.5056 | 0.077* | |
| C27 | 0.4949 (2) | 0.11306 (16) | 0.40575 (18) | 0.0675 (7) | |
| H27 | 0.4161 | 0.1022 | 0.4258 | 0.081* | |
| C28 | 0.4967 (2) | 0.15721 (17) | 0.32982 (19) | 0.0766 (7) | |
| H28 | 0.4164 | 0.1829 | 0.2868 | 0.092* | |
| C29 | 0.8654 (2) | 0.10866 (14) | 0.40970 (14) | 0.0512 (5) | |
| H29 | 0.9203 | 0.0741 | 0.4604 | 0.061* | |
| C30 | 0.9330 (2) | 0.14664 (15) | 0.35584 (15) | 0.0564 (6) | |
| H30 | 0.8818 | 0.1819 | 0.3046 | 0.068* | |
| C31 | 0.6315 (2) | 0.36980 (14) | 0.48376 (15) | 0.0505 (5) | |
| C32 | 0.7617 (2) | 0.34777 (16) | 0.53904 (15) | 0.0635 (6) | |
| H32 | 0.7822 | 0.3193 | 0.5966 | 0.076* | |
| C33 | 0.8624 (2) | 0.37469 (15) | 0.49599 (17) | 0.0628 (6) | |
| H33 | 0.9613 | 0.3678 | 0.5186 | 0.075* | |
| C34 | 0.7873 (3) | 0.41190 (17) | 0.41671 (17) | 0.0739 (7) | |
| H34 | 0.8270 | 0.4361 | 0.3734 | 0.089* | |
| C35 | 0.4923 (2) | 0.35871 (14) | 0.49243 (14) | 0.0522 (5) | |
| H35 | 0.4868 | 0.3295 | 0.5459 | 0.063* | |
| C36 | 0.3707 (2) | 0.38543 (15) | 0.43283 (15) | 0.0565 (6) | |
| H36 | 0.3716 | 0.4150 | 0.3785 | 0.068* | |
| N1 | 0.39430 (19) | 0.88338 (14) | 0.28277 (13) | 0.0603 (5) | |
| N2 | 0.3605 (2) | 0.61778 (13) | 0.21816 (13) | 0.0608 (5) | |
| N3 | 1.19984 (19) | 0.37178 (13) | 0.13825 (14) | 0.0597 (5) | |
| N4 | 0.51390 (19) | 0.11858 (12) | 0.04148 (14) | 0.0589 (5) | |
| N5 | 1.08370 (18) | 0.13472 (13) | 0.37455 (13) | 0.0574 (5) | |
| N6 | 0.23663 (19) | 0.36992 (13) | 0.44974 (14) | 0.0600 (5) | |
| O1 | 0.80031 (15) | 0.92554 (10) | 0.23725 (10) | 0.0586 (4) | |
| O2 | 0.38865 (17) | 0.82959 (14) | 0.34471 (12) | 0.0853 (5) | |
| O3 | 0.29023 (16) | 0.92505 (12) | 0.23200 (12) | 0.0870 (5) | |
| O4 | 0.80683 (15) | 0.57538 (10) | 0.25532 (10) | 0.0614 (4) | |
| O5 | 0.30358 (18) | 0.57248 (13) | 0.26641 (12) | 0.0847 (5) | |
| O6 | 0.29579 (16) | 0.67661 (12) | 0.16014 (11) | 0.0804 (5) | |
| O7 | 0.79022 (15) | 0.33117 (10) | 0.17990 (9) | 0.0588 (4) | |
| O8 | 1.30633 (16) | 0.33720 (12) | 0.19333 (12) | 0.0844 (5) | |
| O9 | 1.20458 (17) | 0.41996 (14) | 0.07243 (12) | 0.0877 (6) | |
| O10 | 0.96849 (15) | 0.10452 (10) | 0.10184 (10) | 0.0629 (4) | |
| O11 | 0.44264 (16) | 0.15312 (12) | −0.03166 (11) | 0.0803 (5) | |
| O12 | 0.46063 (17) | 0.08859 (13) | 0.09852 (12) | 0.0849 (5) | |
| O13 | 0.63096 (15) | 0.16044 (11) | 0.32253 (11) | 0.0711 (5) | |
