Abstract
The yeast (Schizosaccharomyces pombe) SOD2 (Sodium2) gene was introduced into Arabidopsis under the control of the cauliflower mosaic virus 35S promoter. Transformants were selected for their ability to grow on medium containing kanamycin. Southern- and northern-blot analyses confirmed that SOD2 was transferred into the Arabidopsis genome. There were no obvious morphological or developmental differences between the transgenic and wild-type (wt) plants. Several transgenic homozygous lines and wt plants (control) were evaluated for salt tolerance and gene expression. Overexpression of SOD2 in Arabidopsis improved seed germination and seedling salt tolerance. Analysis of Na+ and K+ contents of the symplast and apoplast in the parenchyma cells of the root cortex and mesophyll cells in the spongy tissue of the leaf showed that transgenic lines accumulated less Na+ and more K+ in the symplast than the wt plants did. The photosynthetic rate and the fresh weight of the transgenic lines were distinctly higher than that of wt plants after NaCl treatment. Results from different tests indicated that the expression of the SOD2 gene promoted a higher level of salt tolerance in vivo in transgenic Arabidopsis plants.
A major factor impairing worldwide agricultural productivity is salinity, which is believed to affect nearly one-fifth of the world's irrigated land and resulted in a loss of 10 million ha of otherwise arable land each year (Boyer, 1982; Szaboles, 1987; Flowers and Yeo, 1995; Nelson et al., 1998). To solve the problems caused by salinity in agricultural areas, several approaches have been applied, such as irrigation with fresh water and improving soil drainage. However, these expensive solutions are not always practical. Therefore, the study of plant salt tolerance, with a view to identify and eventually to manipulate the genes involved in salt perception and responses, seems to be a promising approach (Zhu, 2000).
Plant growth depends on mineral nutrients absorbed from the soil by roots. Although Na+ is a major cation present in the soil, it is not considered an essential mineral for most plants. In saline soils, high concentrations of Na+ disrupt the balance of other minerals such as K+, thereby creating osmotic stress and causing secondary problems such as oxidative stress (Zhu, 2001). These adverse effects result in plant growth inhibition and even plant death.
The mechanisms for plant cells to prevent excessive accumulation of Na+ in the cytosol are as follows. First, Na+ entry to plant cells may be restricted by selective ion uptake. However, nonselective cation channels have been proposed to mediate substantial Na+ entry into plant roots (Davenport and Tester, 2000; Demidchik and Tester, 2002). The cloned high-affinity potassium uptake transporter (HKT1) and low-affinity cation transporter (LCT1) affect Na+ permeability when expressed in yeast or oocytes, suggesting that they may also be considered Na+ influx transporters (Rubio et al., 1995; Schachtman et al., 1997; Liu et al., 2000).
Second, internalized Na+ can be stored in vacuoles. Vacuolar compartmentalization is an efficient strategy for plant cells to deal with salt stress because the stored Na+ contributes to osmotic adjustment (Flowers et al., 1977). Vacuolar Na+/H+ antiporters have been cloned from glycophytes (i.e. salt-sensitive plants; Apse et al., 1999; Fukuda et al., 1999; Gaxiola et al., 1999; Darley et al., 2000; Quintero et al., 2000) and halophytes (Hamada et al., 2001; Parks et al., 2002). The Arabidopsis AtNHX1 gene encodes a tonoplast Na+/H+ antiporter and functions in compartmentalizing Na+ into the vacuole (Gaxiola et al., 1999). Overexpression of AtNHX1 improved salt tolerance in Arabidopsis, tomato (Lycopersicon esculentum), and canola (Brassica napus; Apse et al., 1999; Zhang and Blumwald, 2001; Zhang et al., 2001). These transgenic lines were able to grow and produce almost normal amounts of fruits in the presence of 200 mm NaCl, a concentration that inhibits wild-type (wt) plant growth, supporting a role of the vacuolar Na+/H+ antiporter in salt tolerance.
Third, Na+ in the cytosol may be exported back to the growth medium or to apoplastic spaces. Na+/H+ antiporters on the plasma membrane are expected to fulfill this function (Blumwald, 2000; Blumwald et al., 2000). Na+ extrusion through these Na+/H+ antiporters is driven by an inwardly directed proton gradient created by a H+-ATPase. Recently, a plasma membrane Na+/H+ antiporter that is encoded by the Arabidopsis SOS1 gene was identified (Shi et al., 2000), initially as a genetic locus required for salt tolerance in Arabidopsis (Wu et al., 1996). SOS1 functions to control long distance Na+ translocation from root to shoot in Arabidopsis (Shi et al., 2002). Overexpression of SOS1 in Arabidopsis conferred the transgenic plants with increased tolerance to NaCl stress (Shi et al., 2003).
Fourth, the Na+ recirculation from shoots to roots by the phloem sap could be important for salt tolerance, limiting Na+ accumulation in leaves, but this possibility has barely been considered (Munns, 2002). Recently, the functional analysis of AtHKT1 in Arabidopsis showed that Na+ recirculation by the phloem is crucial for salt tolerance (Berthomieu et al., 2003).
