Abstract
Fasciclin-like arabinogalactan proteins (FLAs) are a subclass of arabinogalactan proteins (AGPs) that have, in addition to predicted AGP-like glycosylated regions, putative cell adhesion domains known as fasciclin domains. In other eukaryotes (e.g. fruitfly [Drosophila melanogaster] and humans [Homo sapiens]), fasciclin domain-containing proteins are involved in cell adhesion. There are at least 21 FLAs in the annotated Arabidopsis genome. Despite the deduced proteins having low overall similarity, sequence analysis of the fasciclin domains in Arabidopsis FLAs identified two highly conserved regions that define this motif, suggesting that the cell adhesion function is conserved. We show that FLAs precipitate with β-glucosyl Yariv reagent, indicating that they share structural characteristics with AGPs. Fourteen of the FLA family members are predicted to be C-terminally substituted with a glycosylphosphatidylinositol anchor, a cleavable form of membrane anchor for proteins, indicating different FLAs may have different developmental roles. Publicly available microarray and expressed sequence tag data were used to select FLAs for further expression analysis. RNA gel blots for a number of FLAs indicate that they are likely to be important during plant development and in response to abiotic stress. FLAs 1,2, and 8 show a rapid decrease in mRNA abundance in response to the phytohormone abscisic acid. Also, the accumulation of FLA1 and FLA2 transcripts differs during callus and shoot development, indicating that the proteins may be significant in the process of competence acquisition and induction of shoot development.
Cell-to-cell interactions and communication provide key structural, positional, and environmental signals during plant development. In plant cells, such signals must traverse the cell wall that surrounds the plasma membrane. Plant cell walls are primarily composed of the polysaccharides cellulose, cross-linking glycans, pectins, and some proteins (Cosgrove, 1997) that together form a complex interactive network known as the extracellular matrix (ECM; Bacic et al., 1988; Carpita and Gibeaut, 1993). The nature of the interactions changes during development and is influenced by biotic and abiotic stresses, resulting in altered wall composition and structure (Cassab and Varner, 1988). Cell wall proteins, which generally comprise less than 10% of the dry weight of the primary wall (Bacic et al., 1988), are recognized as critical components in maintaining the physical and biological functions of the plant ECM. Most ECM proteins belong to large families that include enzymes (such as the hydrolases, proteases, glycosidases, peroxidases, and esterases), expansins, wall-associated kinases, and hydroxyproline (Hyp)-rich glycoproteins (Arabidopsis Genome Initiative [AGI], 2000).
Arabinogalactan proteins (AGPs) are a class of Hyp-rich glycoproteins that are highly glycosylated and are abundant in the plant cell wall and plasma membrane (Fincher et al., 1983; Nothnagel, 1997; Bacic et al., 2000; Gaspar et al., 2001; Showalter, 2001). Type II arabinogalactan (AG) polysaccharide chains predominate and are O-glycosidically linked to Hyp residues in the protein backbone, resulting in the total mass of the molecule consisting of 90% to 99% carbohydrate (Nothnagel, 1997; Serpe and Nothnagel, 1999; Bacic et al., 2000). There is increasing evidence that Hyp (arabino) galactosylation depends on the clustered, noncontiguous arrangement of the Hyp residues (Goodrum et al., 2000; Zhou et al., 2002). In contrast, blocks of contiguous Hyp residues, such as those that occur in extensins, are arabinosylated with short oligosaccharides (Kieliszewski and Lamport, 1994; Shpak et al., 2001).
In Arabidopsis, the glycosylphosphatidylinositol (GPI)-anchored AGPs can be divided into four subclasses, the classical AGPs, those with Lys-rich domains, and AG peptides with short protein backbones (Schultz et al., 2002). The fasciclin-like AGPs (FLAs) constitute a fourth distinct subclass of AGPs.
Proteins with variable numbers of fasciclin domains, generally one to four, were first identified in fruitfly (Drosophila melanogaster) and have since been identified in proteins from animals, yeast, bacteria, algae, lichens, and higher plants (Elkins et al., 1990b; Huber and Sumper, 1994; Kawamoto et al., 1998; Gaspar et al., 2001; Paulsrud and Lindblad, 2002; Johnson et al., 2003). Fasciclin domains are 110 to 150 amino acids long and have low sequence similarity. This low sequence similarity may account for the lack of a single consensus sequence for fasciclin domains. However; all fasciclin domains contain two highly conserved regions (H1 and H2) of approximately 10 amino acids each (Kawamoto et al., 1998). Proteins containing fasciclin domains, from a broad spectrum of organisms, have been shown to function as adhesion molecules (Elkins et al., 1990a; Huber and Sumper, 1994; Kawamoto et al., 1998; Kim et al., 2000).
Fasciclin 1 (Fas1) from fruitfly is capable of promoting cell adhesion through homophilic interactions (Elkins et al., 1990a). Fas1 and midline fasciclin from fruitfly have roles in the positioning of axonal pathways (Elkins et al., 1990b; Hu et al., 1998) and synaptic plasticity (Zhong and Shanley, 1995).
Monoclonal antibodies raised against Algal-CAM, a fasciclin-like adhesion protein, prevent appropriate cell-cell contacts of the four-cell Volvox carteri embryo (Huber and Sumper, 1994). The gene for Algal-CAM predicts two fasciclin domains and an extensin-like domain rich in Ser-Pro3-5 with the Pro residues likely to be hydroxylated and subsequently O-glycosylated with short arabinooligosaccharides (Kieliszewski and Lamport, 1994; Kieliszewski et al., 1995; Shpak et al., 2001). The recent identification of a sos5 (salt overly sensitive) mutant in Arabidopsis with an amino acid substitution in the H2 region of FLA4 (Shi et al., 2003) indicates that this domain is important for FLA function.
Twenty-one genes encoding FLAs have been identified in Arabidopsis (Gaspar et al., 2001; Schultz et al., 2002). Compared with other classical AGPs, the FLAs are a relatively heterogeneous group because they can have one or two AGP regions and one or two fasciclin-like domains. Fourteen of the FLAs are predicted to be GPI-anchored. FLAs are one of the many classes of chimeric GPI-anchored proteins that also contain AG glycomodules (Borner et al., 2003). Despite a large number of Arabidopsis proteins predicted to be GPI-anchored (Borner et al., 2002), until recently there was little direct experimental evidence. Experimental support for the GPI-anchoring of FLAs 1, 7, 8, and 10 was recently obtained. A proteomics approach showed that FLAs 1, 7, 8, and 10 are sensitive to phospholipase C cleavage from the plasma membrane (Borner et al., 2003). This suggests that other FLAs predicted to be GPI-anchored will also prove to be GPI-anchored in vivo. GPI anchors in animals are thought to provide properties such as increased lateral mobility in the lipid bilayer, regulated release from the cell surface, polarized targeting to different cell surfaces, and inclusion in lipid rafts (Hooper, 1997; Gaspar et al., 2001; Harris and Siu, 2002). Both Fas1 from fruitfly (Hortsch and Goodman, 1990) and Algal-CAM from V. carteri (Huber and Sumper, 1994) have GPI-anchored variants that may regulate their release from the plasma membrane.
Although FLAs and AGPs may have both short arabinooligosaccharides and large type II AG polysaccharide chains, it is the AG polysaccharides that are most likely to be involved in development. Most experiments that have led to proposed AGP function(s) have used monoclonal antibodies that recognize AGP carbohydrate epitopes or β-glucosyl Yariv reagent (Yariv et al., 1967; Knox et al., 1991; Pennell, 1992). AGPs have been implicated in several roles in plant growth and development such as cell fate determination, somatic embryogenesis, and cell proliferation (Majewski-Sawka and Nothnagel, 2000). Of major interest is the role of AGPs as signal molecules, suggested through use of the JIM8 antibody, which recognizes a set of AGPs that are thought to be involved in somatic embryogenesis in cell cultures (McCabe et al., 1997).
In this study, we have identified all the FLAs from Arabidopsis and divided them into different groups based on their domain structure. Characterization of the proteins show that FLAs precipitate with β-glucosyl Yariv reagent, a reagent that has been used to selectively precipitate AGPs and, hence, define this class of glycoproteins/proteoglycans. We have prioritized the study of selected FLAs based on available genomic resources, focusing on FLA1, FLA2, and FLA8. FLA2 and FLA8 are abundant FLAs, based on the number of expressed sequence tags (ESTs) and mass spectrometry (MS), suggests they are also abundant proteins. These three FLAs also showed interesting developmental and environmental regulation in Arabidopsis Functional Genomics Consortium (AFGC) microarray experiments. We verify these microarray results and show that selected FLAs are likely to be important during plant development and in response to abiotic stress. The putative function of the FLAs and their role in the ECM are discussed.
RESULTS
Structure and Sequence Analysis of the Arabidopsis FLAs
FLA sequences were identified from the annotated Arabidopsis database using hidden markov models (Schultz et al., 2002). Hidden markov models, as used by PSI-BLAST (Altschul et al., 1997) and HMMer (Durbin et al., 1998), are able to find proteins with low identity to the target sequence because they use multiple sequence alignments rather than pair-wise alignments and give higher weightings to the more conserved regions of the protein and lower weightings to the less conserved regions of the protein (Schultz et al., 2002). The annotation of these genes was relatively straightforward as only FLA1 has an intron (743 bp) in the genomic sequence.
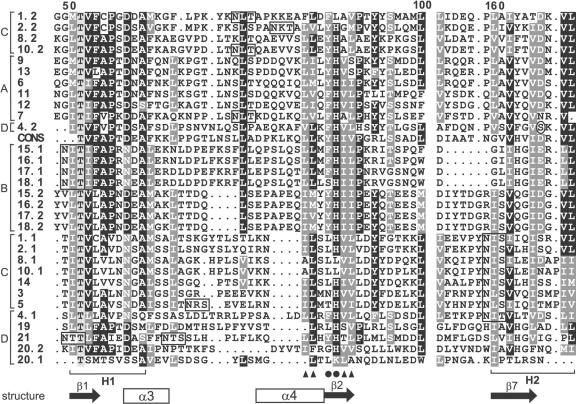
A multiple sequence alignment of all the fasciclin domains of FLAs from Arabidopsis and a consensus sequence (smart00554, see “Materials and Methods”) identified the conserved regions common to all fasciclin domains, called H1 and H2 (Fig. 1). There are currently three different fasciclin-related domains with different protein motif databases using different numbers of fasciclin related molecules to generate their consensus sequences (see “Materials and Methods”). Most of the Arabidopsis FLAs contain other conserved residues such as Leu and Ile near the H1 domain that are thought to be involved in either maintaining the structure of the fasciclin domain and/or cell adhesion (indicated by triangles in Fig. 1; Kim et al., 2000).
