Abstract
Fertilization of the female gametophyte in angiosperm plants initiates a process of coordinated development of embryo, endosperm, and seed coat that ensures the production of a viable seed. Mutant analysis has suggested that communication between the endosperm and the seed coat is an important determinant in this process. In addition, cell groups within the embryo, derived from the apical and from the basal cell, respectively, after zygote division, concertedly establish a functional root meristem, and cells in the apical region of the embryo are hypothesized to repress cell divisions in the basal cell-derived suspensor. The available evidence for these interregional communication events mostly relies on the analysis of mutant phenotypes in Arabidopsis. To provide independent and direct evidence for communication events, we used conditional domain-specific expression of the diphtheria toxin A chain (DTA) in developing Arabidopsis seeds. By using a collection of cell- or tissue-type-specific promoters, we show that the mGAL4:VP16/UAS two-component gene expression allows reliable spatiotemporal and conditional expression of the GFP:GUS reporter and the DTA gene in the developing embryo and endosperm. Expression of DTA in the protoderm of the embryo proper led to excessive proliferation of suspensor cells, sometimes resulting in the formation of secondary embryos. Endosperm-specific expression of DTA caused complete cessation of seed growth, followed by pattern defects in the embryo and embryo arrest. Taken together, the results presented here substantiate the evidence for and underline the importance of interregional communication in embryo and seed development and demonstrate the usefulness of conditional toxin expression as a method complementary to phenotypic analysis of developmental mutants.
Seed development in higher plants is characterized by the coordinated development of distinct tissues. Seed tissues mainly arise from cells and tissues of the female gametophyte that are formed before fertilization. The multinucleate female gametophyte is enclosed by several sporophytic maternal cell layers that constitute the ovule. In angiosperms, double fertilization by fusion of the egg cell and the central cell of the female gametophyte with the two sperm cells delivered by the pollen tube generates the diploid embryo and triploid endosperm, respectively. The embryo and endosperm both develop within the confines of the maternal tissue, now referred to as the seed coat, but each follows a different developmental program (for review on embryo and endosperm development, see Jürgens and Mayer, 1994; Berger, 2003, respectively). Within the embryo, a complex but precise pattern of organs and cell types is laid down from a single cell, the zygote (Jürgens and Mayer, 1994). Genetic controls are required to establish the embryo pattern and to ensure the proper initiation and relative positioning of distinct cell groups and organs, such as the meristems and vasculature. The endosperm first undergoes a series of synchronized nuclear divisions and forms a free-nuclear syncytium divided in three domains. The micropylar endosperm surrounds the developing embryo, the peripheral endosperm fills up most of the seed volume, and a dense nucleocytoplasmic domain, the chalazal endosperm, extends to the nucellar region of the seed. As soon as the embryo reaches the heart stage, endosperm nuclei are partitioned into individual units by cell walls during a process called cellularization. Only a remnant of the endosperm is present in the mature seed as a thin layer that separates the embryo from the seed coat tissues (Berger, 2003). The relative growth of seed coat, endosperm, and embryo must be stringently regulated to secure development of a viable seed.
Several Arabidopsis mutants in seed and endosperm development and embryo pattern formation have been isolated, and detailed phenotypic analysis of a number of them has brought new insights into the importance of communication between different functional/cellular domains—referred to here as interregional communication—in plant seeds and, specifically, in embryos. For example, mutants of the fis (fertilization-independent seed) category and seeds with an excess of maternal genomes resulting from crosses between parents with different ploidy levels show reduced endosperm development and reduced seed size. In some cases, endosperm defects in such genotypes are accompanied by aberrant embryo development (Ohad et al., 1996; Scott et al., 1998). The recent isolation of the haiku mutants reinforces the hypothesis that endosperm development plays an important role in overall seed growth and also has a function in late embryo development (Garcia et al., 2003). The role of the endosperm in early embryo development, however, is as yet unclear.
Phenotypic analysis of the large set of Arabidopsis embryo patterning mutants (e.g. Mayer et al., 1991) has highlighted the importance of interregional communication in the regulation of pattern formation. Analysis of the sus and twin mutants classes, in which the basally located suspensor cells proliferate after cellular defects become visible in the embryo proper (Vernon and Meinke, 1994; Schwartz et al., 1994; Zhang and Somerville, 1997) and of bdl (bodenlos) and mp (monopteros) mutants, which show initial apical cell division defects and subsequent failure to initiate a root meristem from a clonally distinct cell population (Berleth and Jürgens, 1993; Hamann et al., 1999), led to the hypothesis that the apical region of the embryo exerts significant control over the development of the basal region of the embryo and the suspensor.
Although cloning of the BDL (Hamann et al., 2002) and MP (Hardtke and Berleth, 1998) genes has given important insight into the possible mechanism underlying this interregional communication, identification of the TWN2 (Zhang and Somerville, 1997) and SUS1 (Golden et al., 2002) genes has not significantly improved our understanding of the mechanism of embryo-suspensor interactions.
Generally, in the mentioned cases of hypothesized regulatory interactions between cells or domains, it is difficult to conclude that a mutation affects interregional communication, e.g. between the endosperm and the embryo proper, unless the expression of the mutated gene is known to be confined to specific cells or domains. Therefore, independent evidence should come from experiments where a strictly defined set of cells within the seed or embryo is disabled, e.g. by expression of a toxin. Subsequent non-cell-autonomous effects on development then directly show functional interactions. Until now, only a single study has made use of toxin expression in the embryo to study cellular interactions. However, the observed non-cell-autonomous effects were mild and did not provide evidence for repression of suspensor development by the embryo proper (Baroux et al., 2001).
Here, we present an analysis of cellular communication within the seed by local expression of the diphtheria toxin A chain (DTA). This protein is highly toxic (Yamaizumi et al., 1978), and the encoding gene has been used successfully to ablate root cap cells (Tsugeki and Fedoroff, 1999), petal and stamen primordia in flowers (Day et al., 1995), or even entire flowers (Nilsson et al., 1998). To circumvent any expression of the toxin before fertilization, we adopted the mGAL4:VP16/UAS two-component gene expression system. Two-component gene expression technology (transactivation) makes use of two plant lines—one that expresses a heterologous transcription factor under control of a chosen plant promoter and another that contains a gene of interest under control of a silent promoter that is only activated by the heterologous transcription factor. Only upon crossing the two lines, the gene of interest will be expressed in the zygotic tissues.
The mGAL4:VP16/UAS system has been optimized for Arabidopsis (Haseloff, 1999) and has been used by several researchers to express reporter genes or genes of interest during postembryonic stages of development (e.g. Kiegle et al., 2000; Benjamins et al., 2001; Sabatini et al., 2003) or in the endosperm (Boisnard-Lorig et al., 2001). No data were available on its reliability in the embryo; therefore, we first analyzed this aspect in detail. The analysis shows that mGAL4:VP16/UAS technology is reliable in embryos. Here, we use the technology to express DTA toxin in specific tissues in the embryo or in the endosperm, and we provide direct evidence for the previously hypothesized cellular interactions in embryo and seed development.