| O14 | 1.15176 (16) | 0.09038 (12) | 0.44346 (11) | 0.0782 (5) | |
| O15 | 1.13827 (17) | 0.17002 (12) | 0.32070 (12) | 0.0805 (5) | |
| O16 | 0.64420 (16) | 0.41055 (11) | 0.40583 (10) | 0.0709 (5) | |
| O17 | 0.23464 (18) | 0.33222 (13) | 0.52140 (13) | 0.0894 (6) | |
| O18 | 0.12831 (16) | 0.39605 (12) | 0.39036 (11) | 0.0817 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0618 (16) | 0.0695 (15) | 0.0780 (17) | −0.0058 (12) | 0.0427 (14) | 0.0025 (13) |
| C2 | 0.0469 (13) | 0.0728 (15) | 0.0772 (17) | 0.0049 (11) | 0.0246 (13) | 0.0062 (13) |
| C3 | 0.0560 (14) | 0.0666 (14) | 0.0570 (14) | 0.0048 (11) | 0.0171 (12) | 0.0129 (11) |
| C4 | 0.0490 (13) | 0.0499 (12) | 0.0475 (13) | −0.0027 (9) | 0.0190 (10) | 0.0047 (10) |
| C5 | 0.0531 (13) | 0.0529 (12) | 0.0511 (13) | −0.0015 (10) | 0.0195 (11) | 0.0040 (10) |
| C6 | 0.0502 (13) | 0.0580 (13) | 0.0565 (14) | −0.0032 (10) | 0.0215 (11) | 0.0050 (10) |
| C7 | 0.0463 (12) | 0.0480 (12) | 0.0522 (13) | 0.0049 (9) | 0.0142 (10) | 0.0078 (10) |
| C8 | 0.0573 (14) | 0.0591 (14) | 0.0791 (17) | 0.0037 (11) | 0.0264 (13) | 0.0173 (12) |
| C9 | 0.0541 (15) | 0.0621 (15) | 0.0925 (19) | 0.0008 (11) | 0.0344 (14) | 0.0078 (13) |
| C10 | 0.0481 (14) | 0.0652 (15) | 0.0800 (18) | 0.0130 (11) | 0.0144 (12) | 0.0033 (13) |
| C11 | 0.0502 (13) | 0.0460 (12) | 0.0575 (14) | 0.0037 (9) | 0.0158 (11) | 0.0053 (10) |
| C12 | 0.0507 (13) | 0.0602 (13) | 0.0621 (15) | 0.0135 (10) | 0.0207 (12) | 0.0092 (11) |
| C13 | 0.0480 (12) | 0.0481 (12) | 0.0525 (13) | −0.0016 (9) | 0.0210 (10) | 0.0071 (10) |
| C14 | 0.0487 (13) | 0.0608 (13) | 0.0608 (14) | 0.0065 (10) | 0.0156 (11) | 0.0135 (11) |
| C15 | 0.0432 (12) | 0.0628 (14) | 0.0753 (16) | 0.0035 (10) | 0.0208 (12) | 0.0050 (12) |
| C16 | 0.0562 (15) | 0.0691 (15) | 0.0791 (17) | −0.0022 (11) | 0.0378 (13) | 0.0097 (13) |
| C17 | 0.0507 (13) | 0.0516 (12) | 0.0514 (13) | −0.0012 (10) | 0.0223 (10) | 0.0042 (10) |
| C18 | 0.0462 (12) | 0.0584 (13) | 0.0573 (14) | −0.0003 (10) | 0.0226 (11) | 0.0071 (10) |
| C19 | 0.0449 (12) | 0.0456 (12) | 0.0581 (14) | 0.0041 (9) | 0.0161 (11) | 0.0067 (10) |
| C20 | 0.0558 (14) | 0.0641 (14) | 0.0666 (15) | 0.0086 (11) | 0.0244 (12) | 0.0153 (12) |
| C21 | 0.0523 (14) | 0.0609 (14) | 0.0933 (19) | 0.0058 (11) | 0.0343 (14) | 0.0140 (13) |
| C22 | 0.0402 (13) | 0.0669 (15) | 0.097 (2) | 0.0073 (10) | 0.0131 (13) | 0.0213 (14) |