In yeast, SOD2 was identified from Schizosaccharomyces pombe as an Na+/H+ antiporter on the plasma membrane involved in salt tolerance (Jia et al., 1992). Expression of SOD2 in a Saccharomyces cerevisiae strain (RH16.6) lacking any Na+-ATPase activity restores the ability of these cells to export sodium and greatly increases their resistance to both Na+ and Li+ in the medium (Hahnenberger et al., 1996). A homolog of SOD2, NHA1, was cloned from S. cerevisiae and found to contribute to Na+ extrusion, although Na+-ATPases have a dominant role in reducing intracelluar Na+ concentration in the yeast (Prior et al., 1996). Here, we report the introduction of the yeast SOD2 gene into Arabidopsis and evaluate its effect on germination and physiological responses compared with those of wt plants under both stressed and non-stressed conditions. Our results showed that ectopic expression of SOD2 conferred enhanced tolerance to NaCl stress. Moreover, SOD2-expressing plants accumulate less Na+ and more K+ in the symplast than do wt plants under NaCl stress.
RESULTS
Genetic Transformation
Five-week-old Arabidopsis plants were infected with Agrobacterium tumefaciens carrying the SOD2 and the nptII genes in the plasmid pROK-SOD2 by the floral dipping method. Forty-six individual kanamycin resistant plants were obtained from 87,000 seeds. The initial kanamycin-resistant plants were named T1, and the progeny obtained from a T1 transgenic plant (by self-cross) were named T2. Several transgenic homozygous lines (T3) that were all tolerant to kanamycin (30 μg mL-1) were selected, named S04-9, S15-3, S23-2, S20-6, S21-5, S29-1, S30-7, S35-8, etc., and used for molecular and physiological analysis and further experiments. There were no obvious morphological or developmental differences between the transgenic and wt plants.
Molecular Characterization of the Transgenic Plants
T3 kanamycin-tolerant plants were checked by PCR. The amplification was carried out in the presence of 5′ and 3′ primers of SOD2. An intense 1.4-kb band corresponding in size to the SOD2 product was obtained from the kanamycin-tolerant plants, whereas nothing was obtained from wt plants. To confirm the amplification products, PCR-Southern hybridization (data not shown) was performed using the SOD2 gene as the probe. The results showed that all kanamycin-tolerant plants had strong positive signals, and no signal was present in wt plants.
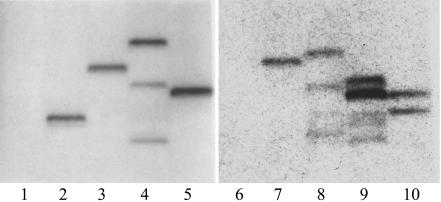
To characterize the copy number of integrated foreign SOD2 gene in the transgenic plants, genomic DNA of T3 plants digested by HindIII was hybridized with the SOD2 probe. Southern-blot analysis showed that all transgenic plants had hybridization signals, and no signal was present in wt plants. The results also showed that four lines (S04-9, S15-3, S20-6, and S21-5) had integrated a single copy of the SOD2 gene, and the other four lines (S23-2, S29-1, S30-7, and S35-8) might carry more than one copy of the transgene (Fig. 1).
Figure 1.
Southern hybridization of DNA from the wt and some transgenic plants. DNA (10 μg) was digested with HindIII, and the resulting fragments were resolved by gel electrophoresis and transferred to a nylon membrane. The membrane was hybridized (65°C) with the gene-specific DNA probe for SOD2. Lanes 1 and 6, wt plants; lanes 2 to 5 and 7 to 10, different transgenic plants (S04-9, S15-3, S23-2, S20-6, S21-5, S29-1, S30-7, and S35-8).
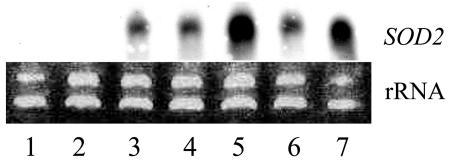
Northern-blot analysis revealed the presence of SOD2 mRNA in T3 plants of several homozygous transgenic lines. The levels of expression among different transgenic lines were different, which might relate with the copy number of the inserted gene. For example, lane 3 shows line S04-9 that contains one copy of SOD2 in its genome, and the expression level of SOD2 is low. However, lane 5 is line S23-2 with more than one copy of the SOD2 gene, so its expression is more than that of line S04-9. The wt showed no hybridization signal in the northern analysis (Fig. 2).
Figure 2.
Expression of SOD2 in wt and some transgenic plants by northern blot. Thirty micrograms (30 μg) of total RNA was analyzed by RNA gel blotting. The blot was hybridized (55°C) with the gene-specific DNA probe for SOD2. The northern blot was exposed to x-ray film for 7 d. The ethidium bromide-stained rRNA band in the agarose gel is shown as a loading control. Lanes 1 and 2, wt plants; lanes 3 to 7, different transgenic plants (S04-9, S15-3, S23-2, S20-6, and S35-8).