Figure 1.
Sequence alignment of part of the fascilcin domains of the Arabidopsis FLAs (1-21) and a consensus sequence (CONS) generated from 78 fasciclin domains from a diverse range of species. Where there are two fasciclin domains in the protein, the domain closest to the N terminus is indicated by .1 and the second by .2. The alignment was generated by ClustalX and manually edited. Conserved and identical residues are shaded dark gray and black, respectively, and similar residues are indicated in light gray. Periods represent gaps introduced by the program for optimal alignment, and the space before the H2 domain corresponds to 52 amino acid residues (or gaps) that show little or no similarity. The H1 and H2 conserved regions characteristic of fasciclin domains are indicated below the alignment. Amino acids thought to be involved in adhesion (Ile, Leu, and Val; Kim et al., 2000, 2002) are indicated by black triangles and YH motifs by black circles. The predicted secondary structure elements of the fasciclin domain based on the crystal structure of fas3 and fas4 domains of fruitfly are indicated below the alignment. These secondary structure elements are in most FLAs, but some, e.g. α4, are not conserved in group B FLAs (see “Discussion”). N-glycosylation motifs are boxed. The amino acid in the H2 region of FLA4 that is substituted in the sos5 mutant (Ser to Phe) is circled. The FLAs are grouped into four groups (A-D) based on phylogenetic analysis and pair-wise sequence comparison (see “Results”), as indicated on the left-hand side of the alignment.
The domain structure for each FLA was determined by identifying the arrangement and number of fasciclin domains, AGP regions, and the presence or absence of a putative GPI anchor signal (Fig. 2). The predicted N-terminal signal sequence was identified as described previously (Schultz et al., 2002), and C-terminal GPI anchor signal sequences were identified using PSORT (Nakai and Horton, 1999) and an improved GPI anchor prediction program for plants (Eisenhaber et al., 2003). Putative AGP regions were identified based on the presence of at least two noncontiguous Pro residues; for example, the sequence (A/S) P(A/S) P. In vivo, these regions are predicted to be Hyp-O-glycosylated and are increasingly being referred to as “glycomodules” (Tan et al., 2003).
Figure 2.
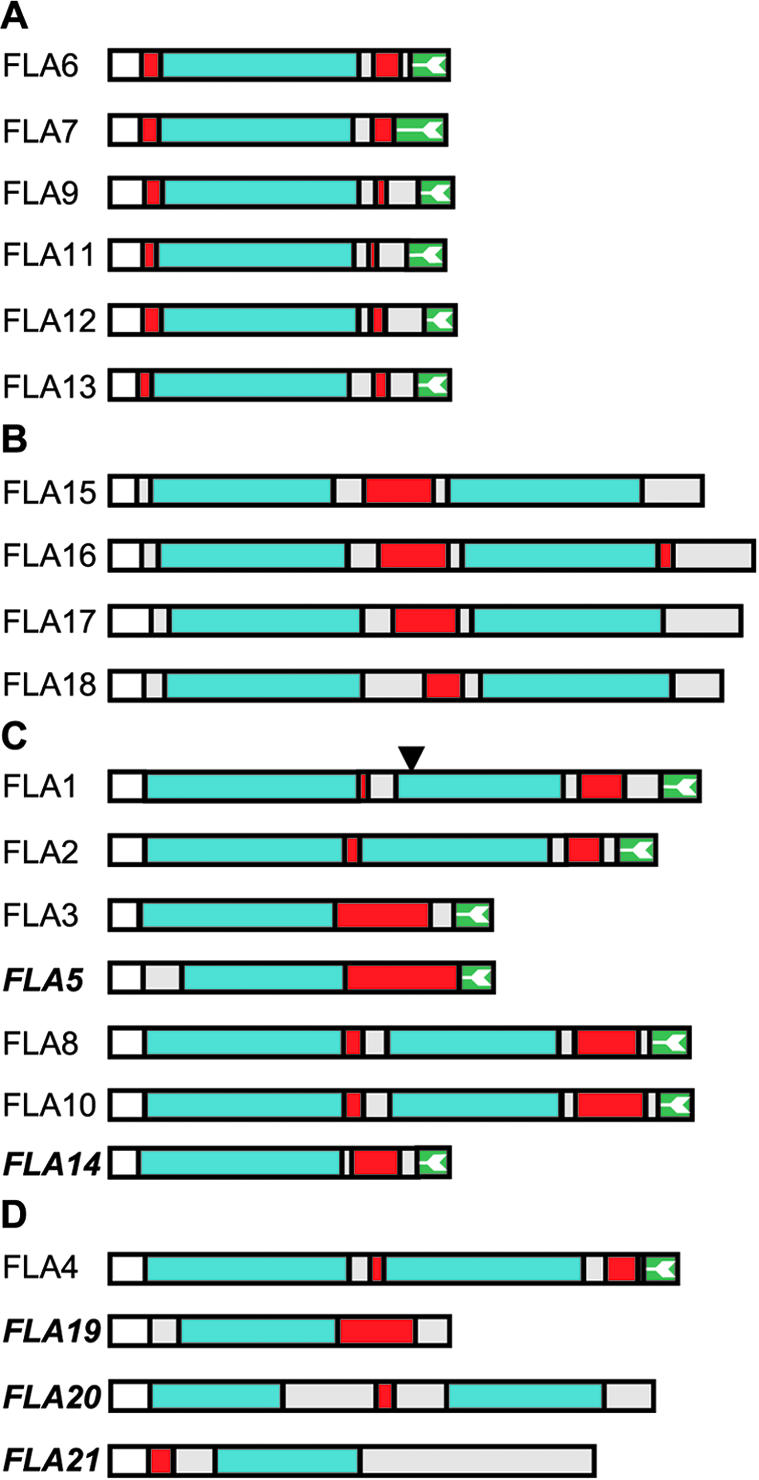
Schematic representation of the Arabidopsis FLAs deduced from DNA sequence (see “Materials and Methods” for accession nos.). The FLAs are grouped into four groups (A-D) based on phylogenetic analysis and pair-wise sequence comparison (see “Results”). The protein backbone of FLAs contains either one or two fasciclin-like domains (blue) and one or two AGP regions (red; Gaspar et al., 2001). Only the gene for FLA1 is predicted to have an intron, the position of which is indicated by a triangle, and where there are no ESTs, FLA names are bold and italicized. FLAs are predicted to contain an N-terminal secretion signal (white), and 14 of the 21 FLAs have a C-terminal signal for addition of a GPI anchor (green with white arrow; Schultz et al., 2002). Additional protein regions are shown in light gray. The longest FLA is FLA16 with 475 amino acids, and the shortest is FLA19 with 248 amino acids.
Due to the low sequence similarity of the FLAs, phylogenetic analysis is problematic (Li, 1997). To classify the FLAs, we used pair-wise sequence similarity, domain structure, and phylogenetic analysis. Taken together, these results indicated that the FLAs can be placed into four groups (Fig. 2). FLAs 6, 7, 9, and 11 to 13 form a group (A) characterized by a single fasciclin domain that is flanked by AGP regions and a C-terminal GPI-anchor signal. FLAs from group A have between 43% and 85% similarity at the amino acid level. The second group (B) comprises four sequences (FLAs 15-18) that have between 81% and 88% similarity. The group B FLAs have two fasciclin domains flanking an AGP region, with FLA16 having an additional small AGP region near its C terminus. None of the FLAs from group B have a C-terminal GPI anchor signal sequence. The third group (C) includes FLAs 1 to 3, 5, 8, 10, and 14. Group C FLAs have a variable domain structure with one or two fasciclin domains, one or two AGP regions, and a C-terminal GPI anchor signal. FLAs 3, 5, and 14 are placed in this group rather than group A because their fasciclin domains are more closely related to the first domain of the other group C FLAs (46%-62% amino acid similarity; Fig. 1). FLAs 4 and 19 to 21 do not appear to have a strong relationship to each other or to any of the other FLAs (33%-48% similarity) and are placed in the final group (D). FLAs that are not likely to be GPI anchored, such as FLAs 15 to 21 (Eisenhaber et al., 2003), have a C-terminal region that is either hydrophilic or shows no other recognizable motifs or structure.
FLAs Contain O-Linked Glycosylation as Predicted
FLAs were originally identified in Arabidopsis as part of our search for AGPs. The presence of clustered, noncontiguous Pro residues, separated by Ala or Ser residues in the protein backbone, suggests that they will contain large AG polysaccharide chains. In addition, all FLAs are predicted to have at least one N-glycosylation site in the fasciclin domains (Fig. 1) and more sites predicted in the other regions of the FLA sequence.
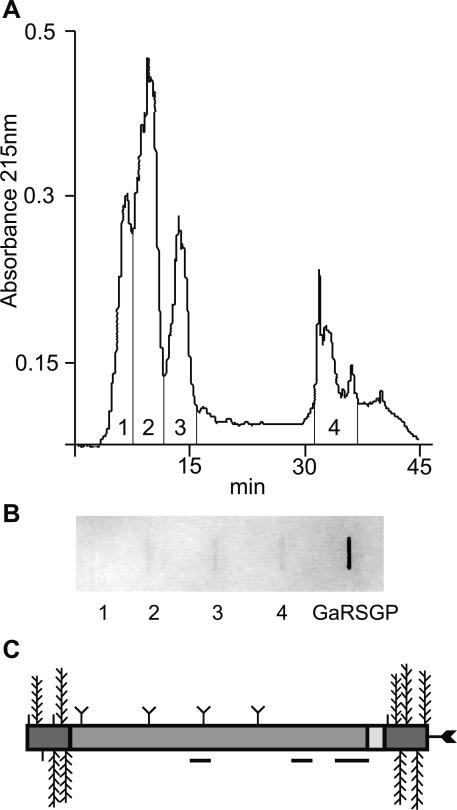
To verify the predicted O- and N-linked glycosylation of FLAs, total AGPs were purified from liquid culture-grown Arabidopsis plants. β-Glucosyl Yariv reagent was used to precipitate proteins containing O-linked AG polysaccharides, and these proteoglycans were then separated by reversed phase (RP)-HPLC as previously described (Fig. 3; Schultz et al., 2000). To identify fractions that contained both O-and N-linked glycans as predicted for FLAs, the RPHPLC fractions were blotted onto a membrane and incubated with a digoxygenin (DIG)-labeled WGA lectin (Goldstein and Hayes, 1978). WGA recognizes chitobiose (β1→4 linked GlcNAc)-containing sugar molecules. Fractions two to four contained AGPs that bound to WGA (Fig. 3). Classical AGPs, not predicted to contain N-glycans by analysis of their protein backbone sequence (Schultz et al., 2000), have been isolated previously from fractions one to three. Fraction four was selected for further analysis because FLAs are likely to be more hydrophobic than the classical AGPs.