| C23 | 0.0449 (12) | 0.0487 (12) | 0.0589 (14) | 0.0041 (9) | 0.0147 (11) | 0.0060 (10) |
| C24 | 0.0408 (12) | 0.0586 (13) | 0.0652 (15) | 0.0022 (10) | 0.0100 (11) | 0.0069 (11) |
| C25 | 0.0444 (12) | 0.0465 (12) | 0.0606 (14) | 0.0040 (9) | 0.0165 (11) | 0.0053 (10) |
| C26 | 0.0591 (15) | 0.0654 (15) | 0.0714 (16) | 0.0058 (11) | 0.0258 (13) | 0.0098 (12) |
| C27 | 0.0523 (14) | 0.0587 (14) | 0.099 (2) | 0.0009 (11) | 0.0341 (14) | 0.0041 (13) |
| C28 | 0.0416 (13) | 0.0735 (16) | 0.110 (2) | 0.0120 (11) | 0.0151 (14) | 0.0239 (15) |
| C29 | 0.0441 (12) | 0.0502 (12) | 0.0580 (14) | 0.0041 (9) | 0.0140 (10) | 0.0055 (10) |
| C30 | 0.0418 (12) | 0.0607 (13) | 0.0644 (15) | 0.0063 (10) | 0.0130 (11) | 0.0113 (11) |
| C31 | 0.0547 (14) | 0.0463 (12) | 0.0535 (14) | −0.0002 (10) | 0.0212 (11) | 0.0068 (10) |
| C32 | 0.0570 (14) | 0.0734 (15) | 0.0646 (15) | 0.0101 (11) | 0.0241 (13) | 0.0195 (12) |
| C33 | 0.0493 (13) | 0.0651 (14) | 0.0773 (17) | 0.0053 (11) | 0.0241 (13) | 0.0112 (12) |
| C34 | 0.0607 (16) | 0.0906 (18) | 0.0818 (19) | −0.0017 (13) | 0.0385 (14) | 0.0167 (14) |
| C35 | 0.0546 (13) | 0.0515 (12) | 0.0549 (14) | 0.0029 (10) | 0.0232 (11) | 0.0059 (10) |
| C36 | 0.0501 (13) | 0.0594 (13) | 0.0638 (15) | −0.0009 (10) | 0.0233 (12) | 0.0082 (11) |
| N1 | 0.0445 (11) | 0.0722 (13) | 0.0635 (13) | −0.0022 (9) | 0.0162 (10) | 0.0023 (10) |
| N2 | 0.0531 (12) | 0.0695 (13) | 0.0664 (13) | 0.0092 (10) | 0.0277 (10) | 0.0066 (10) |
| N3 | 0.0474 (11) | 0.0686 (12) | 0.0667 (13) | 0.0013 (9) | 0.0232 (10) | 0.0070 (10) |
| N4 | 0.0449 (11) | 0.0592 (12) | 0.0732 (14) | 0.0007 (9) | 0.0194 (10) | 0.0058 (10) |
| N5 | 0.0436 (11) | 0.0605 (12) | 0.0689 (14) | 0.0040 (9) | 0.0186 (10) | 0.0046 (10) |
| N6 | 0.0503 (12) | 0.0578 (12) | 0.0761 (14) | 0.0038 (9) | 0.0257 (11) | 0.0066 (10) |
| O1 | 0.0573 (9) | 0.0610 (9) | 0.0617 (10) | 0.0035 (7) | 0.0243 (8) | 0.0130 (7) |
| O2 | 0.0609 (11) | 0.1205 (14) | 0.0839 (13) | 0.0062 (10) | 0.0338 (9) | 0.0390 (11) |
| O3 | 0.0484 (10) | 0.1061 (13) | 0.1021 (13) | 0.0130 (9) | 0.0155 (9) | 0.0318 (10) |
| O4 | 0.0534 (9) | 0.0672 (10) | 0.0656 (10) | 0.0106 (7) | 0.0203 (8) | 0.0171 (8) |
| O5 | 0.0710 (11) | 0.1055 (13) | 0.0971 (13) | 0.0118 (10) | 0.0522 (10) | 0.0297 (10) |
| O6 | 0.0586 (10) | 0.1003 (13) | 0.0887 (12) | 0.0279 (9) | 0.0290 (9) | 0.0350 (10) |