Overexpression of SOD2 Improved Seed Germination
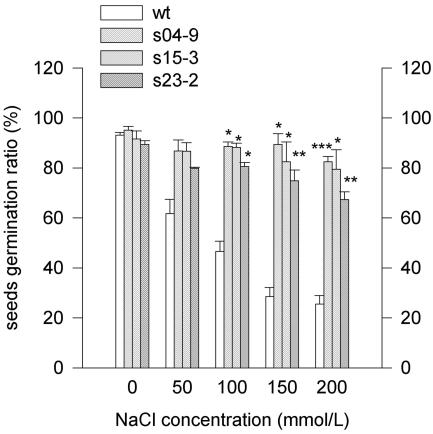
In Arabidopsis, salt sensitivity is most evident at the seed germination and seedling stages. The seeds of Arabidopsis (ecotype C24) were planted on a filter paper saturated with 80 mm NaCl and incubated at 4°C for 2 d, then moved to room temperature. Seed germination was greatly impaired at salt concentrations at or above 75 mm NaCl (Xiong and Zhu, 2002). To test for the ability to germinate under stress conditions, seeds from wt and several homozygous lines were surface sterilized and placed on Murashige and Skoog medium containing different levels of NaCl (0, 50, 100, 150, and 200 mm NaCl). On the Murashige and Skoog medium (0 mm NaCl), all the germination rates were around 95%. On the medium with 200 mm NaCl, the germination rate of wt was 25.5%, whereas the germination rates of T3 transgenic plants' seeds of lines S04-9, S15-3, and S23-2 were 82.5%, 79.5%, and 67.4%, respectively (Fig. 3).
Figure 3.
Germination of wt and some transgenic plants after 7 d sown on Murashige and Skoog medium with different concentrations of NaCl (0, 50, 100, 150, and 200 mm). Results as means ± se (n = 3). *, **, and ***, Significantly different from the wt at P < 0.05, 0.01, and < 0.001, respectively, by Student's t test.
Overexpression of SOD2 Enhanced Early Seedling Tolerance under Salt Stress
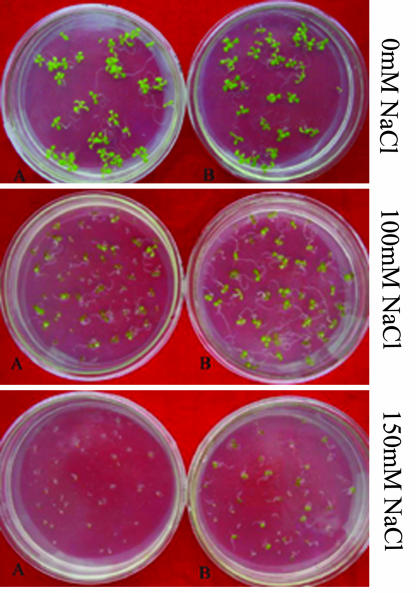
Expression of SOD2 in an S. cerevisiae strain lacking any Na+-ATPase activity (RH16.6) restores the ability of these cells to export sodium and greatly increases their resistance to both Na+ and Li+ in the medium (Hahnenberger et al., 1996). To determine whether SOD2-expressing plants have elevated salt tolerance during early seedling development, seeds from the homozygous SOD2-expressing line S04-9 were germinated on Murashige and Skoog media containing different levels of NaCl. After 15 d of cultivation on Murashige and Skoog medium with 100 mm NaCl, wt seedlings showed growth inhibition, whereas the transgenic plants could continue their growth normally. Also, wt roots were shorter than those of the transgenic line. The wt plants turned yellow and ceased growth on Murashige and Skoog medium with 150 mm NaCl after 15 d of cultivation, but the transgenic plants turned green and could grow slowly (Fig. 4).
Figure 4.
Photograph for wt (A) and transgenic plants of line S04-9 (B) after 15 d of cultivation on Murashige and Skoog + 0mm NaCl, Murashige and Skoog +100 mm NaCl medium, and Murashige and Skoog + 150 mm NaCl medium.
To obtain further evidence that overexpression of SOD2 conferred resistance to salt stress, we measured the root growth of S04-9 and wt plants under salt stress conditions. Seeds of S04-9 and wt were grown on Murashige and Skoog medium for 4 d and transferred onto Murashige and Skoog plates with different levels of NaCl and LiCl. As shown in Table I, transgenic line S04-9 showed nearly the same root growth as wt under normal conditions. However, under salt stress conditions, the transgenic line S04-9 showed a somewhat lower degree of inhibition than did wt plants. These results indicate that overexpression of SOD2 results in enhanced early seedling tolerance to salt stress.
Table I.