Figure 3.
Separation of AGPs and FLAs by RP-HPLC and detection of N-glycans using wheat germ agglutinin (WGA). A, Separation of β-glucosyl Yariv precipitated proteoglycans into four fractions by RP-HPLC. B, Slot blot representing approximately 40 μg of total carbohydrate from four fractions of proteoglycans and 250 μg of a control, galactose-rich stylar glycoprotein (Sommer-Knudsen et al., 1996), a known N-linked glycoprotein from Nicotiana alata, were detected for their ability to bind WGA. Fractions two to four and galactose-rich stylar glycoprotein bind to WGA, indicating the presence of N-glycans. C, Schematic representation of the predicted structure of native FLA7 after processing and posttranslational modifications. Horizontal lines below the protein backbone represent peptides sequenced from a trypsin digest of fraction four (Tables I and II). Posttranslational modifications include Pro residues hydroxylated to Hyp and O-linked sugars added to these Hyp residues (vertical lines, arabinooligosaccharides; feathers, type II AG polysaccharide chains) in the AGP regions (dark gray). Predicted N-glycosylation sites in the fasciclin domain (medium gray) are indicated by a Y shape and a C-terminal GPI anchor by an arrow.
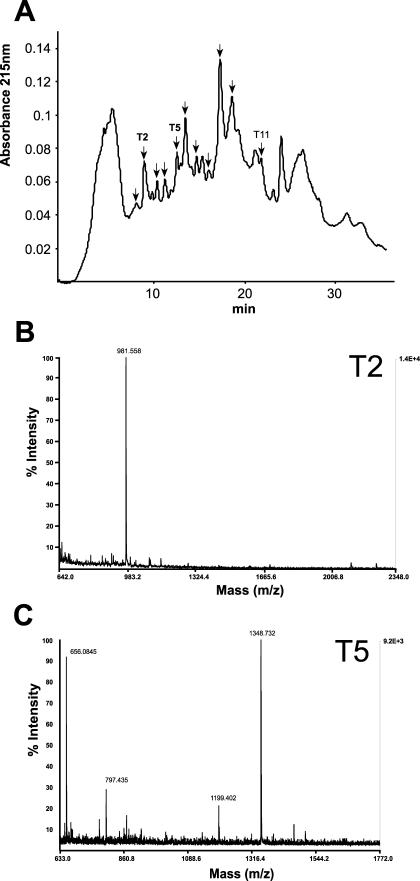
Fraction four was digested with trypsin, and the resulting peptides (Fig. 4) were first analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) MS. MALDI-TOF MS was used because fraction four likely contained a mixture of FLAs. Ions of molecular mass were obtained that correspond to those predicted for the trypsin-digested peptides of FLA2, FLA7, and FLA8 (Table I). N-terminal Edman sequencing of two different peaks (Fig. 4) identified three peptides that matched the FLA7 protein backbone (Fig. 3C) and one peptide matching an ENOD-like protein (Lin et al., 1999; Table II). This particular ENOD (GenBank accession no. AAD23007) has an AG glycomodule (SPAPSP), so it is not surprising that it precipitates with β-glucosyl Yariv reagent. The ENOD-like protein is also predicted to have two N-glycosylation sites (Table II). N-terminal sequencing of one of the FLA7 peptides (NPPLSNLTK) provided direct evidence that this FLA is N-glycosylated due to the blank cycle at the Asn residue in cycle 6. FLAs are predicted to contain two to eight N-glycosylation sites. FLA7 is predicted to have four (Fig. 3C). FLA7 is also predicted to contain up to nine type II AG polysaccharide chains and five arabinooligosacharides (Fig. 3C). The presence of FLAs 2, 7, and 8 in β-glucosyl Yariv precipitated material confirms the presence of Hyp-O-glycosylation in these FLAs.
Figure 4.
Separation of trypsin digested fraction four (see Fig. 3) by RP-HPLC (A) and representative examples of analysis of two fractions by MALDI-TOF MS (B and C). A, Separation of trypsin-digested proteoglycans in fraction four (see Fig. 3) by RP-HPLC. Fractions indicated by arrows were analyzed by MALDI-TOF MS (Table I), and fractions T2, T5, and T11 were used for N-terminal Edman sequencing (see Table II). B, MALDI-TOF MS spectrum of fraction T2 from RP-HPLC profile of peptide digest. The ion at m/z 981.558 matches the expected mass of a tryptic peptide of FLA7, FTDVSGTVR (981.501 Da). C, MALDI-TOF spectrum of fraction T5 from RP-HPLC profile of peptide digest. The ion at m/z 1,348.732 matches the expected mass of a tryptic peptide of FLA7, STDPVAVYQVNR (1,348.686 Da).
Table I.
Peptides corresponding to three FLAs obtained from a trypsin digest of hydrophobic fraction four of β-glucosyl Yariv precipitated proteoglycans (see Fig. 3)
| Protein | Peptide Sequencea | Trypsin Fractionb | Position in Protein | Predictedc | Mass Observedd | Difference |
|---|---|---|---|---|---|---|
| FLA7 | (F)VPKDDAFK | T3 | 88-95 | 919.489 | 919.47 | 0.019 |
| FLA7 | (K)FTDVSGTVRe | T2 | 149-157 | 981.501 | 981.558 | −0.057 |
| FLA7 | (K)NPPLSNLTKe | T2 | 99-107 | 983.557 | 983.569 | −0.012 |
| FLA7 | (F)STDPVAVYQVNRe | T5 | 167-184 | 1,348.686 | 1,348.732 | −0.046 |
| FLA2 | (K)TALVLY | T9 | 250-255 | 679.403 | 679.421 | −0.018 |
| FLA2 | (K)FMPKFK | T4, T5 | 237-242 | 797.438 | 797.435 | 0.003 |
| FLA2 | (F)QSTGSATGTSGY | T1 | 117-128 | 1,099.454 | 1,099.496 | −0.042 |
| FLA2 | (Y)SLYQIRNILSLHVLVDYF | T11 | 80-97 | 2,193.207 | 2,193.259 | −0.052 |
| FLA8 | (K)LADEINSR | T3 | 50-57 | 917.47 | 917.486 | −0.016 |
| FLA8 | (Y)HALAEYKPK | T1 | 256-262 | 1,056.584 | 1,056.51 | 0.074 |
| FLA8 | (K)TFANLLVSSGVLK | T5 | 201-213 | 1,348.784 | 1,348.717 | 0.067 |
| ENOD likef | (K)LSLVVISPR | T2 | 121-129 | 983.625 | 983.668 | −0.043 |
a Theoretical digest of FLA protein backbone with “msdigest” (http://prospector.ucsf.edu) was performed. Peptides shown had close agreement between predicted (c) and observed (d) masses. The first amino acid is predicted (parentheses) because it is the residue important for defining the trypsin cleavage site, but is not observed in the peptide. b Trypsin-digested fraction four of hydrophobic AGPs (see Fig. 3) was separated by RP-HPLC into 11 fractions (see Fig. 4A). c Predicted molecular mass of the FLA peptides. d Observed molecular mass inferred by MALDI-TOF MS of a trypsin-digested fraction of hydrophobic AGPs (see Fig. 4). e Peptide sequence was also determined using Edman degradation chemistry. f Early nodulin-like protein from Arabidopsis (GenBank accession no. AAD23007). Another peptide (residues 77-96) for this ENOD-like protein was sequenced by N-terminal sequencing (Table II).
Table II.
N-terminal peptide sequence derived from a trypsin digest of fraction four of β-glucosyl Yariv precipitated proteoglycans (see Fig. 3)
| Protein | Peptide Sequencea | Trypsin Fractionb | Position in Protein |
|---|---|---|---|
| FLA7 | NPPLSXLTK | T2 | 99-107 |
| FLA7 | FTDVSGTVR | T2 | 149-157 |
| FLA7 | STDPVAVYQVNR | T5 | 167-184 |
| ENOD likec | EAYNSCXTTNPLAXYTDG | T5 | 77-96 |
a Peptide sequence was determined using Edman degradation chemistry. X indicates that no amino acid was detected in this cycle, and underlined amino acids indicate predicted N-glycosylation sites. b Trypsin-digested fraction four of hydrophobic AGPs (see Fig. 3) was separated by RP-HPLC into 11 fractions (see Fig. 4A). c Early nodulin-like protein from Arabidopsis (GenBank accession no. AAD23007).