| O7 | 0.0507 (9) | 0.0679 (10) | 0.0616 (10) | 0.0032 (7) | 0.0220 (7) | 0.0174 (8) |
| O8 | 0.0466 (9) | 0.1086 (13) | 0.0965 (13) | 0.0181 (9) | 0.0180 (9) | 0.0305 (10) |
| O9 | 0.0608 (11) | 0.1255 (15) | 0.0889 (13) | 0.0044 (10) | 0.0385 (10) | 0.0417 (11) |
| O10 | 0.0447 (9) | 0.0705 (10) | 0.0728 (11) | 0.0043 (7) | 0.0161 (8) | 0.0234 (8) |
| O11 | 0.0480 (9) | 0.1092 (13) | 0.0791 (12) | 0.0135 (9) | 0.0119 (9) | 0.0261 (10) |
| O12 | 0.0587 (11) | 0.1081 (14) | 0.0985 (14) | −0.0004 (9) | 0.0386 (10) | 0.0290 (11) |
| O13 | 0.0428 (9) | 0.0858 (11) | 0.0836 (12) | 0.0072 (8) | 0.0167 (8) | 0.0322 (9) |
| O14 | 0.0495 (9) | 0.1025 (13) | 0.0824 (12) | 0.0191 (9) | 0.0184 (9) | 0.0287 (10) |
| O15 | 0.0602 (10) | 0.1047 (13) | 0.0902 (13) | 0.0043 (9) | 0.0414 (10) | 0.0295 (10) |
| O16 | 0.0574 (10) | 0.0928 (12) | 0.0663 (11) | 0.0032 (8) | 0.0235 (8) | 0.0250 (9) |
| O17 | 0.0698 (12) | 0.1143 (14) | 0.0977 (14) | 0.0103 (10) | 0.0431 (11) | 0.0430 (11) |
| O18 | 0.0482 (10) | 0.1026 (13) | 0.0895 (13) | 0.0134 (9) | 0.0139 (9) | 0.0155 (10) |
Geometric parameters (Å, °)
| C1—C2 | 1.321 (3) | C21—H21 | 0.9300 |
| C1—O1 | 1.362 (2) | C22—O10 | 1.368 (3) |
| C1—H1 | 0.9300 | C22—H22 | 0.9300 |
| C2—C3 | 1.397 (3) | C23—C24 | 1.320 (3) |
| C2—H2 | 0.9300 | C23—H23 | 0.9300 |
| C3—C4 | 1.342 (3) | C24—N4 | 1.434 (2) |
| C3—H3 | 0.9300 | C24—H24 | 0.9300 |
| C4—O1 | 1.362 (2) | C25—C26 | 1.343 (3) |
| C4—C5 | 1.425 (3) | C25—O13 | 1.353 (2) |
| C5—C6 | 1.317 (3) | C25—C29 | 1.422 (3) |
| C5—H5 | 0.9300 | C26—C27 | 1.405 (3) |
| C6—N1 | 1.438 (3) | C26—H26 | 0.9300 |
| C6—H6 | 0.9300 | C27—C28 | 1.319 (3) |
| C7—C8 | 1.336 (3) | C27—H27 | 0.9300 |
| C7—O4 | 1.368 (2) | C28—O13 | 1.361 (3) |
| C7—C11 | 1.424 (3) | C28—H28 | 0.9300 |
| C8—C9 | 1.398 (3) | C29—C30 | 1.317 (3) |
| C8—H8 | 0.9300 | C29—H29 | 0.9300 |
| C9—C10 | 1.327 (3) | C30—N5 | 1.431 (2) |
| C9—H9 | 0.9300 | C30—H30 | 0.9300 |
| C10—O4 | 1.360 (2) | C31—C32 | 1.334 (3) |
| C10—H10 | 0.9300 | C31—O16 | 1.362 (2) |
| C11—C12 | 1.320 (3) | C31—C35 | 1.423 (3) |
| C11—H11 | 0.9300 | C32—C33 | 1.399 (3) |
| C12—N2 | 1.436 (3) | C32—H32 | 0.9300 |
| C12—H12 | 0.9300 | C33—C34 | 1.317 (3) |
| C13—C14 | 1.341 (3) | C33—H33 | 0.9300 |
| C13—O7 | 1.365 (2) | C34—O16 | 1.364 (3) |
| C13—C17 | 1.426 (3) | C34—H34 | 0.9300 |
| C14—C15 | 1.401 (3) | C35—C36 | 1.314 (3) |
| C14—H14 | 0.9300 | C35—H35 | 0.9300 |