Overexpression SOD2 impacts root growth under NaCl and LiCl stress
| Stress
|
Effect on Root Growth
|
|
|---|---|---|
| wt | S04-9 | |
| 0 mm NaCl (mm week−1) | 22.2 (2.52) | 23.8 (2.17) |
| 100 mm NaCl (mm week−1) | 11.3 (0.56) | 15.3 (1.45)** |
| Relative growth (%) | 50.9 | 64.3 |
| 150 mm NaCl (mm week−1) | 3.4 (0.16) | 6.3 (0.47)*** |
| Relative growth (%) | 15.3 | 26.4 |
| 0 mm LiCl (mm week−1) | 22.2 (2.52) | 23.8 (2.17) |
| 10 mm LiCl (mm week−1) | 12.1 (1.02) | 18.7 (1.78)* |
| Relative growth (%) | 54.5 | 78.6 |
| 20 mm LiCl (mm week−1) | 1.6 (0.22) | 3.6 (0.7)** |
| Relative growth (%) | 7.2 | 15.1 |
wt and S04-9 plants were grown on Murashige and Skoog plates for 4 d and transferred onto Murashige and Skoog plates containing the indicated concentrations of NaCl and LiCl. The root growth of 10 plants was measured 7 d after transfer in each of three independent experiments. Results as means ± SE (n =10). *, **, and ***, Significantly different from the wt at P < 0.05, 0.01, and P < 0.001, respectively, by Student's t test.
Effect of Constitutive SOD2 Expression on the Photosynthetic Rate (Pn), Fresh Weight, Dry Weight, and Pro Content
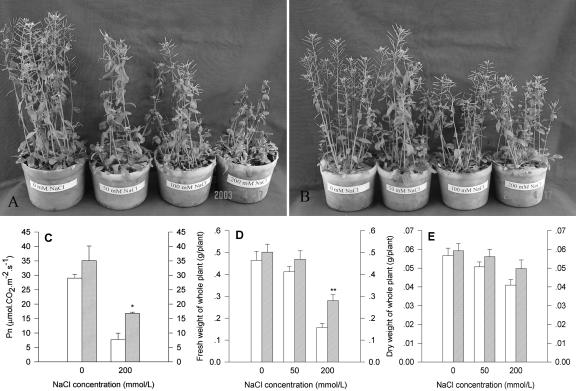
Progeny of T3 homozygous kanamycin-tolerant plants (T4 generation) of lines S04-9 and S23-2 and wt plants were cultivated under salt or non-salt conditions. wt plants displayed progressive chlorosis, reduced leaf size, and a general growth inhibition when watered with NaCl-containing solution. These inhibitory effects increased progressively with increasing NaCl concentration in the watering solution, and the wt plants ceased to flower and died later (Fig. 5A). The transgenic plants of line S04-9 were affected slightly and could survive up to 200 mm NaCl (Fig. 5B). Similar phenomena were also observed in the progeny of line S23-2 (data not shown). After finishing the above treatment, the leaves of the plants were used for measuring Pns. Figure 5C showed that the Pn was decreased with the NaCl treatment, but the Pn of the transgenic plants was almost twice as great as that of the wt plants.
Figure 5.
Increased resistance to NaCl stress of transgenic plants overexpressing SOD2. Phenotypes of wt (A) and T4 transgenic plants of line S04-9 (B) after the NaCl treatment. Left to right, Concentrations of NaCl were 0, 50, 100, and 200 mm, respectively. wt plants and transgenic plans were grown on soil and watered every 3 d until they were subjected to NaCl stress. The Pn (C) of the leaf, fresh weight (D), and dry weight (E) of whole plant were measured when the NaCl treatment was finished. White and cross-hatched line bars, wt and S04-9, respectively. * and **, Significantly different from the wt at P < 0.05 and 0.01, respectively, by Student's t test.
The above plants were harvested for measuring fresh weight and dry weight. As shown in Figure 5 (D and E), NaCl treatment significantly decreased the fresh weight and dry weight of wt plants, but the treatment did not markedly affect the dry weight of the transgenic plants. Both the fresh weight and dry weight of the transgenic plants were higher than that of the wt plants.
To determine whether SOD2 overexpression impacted the Pro content, we measured the Pro content both in transgenic line and wt plants. No significant difference in Pro content was observed between transgenic line and wt plants, although the Pro content was a little lower in transgenic line than wt plants (data not shown).
Transgenic Plants Accumulated Less Na+ and More K+ Than wt Plants
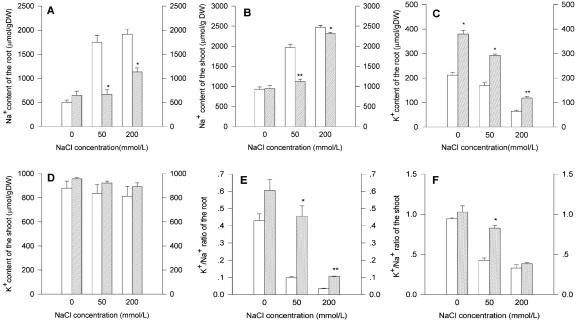
We analyzed the ion content of the transgenic and wt plants before and after exposure to NaCl stress. We analyzed Na+ and K+ because these ions are known to play important roles in plants under NaCl stress. Without NaCl stress, Na+ contents were nearly the same in both wt and transgenic plants. As expected, NaCl treatment increased cellular Na+ levels in both transgenic and wt plants, but the Na+ content in the transgenic line was distinctly lower than in wt (Fig. 6, A and B).