Expression of FLAs in Various Tissue Types
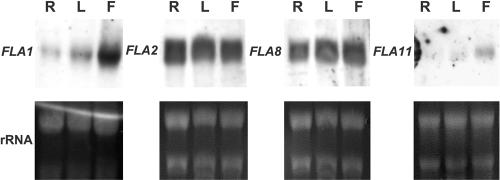
We have adopted a systematic approach to obtaining and evaluating the publicly available Arabidopsis resources to help us select specific subsets of genes for targeted experimental approaches (Schultz et al., 2002). Using this approach, an “expression summary” of each FLA gene was obtained from the Arabidopsis Gene Index (Table III; Schultz et al., 2002). There are currently ESTs for all FLAs except FLA5,FLA14, and FLAs19 to 21. FLA2 and FLA8 have the most ESTs, with 46 and 45 ESTs, respectively, identified in EST libraries. Some FLAs appear to have more restricted expression; for example, FLA3 has ESTs only in silique and flower tissue libraries (Table III). Eleven of the 21 FLAs are represented on the now complete AFGC microarray (Table IV; Schultz et al., 2002). Experiments conducted with the AFGC array include tissue comparisons that give an indication of expression levels in different plant organs. Results from microarrays suggest that many FLAs are more abundant in flowers than in other tissues (Table IV). To confirm the EST summary and microarray information, RNA gel blots were performed for four of the FLA genes, FLA1, FLA2, FLA8, and FLA11 (Fig. 5). FLA1 is more abundant in flowers, which is consistent with the microarray signal ratio of 23:1 obtained by comparing mRNA abundance in flowers and leaves. In a microarray comparison of flowers and leaves FLA2 and FLA8 had a signal ratio of approximately 8:1; however, in RNA gel blots (Fig. 5) expression levels were equally abundant in all tissues tested. In contrast, for FLA1 and FLA11, the results for RNA gel blots and the microarray were consistent with transcripts being more abundant in flowers than leaves and higher in leaves than roots. The discrepancy between the microarray data and RNA gel blots may be due to different growth conditions and suggests that within a given organ type, e.g. flower, the precise developmental timing and growth conditions can have profound effects.
Table III.
EST expression summary for Arabidopsis FLAsa
| Description
|
Total ESTs in Library
|
Library
|
FLA
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1b | 2b | 3 | 4 | 6b | 7b | 8b | 9b | 10 | 11b | 12b | 13 | 15b | 16b | 17 | 18b | |||
| Various developmental stages | 28,252 | #A6B | 2 | 2 | — | — | — | — | — | 3 | — | 2 | — | 2 | — | 8 | — | 6 |
| Mixed tissues | 27,631 | NH11 | 5 | 3 | — | — | 4 | 9 | 9 | 8 | — | 1 | 3 | 2 | 3 | 2 | — | — |
| Roots | 17,573 | 5336 | 3 | 20 | — | — | — | 9 | 6 | 10 | 2 | 8 | 1 | — | 1 | 2 | — | 1 |
| Green siliques | 12,589 | 5339 | 2 | 2 | — | — | — | 2 | 7 | 1 | — | 2 | 1 | 1 | — | — | — | — |
| Aboveground organs (2 to 6 weeks) | 12,264 | 5335 | 1 | 7 | — | — | — | 2 | 11 | — | — | — | — | — | — | 2 | — | — |
| Developing seed | 10,800 | 5564 | 3 | 1 | — | — | — | — | 2 | — | — | — | — | — | 3 | 2 | — | — |
| Flower buds | 5,719 | 5337 | — | 3 | — | — | — | — | 5 | — | — | 1 | — | — | 1 | — | — | 1 |
| Rosette plants | 3,376 | #A69 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 2 |
| Aboveground organs (2 to 6 weeks) | 3,346 | 4063 | 1 | 2 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Ohio clone set | 3,013 | 2339 | 1 | — | — | — | 1 | 1 | 1 | 1 | — | — | — | — | — | 1 | — | — |
| Various developmental stages | 2,607 | #A60 | — | — | — | — | — | 1 | — | — | — | — | — | — | 2 | — | — | — |
| Root | 2,514 | #A5T | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 | — |
| Unknown | 2,372 | #A5V | — | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — |
| Mixture silique and flower | 1,867 | #A62 | — | — | 2 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Rosette plants | 1,720 | #A66 | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | — | — |
| Mixture silique and flower | 1,534 | #A5U | — | — | 1 | — | — | — | — | — | 2 | 2 | — | — | — | — | — | — |
| Seedling (3 d) | 1,503 | NH18 | — | 2 | — | — | — | 1 | 2 | 4 | — | — | — | 1 | — | — | — | — |
| Liquid-cultured seedlings | 1,135 | 5338 | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Cycling cells suspension culture | 897 | NH02 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 |
| Three-leaf rosette | 773 | 2341 | — | 1 | — | — | — | — | — | — | — | 1 | — | — | — | — | — | 1 |
| Seedling hypocotyl (3 d) | 724 | NH28 | — | 1 | — | — | — | — | — | 3 | — | — | — | — | — | — | — | — |
| Various developmental stages | 693 | #A68 | — | — | — | — | — | 2 | 2 | 1 | — | — | 2 | — | — | — | — | 1 |
| Seedling hypocotyl | 453 | NH25 | — | — | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — |
| Green silique | 389 | NH05 | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | — |
| Inflorescence | 347 | 2334 | — | — | — | — | — | — | — | — | — | — | — | — | 1 | — | — | — |
| Salt-induced transcripts | 148 | 6523 | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| In vitro-etiolated seedlings (5 d) | 124 | NH19 | — | — | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — |
| Whole seedling | 37 | NH39 | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — | — | 1 |
| Total | 18 | 46 | 3 | 1 | 5 | 28 | 45 | 33 | 5 | 18 | 7 | 6 | 11 | 17 | 1 | 14 | ||
a Obtained from The institute for Genomic Research Web site (January 2003). There are no ESTs for FLA5, FLA14, FLA19, FLA20, and FLA21. bThese FLAs have ESTs on the AFGC microarray (Schultz et al., 2002).
Table IV.
Selected microarray experiments for FLA genesa
| Experiment Identificationb | Descriptionc | FLA1 | FLA2a | FLA2b | FLA6 | FLA7 | FLA8 | FLA9a | FLA9bd | FLA11 | FLA12 | FLA15a | FLA15b | FLA16 | FLA18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratio of Channel 2:Channel 1e | |||||||||||||||
| 2370 | Leaves to flowers | 0.06 | 0.165 | 0.172 | 0.75 | 0.303 | 0.213 | 0.989 | 0.258 | 0.271 | 0.279 | 0.575 | 0.24 | 0.313 | 0.657 |
| 2371 | Flowers to leaves | 22.828 | 8.239 | 5.388 | 0.51 | 3.114 | 8.848 | 1.48 | 6.645 | 3.109 | 4.745 | 3.3 | 3.564 | 4.045 | 2.819 |
| 11991 | Callus-inducing medium (CIM) to root | NAf | 0.235 | 0.182 | 0.395 | NA | 0.667 | 0.446 | 0.884 | 0.389 | 0.469 | 0.224 | 0.399 | 0.355 | 0.517 |
| 11992 | Root to CIM | NA | 1.885 | 7.567 | 1.326 | NA | 1.933 | 1.2 | 0.802 | 2.346 | 0.99 | 1.768 | 1.352 | 1.227 | 0.844 |
| 12106 | Shoot-inducing medium (SIM; after CIM) to root | NA | 0.688 | 0.378 | 0.978 | 0.501 | 0.992 | 0.638 | 1.187 | 0.45 | 1.254 | 0.414 | 0.581 | 0.796 | 0.935 |
| 12107 | Root to SIM | NA | 1.899 | 1.715 | 1.479 | NA | 0.821 | 2.703 | 0.846 | 2.366 | 1.442 | 2.683 | 1.407 | 1.598 | 1.448 |
| 11757 | Wild type to abi1-1g | 1.293 | 1.645 | LOW | 1.63 | 0.954 | 0.642 | 1.068 | 0.814 | 1.049 | 1.728 | 1.403 | 1.355 | 1.592 | 1.095 |
| 11895 | abi1-1 to wild type | 1.034 | 0.62 | 6.735 | 0.227 | 1.852 | 3.709 | 0.297 | 1.977 | 1.878 | 0.257 | 0.823 | 0.614 | 0.159 | 0.572 |
| 5197 | 30 min of Antimycin A (AA) to control | 1.021 | 2.296 | 2.935 | 1.34 | 1.447 | 1.198 | 1.563 | 1.425 | 0.832 | 1.172 | 1.45 | 1.328 | 1.393 | 3.143 |
| 5200 | Control to 30 min of AA | 1.018 | 0.482 | 0.174 | 0.872 | 0.726 | 1.109 | 0.671 | 1.192 | 0.799 | 1.111 | 0.582 | 0.803 | 0.634 | 0.371 |
| 5198 | 4 h of AA to control | 0.643 | 1.766 | 2.272 | 1.383 | 0.938 | 0.976 | 1.336 | 1.283 | 0.593 | 1.355 | 1.194 | 1.484 | 1.16 | 1.889 |
| 5201 | Control to 4 h of AA | 2.038 | 0.799 | 0.331 | 0.915 | 0.683 | 1.359 | 0.752 | 0.885 | 1.218 | 0.767 | 0.762 | 0.905 | 0.882 | 0.601 |
a This is a selection of “significant” experiments for 11 of the FLAs from a large spreadsheet containing all of the AFGC experiments (rows) and as many genes as desired (e.g. columns 3, 4, n) obtained January of 2003. b The experiment identification is the one used by AFGC. c Descriptions have been modified to be more informative and are reported as the ratio of channel 2 to channel 1. d A third EST representing FLA9 (172M22T7) on the AFGC microarray is not shown. e Values considered significant (i.e. less than 0.5 or greater than 2) are bold. f NA, Not available. g abi1-1, abscisic acid 1 mutant.
Figure 5.
Expression of FLA genes in roots, leaves, and flowers of Arabidopsis ecotype Wassilewskija (Ws-2). RNA gel blots were hybridized with gene-specific probes for FLA1, FLA2, FLA8, and FLA11. Exposure times were 3 h for FLA1 and FLA11 and 45 min for FLA2 and FLA8. The staining of rRNAs with ethidium bromide in the lower panels shows the loading in each sample. R, Roots; L, leaves; F, flowers.