RESULTS
Plant Lines for Conditional Domain-Specific Expression in Seed or Embryo
An experimental strategy for cell ablation by expressing the DTA cytotoxin in limited domains of the developing seed needs to fulfill two important criteria. First, it requires a selected set of gene promoters with a well-defined spatiotemporal expression specificity in the seed; second, the expression must be conditional, i.e. expression of DTA must not take place during generation of lines or at any time before but should be readily activated upon fertilization. As for the first requirement, we have selected a set of previously described promoters that provide domain-specific expression in the embryo or the endosperm. The Arabidopsis RPS5A (RIBOSOMAL PROTEIN S5A) promoter is strongly expressed in dividing cells, starting as early as the one-cell embryo stage (Weijers et al., 2001a). The Arabidopsis LTP1 (LIPID TRANSFER PROTEIN 1) promoter marks the L1 layer of all newly formed organs and is preferentially expressed in the apical domain of the embryo (Thoma et al., 1994). The synthetic auxin-responsive DR5(7x) promoter has an expression peak in the central root cap cells of the torpedo stage Arabidopsis embryo (Sabatini et al., 1999). The enhancer-trapped mGAL4:VP16 gene (see below) in line KS22I was used as a tool to drive expression in the developing endosperm (Boisnard-Lorig et al., 2001).
To obtain conditional expression of DTA, we adopted the mGAL4:VP16/UAS two-component gene expression system (Fig. 1A; Haseloff, 1999). ACT lines were generated that harbor the mGAL4:VP16 coding sequence coupled to the RPS5A, LTP1, or DR5(7x) promoters. For all ACT constructs, lines were obtained with a single T-DNA insertion that did not display abnormal embryo- or seedling development at frequencies exceeding those in wild type (Table I). The mGAL4:VP16 mRNA was easily detected in ACT RPS5A, ACT 35S (pCaMV35S::mGAL4:VP16; J. Haseloff, unpublished data), ACT LTP1 (Fig. 1B), and ACT DR5(7x) (not shown) seedlings. In addition, the mGAL4:VP16 protein was detectable in extracts from ACT 35S seedlings (Fig. 1C).
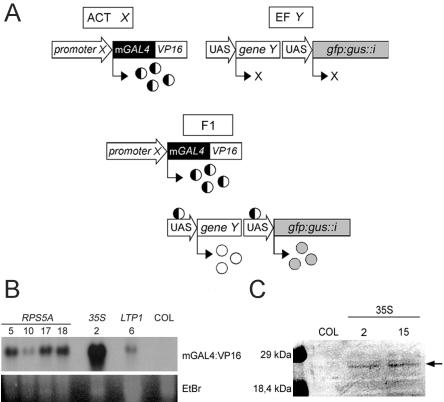
Figure 1.
Expression of mGAL4:VP16 in Arabidopsis. A, mGAL4: VP16/UAS transactivation approach: A promoter of interest (promoter X) is fused to the mGAL4:VP16 coding sequence in activator (ACT) line X. The mGAL4:VP16 protein is then produced only in cells in which promoter X is active. In effector (EF) line Y, both a target gene (gene Y) and eGFP:GUS::intron are controlled by the GAL4: VP16-responsive UAS promoter (UAS; contains five GAL4-binding sites placed upstream of the -47 CaMV 35S promoter). No transcription of these genes occurs in the absence of GAL4:VP16. Upon crossing ACT X and EF Y, transactivation of Y and GGi only occurs in those cells of F1 (and F2) progeny in which promoter Y is active. B, mGAL4:VP16 mRNA accumulation in seedlings of the ACT RPS5A (nos. 5, 10, 17, and 18), ACT 35S#2 and ACT LTP1#8 lines. Columbia (Col), Wild-type control. Twenty micrograms of total RNA was loaded for ACT LTP1 and 10 μg for all other samples. Ethidium bromide-stained gel to show loading differences. C, Western blot incubated with anti-VP16 antibody shows the expected band of 26 kD in total protein extracts of ACT 35S seedlings (30 μg of protein) but not in wild type (Col).
Table I.
Embryo and seedling pattern defects in ACT and EF lines
| Line
|
Altered pattern
|
Segregation of T-DNA in T2a | ||
|---|---|---|---|---|
| Generationb | Seedlingsc | Embryosd | (R:S [X2]) | |
| Columbia wild type | - | 0.7% (2/294) | 1.8% (4/223) | - |
| ACT RPS5A no. 5 | T4 | 0.5% (1/194) | 0.6% (1/166) | 61:21 (0.02) |
| ACT RPS5A no. 10 | T3 | <1.6% (0/63) | - | 51:22 (0.98) |
| ACT RPS5A no. 17 | T2 | <0.2% (0/561) | 1.5% (3/194) | 48:13 (0.44) |
| ACT RPS5A no. 18 | T2 | <0.1% (0/986) | - | 49:18 (0.12) |
| ACT LTP1 no. 8 | T4 | 0.2% (2/1,122) | 0.9% (1/108) | - |
| ACT LTP1 no. 9 | T4 | 0.6% (2/355) | - | - |
| EF DTA no. 6 | T4 | - | 2.1% (7/325) | 55:18 (0.08) |
| EF GGi no. 15 | T5 | 0.9% (1/109) | 1.0% (1/104) | - |
a Segregation of the resistance marker on the T-DNA construct in T2 generation seedlings. The ratio is expressed as resistant (R):sensitive (S) seedlings. The observed ratios were tested for goodness of fit with the 3:1 ratio expected for a single T-DNA insertion locus in the X2 test (X2 < 3.84; P > 0.05). b Lines that were analyzed in the T3 or T4 generations are homozygous for the T-DNA insertion. c Altered seedling pattern is defined as changes in the no. of cotyledons, fusion of cotyledons, the absence of hypocotyl or root, or other clear malformations in 7-d-old seedlings germinated on nutrient medium, with the addition of kanamycin (ACT lines) or phosphinotricin (EF lines). Nos. in parentheses indicate the no. of seedlings or embryos with a pattern defect per no. of individuals analyzed. d Embryo phenotypes were analyzed at the globular to torpedo stage. Pattern defects in embryos are defined as obvious changes in the pattern of cell divisions relative to that described in Jürgens and Mayer (1994). Small variations in embryo size or developmental stage were not included in this analysis, but obvious growth retardation was scored.