| C15—C16 | 1.332 (3) | C36—N6 | 1.435 (3) |
| C15—H15 | 0.9300 | C36—H36 | 0.9300 |
| C16—O7 | 1.361 (2) | N1—O2 | 1.217 (2) |
| C16—H16 | 0.9300 | N1—O3 | 1.224 (2) |
| C17—C18 | 1.323 (3) | N2—O5 | 1.223 (2) |
| C17—H17 | 0.9300 | N2—O6 | 1.226 (2) |
| C18—N3 | 1.431 (3) | N3—O9 | 1.220 (2) |
| C18—H18 | 0.9300 | N3—O8 | 1.223 (2) |
| C19—C20 | 1.341 (3) | N4—O11 | 1.221 (2) |
| C19—O10 | 1.368 (2) | N4—O12 | 1.223 (2) |
| C19—C23 | 1.419 (3) | N5—O15 | 1.216 (2) |
| C20—C21 | 1.407 (3) | N5—O14 | 1.227 (2) |
| C20—H20 | 0.9300 | N6—O17 | 1.222 (2) |
| C21—C22 | 1.325 (3) | N6—O18 | 1.222 (2) |
| C2—C1—O1 | 111.4 (2) | C24—C23—C19 | 125.8 (2) |
| C2—C1—H1 | 124.3 | C24—C23—H23 | 117.1 |
| O1—C1—H1 | 124.3 | C19—C23—H23 | 117.1 |
| C1—C2—C3 | 106.0 (2) | C23—C24—N4 | 120.9 (2) |
| C1—C2—H2 | 127.0 | C23—C24—H24 | 119.5 |
| C3—C2—H2 | 127.0 | N4—C24—H24 | 119.5 |
| C4—C3—C2 | 107.74 (19) | C26—C25—O13 | 108.90 (18) |
| C4—C3—H3 | 126.1 | C26—C25—C29 | 132.1 (2) |
| C2—C3—H3 | 126.1 | O13—C25—C29 | 119.0 (2) |
| C3—C4—O1 | 109.20 (18) | C25—C26—C27 | 107.9 (2) |
| C3—C4—C5 | 131.5 (2) | C25—C26—H26 | 126.0 |
| O1—C4—C5 | 119.26 (19) | C27—C26—H26 | 126.0 |
| C6—C5—C4 | 125.6 (2) | C28—C27—C26 | 105.5 (2) |
| C6—C5—H5 | 117.2 | C28—C27—H27 | 127.2 |
| C4—C5—H5 | 117.2 | C26—C27—H27 | 127.2 |
| C5—C6—N1 | 120.4 (2) | C27—C28—O13 | 111.4 (2) |
| C5—C6—H6 | 119.8 | C27—C28—H28 | 124.3 |
| N1—C6—H6 | 119.8 | O13—C28—H28 | 124.3 |
| C8—C7—O4 | 109.22 (18) | C30—C29—C25 | 126.3 (2) |
| C8—C7—C11 | 131.9 (2) | C30—C29—H29 | 116.9 |
| O4—C7—C11 | 118.83 (19) | C25—C29—H29 | 116.9 |
| C7—C8—C9 | 107.9 (2) | C29—C30—N5 | 121.3 (2) |
| C7—C8—H8 | 126.0 | C29—C30—H30 | 119.3 |
| C9—C8—H8 | 126.0 | N5—C30—H30 | 119.3 |
| C10—C9—C8 | 106.0 (2) | C32—C31—O16 | 108.79 (19) |
| C10—C9—H9 | 127.0 | C32—C31—C35 | 132.6 (2) |
| C8—C9—H9 | 127.0 | O16—C31—C35 | 118.58 (19) |
| C9—C10—O4 | 111.2 (2) | C31—C32—C33 | 108.6 (2) |
| C9—C10—H10 | 124.4 | C31—C32—H32 | 125.7 |
| O4—C10—H10 | 124.4 | C33—C32—H32 | 125.7 |
| C12—C11—C7 | 126.0 (2) | C34—C33—C32 | 105.3 (2) |
| C12—C11—H11 | 117.0 | C34—C33—H33 | 127.4 |
| C7—C11—H11 | 117.0 | C32—C33—H33 | 127.4 |
| C11—C12—N2 | 120.57 (19) | C33—C34—O16 | 111.7 (2) |
| C11—C12—H12 | 119.7 | C33—C34—H34 | 124.1 |
| N2—C12—H12 | 119.7 | O16—C34—H34 | 124.1 |
| C14—C13—O7 | 109.09 (18) | C36—C35—C31 | 126.8 (2) |