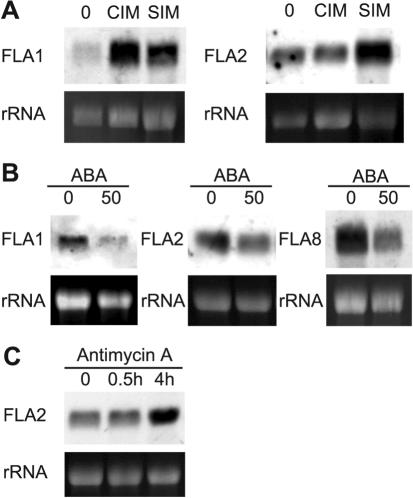
RNA gel blot analyses (Fig. 6) were also used to verify development and stress experiments that resulted in significant (i.e. less than 0.5 or greater than 2) and/or borderline changes in FLA expression on the microarrays (Table IV; Schultz et al., 2002). In the first experiment, Arabidopsis roots from 14-d-old plants were cut into segments and transferred first to CIM and then to SIM according to the method of Cary et al. (2001). RNA gel-blot analyses suggested FLA2 transcripts were not down-regulated after CIM treatment (Fig. 6A) as indicated by the microarray experiments (Table IV). FLA2 transcripts do, however, accumulate after SIM incubation, consistent with the microarray expression profile for this gene (Table IV; http://www.bioinformatics.iastate.edu/howell). FLA1 is the most closely related to FLA2 at the amino acid level (65%); therefore, FLA1 transcript levels were also determined to see if it was regulated in a similar way. Unlike FLA2, FLA1 transcripts increased in abundance after CIM treatment and remained high when grown on SIM.
Figure 6.
Expression of FLAs in response to a variety of hormones and stress treatments. A, RNA from 2-week-old roots (0), root segments incubated for 4 d on CIM and SIM for 14 d. B, RNA extracted from 11-d-old seedlings grown on 0 or 50 μm abscisic acid (ABA) for 2 d. C, RNA from leaves incubated for 0 or 30 min or 4 h on a solution containing the inhibitor of mitochondrial electron transport, AA. RNA gel blots were hybridized with gene-specific probes for FLA1, FLA2, and FLA8. Exposure times were 2 h, except for the AA blot, which was a 1 h exposure. The staining of rRNAs with ethidium bromide in the lower panels shows the loading in each sample.
In the second experiment, we looked at the expression of FLA1, FLA2, and FLA8 in response to ABA (Fig. 6B). Microarray experiments found that transcript accumulation of FLA2 and FLA8 was higher in the ABA-insensitive mutant abi1-1 compared with wild-type (Table IV), suggesting that they are normally down-regulated by an ABA-dependent pathway. Consistent with this prediction, both FLA2 and FLA8 were down-regulated in the presence of ABA (Fig. 6B). FLA1 transcript levels were not significantly altered on the microarray experiments in the abi1-1 mutant compared with wild type (Table IV). RNA gel-blot analysis shows that, like FLA2 and FLA8, it is down-regulated in the presence of ABA (Fig. 6B).
The hormone ABA is implicated in both development and stress pathways (Leung and Giraudat, 1998). Additional experiments are required to determine if ABA regulation of FLAs is important for development, stress responses, or both. For FLA2, there is some evidence that it is involved in both, although whether this is related to ABA is unknown. The regulation of FLA2 by CIM and SIM experiments suggests that FLA2 is involved in development. Microarray data indicates FLA2 is also regulated by stress responses in the experiments to inhibit mitochondrial electron transport using AA (Yu et al., 2001). Leaves treated with AA caused a slight (approximately 2-fold) up-regulation in FLA2 mRNA accumulation after 4 h, with no obvious change after treatment for 30 min (Fig. 6C). Overall, the RNA gel-blot experiments presented here generally confirm the AFGC results for the FLA genes, indicating that FLAs are regulated both developmentally and during stress responses.
DISCUSSION
FLAs Represent a Complex Multigene Family of Proteoglycans
The practical completion of the Arabidopsis genome facilitated the identification of 21 members of the FLA family. The first FLA, FLA8, was identified based on the presence of a predicted AGP-like region. Subsequent identification of fasciclin-like domains in this sequence led to the definition of a new subgroup of the AGP family (Schultz et al., 2000). Identification of the additional 20 FLAs in the Arabidopsis genome revealed that the FLA gene family is surprisingly diverse. Based on overall domain structure of the protein and sequence similarity of the fasciclin domains, the 21 FLAs can be divided into four groups (Fig. 2). At the sequence level, the predicted proteins have low similarity except for members of group B (81%-88% similarity) and two apparent gene duplications resulting in FLA8 and FLA10 with 88% similarity and FLA9 and FLA13 with 85% similarity. FLAs from the same group have similar domain structures and generally greater than 45% amino acid similarity.
The unique feature of all FLAs is the presence of putative cell adhesion domain(s) adjacent to putative AG glycosylation regions. Glycosylation requires the posttranslational modification of Pro residues to Hyp and the type of glycosylation relies on the contiguous or noncontiguous nature of these Hyp residues. The Hyp contiguity hypothesis suggests that blocks of contiguous Hyp residues, such as those occur in extensins, are arabinosylated with short oligosaccharides (Kieliszewski and Lamport, 1994; Shpak et al., 2001) and that Hyp (arabino) galactosylation occurs on the clustered noncontiguous Hyp residues commonly found in AGPs (Goodrum et al., 2000).
We confirmed that FLA7 is O-glycosylated like other AGPs because three peptides that match FLA7 were obtained from a tryptic digest of a hydrophobic fraction of β-glucosyl Yariv precipitated material (Fig. 3; Table I). The length of the predicted AGP region(s) in the protein backbones of FLAs is highly variable. The smallest is only six amino acids (FLA11), with three potential sites for O-glycosylation of AG chains in the APAPAP glycomodule (Shpak et al., 1999). The longest is 68 amino acids with 11 potential O-glycosylation sites (FLA5). A number of the FLAs have Ser-Pro-2 motifs, and FLAs 3 to 5 have blocks of contiguous Pro-3-5 residues.
Initially, it was thought the O-linked AG polysaccharides were of most interest because many experiments using antibodies that recognize carbohydrate epitopes on AGPs implicated AGPs in a variety of roles in plant growth and development such as cell fate, somatic embryogenesis, and cell proliferation (for review, see Majewski-Sawka and Nothnagel, 2000). More recent findings have shown that the embryogenic potential of somatic cells in culture can increase in the presence of AGPs containing N-glycans (Domon et al., 2000; van Hengel et al., 2001; van Hengel et al., 2002). Because N-glycosylation is not a feature of classical AGPs, an interesting possibility is that these bioactive AGPs are FLAs because FLAs are predicted to contain two to eight N-glycosylation sites. Alternatively, it could be the ENOD-like protein or one of the approximately 70 other GPI-anchored proteins containing AG glycomodules in their mature form (Borner et al., 2002, 2003), provided they also contain N-linked glycans. The classical AGPs and AG peptides have been excluded from the 100 proteins predicted to contain AG glycomodules because they do not contain sites for N-linked glycosylation. Borner et al. (2003) found that a surprisingly large number of GPI-anchored proteins (40%) contain AG glycosylation motifs. Therefore, if FLAs are involved in somatic embryogenesis and other developmental processes, it is likely that, in addition to the AGP domains, the fasciclin domains are important for the function of the molecule. The control of developmental or environmental responsiveness of FLAs could occur via one of three mechanisms: (a) by de novo synthesis of the appropriate FLA, (b) by modification of the carbohydrate component to change the FLA structure (van Hengel et al., 2001; van Hengel et al., 2002), or (c) by release of the FLA from the plasma membrane by GPI anchor cleavage (Schultz et al., 1998; Borner et al., 2002, 2003).
FLAs Are Differentially Regulated during Shoot Development
Another potential developmental role where FLAs may be important is the production of new shoot and root meristems. Shoot organogenesis induced in tissue culture has been described as comprising three phases: (a) competence acquisition, (b) induction, and (c) differentiation, based on the changing hormone requirements for each phase (Christianson and Warnick, 1983, 1984, 1985). Arabidopsis root explants require an initial incubation on CIM to gain competence to respond to morphogenic signals (Valvekens et al., 1988). Mutants with increased shoot regeneration properties support the notion that competency to produce roots, green callus, or shoots is related (Cary et al., 2001).
RNA gel-blot analysis of FLA1 and FLA2 after CIM and SIM treatment showed that transcripts for the two genes were induced during this developmental process. FLA1 mRNA accumulation was noticeably up-regulated after 4 d on CIM, and this high level was maintained during the SIM treatment. There was no obvious change in FLA2 expression on CIM, but a significant increase was observed after SIM treatment (Fig. 6).
It is possible that both FLAs are involved in different steps of the same developmental pathway. The earlier response of FLA1 suggests it is involved in the competence acquisition stage where cells are thought to “dedifferentiate” and proliferate to produce callus (Cary et al., 2001). FLA2 may be involved in the second induction phase, where the developmental fate of the regenerating tissue is specified by the hormonal composition of the medium. Based on the microarray data from Stephen Howell's laboratory, a Web site has been established that enables the expression profile of your favorite gene to be compared with other genes regulated by CIM and SIM (http://bioinformatics.iastate.edu/howell/; Che, 2002). FLA2 appears to have a similar profile to a hypothetical protein with putative oligopeptide transporter activity (At4g27730), a putative protein kinase (At2g32800), and an Myb-related transcription factor (At5g57620; Che, 2002). There are no available data for FLA1 on the microarray; therefore, a comparison is not possible. Two groups of mutants, the srd (shoot regeneration defective; Yasutani et al., 1994) and ire (increased organ regeneration; Cary et al., 2001), have been isolated based on their ability to reduce or enhance shoot and/or root organogenesis (Cary et al., 2002). Mutants in FLA1 and FLA2 may enable us to further explore the role of these proteins in organogenesis.
Expression of Some FLAs Is Regulated by ABA
Because it appears that most FLA mutants will not have a phenotype when grown under standard conditions (K.L. Johnson, A. Bacic, and C.J. Schultz, unpublished data), understanding the environmental and developmental stimuli that control the expression of each FLA may assist in uncovering phenotypes. The AFGC microarray data provides an excellent resource to begin the process of characterizing FLA expression (Table IV). RNA gel blots were performed to verify some of the results from the microarray. Microarray analysis comparing the abi1-1 mutant with the wild-type seedlings indicated that FLA2 and FLA8 are more abundantly expressed in the abi1-1 mutant. We confirmed this result, showing that FLA2 and FLA8 were both down-regulated through an ABA-dependent pathway (Fig. 6). FLA1 transcript levels were also down-regulated in the presence of ABA, in contrast to no significant change on the microarray.