Separate EF DTA plant lines were generated containing the DTA gene driven by the mGAL4:VP16-responsive UAS promoter (Fig. 1A). As a control, EF GGi lines containing a UAS::GFP:GUS reporter gene were generated. No GUS activity could be detected in 30 independent EF GFP:GUS lines, and the development of all EF DTA lines (n = 28; Table I; data not shown) was indistinguishable from that of wild-type Arabidopsis plants. These results, together with those of Bougourd et al. (2000), show that the UAS promoter is inactive in the absence of mGAL4:VP16 and, therefore, not a target of endogenous transcription factors.
Reliable Domain-Specific Reporter Gene Expression after mGAL4:VP16/UAS Transactivation
To first assess the reliability of cell-specific transactivation in the seed, homozygous EF GGi plants were crossed with the homozygous ACT lines and with the KS022I enhancer trap line. The ACT lines were consistently used as female parents in crosses with EF lines because we observed that this provided the strongest transactivation during early embryogenesis (Weijers et al., 2001b).
Transactivation of the UAS::GFP:GUS gene in RPS5A»GGi F1 (the notation promoter Y » gene X is used to distinguish transactivation from direct promoter Y::gene X fusions) seeds was detectable in the embryo and endosperm in the one-cell embryo stage (Fig. 2A), and the intensity of GUS staining increased until the heart stage (Fig. 2, A-D). Although the expression level in RPS5A»GGi embryos was much stronger than in RPS5A::GUS reporter lines (compare Fig. 2C with Weijers et al., 2001a), the spatiotemporal pattern did not differ.
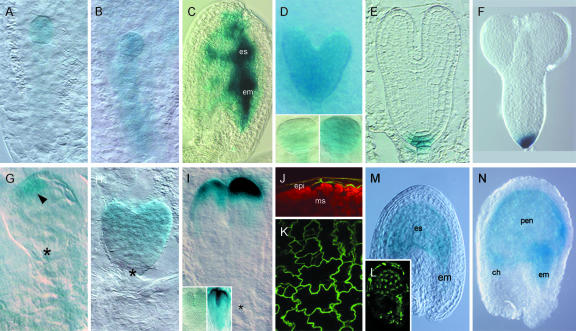
Figure 2.
Cell type-specific transactivation in the Arabidopsis embryo and endosperm. A to D, RPS5A#5»GGi#15 F1 embryos. A, One-cell stage embryo showing β-glucuronidase (GUS) activity in the apical cell. B, Two-cell stage embryo showing GUS activity both in the apical and the basal cell. C, Strong GUS activity in globular stage embryo and endosperm. D, Global GUS activity in a globular to heart stage embryos. Insets, Globular embryos from the same silique that were stained for the same time. E, GUS activity in a median section through a torpedo stage DR5(7x)::GFP:GUS#15 T3 embryo. F, GUS activity in a DR5(7x)#3»GGi#15 F1 embryo. G to K, LTP1#8»GGi#15 F1 embryos (G-I) and seedlings (J and K). G, GUS activity in a single protodermal cell (arrowhead) in a dermatogen stage embryo. H, Preferential apical staining in a heart stage embryo. I, GUS activity in the L1 of the cotyledons in a torpedo stage embryo. Insets, Two embryos from the same silique that were stained for the same time. J, Confocal cross section showing green fluorescent protein (GFP):GUS fluorescence in the epidermis of a leaf primordium. Red fluorescence is chlorophyll. K, Dermal optical section of a cotyledon. L, Endosperm-specific mGFP5-ER fluorescence in a developing KS22I seed. Note the fluorescence in the seed coat. M and N, GUS activity in KS22I»GGi#15 F1 seeds 2 (M) and 4 (N) days after pollination (dap). Staining times were 4 h in (A-D) and 16 h in (E-I). The asterisk in G to I marks the position of the hypophysis or root pole. es, Endosperm; em, embryo; epi, epidermis; ms, mesophyll; ch, chalazal pole; pen, peripheral endosperm.
As expected for an L1 layer-specific promoter, GUS activity in LTP1»GGi F1 embryos was detectable in protodermal cells in dermatogen stage embryos (Fig. 2G). Subsequently, the expression spread to cover the entire apical region of the heart stage embryo (Fig. 2H), after which the GUS signal became exclusively confined to the L1 layer of the cotyledons (Fig. 2I). In F1 seedlings, the GUS pattern did not differ from the reported LTP1::GUS pattern (Thoma et al., 1994; data not shown), and analysis of GFP fluorescence revealed strict epidermal specificity (Fig. 2, J and K).
Similarly, GFP:GUS transactivation in DR5(7x)»GGi (Fig. 2F) embryos mirrored the direct promoter::GUS fusion activity (Fig. 2E). In KS22I»GGi F1 plants, we observed endosperm specific expression of both GFP: GUS on the EF GGi construct (Fig. 2, M and N) and mGFP5-ER on the enhancer trap construct (Fig. 2L), indicating coregulation of both UAS-controlled reporter genes. In summary, ACT line-driven UAS::GGi expression conserved the spatiotemporal expression of each promoter and provided stronger GGi expression than a corresponding direct promoter::GGi fusion. Notably, for reasons that we do not know, a significant variation in gene expression levels was always observed between sibling embryos (Fig. 2, D and I, insets).
Protoderm Ablation Reveals Apical Control of Basal Embryo Development
A striking example of interregional communication in embryo development is the hypothesized suppression of cell divisions in the suspensor and the regulation of hypophysis division by the embryo proper (Schwartz et al., 1994; Vernon and Meinke, 1994; Zhang and Somerville, 1997; Jürgens, 2001). A previous study has challenged the hypothesis of apical control of basal embryo development (Baroux et al., 2001). Upon LTP1-driven expression of the BARNASE (Bacillus amyloliquefaciens ribonuclease) gene, embryos showed defects in the basal tier of the embryo proper and in hypophysis division. Surprisingly, however, no signs for deregulation of suspensor development were found. Possibly, the effectiveness of BARNASE-induced cell lethality (Baroux et al., 2001) was not sufficient to induce such defects in the embryo proper that would allow the detection of embryo-suspensor interregional communication. Alternatively, it may be that the defects in suspensor development observed in Arabidopsis sus and twn mutants are not a result of improper regulation by the embryo proper. Here, we used the ACT LTP1 and EF DTA lines to challenge this hypothesis.
First, we confirmed that the UAS::DTA gene encodes a functional toxin by analyzing RPS5A»DTA F1 embryos. Cell ablation was visible in Schiff reagent-stained ovules shortly after fertilization as bright nuclear fluorescence and degeneration of the young embryo and endosperm (Fig. 3B). To confirm that the DTA protein works cell autonomously, LTP1»DTA embryos were generated and analyzed at early developmental stages. The first obvious aberrations were found during the 16-cell stage. At this stage, protodermal cells but not inner cells showed the hallmarks of cell ablation, ranging from strong uniform nuclear fluorescence (not shown) to the presence of nucleus-free cellular remnants (Fig. 3H).