Figure 6.
The Na+ and K+ content in the tissues of wt (white bars) and the T4 transgenic plants of line S04-9 (cross-hatched line bars) after 50 and 200 mm NaCl treatment. The ion contents were determined by atomic absorption spectrophotometry. A, Na+ content of the root; B, Na+ content of the shoot; C, K+ content of the root; D, K+ content of the shoot; E, K+ to Na+ ratio of the root; F, K+ to Na+ ratio of the shoot. Results as means ± se (n = 3). * and **, Significantly different from the wt at P < 0.05 and < 0.01, respectively, by Student's t test.
Without NaCl stress, the K+ content was similar in the shoots of wt and transgenic plants but surprisingly different in the roots. NaCl treatment decreased the K+ content of both types of roots, but at all levels, it was clearly higher in the roots of transgenic lines than in those of wt (Fig. 6C). NaCl treatment did not affect K+ levels in either type of plants (Fig. 6D). The ratio of K+ to Na+ was decreased with the increasing concentration of NaCl in both roots and shoots (Fig. 6, E and F). However, it was higher in the transgenic line than in the wt plants under salt-stressed conditions, except for the ratio in shoots watered at 200 mm NaCl.
Reduced Na+ Accumulation in the Symplast of the Plants Overexpressing SOD2
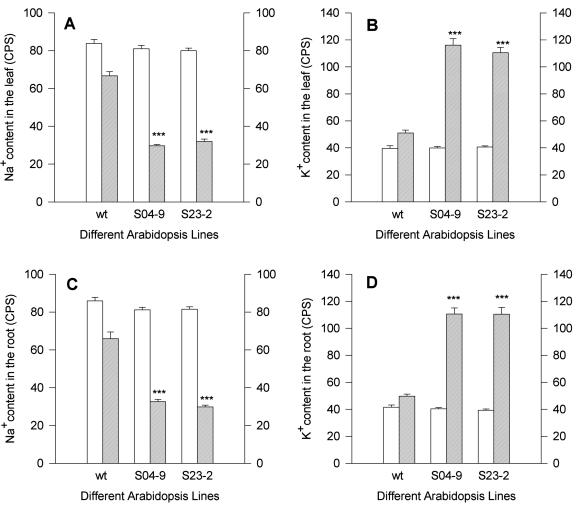
Under severe salt stress, SOD2 functions to export Na+ to limit accumulation of Na+ in yeast cells. To determine if overexpression of SOD2 reduces Na+ accumulation in Arabidopsis, the Na+ content in transgenic plants overexpressing SOD2 and wt plants was examined. In response to 100 mm NaCl treatment for up to 6 d, both S04-9 and S23-2 plants accumulated less Na+ in the symplast of the leaf and root than in the apoplast. However, the Na+ content of the symplast in wt plants was obviously more than that in transgenic plants after the NaCl treatment (Fig. 7, A and C). The content of K+ in the symplast of transgenic plants was significantly higher than that in wt plants (Fig. 7, B and D). These results suggested that overexpression of SOD2 might increase Na+ export and decrease Na+ accumulation in the symplast both in the root and leaf.
Figure 7.
X-ray microanalysis of Na+ and K+ content in the apoplast and symplast of wt and the T4 transgenic plants of lines S04-9 and S23-2 after 100 mm NaCl treatment for 6 d. A, Na+ content in the leaf; B, K+ content in the leaf; C, Na+ content in the root; D, K+ content in the root. White bars and cross-hatched line bars, Apoplast and symplast, respectively. Results as means ± se (n = 7). ***, Significantly different from the wt at P < 0.001 by Student's t test. CPS, X-ray counts per second.
DISCUSSION
Soil salinity is a prevalent abiotic stress for crop plants. Excess salts in the soil solution interfere with mineral nutrition and water uptake and lead to the undue accumulation of toxic ions. Plant growth under salt stress depends, among other concomitant processes, on the reestablishment of proper cellular ion homeostasis. Low cytosolic Na+ content is preserved by the concerted interplay of regulated ion uptake, vacuolar compartmentation, and active extrusion to the extracellular milieu (Blumwald et al., 2000).