The down-regulation of FLAs1,2, and 8 is consistent with a role for these FLAs in stress responses. ABA is known to be an essential mediator of plant responses to water loss by reducing stomatal aperture and in root morphogenetic responses to drought (Giraudat et al., 1994; Vartanian et al., 1994). Molecular studies of ABA-deficient mutants show they are also affected in the regulation of genes by drought, salt, or cold stress (Leung and Giraudat, 1998). Interestingly, a pine (Pinus taeda) FLA, PtX14A9, the likely ortholog of FLA11, is preferentially expressed in the xylem, and although it showed no response to ABA, it was down-regulated by drought stress (No and Loopstra, 2000). This suggests FLAs can have different responses to plant hormones and that they are likely to be involved in general stress responses.
Regulation of FLA2 by AA
A set of genes thought to be involved in general stress responses are up-regulated by AA, which acts to inhibit the mitochondrial electron transport, inducing transcription of genes for alternative oxidase (Yu et al., 2001). Many genes affected by AA treatment are also affected by other stresses, including aluminum stress (Schultz et al., 2002). In addition to FLA2 and FLA18, a number of classical AGPs (AGP1 and AGP2), Lys-rich AGP18, and AG peptide AGP12 are up-regulated by AA treatment in the microarray experiments (Schultz et al., 2002). RNA gel blots confirmed FLA2 was up-regulated in leaves after 4 h treatment of AA. Nuclear-encoded genes up-regulated by AA treatment encode proteins with a wide range of functions, from signal transduction and biosynthetic enzymes to stress and defense response genes (Yu et al., 2001). It is not known how FLAs are involved in plant responses to mitochondrial-nuclear communication. It is possible that FLAs belong to a set of genes involved in general stress responses from common signal transduction pathways (Yu et al., 2001).
Structural Features of FLAs. How Could the Fasciclin Domain Facilitate Cell Adhesion?
If FLAs are involved in development and stress responses, it raises the question of how the putative cell adhesion domain functions. Fasciclin domains of Arabidopsis FLAs include the highly conserved H1 and H2 regions that characterize fasciclin domains from all species (Fig. 1). Although the H1 and H2 domains are highly conserved, they are not directly involved in cell adhesion (Kim et al., 2000; Kim et al., 2002). This evidence comes from studies of human (Homo sapiens) adhesion molecules containing fasciclin domains, where they have used in vivo experiments with synthetic peptides containing H1 and H2 to show that they did not compete in binding assays, whereas other peptides did.
Fasciclin 1 (Fas1) from fruitfly is known to be capable of homophilic adhesion (Elkins et al., 1990a). Surprisingly, even with a crystal structure for Fas1 of fruitfly, the cell adhesion mechanism is still poorly understood (Clout et al., 2003). The three-dimensional structure of a polypeptide containing domains 3 and 4 of Fas1 has, nevertheless, located conserved amino acids that form hydrogen bonds essential for the correct folding of the fasciclin domain. These include a number of conserved Ile and Leu residues in regions flanking the H1 domain that are also observed in all Arabidopsis FLAs (Fig. 1). Although these residues are conserved, they are more likely to be involved in maintaining the folded state rather than in direct cell adhesion interactions (Clout et al., 2003).
Unfortunately, the low similarity between the Arabidopsis FLAs and Fas1 means that it is not possible to predict the three-dimensional structure of FLAs using automated threading techniques (http://swissmodel-.expasy.org). Preliminary analysis using secondary structure predictions such as “PredictProtein” (http://www.embl-heidelberg.de/predictprotein/) shows that the secondary structure of most Arabidopsis FLAs is similar to that determined by the crystal structure of fasciclin domains 3 and 4 in fruit-fly (Fig. 1). α helix 4, however, is not conserved in FLAs 15 to 19 because of the presence of Pro residues, which are helix breakers (Fig. 1).
The best clues to identifying the residues involved in cell adhesion comes from studies using interfering peptides based on the big-h3 proteins from humans. These studies have identified both DI and YH motifs thought to interact with different integrins and mediate human corneal cell adhesion (Kim et al., 2000) and fibroblast adhesion (Kim et al., 2002), respectively.
The DI motif is present only in the group B FLAs. The YH motif is more conserved and is found in 11 of the FLAs, with an additional nine FLAs having the conserved amino acids FH. In human cell cultures, the DI and YH motifs are important for interactions with other components of the ECM, such as integrins. Integrins are animal ECM receptors that mediate signals to intercellular kinases (van der Flier and Sonnenberg, 2001). Some of the FLAs are likely to be attached to the plasma membrane through a GPI anchor and may interact with receptor-like kinases, such as the wall-associated kinases (Gens et al., 2000). The localization of FLAs at the plasma membrane suggests they could have a signaling role.
Interactions at the Plasma Membrane
GPI-anchored adhesion molecules, including Fas1 from fruitfly, are proposed to be recruited into membrane rafts to enable more stable interactions between adhesion complexes (Harris and Siu, 2002). The interactions between individual cell adhesion molecules in lipid rafts are thought to be weak. This could explain why the crystal structure of the fasciclin domains from fruitfly failed to identify any obvious cell adhesion domains. Strong interactions are achieved by the clustering of many cell adhesion molecules, within and between cells. The enlarged rafts that result are thought to mediate signaling and recruit the cytoskeleton to stabilize cell-cell adhesion. This suggested role of fasciclins in signaling is supported by double mutant combinations of a non-receptor Tyr kinase and fasciclin proteins from fruit-fly that show more severe defects than single loss-of-function mutants (Elkins et al., 1990b; Hu et al., 1998).
The same 14 FLAs are predicted to be GPI anchored using two recently revised GPI anchor prediction programs for plants (Borner et al., 2003; Eisenhaber et al., 2003). A brief description of the mechanistic differences between these two prediction methods is described in “Materials and Methods.” The two programs (Borner et al., 2003; Eisenhaber et al., 2003) identify more GPI-anchored FLAs, i.e. FLA11 and FLA14, than the PSORT program (Nakai and Horton, 1999) used previously (Schultz et al., 2002). The new Big-PI plant predictor is available at http://mendel.imp.univie.ac.at/gpi/plant-server.html and has the advantage of also predicting the cleavage site residue (ω; Eisenhaber et al., 2003). FLA17 is possibly a Type 1a membrane protein with a putative transmembrane domain of 20 amino acids at the C-terminal, and the remaining six FLAs (15, 16 and 18-21) are likely to be secreted. Four of the GPI-anchored FLAs (FLAs 1, 7, 8, and 10) were shown to be sensitive to phosphatidylinositol-specific phospholipase C cleavage (Borner et al., 2003), indicating some FLAs may be released into the ECM.
Fas1 from fruitfly (McAllister et al., 1992) and Algal-CAM from V. carteri (Huber and Sumper, 1994) have GPI-anchored variants that can be released from the membrane surface via cleavage of the lipid anchor. Unlike Fas1 and Algal-CAM, there is no opportunity for alternate splicing of FLAs because there is only one known intron (in FLA1). Plants may have evolved separate genes for GPI-and non-GPI-anchored FLAs. One possibility is that the secreted forms control cell expansion in the primary cell wall, whereas GPI-anchored forms might maintain integrity of the plasma membrane during cell expansion.
Mutants will play an important role in determining the role of GPI anchoring in FLA function. A mutant in FLA4 that is sos5 (salt overly sensitive) has been identified recently (Shi et al., 2003). The sos5 (fla4) mutant displayed root swelling and root growth arrest when grown on a high-salt medium. A root swelling phenotype was also observed for sos5 (fla4) roots grown without salt; however, this phenotype is conditional. Roots grown without salt displayed swelling only when they grew on the surface of the agar (Shi et al., 2003). The conditionality of the root swelling phenotype suggests a role for FLA4 as a non-covalent cross-linker of plant cell walls. We propose that the fasciclin domains of FLA4 are involved in ionic interactions, most likely with other FLAs, such that they form a network at either the plasma membrane, or in the cell wall that controls the rate of cell expansion. The alteration of FLA4 in sos5 mutants allows uncontrolled expansion unless there is an external pressure such as provided by the agar.
Conclusion
We have shown that FLAs are regulated by both development and stress. The use of mutants will determine the importance of FLAs during these processes and establish if FLAs function as cell adhesion molecules. A number of FLA mutants have already been identified in the various resources of insertion mutants (Schultz et al., 2002). Although the characterization of these may prove difficult due to the potential for redundancy in large gene families (Pickett and Meeks-Wagner, 1995), EST expression summaries and microarray data will be useful in determining what double mutant combinations and developmental and stress experiments may be successful in uncovering phenotypes.
The conservation of the fasciclin domains across species provides an exciting opportunity to study the dynamic nature of plant and animal cells. The putative role of FLAs as cell adhesion molecules suggests that they are vital components of the interactions that occur in the plant ECM. The formation of membrane rafts is one proposed mechanism that GPI-anchored molecules can facilitate signaling and stabilize adhesion (Harris and Siu, 2002). In plant cells, the important “adhesion” events are likely to be between molecules at the plasma membrane and within the primary cell wall. Study of the potential of FLAs to interact with themselves and receptor-like kinases or other components of the ECM will help us to better understand the role of FLAs in plant growth and development.