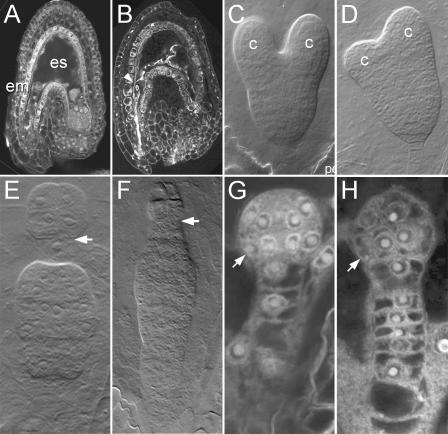
Figure 3.
DTA expression in domains of the Arabidopsis embryo. A, C, and G, Wild-type two-cell (A), heart (C), and dermatogen stage (G) embryos. B, RPS5A#5»DTA#6 F1 seed containing only remnants of embryo and endosperm. D, Heart stage LTP1#8»DTA#6 F1 embryos with normal pattern but defective cotyledons. E and F, Globular stage LTP1#8»DTA#6 F1 embryos showing arrest of the embryo proper and proliferation of the suspensor. The clonal boundary of the first embryonic cell division is marked by an arrow. H, Dermatogen stage LTP1#8»DTA#6 F1 embryo showing ablation of the protoderm (arrow, compare with wild-type embryo in G). Note that the number of suspensor cells is doubled in the LTP1#8»DTA#6 embryo. Em, Embryo; es, endosperm; c, cotyledon.
Cell ablation in LTP1»DTA embryos showed significant variation in timing, and in the severity of the induced defects. Extreme defects were observed in 8.7% of the analyzed embryos (n = 92), including complete protoderm ablation during the 16-cell stage (Fig. 3, E and F), whereas in the remaining embryos, the defects originated from post-globular ablation events (Fig. 3D). Interestingly, preglobular ablation of the protoderm induced excessive divisions in the suspensor (Fig. 3, E, F, and H). In one class of embryos, excessive divisions were in appropriate orientation, which merely anticipated later suspensor divisions (Fig. 3H). In another class of embryos, suspensor cells were more obviously released from repression because a dense cell group developed in the middle of the suspensor. Often, these cell groups attained division patterns common to the embryo (Fig. 3, E and F). In the remaining embryos, cellular defects suggested the ablation events to have taken place after the globular to heart stage transition. In these embryos, the ablation events resulted in strong defects in cotyledon development. Embryos showed only rudimentary cotyledons at the apical end of the axis. Surprisingly, such embryos had no visible defects in root pole patterning (Fig. 3D), which indicates that the apical control over suspensor development, evident at early stages, loses its effect after transition to the heart stage.
The sites that are marked by cell ablation in post-globular LTP1»DTA embryos are exactly those where LTP1 promoter activity is reported by GUS activity in LTP1»GGi embryos. Moreover, in contrast to DR5(7x)»DTA embryos (Friml et al., 2003), no signs of cell ablation were found in LTP1»DTA suspensor cells or in the hypophysis, This indicates that the aberrant basal cell divisions after early ablation events in the embryo proper are not a result of local DTA expression but are induced by a lack of “controlling” signals from the apical cell lineage.
Endosperm Ablation Reveals Its Role in Seed Size Regulation and Embryo Patterning
Several Arabidopsis mutants and transgenic lines and seeds resulting from crosses between parent plants with different ploidies show reduced endosperm development and reduced seed size (Scott et al., 1998; Luo et al., 2000; Garcia et al., 2003). The reduction in seed size in these cases is zygotically determined, suggesting that the regulation of seed size is controlled by the endosperm. This regulatory interaction between the endosperm, embryo, and seed coat represents another example of interregional communication within the seed. Because the mutant genotype may not only affect the endosperm, it is difficult to draw unequivocal conclusions as for the role of endosperm in seed growth and embryo development. To obtain such direct evidence, we analyzed seed growth and development in seeds that express DTA under control of the endosperm-expressed mGAL4:VP16 activator in enhancer trap line KS22I.
Based on fluorescence of the UAS::mGFP5-ER gene present on the mGAL4:VP16 enhancer trap T-DNA, KS22I enhancer expression commences after the second round of nuclear divisions (M. Ingouff and F. Berger, personal communication). In accordance with this expression pattern, endosperm development arrested almost immediately in KS22I»DTA seeds, and the endosperm soon degenerated, leaving only a few remnant cellular structures (Fig. 4, E-G). The most striking phenotype of KS22I»DTA F1 seeds was the complete cessation of seed expansion. Wild-type seeds underwent rapid and continuous growth shortly after fertilization (Fig. 4, A-D), whereas seed expansion was impaired in 57% (47/83) of the KS22I»DTA seeds analyzed 2 dap, in 56% (46/83) at 4 dap, and in 97% (131/135) at 6 dap (Fig. 4, E-H). This result probably reflects the variability in the onset of DTA expression in independent F1 seeds, but it also shows that the induced effect is fully penetrant at stages when the phenotype is unambiguously identifiable (6 dap, under these experimental conditions more or less corresponding to wild-type seeds with heart stage embryos). The absence of seed growth upon genetic endosperm ablation presents direct evidence that development of the endosperm is stringently required for seed growth. Similar results were obtained with enhancer trap line M003B (F. Berger and J. Haseloff, unpublished data) that also provides expression of mGAL4:VP16 in the endosperm (data not shown).
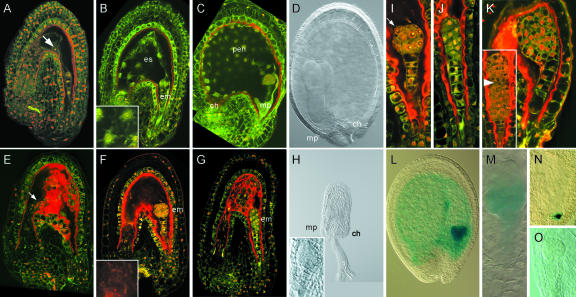
Figure 4.