Salinity tolerance clearly depends on sustaining the cytosolic environment, limiting Na+ accumulation, and maintaining K+ concentration. In this work, the transgenic Arabidopsis plants showed a tendency to accumulate less Na+ under saline condition than wt plants, and the content of Na+ was significantly lower in the roots of transgenic lines than in wt plants (Fig. 6, A and B). Meristematic cells are known to lack large vacuoles for Na+ compartmentation; thus, Na+ extrusion to the external medium by epidermal cells might be crucial to protect the root meristem. Moreover, the Na+ transported to the shoot was decreased because of Na+ extrusion, which also can protect the plants. More importantly, compared with the ion content of the wt plants, the content of Na+ in the symplast of the transgenic lines was significantly decreased, and the K+ content was higher (Fig. 7), which indicated SOD2 function to export Na+ in the transgenic plants. Export of Na+ from the symplast (the continuum of cytoplasm and plasmodesmata) to the apoplast (dead cells, empty space surrounding the symplast, and the wall matrix), reduces the damage of the toxic ion to the cell to some extent and, thus, is an important strategy for cell survival. The fresh weight, dry weight, and Pn of the transgenic plants under salt stress were higher than those of the wt plants, illustrating fully the beneficial effects of expression of SOD2. Therefore, the results reported here showed that this Na+ export strategy in the transgenic plants worked to decrease the symplast Na+ content, keep higher K+ content in the symplast, and increase salt tolerance in plants.
There have been only a few studies aimed at increasing salt tolerance by helping plants reestablish homeostasis under stress (Shi et al., 2003). Calcineurin and HAL1 were regulators of intracellular K+ and Na+ homeostasis in yeast S. cerevisiae (Gaxiola et al., 1992; Nakamura et al., 1993). Ectopic expression of these yeast regulatory genes improved salt tolerance in transgenic plants (Pardo et al., 1998; Gibert et al., 2000; Yang et al., 2001). Very recently, overexpression of a plasma membrane Na+/H+ antiporter gene (ApNhaP) in a freshwater cyanobacterium, a photosynthetic organism, was shown to so dramatically enhance the salt tolerance that the transgenic cyanobacterium could grow in seawater (Waditee et al., 2002). In the fission yeast S. pombe, loss-of-function mutations in the plasma membrane Na+/H+ antiporter gene SOD2 decreased salt tolerance, whereas overexpression of SOD2 substantially increased Na+ export capacity and conferred salt tolerance (Jia et al., 1992).
Soil salinity is a major factor in reducing plant growth and productivity. One strategy for improving the salt tolerance of a plant is to increase the production of small osmolytes or stress proteins that protect or reduce damage caused by salt stress (Zhu, 2001). The results showed that the transgenic lines accumulated less Pro than wt plants. Maybe the cytosolic Na+ was extruded into apoplastic space by SOD2, so it reduced the damnification to the cell, and the plants did not need to increase Pro to protect the plants, or maybe the transgenic plants were expected to have enhanced accumulation of other solutes in the vacuole and cytosol to compensate for the increased Na+ export from cells.
This is the first report, to our knowledge, that a yeast plasma membrane Na+/H+ antiporter is ectopically expressed in high plants. In this work, the content of Na+ in the symplast of transgenic lines was lower than wt, whereas the content of K+ in transgenic lines was higher than wt. Therefore, the transgenic plants might use the activity of SOD2 to export Na+ back to the growth medium or to apoplastic spaces, thereby reducing the content of Na+ and improving salt tolerance. The results indicate that overexpression of the SOD2 gene can promote a high level of salt tolerance in transgenic Arabidopsis plants.
MATERIALS AND METHODS
Plasmid Construction and Plant Transformation
The SOD2 open reading frame was amplified using the Schizosaccharomyces pombe cDNA as template with primers 5′-CATCGATGGGCTGGAGACAACT-3′ 5′-CTGGATCCTAAACGTAATCTTCC-3′. PCR conditions were as follows: 94°C for 3 min, followed by 35 cycles of 94°C for 45 s, 55°C for 45 s, 72°C for 1 min and 30 s, and 72°C for 10 min. SOD2 PCR products were confirmed by sequence analysis on an Applied Biosystem 373 Automated DNA sequencer (ABI/Perkin-Elmer, Foster City, CA). The open reading frame region of SOD2 is 1,404 bp. This SOD2 PCR product was inserted into binary plant vector pROK2, between the cauliflower mosaic virus 35S promoter and octopine synthase terminator. The resulting plasmid, named pROK-SOD2, was mobilized to Agrobacterium tumefaciens strain GV3101 and used for plant transformation. Arabidopsis adult plants (5 weeks old) were infected with the A. tumefaciens by floral dipping method (Clough and Bent, 1998) and grown in the greenhouse. These seeds collected were screened in Murashige and Skoog medium supplemented with 30 μg mL-1 kanamycin.
Southern and Northern Analysis
Southern- and northern-blotting experiments were performed using approximately 10 μg of genomic DNA and 30 μg of total RNA per track. Genomic DNA was isolated from transgenic lines and wt Arabidopsis as described by Murray and Thompson (1980) and digested with HindIII, then transferred to a Hybond N+ nylon membrane (Amersham, Buckingham-shire, UK). Southern hybridizations (65°C) were carried out as described by Sambrook et al. (1989). DNA probes were labeled with [32P]dCTP according to the manufacturer's instruction (Sigma, St. Louis). Total RNA was extracted from transgenic lines and wt Arabidopsis according to the method of Chomczynski and Sacci (1987). The probes and method of hybridization (55°C) are the same with Southern blots.