| C14—C13—C17 | 131.9 (2) | C36—C35—H35 | 116.6 |
| O7—C13—C17 | 119.01 (18) | C31—C35—H35 | 116.6 |
| C13—C14—C15 | 108.30 (19) | C35—C36—N6 | 121.4 (2) |
| C13—C14—H14 | 125.8 | C35—C36—H36 | 119.3 |
| C15—C14—H14 | 125.8 | N6—C36—H36 | 119.3 |
| C16—C15—C14 | 105.28 (19) | O2—N1—O3 | 123.55 (19) |
| C16—C15—H15 | 127.4 | O2—N1—C6 | 120.06 (19) |
| C14—C15—H15 | 127.4 | O3—N1—C6 | 116.38 (19) |
| C15—C16—O7 | 111.6 (2) | O5—N2—O6 | 122.62 (19) |
| C15—C16—H16 | 124.2 | O5—N2—C12 | 117.32 (18) |
| O7—C16—H16 | 124.2 | O6—N2—C12 | 120.07 (19) |
| C18—C17—C13 | 126.3 (2) | O9—N3—O8 | 122.83 (19) |
| C18—C17—H17 | 116.9 | O9—N3—C18 | 119.96 (18) |
| C13—C17—H17 | 116.9 | O8—N3—C18 | 117.21 (19) |
| C17—C18—N3 | 120.6 (2) | O11—N4—O12 | 122.61 (19) |
| C17—C18—H18 | 119.7 | O11—N4—C24 | 120.2 (2) |
| N3—C18—H18 | 119.7 | O12—N4—C24 | 117.14 (19) |
| C20—C19—O10 | 109.30 (18) | O15—N5—O14 | 123.11 (18) |
| C20—C19—C23 | 132.3 (2) | O15—N5—C30 | 117.66 (19) |
| O10—C19—C23 | 118.35 (19) | O14—N5—C30 | 119.2 (2) |
| C19—C20—C21 | 107.7 (2) | O17—N6—O18 | 122.96 (19) |
| C19—C20—H20 | 126.2 | O17—N6—C36 | 119.78 (19) |
| C21—C20—H20 | 126.2 | O18—N6—C36 | 117.3 (2) |
| C22—C21—C20 | 106.1 (2) | C1—O1—C4 | 105.67 (17) |
| C22—C21—H21 | 127.0 | C10—O4—C7 | 105.69 (17) |
| C20—C21—H21 | 127.0 | C16—O7—C13 | 105.73 (16) |
| C21—C22—O10 | 111.2 (2) | C22—O10—C19 | 105.77 (18) |
| C21—C22—H22 | 124.4 | C25—O13—C28 | 106.26 (18) |
| O10—C22—H22 | 124.4 | C31—O16—C34 | 105.61 (17) |
| O1—C1—C2—C3 | 0.3 (3) | C35—C31—C32—C33 | −179.9 (2) |
| C1—C2—C3—C4 | −0.4 (3) | C31—C32—C33—C34 | 0.1 (3) |
| C2—C3—C4—O1 | 0.4 (2) | C32—C33—C34—O16 | −0.1 (3) |
| C2—C3—C4—C5 | −179.7 (2) | C32—C31—C35—C36 | 178.6 (2) |
| C3—C4—C5—C6 | −179.2 (2) | O16—C31—C35—C36 | −1.2 (3) |
| O1—C4—C5—C6 | 0.7 (3) | C31—C35—C36—N6 | 179.89 (18) |
| C4—C5—C6—N1 | 179.79 (18) | C5—C6—N1—O2 | −2.8 (3) |
| O4—C7—C8—C9 | 0.0 (2) | C5—C6—N1—O3 | 178.35 (19) |
| C11—C7—C8—C9 | −179.6 (2) | C11—C12—N2—O5 | 178.7 (2) |
| C7—C8—C9—C10 | −0.1 (3) | C11—C12—N2—O6 | −1.9 (3) |
| C8—C9—C10—O4 | 0.1 (3) | C17—C18—N3—O9 | 2.5 (3) |
| C8—C7—C11—C12 | 177.2 (2) | C17—C18—N3—O8 | −178.20 (19) |
| O4—C7—C11—C12 | −2.4 (3) | C23—C24—N4—O11 | −1.3 (3) |
| C7—C11—C12—N2 | −179.95 (18) | C23—C24—N4—O12 | 178.77 (19) |
| O7—C13—C14—C15 | −0.2 (2) | C29—C30—N5—O15 | 178.5 (2) |