MATERIALS AND METHODS
Sequence Analysis
FLA sequences were identified from the annotated Arabidopsis database using hidden markov models (Schultz et al., 2002). The locus identity numbers assigned by the AGI (2000) for the FLA genes are: FLA1, At5g55730; FLA2, At4g12730; FLA3, At2g24450; FLA4, At3g46550; FLA5, At4g31370; FLA6, At2g20520; FLA7, At2g04780; FLA8, At2g45470; FLA9, At1g03870; FLA10, At3g60900; FLA11, At5g03170; FLA12, At5g60490; FLA13, At5g44130; FLA14, At3g12660; FLA15, At3g52370; FLA16, At2g35860; FLA17, At5g06390; FLA18, At3g11700; FLA19, At1g15190; FLA20, At5g40940; and FLA21, At5g06920. SignalP (www.cbs.dtu.dk/services/SignalP/#submission) was used to determine the predicted length of the N-terminal signal sequence (Nielsen et al., 1997; Schultz et al., 2002). Original prediction for the GPI anchoring of FLAs (Schultz et al., 2002) was performed using PSORT (http://psort.nibb.ac.jp/form.html; Nakai and Horton, 1999). Additional FLAs, i.e. FLA11 and FLA14, were predicted as GPI anchored using a new program, big-PI plant predictor (http://mendel.imp.univie.ac.at/gpi/plant_server.html; Eisenhaber et al., 2003). The big-PI plant predictor assumes that the C-terminal signal for addition of a GPI anchor consists of four necessary elements: (a) a polar and flexible linker region of about 11 residues (ω - 11... ω - 1), (b) a region of small residues (ω-1... ω + 2) with the ω-site, (c) a spacer region (ω + 3... ω + 9) of moderately polar residues, and (d) a tail beginning with ω + 9 or ω + 10 up to the C-terminal end with a long, sufficiently hydrophobic stretch (Eisenhaber et al., 1998, 2003). The big-PI method differs from another plant-specific method, recently revised by Borner et al. (2003), in that it considers some of the mature protein (i.e. ω - 11... ω-1), in addition to the cleavage site residue (ω) and the C-terminal cleaved portion of the GPI signal. Other differences between the programs are detailed by Eisenhaber et al. (2003). Profilescan (http://hits.isb-sib.ch/cgi-bin/PFSCAN) was used to identify the length of the fasciclin-like domains (INTERPRO; IPR000782) and potential N-glycosylation sites by using all four databases. Profilescan aligned the Arabidopsis FLAs to consensus sequences based on the bigh3 domain (PROSITE; PS50213), the fasciclin domain (PFAM; PF02469; 13 proteins), and FAS1 (SMART; SM00554; 78 proteins) sequences of approximately 140 amino acids in length. The first fasciclin domain of six of the FLAs (1-3, 5, 8, and 10) is predicted to be only 70 amino acids in length when aligned to the bigh3 domain. Although this is possible, alignment of these domains with the FAS1 consensus sequence made up of 79 proteins (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=smart00554&version=v1.62) shows these FLAs have conserved H2 domains at the C-terminal end of the domains. AGP regions were identified based on rules established by Schultz et al. (2002).
Sequences were aligned using ClustalX program (Thompson et al., 1994) and manually edited. The full-length alignment can be accessed on-line as supplementary data. An alignment of the full-length protein sequence in a NEXUS matrix was analyzed using PAUP 4.01 (Swofford, 1999). Parsimony searches were performed using a heuristic search with 100 resampling replicates for each analysis. Pair-wise sequence alignments were performed using the Gap program (Genetics Computer Group, Madison, WI) in BioManager (http://www.angis.org.au). The parameters used were: a BLOSUM62 scoring matrix, gap creation and gap extension penalties of six and one, respectively, and no penalty for gaps extensions longer than 12.
Protein Extraction and Detection of N-Linked Glycosylation
AGPs were extracted from liquid culture-grown Arabidopsis tissue (Ws-2 strain CS2360, Arabidopsis Biological Research Center, Columbus OH) and separated by RP-HPLC as described previously (Schultz et al., 2000). Colorimetric analysis (Dubois et al., 1956) was used to determine total carbohydrate in RP-HPLC fractions of AGPs. Carbohydrate (40 μg) from the four fractions and 250 μg of GaRGSP (Sommer-Knudsen et al., 1996) were blotted onto a nitrocellulose membrane and probed for N-glycans using DIG-labeled WGA (Boehringer Mannheim/Roche, Germany) according to the manufacturer's instructions. Fraction four from RP-HPLC (Fig. 3) was digested with trypsin and the resulting peptides separated by RP-HPLC (Schultz et al., 2000) using a Brownlee Aquapore RP-300 column (C18, 2.1 × 100 mm, Perkin-Elmer Applied Biosystems, Foster City, CA) with a linear gradient of 0% to 100% (w/v) in 60 min from Solvent A (0.1% [w/v] trifluoracetic acid [TFA]) to Solvent B (80% [w/v] acetonitrile in 0.1% [w/v] TFA). Peptides were N-terminal sequenced using Edman degradation chemistry on a model LF 3400 Beckman sequencer (Beckman Instruments, Fullerton, CA) with on-line analysis on a Beckman system gold HPLC.
MALDI-TOF MS
Fractions were analyzed using MALDI performed on a Voyager-DE STR mass spectrometer from Perkin-Elmer Applied Biosystems, equipped with a 337-nm N2 laser. Parent ion masses were measured in reflector/delayed extraction mode, with accelerating voltage of 20 kV, grid voltage of 74%, and a 74-ns delay. Positive polarity was used. Fifty scans were averaged per sample, and spectra were subject to two-point external calibration using the appropriate standards. The external standard used were bradykinin, angiotensin I, ACTH (1-17) with molecular weights of 1,060.5692, 1,296.6853, and 2,093.0867, respectively, and ACTH (18-39) with molecular masses of 1,060.5692, 1,296.6853, and 2,465.1989, respectively. The matrix used was α-cyano-4-hydroxycinnamic acid (saturated solution in 50% [w/v] acetonitrile with 0.1% [w/v] TFA). Predicted masses were determined using “ms-digest” (http://prospector.ucsf.edu) selecting the slymotrypsin digest with no missed cleavages. Other options were unmodified Cys residues, hydrogen at the N terminus, and free acid at the C terminus.
Analysis of AFGC Microarray Information
A detailed description of the array data reformatting procedure is given by Schultz et al. (2002), although the Web site addresses have changed as follows. The AGI locus (e.g. At5g10430) was used to determine which FLA ESTs were on the AFGC array. These data are accessible from The Arabidopsis Information Resource (http://www.Arabidopsis.org/tools/bulk/microarray/index.html).
Information about each experiment (i.e. abstracts and RNA information) can be accessed from the TAIR microarray experiments site (http://www.Arabidopsis.org/servlets/Search?type=expr&search_action=new_search). An alternate way to access all of the experiments is from the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD). The PERL program (AFGC microarray reformatting tool) is available by contacting the corresponding author.
RNA Gel-Blot Analysis
RNA gel-blot analysis was performed using DIG probes as described previously (Schultz et al., 2000). Arabidopsis seedlings (14 d) grown on Murashige and Skoog plates and liquid culture were used to obtain shoot (leaf) and root tissue, respectively. Open and closed flower buds were removed from approximately 8-week-old plants, grown in growth chambers (21°C, 10 h of light; 150 μmol m-2 s-1). RNA was extracted using TRIZOL reagent (Life Technologies/Gibco-BRL, Cleveland) according to the manufacturer's instructions with the addition of a high-salt solution (0.8 m sodium citrate and 1.2 m NaCl) in the isopropanol precipitation step used for root tissue. Total RNA (10 μg) was electrophoresed through 1.5% (w/v) agarose gels containing formaldehyde and transferred to nylon membrane as previously described (Schultz et al., 1997). Single-stranded DIG-labeled probes were prepared using a two-stage PCR protocol (Schultz et al., 1997). For each gene the first (non-labeling) PCR round included the appropriate forward and reverse primers (see below). In the second (labeling) round of PCR, only the reverse primer was used. The sequence of primers are as follows: FLA1-F, 5′-CTCTCCCTCCACGTCCTTTTAGATTACTT-3′; FLA1-R, 5′-AGTCGCATATATAGCTAAAGGCTGCTCAT-3′; FLA2-F, 5′-ATCCTCTCTAACGGATACCACTCTACCAA; FLA2-R, 5′-GCGTTCTTAGGCTTTTTCTTACTCGACTT-3′; FLA8-F, 5′-AATTCGATCTAACGACCTCTACC-3′; FLA8-R, 5′-CAAACTCAACAACACATAACCAC-3′; FLA11-F, 5′-CCTTCAGGTCCAACGAACAT-3′; and FLA11-R, 5′-AAACCCGAAT CCAGTCCTCT-3′. Probe sizes amplified were: FLA1, 702 nucleotides; FLA2, 836 nucleotides; FLA8, 572 nucleotides; and FLA11, 593 nucleotides. Hybridization and chemiluminescent detection of DIG-labeled probes was as previously described (Schultz et al., 1997).
Plant Material for Plate-Based Assays
Ws-2 seeds were surface sterilized with 12% (v/v) sodium hypochlorite for 5 min, rinsed with sterile water, and transferred in 0.8% (w/v) SeaPlaque agarose (Cambrex Bio Science, Rockland, ME) to sterile Murashige and Skoog plates (1× Murashige and Skoog [Life Technologies/Gibco-BRL], 3% [w/v] Suc, and 0.8% [w/v] agar). Plates were incubated at 4°C for 3 d, then placed in a chamber for 16 h of light and 8 h of dark, with day temperature of 22°C and a night temperature 16°C for 12 to 14 d.
Plant Material for Callus Induction/Shoot Development
Ws-2 seeds plated on Murashige and Skoog plates were grown for 13 d with 12 h of light at 25°C and 12 h of dark at 17°C. Callus induction and shoot development were performed according to the method of Cary et al. (2001). Approximately 10 root sections of 5 to 10 mm were placed onto three replicate plates of CIM containing 2.2 mm 2,4-dichlorophenoxyacetic acid and 0.2 mm kinetin in a 20°C chamber for 16-h-light and 8-h-dark cycle. After incubation on CIM for 4 d, root explants were transferred to plates containing SIM with 0.9 mm 3-indoleacetic acid and 5 mm isopentenyladenine for a further 14 d. Root tissue was collected for RNA extraction before transfer to CIM, after 4 d on CIM and 14 d on SIM.
Plant Material for ABA Treatment
ABA treatments were based on the method of Söderman et al. (2000). Seedlings were grown on Murashige and Skoog plates (1× Murashige and Skoog [Life Technologies/Gibco-BRL], 3% [w/v] Suc, and 0.8% [w/v] agar) for 11 d; then, five plants were transferred to fresh Murashige and Skoog medium (1× Murashige and Skoog, 2% [w/v] Suc, and 0.8% [w/v] agar) containing either 0 or 50 μm ABA (four replicates each treatment) for an additional 2 d before harvest (Söderman et al., 2000). Tissue for RNA isolation was snap frozen in liquid nitrogen and stored at -70°C until extraction.