Growth Parameters and Germination Studies
Arabidopsis (ecotype Landsberg erecta) plants were grown in the greenhouse under a 16-h-light and 8-h-dark cycle at 22°C. For germination study, wt and transgenic surface-sterilized Arabidopsis seeds were sown in petri dishes containing 30 mL of Murashige and Skoog medium (Murashige and Skoog [Murashige and Skoog, 1962] + 3% [w/v] Suc), Murashige and Skoog + 50 mm NaCl, Murashige and Skoog +100 mm NaCl, Murashige and Skoog + 150 mm NaCl and Murashige, and Skoog + 200 mm NaCl. Approximately 100 seeds were placed on each 9-cm petri dish. Germination was scored after 7 d of germination under conditions as described above.
Growth Measurement
For stress treatments and growth measurement, 4-d-old seedlings from vertical plates in Murashige and Skoog medium were transferred and placed with roots pointing downward onto vertical agar plates supplemented with different levels of NaCl and LiCl. Each plate contained 10 wt and transgenic seedlings. Three replicate plates were used for each treatment. Increases in root length were measured 7 d after transfer in each of the three independent experiments.
Determination of the Total K+ and Na+ Contents
Transgenic T4 plants from the lines of single or multiple copy insertion of SOD2 were evaluated for salt tolerance. The seeds of these lines were sown in 9-cm plastic pots filled with a 2:1:1 (v/v) mixture of soil:perlite:vermiculite, and the plants were grown in a greenhouse with a photoperiod of 16 h of light and 8 h of darkness, a temperature of 25°C/20°C, and a relative humidity of 60%/80%. Four-week-old seedlings were subjected to salt stress. We applied 60 mL of Hoagland solution to each pot every other day over the 16-d watering treatment. The control group received no NaCl supplementation, and the other three groups were watered with Hoagland solutions supplemented with NaCl. The concentrations of NaCl supplementation were increased stepwise by 50 mm every 4 d for each group to the indicated maximum. The roots and shoots of plants were harvested at the end of the salt treatment. Dry weight was measured after 48 h at 70°C. The samples were digested with HNO3, and K+ and Na+ contents were determined using an atomic absorption spectrophotometer (Z-8000, Hitachi, Tokyo) according to Wang and Zhao (1995). Values are the mean ± se (n = 3).
Measurement of the Pn, Fresh Weight, and Dry Weight
When the above NaCl treatment was finished, plant growth was determined by measuring Pn, fresh weight, and dry weight. The Pn was determined using the automatic photosynthetic measuring apparatus (Ciras-2, PP Systems, Hitchin, Hertfordshire, UK). The fresh weight of each individual whole plant was measured immediately after the harvest. Dry weight was measured after 48 h at 70°C.
X-Ray Microanalysis of K+ and Na+ Content of the Symplast and Apoplast
Transgenic T4 plants were grown under a short-day cycle (8 h of light and 16 h of dark). After 3 weeks, we watered the plants with different concentrations of NaCl every other day. Both the wt plants and transgenic plants received the same concentration of NaCl, every 2 d for four times, from 25 to 100 mm (final concentration). After the six times treatment with 100 mm NaCl, the leaf (0.5 cm2) and the root (1 cm) were harvested and immediately stored in liquid nitrogen in a copper bag. The materials were used to analyze Na+ and K+ contents in the symplast and apoplast according to the method of Fritz (1989) and Li et al. (1991). The mesophyll cells of the spongy tissue in the leaf and the parenchyma cells of the cortex in the root were used for the analysis. For each tissue, seven measurements were carried out. The results are expressed as x-ray counts per second of an element peak after background subtraction.
Measurement of Pro Content
For Pro content measurement, 2 g of fresh leaves was ground in 10 mL of 5% (v/v) acetic acid, and the homogenate was diluted to 50 mL with distilled water. Pro concentration was determined as described by Troll and Lindsley (1955). Values are the means ± se (n = 3).
Acknowledgments
We thank Dr. Jian-kang Zhu and Dr. Ray Wu for helpful revision of the manuscript and Dr. Huazhong Shi for providing helpful suggestions.