Consequences of endosperm ablation on seed development. A to D, Wild-type seed development. A, Seed containing a two-celled embryo (arrow marks endosperm nucleus). B, Seed containing an eight-celled embryo. The inset shows a magnification of endosperm nuclei with surrounding cytoplasm and organelles. C, Seed containing a globular stage embryo. The endosperm domains (ch, pen, and mp) are clearly recognizable along the anterio-posterior axis of the seed. D, Seed containing a torpedo stage embryo. E to K, KS22I»DTA#6 F1 seed development. E, Seed of a stage comparable with A. Note the intense red fluorescence and the tracheid-like structure in the degraded endosperm (arrow). The embryo is not visible in this optical section. F, Seed containing a globular stage embryo. The endosperm is absent, as shown by a magnification of the endosperm cavity in the inset. G, Seed containing a globular stage embryo that is directly surrounded by seed coat tissues. The seed starts to collapse at this stage, which is visible because the seed coat tissues are in the same focal plane as the embryo. H, Collapsed seed containing a “heart stage” embryo. Magnification and stage are the same as in D. Inset, Magnification of the micropylar region containing the embryo. I, Higher magnification of the embryo in F shows a normal embryo pattern, except for a slight bulging of the epidermal cells (arrow). J, Higher magnification of the embryo in G, which shows severe lateral compression, indicating aberrant patterning. K, Heart stage embryos enclosed by seed coat tissues show abnormal patterns of cell division. Note that a structure resembling an embryo proper develops in the suspensor of the embryo in the inset (arrowhead marks the boundary between two embryo structures). L and M, RPS5A::GFP:GUS#20 activity in phenotypically wild-type (L) and KS22I»DTA#6 F1 sibling seeds (M). Note that GUS activity is still detectable in the embryo in (M) but only in the basal domain that does not yet show signs of physical obstruction. The KS22I line was heterozygous for the transgene. N and O, DR5(7x)::GFP:GUS#13 activity in wild-type (N) and phenotypically affected (O) sibling KS22I»DTA embryos. The embryo in O is delayed and does not show GUS activity. Seeds were stained for GUS activity for 4 h. es, Endosperm; em, embryo; ch, chalazal pole; pen, peripheral endosperm; mp, micropylar pole.
In addition to the defects in seed growth, embryos in KS22I»DTA F1 seeds showed defects in growth and cell division patterns. However, less than 30% of the embryos (10/38 at preglobular stage and 8/46 at globular stage) were affected. Preglobular embryos displayed swollen protodermal cells (Fig. 4I) or altered cell division planes (Fig. 4J), but embryos never showed any sign of cell death. Several embryo phenotypes, such as presence of two stacked embryo proper structures (Fig. 4K, inset), were found that indicated defects originating from early preglobular stages. This suggests that the presence of a developing endosperm is at least to some extent required for proper early embryo development.
In the absence of endosperm development, KS22I»DTA post-globular stage embryos became directly enclosed by the seed coat tissues, and, presumably as a result of this physical obstruction, embryos showed heavily distorted division patterns (Fig. 4K), and seeds collapsed after transition stage (Fig. 4H) and did not germinate (not shown). Cells within “arrested” embryos showed reduced expression of the RPS5A::GUS marker (Weijers et al., 2001a; Fig. 4M) that is normally active in dividing cells (Fig. 4L). Interestingly, expression of the RPS5A::GUS marker always ceased first in the apical cells that are in direct contact with the seed coat. Because the phenotypes at the basal pole of KS22I»DTA embryos resembled those of embryos with defects in auxin response or transport (Mayer et al., 1991; Berleth and Jürgens, 1993; Hamann et al., 1999; Hobbie et al., 2000), the DR5(7x)::GUS reporter was crossed into the KS22I line. Upon crossing KS22I;DR5(7x)::GUS lines with EF DTA lines, F1 embryos consistently failed to activate the DR5(7x)::GUS marker (Fig. 4O), which is normally active in the hypophyseal cell group from the mid-heart stage on (Fig. 4N), suggesting that arrested embryos fail to establish the auxin maximum that is associated with patterning of the root meristem (Sabatini et al., 1999). Whether this reflects a defect in auxin-related processes or that the absence of the auxin response maximum is merely the result of auxin-independent defects in embryo patterning due to the endosperm ablation cannot be distinguished.
Our analysis provides direct evidence for the previously suggested role for endosperm development in promoting seed growth and facilitating proper embryo development (Ohad et al., 1996; Scott et al., 1998; Luo et al., 2000; Garcia et al., 2003). The results clearly show that endosperm development is not required per se for proper embryo development during the early preglobular stages because only a part of the nonenlarged seeds contain defective embryos. Whether the patterning defects observed in KS22I»DTA embryos at later stages are caused by the absence of the endosperm or merely by the physical constraints of the underdeveloped integument layers awaits further studies.
DISCUSSION
Intercellular communication in plants, with the central role of protein or peptide movement in root radial patterning (Nakajima et al., 2001), floral patterning (Sessions et al., 2000), and stem cell homeostasis in the shoot meristem (Lenhard and Laux, 2003) as recent examples, attracts increasing attention. Cellular interactions take place during postembryonic and embryo development, where, for example, embryonic root initiation depends on signaling across the clonal boundary set during the first cell division. Previously, communication had been proposed between embryo proper and suspensor and between embryo, endosperm, and seed coat tissues. In most cases, models were based on the phenotypes of developmental mutants; thus, direct evidence for these communication events was still lacking. Here, we provide direct evidence for and confirm the presence of all previously proposed communication and regulatory interactions in the seed by local expression of the DTA toxin in domains of the developing seed or embryo.
mGAL4:VP16/UAS Transactivation in Developing Arabidopsis Seed
GAL4-based transactivation technologies are routine methods in several species, including fruitfly (Drosophila melanogaster), zebra fish (Danio rezio), and Xenopus laevis (Brand and Perrimon, 1993; Scheer and Campos-Ortega, 1999; Hartley et al., 2002), to spatially regulate expression of transgenes. GAL4 components have been optimized for Arabidopsis (Haseloff, 1999), and examples of their successful application have been reported previously (Kiegle et al., 2000; Benjamins et al., 2001; Boisnard-Lorig et al., 2001; Sabatini et al., 2003). However, unlike the Lac operator/repressor system (Moore et al., 1998) that was tested for embryo-specific expression of GUS or the BARNASE toxin in Arabidopsis embryos (Baroux et al., 2001), a systematic analysis of the use of the mGAL4:VP16/UAS system for conditional region-specific expression in developing embryos and seeds has not been presented.
Our analysis using three different cell type-specific promoters and an endosperm-specific GAL4 enhancer trap line shows that transactivation of GUS is reliable in that the expression patterns mimic those seen in direct promoter-GUS fusion lines. In addition, reliable cell type-specific transactivation has been observed with three other promoters (D. Weijers and R. Offringa, unpublished data). It is particularly noteworthy that all of the promoters used here (RPS5A, LTP1, DR5, and KS22I) are also active in sporophytic maternal tissues. Nonetheless, two-component gene expression technology ensures the expression of GUS (or any other gene) only in the F1 zygotic tissues, thereby circumventing the sporophytic expression pattern of the chosen promoter.