| C17—C13—C14—C15 | −179.2 (2) | C29—C30—N5—O14 | −1.9 (3) |
| C13—C14—C15—C16 | 0.3 (2) | C35—C36—N6—O17 | 0.9 (3) |
| C14—C15—C16—O7 | −0.3 (3) | C35—C36—N6—O18 | −179.32 (19) |
| C14—C13—C17—C18 | 179.2 (2) | C2—C1—O1—C4 | −0.1 (2) |
| O7—C13—C17—C18 | 0.3 (3) | C3—C4—O1—C1 | −0.2 (2) |
| C13—C17—C18—N3 | −178.25 (18) | C5—C4—O1—C1 | 179.84 (17) |
| O10—C19—C20—C21 | 0.4 (2) | C9—C10—O4—C7 | 0.0 (2) |
| C23—C19—C20—C21 | −177.6 (2) | C8—C7—O4—C10 | 0.0 (2) |
| C19—C20—C21—C22 | −0.5 (2) | C11—C7—O4—C10 | 179.69 (18) |
| C20—C21—C22—O10 | 0.3 (3) | C15—C16—O7—C13 | 0.2 (2) |
| C20—C19—C23—C24 | 174.8 (2) | C14—C13—O7—C16 | 0.0 (2) |
| O10—C19—C23—C24 | −3.0 (3) | C17—C13—O7—C16 | 179.15 (18) |
| C19—C23—C24—N4 | −179.47 (18) | C21—C22—O10—C19 | −0.1 (2) |
| O13—C25—C26—C27 | 0.2 (2) | C20—C19—O10—C22 | −0.2 (2) |
| C29—C25—C26—C27 | 179.0 (2) | C23—C19—O10—C22 | 178.11 (17) |
| C25—C26—C27—C28 | −0.1 (3) | C26—C25—O13—C28 | −0.3 (2) |
| C26—C27—C28—O13 | −0.1 (3) | C29—C25—O13—C28 | −179.25 (19) |
| C26—C25—C29—C30 | −175.2 (2) | C27—C28—O13—C25 | 0.3 (3) |
| O13—C25—C29—C30 | 3.4 (3) | C32—C31—O16—C34 | 0.1 (2) |
| C25—C29—C30—N5 | −179.79 (18) | C35—C31—O16—C34 | 179.92 (18) |
| O16—C31—C32—C33 | −0.1 (2) | C33—C34—O16—C31 | 0.0 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···O10i | 0.93 | 2.58 | 3.382 (3) | 145 |
| C16—H16···O13 | 0.93 | 2.60 | 3.370 (3) | 141 |
| C26—H26···O14ii | 0.93 | 2.59 | 3.281 (3) | 131 |
| C28—H28···O8iii | 0.93 | 2.59 | 3.405 (3) | 146 |
| C34—H34···O4 | 0.93 | 2.59 | 3.383 (3) | 143 |
Symmetry codes: (i) x, y+1, z; (ii) −x+2, −y, −z+1; (iii) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RZ2466).
References
- Ballini, R. & Bosica, G. (2005). Chem. Rev.105, 933–971. [DOI] [PubMed]
- Bruker (2001). SAINT-Plus and SMART Bruker AXS, Inc., Madison, Wisconsin, USA.
- Ono, N. (2005). The Nitro Group in Organic Synthesis Weinheim: Wiley-VCH.
- Ranu, B. C. & Banerjee, S. (2005). Org. Lett.7, 3049–3052. [DOI] [PubMed]
- Sheldrick, G. M. (1997). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Valerga, P., Puerta, M. C., Rodríguez Negrín, Z., Castañedo Cancio, N. & Palma Lovillo, M. (2009). Acta Cryst. E65, o1979. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053681002492X/rz2466sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681002492X/rz2466Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report