This work was supported by the National Natural Science Foundation of China (grant no. 39980022) and the National Key Fundamental Research Program of China (no. G1999011700).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026062.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256-1258 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozomi N, Oiki S, Yamada K, Cellier F et al. (2003) Fountional analysis of AtNKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12: 431-434 [DOI] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP (2000) Sodium transport in plant cells. Biochim Biophys Acta 1465: 140-151 [DOI] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218: 443-448 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacci N (1987) Single-step method of RNA isolation by acid guanidiumthiocyanate-phenol-chlorofrom extraction. Anal Biochem 162: 156-159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Darley CP, van Wuytswinkel OC, van der Woude K, Mager WH, de Boer AH (2000) Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem J 351: 241-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Tester M (2000) A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol 122: 823-834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Tester M (2002) Sodium fluxes through non-selective cation channels in the plasma membrane of protoplasts from Arabidopsis thaliana roots. Plant Physiol 128: 379-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Troke PF, Yeo AR (1977) The mechanism of salt tolerance in halophytes. Annu Rev Plant Physiol 28: 89-121 [Google Scholar]
- Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants: where next? Aust J Plant Physiol 22: 875-884 [Google Scholar]
- Fritz E (1989) X-ray microanalysis of diffusible elements in plant cells after freeze-drying, pressure-infiltration with ether and embedding in plastic. Scanning Microscopy 3: 517-526 [Google Scholar]
- Fukuda A, Nakamura A, Tanaka Y (1999) Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim Biophys Acta 1446: 149-155 [DOI] [PubMed] [Google Scholar]
- Gaxiola R, de Larrinoa IF, Villalba JM, Serrano R (1992) A novel and conserved salt-induced protein is an important determinant of salt tolerance in yeast. EMBO J 11: 3157-3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96: 1480-1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert C, Rus AM, Bolarin MC, Lopez-Coronado JM, Arrillaga I, Montesinos C, Caro M, Serrano R, Morena V (2000) The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiol 123: 393-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger KM, Jia Z, Young PG (1996) Functional expression of the Schizosaccharomyces pombe Na+/H+ antiporter gene, sod2, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 5031-5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T (2001) Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Mol Biol 46: 35-42 [DOI] [PubMed] [Google Scholar]
- Jia ZP, McCullough N, Martel R, Hemmingsen S, Young PG (1992) Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J 11: 1631-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Fritz E, Li T, Hüttermann A (1991) X-ray microanalysis of ion contents in roots of Populus maximowiczii grown under potassium and phosphorus deficiency. J Plant Physiol 138: 180-185 [Google Scholar]
- Liu W, Schachtmann DP, Zhang W (2000) Partial deletion of a loop region in the high affinity K+ transporter HKT1 changes ionic permeability leading it increased salt tolerance. J Biol Chem 275: 27924-27932 [DOI] [PubMed] [Google Scholar]
- Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239-250 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473-497 [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant cDNA. Nucleic Acids Res 8: 4321-4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T (1993) Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J 12: 4063-4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Shen B, Bohnert HJ (1998) Salinity tolerance: mechanisms, models and the metabolic engineering of complex traits. In JK Setlow, ed, Genetic Engineering, Principles and Methods, Vol 20. Plenum Press, New York, pp 153-176 [DOI] [PubMed] [Google Scholar]
- Pardo JM, Reddy MP, Yang S, Maggio A, Huh GH, Matsumoto T, Coca MA, Paino-D'Urzo M, Koiwa H, Yun DJ et al. (1998) Stress-signaling through Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proc Natl Acad Sci USA 95: 9681-9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks GE, Dietrich MA, Schumaker KS (2002) Increased vacuolar Na+/H+ exchange activity in Salicornia bigelovii Torr. in response to NaCl. J Exp Bot 53: 1055-1065 [DOI] [PubMed] [Google Scholar]
- Prior C, Potier S, Souciet JL, Sychrova H (1996) Characterization of NHA1 gene encoding a Na+/H+ antiporter of the yeast Saccharomyces cerevisiae. FEBS Lett 387: 89-93 [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Blatt MR, Pardo JM (2000) Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett 471: 224-228 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassman W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660-1663 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor laboratory Press, Cold Spring Harbor, NY
- Schachtman DP, Kumar R, Schroeder JI, Marsh L (1997) Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc Natl Acad Sci USA 94: 11079-11084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896-6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) Role of SOS1 as a plasma membrane Na+/H+ antiporter that controls long distance Na+ transport in plant. Plant Cell 14: 465-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wu SJ, Zhu JK (2003) Overexpression of a Plasma membrane Na+/H+ antiporter improves salt tolerance in Arabidopsis. Nat Biotechnol 21: 81-85 [DOI] [PubMed] [Google Scholar]
- Szaboles I (1987) The global problems of salt-affected soils. Acta Agron Hung 36: 159-172 [Google Scholar]
- Troll W, Lindsley J (1955) Proline content determination in plant tissues. J Biol Chem 215: 655-660 [PubMed] [Google Scholar]
- Waditee R, Hibino T, Nakamura T, Incharoensakdi A, Takabe T (2002) Overexpression of a Na+ /H+ antiporter confers salt tolerance on a freshwater cyanobacterium, making it capable of growth in sea water. Proc Natl Acad Sci USA 99: 4109-4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BS, Zhao KF (1995) Comparison of extractive methods of Na+, K+ in wheat leave. Plant Physiol Commun 31: 50-52 [Google Scholar]
- Wu SJ, Lei D, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong LM, Zhu JK (2002) Salt tolerance. In C. Somerville and E. Meyerowitz, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD
- Yang SX, Zhao YX, Zhang Q, He YK, Zhang H, Luo D (2001) HAL1 mediate salt adaptation in Arabidopsis thaliana. Cell Res 11: 142-148 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765-768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832-12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124: 941-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6: 66-71 [DOI] [PubMed] [Google Scholar]