One very striking observation was that embryos showed variable levels of transactivated gene expression regardless of which set of homozygous ACT and EF parent lines was used for crossings. This was obvious when GUS activity was monitored but exaggerated when the DTA toxin was expressed. Variability in onset of transactivated gene expression also has been reported for the pOp/LhG4 system (Baroux et al., 2001). Concentrations of the cytosine-methylation inhibitor 5-azacytidine that are sufficient to derepress UAS-dependent expression in tobacco (Nicotiana tabacum; Gälweiler et al., 2000) leave the variability and the maximal gene expression level unaffected in Arabidopsis (D. Weijers and R. Offringa, unpublished data). Therefore, it is unlikely that methylation plays a significant role in the variability of gene expression. An alternative explanation is that GAL4/UAS transactivation enhances naturally occurring fluctuations in gene expression levels. Along these lines, it is interesting to note that real-time measurement of transgene expression, as monitored by promoter-luciferase fusions, showed that gene expression in itself is a highly stochastic process (van Leeuwen et al., 2001). The amplification of expression levels by GAL4/UAS, superimposed on this stochastic behavior of transgenes, could easily explain the variability in gene expression. Nonetheless, in this system, the variability in gene expression is advantageous in that it can be visualized by coregulated reporter gene expression and, therefore, allows one to create dose-response relationships for transgene expression without the need to analyze a multitude of transgenic lines.
For the mGAL4:VP16/UAS system to be generally applicable in studies on plant development, it is important that transgenic lines do not show phenotypic effects in the absence of transactivation. That is, the mGAL4:VP16 and UAS components should not by themselves interfere with the process under study. As a previous study indicated (Bougourd et al., 2000), we show that in seeds, the UAS promoter is inactive in the absence of GAL4 because UAS::GFP:GUS lines do not show any GUS activity and UAS::DTA plants are wild type but conditionally lethal in the presence of mGAL4:VP16. Expression of a fusion between GAL4:VP16 and the rat (Rattus norvegicus) glucocorticoid receptor domain in Arabidopsis was shown to cause stress-related molecular and developmental phenotypes upon treatment with dexamethasone (Kang et al., 1999). In this study, we focused our analysis on embryo patterning and found it to be unaffected in all mGAL4:VP16-expressing lines. In the course of our work, ACT 35S lines were generated that are phenotypically normal and show appreciable postembryonic transactivation levels (Y. Xiong, D. Weijers, and R. Offringa, unpublished data). This corroborates previous observations that mGAL4:VP16 expression does not affect postembryonic Arabidopsis development (Kiegle et al., 2000; Boisnard-Lorig et al., 2001; Sabatini et al., 2003).
DTA-Mediated Ablation Shows Apical Control of Basal Embryo Development
The first zygotic division in Arabidopsis embryogenesis is asymmetric and yields two cells with distinct fates. One cell produces most of the mature embryo, whereas the other cell gives rise to a filamentous structure, the suspensor, which in the early stages connects the embryo to the maternal seed coat tissues. During the globular stage, the uppermost suspensor cell joins the developing pro-embryo and gives rise to the distal root meristem region, including the quiescent center. Thus, cell lineages that are clonally separated during the first division have to coordinate their development to collectively initiate a root meristem. Detailed analysis of the mp and bdl mutants showed that the failure to initiate a root meristem in these mutants is preceded by defects in specification of the apical zygote daughter cell. In addition, the MP and BDL mRNAs are detected in the embryo proper but not in the hypophysis (Hamann et al., 2002), confirming the idea that hypophysis specification and division requires gene activities in the adjacent pro-embryo. Earlier studies on the sus and twn classes of Arabidopsis mutants had suggested that repression of embryonic potential in the suspensor requires a vital and functional pro-embryo. An elegant study by Baroux et al. (2001) showed that expression of BARNASE in the pro-embryo caused hypophysis defects similar to those found in mp and bdl mutants. However, they did not find indications for derepression of suspensor division activity, which challenged the concept of active repression of the suspensor by the embryo proper. In our analyses, expression of DTA in the LTP1 expression domain resulted in the phenotypes reported by Baroux et al. (2001) and in derepression of suspensor division. The stronger phenotypes observed in our study can be explained by higher expression levels of the mGAL4:VP16-UAS system in the embryo as compared with the pOp/LhG4 transactivation system. Alternatively, the toxicity of DTA used in our experiments may be higher than that of BARNASE used by Baroux and coworkers.
After protoderm ablation in preglobular LTP1»DTA embryos, ectopic cell divisions in the suspensor were either ordered in a linear file, superficially random, or in embryo-like patterns. This result provides strong direct evidence for the hypothesis that the pro-embryo represses suspensor division. Apparently, two important aspects of embryo development, the initiation of the root meristem at the basal pole of the embryo axis and maintenance of the initial fate specification upon the first asymmetric division of the zygote, requires pro-embryo activity. Interestingly, ablation of cotyledons did not affect root pole patterning, which is in accordance with the observation that the root meristem is autonomous at this stage (Schiavone and Racusen, 1990) and with the phenotype of the Arabidopsis gurke mutant that lacks cotyledons but shows normal basal pole patterning (Torres-Ruiz et al., 1996). In conclusion, the communication across the clonal apical-basal lineage boundary is essential for the establishment but not for the maintenance of the embryo pattern.
The Role of Endosperm in Seed Development
Our ablation studies also support the proposed role of the endosperm in seed growth (Ohad et al., 1996; Scott et al., 1998; Garcia et al., 2003). Although it is impossible to draw mechanistic conclusions from these experiments, the advantage of our expression system is that the cytological consequences of DTA-mediated ablation are unambiguous; therefore, it is justified to state that only the endosperm is manipulated. In other studies based on mutants or crosses between plants with different ploidy levels, it cannot be excluded that the effects on embryo and seed development are indirectly caused by the genetic background, rather than by the (lack of) endosperm development. Therefore, our experiments provide more direct evidence for a causal relationship between endosperm development and seed expansion. The possibility that endosperm ablation inhibits embryo development, which in turn inhibits seed growth, can be excluded because the arrest in seed growth occurs very early after the onset of KS22I expression, whereas embryo growth is not severely hampered at these stages.
Although previous reports have postulated and shown a role for endosperm in growth of the embryo (Scott et al., 1998; Garcia et al., 2003), it remained unclear as to whether the endosperm functions in regulating embryo development before the heart stage. Given the fact that a limited percentage of KS22I»DTA embryos showed embryo defects that originated in early embryogenesis, it seems that the endosperm is at least to some extent involved in early embryo development. However, these defects were variable and pleiotropic; therefore, it seems unlikely that the endosperm regulates a specific embryo patterning event. If the endosperm nurtures the young embryo, then endosperm ablation could lead to a lack of sufficient nutrients, thereby causing pleiotropic embryo defects. Alternatively, early endosperm ablation might lead to early loss of mechanical buffering of the seed coat, thereby leading to early physical obstruction of the developing embryo. The use of a GAL4 line that gives even earlier expression during endosperm development may provide insight into the function of the endosperm in early embryo development. Alternatively, this problem could be approached by embryo-specific complementation of a mutation that affects both embryo and endosperm development.
MATERIALS AND METHODS
Plant Lines, Growth, and Crosses
Arabidopsis plants were grown as previously described (Weijers et al., 2001a). ACT lines were germinated on medium supplemented with 25 mg L-1 kanamycin (Duchefa, Haarlem, The Netherlands), and EF lines were germinated on medium supplemented with 15 mg L-1 phosphinotricin (Duchefa). The KS22I line (C24 ecotype; Boisnard-Lorig et al., 2001) was introgressed into Col by two backcrosses. The RPS5A::GFP:GUS and DR5(7x)::GFP:GUS lines have been described previously (Benjamins et al., 2001; Weijers et al., 2001a).
Genetic crosses were performed as described by Weijers et al. (2001a) using homozygous ACT lines as female parents and homozygous EF lines as a male parents. The RPS5A::GFP:GUS and DR5(7x)::GFP:GUS lines were crossed with KS22I, and F1 plants were selected that contained both T-DNAs. These lines were crossed subsequently with EF DTA lines. Two independent transgenic lines were used for each construct.
DNA Cloning
DNA cloning was performed following standard procedures (Sambrook et al., 1989) using Escherichia coli strain DH5α. The Arabidopsis LTP1 promoter was isolated as a 1,150-bp HinDIII/BamHI fragment from pMT121 (Toonen et al., 1997) and used to replace the CaMV 35S promoter in pBIN 35S::mgal4:vp16 (J. Haseloff, unpublished data; here named ACT 35S; pSDM1600). The resulting construct was named ACT LTP1 (pSDM7019). The ACT DR5(7x) construct (pSDM7028) was generated by introducing a 100-bp HinDIII/SalI fragment containing seven tandem DR5 repeats (Ulmasov et al., 1997) from pSDM6215 (R. Offringa, unpublished data) into construct pSDM7027, which contains a -47 35S promoter upstream of mGAL4: VP16. For generating the ACT RPS5A construct (pSDM7040), the 35S promoter in ACT 35S was replaced with a HinDIII/BamHI fragment from AtRPS5A::GGi (Weijers et al., 2001a; pSDM7041) containing the AtRPS5A promoter.
The UAS promoter was excised as a 200-bp SphI/BamHI fragment from pBIN 35S::mgal4:vp16-UAS::mgfp5-ER (J. Haseloff, unpublished data) and cloned into pIC20H, resulting in pIC UAS (pSDM7000). Subsequently, the transcriptional terminator from the NOS (nopaline synthase) gene was isolated as a 300-bp EcoRI-KpnI fragment from pMOG690 (Romano et al., 1991) and ligated into pSDM7000 to yield pIC UAS-tNOS (pSDM7022). A 2.7-kb NcoI-EcoRI fragment from pMP3625 (Quaedvlieg et al., 1998) containing the egfp/gusA::intron (GGi) fusion gene was cloned into pSDM7022 to yield pIC UAS::GGi::tNOS (pSDM7003). A 3.2-kb HinDIII-SacI fragment containing UAS::GGi::tNOS was then inserted in the T-DNA region of pGPTV-BAR (Becker et al., 1992) to yield EF GGi (pSDM7006). The DT-A coding sequence from pTH1 (Breitman et al., 1987) was isolated as an 800-bp BamHI-KpnI fragment from pSDM6023 (R. Offringa, unpublished data) and cloned into pSDM7000 to result in pIC UAS::DTA::t35S (pSDM7020). A 1.3-kb HinDIII fragment from pSDM7020 was cloned into pSDM7006 to yield EF DTA (pSDM7021). The use of BamHI for cloning removed the original translation start codon from the DTA coding sequence. Although this leads to the production of a truncated DT-A protein from the next in-frame ATG codon 15 downstream from the original translation start, our study shows that the resulting protein is still active in cell ablation.
Transformation of Constructs and Selection of Lines
All binary vector constructs were introduced into Agrobacterium tumefaciens LBA1115, and Col plants were transformed by floral dip (Clough and Bent, 1998). The ACT DR5(7x) construct was transformed into EF GGi#15 lines directly. Primary transformants were selected on medium containing 100 mg L-1 timentin and 30 mg L-1 phosphinotricin or 70 mg L-1 kanamycin, respectively, and inspected for growth aberrations during their further development. In the T2 generation, lines were selected that showed a 3:1 segregation ratio for the transgene. To preselect EF GGi lines, T2 seedlings were infected with A. tumefaciens containing the ACT 35S construct, and GUS activity was scored 3 d later. Lines with the highest staining frequency and intensity were used. ACT lines were selected by crossing T2 plants with EF GGi#15 plants and monitoring GUS activity in heart stage F1 embryos. Homozygous lines were selected in the T3 generation.
Blotting and Hybridization
Total RNA was isolated from seedlings as described (Weijers et al., 2001a). A 600-bp BamHI-SacI fragment from ACT 35S spanning the mGAL4: VP16 coding sequence was used as a probe. For protein isolation, 50 to 100 mg of seedling tissue was quickly frozen in liquid nitrogen and ground to a fine powder. After incubation in an equal volume of extraction buffer (50 mm Na-phosphate [pH 7.2], 5 mm dithiothreitol, 5% [v/v] glycerol, 10 mm EDTA, and 0.1% [v/v] Triton X-100) for 20 min on ice and centrifugation, the supernatant was used as a protein extract. Protein concentration in total extracts was quantified using a Bradford assay. Thirty micrograms of protein was separated on a 7.5% (w/v) SDS-polyacrylamide gel according to standard procedures. Protein was then blotted onto Immobilon-P membranes (Millipore, Billerica, MA). Blots were incubated with α-GAL4-DBD (sc-510; Santa Cruz Biotechnology, Santa Cruz, CA) or α-VP16 (1-21; sc-7545; Santa Cruz Biotechnology) as primary antibody. An alkaline phosphatase-linked conjugate was used as a secondary antibody, and detection of phosphatase activity was performed according to the manufacturer's procedures.
GUS Activity Assays and Microscopy
Histochemical staining of plant tissues and phenotypic embryo analysis was performed as described by Weijers et al. (2001a). Staining times are indicated where appropriate. For analysis of GFP fluorescence, seedlings were mounted in 10% (v/v) glycerol and viewed on an Axioplan microscope (Zeiss, Jena, Germany) equipped with a Bio-Rad MRC1024 confocal microscope (Bio-Rad Laboratories, Hercules, CA). The 488-nm laser line from the Kr/Ar laser was used to excite GFP, and the fluorescent signal was detected through a 510-nm bandpass filter. Images were collected in Laser-sharp software (Bio-Rad Laboratories). Confocal laser scanning microscopy on whole-mount seeds was performed as described (Sørensen et al., 2001). All images were recorded using a DKC5000 camera and digital recorder (Sony, Tokyo) and compiled in Adobe Photoshop 5.5 (Adobe Systems, Mountain View, CA).
Acknowledgments
The authors thank Jim Haseloff for generously sharing the mGAL4: VP16/UAS constructs before publication; Fred Berger and Jim Haseloff for providing the KS22I and M003B lines; Sacco de Vries and Herman Spaink for providing DNA clones; Quirien Boone, Nick Wierckx, Olivier Meurette, and Ab Quint for invaluable technical assistance; Peter Hock for digital artwork; Enrico Scarpella, Haico van Attikum, and René Benjamins for helpful discussions; and Fred Berger, Jiri Friml, and Niko Geldner for helpful comments on the manuscript.
This work was supported by the Research Council for Earth and Life Sciences (ALW), with financial aid from the Netherlands Organisation for Scientific Research (NWO).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.030692.
References
- Baroux C, Blainvillain R, Moore IR, Gallois P (2001) Transactivation of BARNASE under the AtLTP1 promoter affects the basal pole of the embryo and shoot development of the adult plant in Arabidopsis. Plant J 28: 503-515 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195-1197 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas PJJ, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057-4067 [DOI] [PubMed] [Google Scholar]
- Berger F (2003) Endosperm: the crossroad of seed development. Curr Opin Plant Biol 6: 42-50 [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575-587 [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougourd S, Marrison J, Haseloff J (2000) An aniline-blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J 24: 543-550 [DOI] [PubMed] [Google Scholar]
- Brand A, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401-415 [DOI] [PubMed] [Google Scholar]
- Breitman ML, Clapoff S, Rossant J, Tsui LC, Glode LM, Maxwell IH, Bernstein A (1987) Genetic ablation: targeted expression of a toxin gene causes microphtalmia in transgenic mice. Science 238: 1563-1565 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Day CD, Galgoci BF, Irish VF (1995) Genetic ablation of petal and stamen primordia to elucidate cell interactions during floral development. Development 121: 2887-2895 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature (in press) [DOI] [PubMed]
- Gälweiler L, Conlan RS, Mader P, Palme K, Moore I (2000) The DNA-binding activity of Gal4 is inhibited by methylation of the Gal4 binding site in plant chromatin. Plant J 23: 143-157 [DOI] [PubMed] [Google Scholar]
- Garcia D, Saingery V, Chambrier P, Mayer U, Jürgens G, Berger F (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131: 1661-1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) SHORT INTEGUMENTS/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130: 808-822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin-response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610-1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387-1395 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley KO, Nutt SL, Amaya E (2002) Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc Natl Acad Sci USA 99: 1377-1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58: 139-151 [DOI] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M (2000) The axr6 mutants of Arabidopsis define a gene involved in auxin response and early development. Development 127: 23-32 [DOI] [PubMed] [Google Scholar]
- Jürgens G (2001) Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J 20: 3609-3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Mayer U (1994). Arabidopsis. In JBL Bard, ed, Embryos, Color Atlas of Development. Wolffe Publishing, London, pp 7-22
- Kang H-G, Fang Y, Singh KB (1999) A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defence-related genes. Plant J 20: 127-133 [DOI] [PubMed] [Google Scholar]
- Kiegle E, Moore CA, Haseloff J, Tester MA, Knight MR (2000) Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J 23: 267-278 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Laux T (2003) Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130: 3163-3173 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637-10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Torrez Ruiz RA, Berleth T, Miséra S, Jürgens G (1991) Mutations affecting body organisation in the Arabidopsis embryo. Nature 353: 402-407 [Google Scholar]
- Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95: 376-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307-311 [DOI] [PubMed] [Google Scholar]
- Nilsson O, Wu E, Wolfe DS, Weigel D (1998) Genetic ablation of flowers in transgenic Arabidopsis. Plant J 15: 799-804 [DOI] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL (1996) A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA 93: 5319-5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg NEM, Schlaman HRM, Admiraal PC, Wijting SE, Stougaard J, Spaink HP (1998) Fusions between green fluorescent protein and β-glucuronidase as sensitive and vital bifunctional reporters in plants. Plant Mol Biol 37: 715-727 [DOI] [PubMed] [Google Scholar]
- Romano CP, Hein MB, Klee HJ (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev 5: 438-446 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463-472 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scheer N, Campos-Ortega JA (1999) Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev 80: 153-158 [DOI] [PubMed] [Google Scholar]
- Schiavone FM, Racusen RH (1990) Microsurgery reveals regional capabilities for pattern reestablishment in somatic carrot embryos. Dev Biol 141: 211-219 [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW (1994) Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235-3245 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329-3341 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D (2000) Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779-782 [DOI] [PubMed] [Google Scholar]
- Sørensen MB, Chaudhury AM, Robert H, Bancharel E, Berger F (2001). Polycomb group genes control pattern formation in plant seed. Curr Biol 11: 277-281 [DOI] [PubMed] [Google Scholar]
- Thoma S, Hecht U, Kippers A, Botella J, de Vries S, Somerville C (1994) Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol 105: 35-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen MA, Verhees JA, Schmidt ED, van Kammen A, de Vries SC (1997) AtLTP1 luciferase expression during carrot somatic embryogenesis. Plant J 12: 1213-1221 [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Lohner A, Jürgens G (1996) The GURKE gene is required for normal organization of the apical region in the Arabidopsis embryo. Plant J 10: 1005-1016 [DOI] [PubMed] [Google Scholar]
- Tsugeki R, Fedoroff NV (1999) Genetic ablation of root cap cells in Arabidopsis. Proc Natl Acad Sci USA 96: 12941-12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963-1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Ruttink T, Borst-Vrenssen AW, van der Plas LH, van der Krol AW (2001) Characterization of position-induced spatial and temporal regulation of transgene promoter activity in plants. J Exp Bot 52: 949-959 [DOI] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW (1994) Embryonic transformation of the suspensor in twin polyembryonic mutant of Arabidopsis. Dev Biol 165: 566-573 [DOI] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken R-J, Quint A, Hooykaas P, Offringa R (2001a) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in A Ribosomal Protein S5 gene. Development 128: 4289-4299 [DOI] [PubMed] [Google Scholar]
- Weijers D, Geldner N, Offringa R, Jürgens G (2001b) Paternal gene activity during Arabidopsis embryogenesis. Nature 414: 709-710 [DOI] [PubMed] [Google Scholar]
- Yamaizumi M, Mekada E, Uchida T, Okada Y (1978) One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell 15: 245-250 [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Somerville CS (1997) Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Proc Natl Acad Sci USA 94: 7349-7355 [DOI] [PMC free article] [PubMed] [Google Scholar]