AA Treatment
AA experiments were performed based on the method of Yu et al. (2001). Arabidopsis Ws-2 leaves were harvested from 5-week-old soil-grown plants grown in growth chambers (21°C, 10 h of light, 150μmol m-2 s-1) for 21 d after transfer from Murashige and Skoog plates. For AA treatment, five leaves were floated (with the top surface of the leaf facing up) in 1/10-strength Murashige and Skoog medium in the dark with 10 μm AA (Sigma) for 30 min and 4 h. Leaves were frozen in liquid nitrogen and used for RNA isolation. Control leaves (0 h) were taken from plants and frozen without incubation.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We are grateful to The Arabidopsis Information Resource for providing access to the microarray data. We thank Kris Ferguson for the MS and N-terminal Edman sequencing analysis and are grateful to Edward Newbigin for critical comments on this manuscript.
This work was supported by the Australian Research Council (Large Grant no. A10020017) and by the University of Melbourne (research scholarship to K.L.J.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.031237.
References
- AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Altschul S, Madden T, Schäffer A, Hang J, Hang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacic A, Currie G, Gilson P, Mau S-L, Oxley D, Schultz CJ, Sommer-Knudsen J, Clarke AE (2000) Structural classes of arabinogalactan-proteins. In EA Nothnagel, A Bacic, AE Clarke, eds, Cell and Developmental Biology of Arabinogalactan-Proteins. Kluwer Academic/Plenum Publishers, New York, pp 11-23
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In J Priess, ed, The Biochemistry of Plants, Vol 14. Academic Press, New York, pp 297-371 [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Cary AJ, Che P, Howell SH (2002) Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J 32: 867-877 [DOI] [PubMed] [Google Scholar]
- Cary A, Uttamchandani SJ, Smets R, Van Onckelen HA, Howell SH (2001) Arabidopsis mutants with increased organ regeneration in tissue culture are more competent to respond to hormonal signals. Planta 213: 700-707 [DOI] [PubMed] [Google Scholar]
- Cassab GI, Varner JE (1988) Cell wall proteins. Annu Rev Pl Physiol Pl Mol Biol 39: 321-353 [DOI] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH (2002) Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2271-2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson ML, Warnick DA (1983) Competence and determination in the process of in vitro shoot organogenesis. Dev Biol 95: 288-293 [DOI] [PubMed] [Google Scholar]
- Christianson ML, Warnick DA (1984) Phenocritical times in the process of in vitro shoot organogenesis. Dev Biol 101: 382-390 [DOI] [PubMed] [Google Scholar]
- Christianson ML, Warnick DA (1985) Temporal requirement for phytohormone balance in the control of organogenesis in vitro. Dev Biol 112: 494-497 [Google Scholar]
- Clout NJ, Tisi D, Hohenester E (2003) Novel fold revealed by the structure of a FAS1 domain pair from the insect cell adhesion molecule Fasciclin I. Structure 11: 197-203 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13: 171-201 [DOI] [PubMed] [Google Scholar]
- Domon J-M, Neutelings G, Roger D, David A, David H (2000) A basic chitinase-like protein secreted by embryogenic tissues of Pinus caribaea acts on arabinogalactan-proteins extracted from the same cell lines. J Plant Physiol 156: 33-39 [Google Scholar]
- Dubois M, Gillies KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for the determination of sugars and related substances. Anal Chem 28: 350-356 [Google Scholar]
- Durbin R, Eddy S, Krogh A, Mitchison G (1998) Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, UK
- Eisenhaber B, Bork P, Eisenhaber F (1998) Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng 11: 1155-1161 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, J SC, Borner GHH, Dupree P, Eisenhaber F (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691-1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins T, Hortsch M, Bieber AJ, Snow PM, Goodman CS (1990a) Drosophila fasciclin I is a novel homophilic adhesion molecule that along with fasciclin III can mediate cell sorting. J Cell Biol 110: 1825-1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins T, Zinn K, McAllister L, Hoffmann FM, Goodman CS (1990b) Genetic analysis of a Drosophila neural cell adhesion molecule: interaction of fasciclin I and abelson tyrosine kinase mutations. Cell 60: 565-575 [DOI] [PubMed] [Google Scholar]
- Fincher GB, Stone BA, Clarke AE (1983) Arabinogalactan-proteins: structure, biosynthesis and function. Annu Rev Plant Physiol 34: 47-70 [Google Scholar]
- Gaspar YM, Johnson KL, McKenna JA, Bacic A, Schultz CJ (2001) The complex structures of arabinogalactan-proteins and the journey towards a function. Plant Mol Biol 47: 161-176 [PubMed] [Google Scholar]
- Gens JS, Fujiki M, Pickard BG (2000) Arabinogalactan-protein and wall-associated kinase in a plasmalemmal reticulum with specialized vertices. Protoplasma 212: 115-134 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J, Morris P-C, Bouvier-Durond M, Vartanian N (1994) Current advances in abscisic acid action and signaling. Plant Mol Biol 26: 1557-1577 [DOI] [PubMed] [Google Scholar]
- Goldstein IJ, Hayes CE (1978) The lectins: carbohydrate-binding proteins of plants and animals. Adv Carbohydr Chem Biochem 35: 127-340 [DOI] [PubMed] [Google Scholar]
- Goodrum LJ, Patel A, Leykam JF, Kieliszewski MJ (2000) Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54: 99-106 [DOI] [PubMed] [Google Scholar]
- Harris TJC, Siu C-H (2002) Reciprocal raft-receptor interactions and the assembly of adhesion complexes. BioEssays 24: 996-1003 [DOI] [PubMed] [Google Scholar]
- Hooper NM (1997) Glycosylphosphatidylinositol anchored membrane enzymes. Clin Chim Acta 266: 3-12 [DOI] [PubMed] [Google Scholar]
- Hortsch M, Goodman CS (1990) Drosophila fasciclin I, a neural cell adhesion molecule, has a phosphatidylinositol lipid membrane anchor that is developmentally regulated. J Biol Chem 265: 15104-15109 [PubMed] [Google Scholar]
- Hu S, Sonnenfeld M, Stahl S, Crews ST (1998) Midline fasciclin: A Drosophila fasciclin-I-related membrane protein localized to the CNS midline cells and trachea. J Neurobiol 35: 77-93 [DOI] [PubMed] [Google Scholar]
- Huber O, Sumper M (1994) Algal-CAMs: isoforms of a cell adhesion molecule in embryos of the alga Volvox with homology to Drosophila fasciclin I. EMBO J 13: 4212-4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Schultz CJ, Bacic A (2003) Non-enzymatic cell wall (glyco)proteins. In JK Rose, ed, The Plant Cell Wall, Vol 8. Blackwell Publishing, Oxford, UK, pp 111-154 [Google Scholar]
- Kawamoto T, Noshiro M, Shen M, Nakamasu K, Hashimoto K, Kawashima-Ohya Y, Gotoh O, Kato Y (1998) Structural and phylogenetic analyses of RGD-CAP/βig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim Biophys Acta 1395: 288-292 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J 5: 157-172 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, O'Neill M, Leykam J, Orlando R (1995) Tandem mass spectrometry and structural elucidation of glycopeptides from a hydroxyproline-rich plant cell wall glycoprotein indicate that contiguous hydroxyproline residues are the major sites of hydroxyproline O-arabinosylation. J Biol Chem 279: 2541-2549 [DOI] [PubMed] [Google Scholar]
- Kim J-E, Jeong H-W, Nam J-O, Lee B-H, Choi J-Y, Park R-W, Park JY, Kim I-S (2002) Identification of motifs in the fasciclin domains of the transforming growth factor-β-induced matrix protein βig-h3 that interact with the αvβ5 integrin. J Biol Chem 277: 46159-46165 [DOI] [PubMed] [Google Scholar]
- Kim J-E, Kim S-J, Lee B-H, Park R-W, Kim K-S, Kim I-S (2000) Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-β-induced gene, βig-h3. J Biol Chem 275: 30907-30915 [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, Peart M, Cooper C, Roberts K (1991) Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J 1: 317-326 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Pl Mol Biol 49: 199-222 [DOI] [PubMed] [Google Scholar]
- Li W-H (1997) Molecular Phylogenetics: Methods. Sinauer Associates, Inc., Sunderland, MA
- Lin XY, Kaul SS, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M et al. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402: 761-768 [DOI] [PubMed] [Google Scholar]
- Majewski-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan-proteins in plant development. Plant Physiol 122: 3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister L, Goodman CS, Zinn K (1992) Dynamic expression of the cell adhesion molecule fasciclin I during embryonic development in Drosophila. Development 115: 267-276 [DOI] [PubMed] [Google Scholar]
- McCabe PF, Valentine TA, Forsberg LS, Pennell RI (1997) Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell 9: 2225-2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34-36 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne Gv (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1-6 [DOI] [PubMed] [Google Scholar]
- No E-G, Loopstra CA (2000) Hormonal and developmental regulation of two arabinogalactan-proteins in xylem of loblolly pine (Pinus taeda). Physiol Plant 110: 524-529 [Google Scholar]
- Nothnagel EA (1997) Proteoglycans and related components in plant cells. Int Rev Cytol 174: 195-291 [DOI] [PubMed] [Google Scholar]
- Paulsrud P, Lindblad P (2002) Fasciclin domain proteins are present in Nostoc symbionts of lichens. Appl Environ Microbiol 68: 2036-2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI (1992) Cell surface arabinogalactan proteins, arabinogalactans and plant development. In AJ Callow, JR Green, eds, Prospectives in Plant Cell Recognition, Vol. 48. Cambridge University Press, Cambridge, pp 105-121 [Google Scholar]
- Pickett FB, Meeks-Wagner DR (1995) Seeing double: appreciating genetic redundancy. Plant Cell 7: 1347-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Gilson P, Oxley D, Youl JJ, Bacic A (1998) GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci 3: 426-431 [Google Scholar]
- Schultz CJ, Hauser K, Lind JL, Atkinson AH, Pu Z-Y, Anderson MA, Clarke AE (1997) Molecular characterisation of a cDNA sequence encoding the backbone of a style-specific 120kDa glycoprotein which has features of both extensins and arabinogalactan-proteins. Plant Mol Biol 35: 833-845 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Johnson K, Currie G, Bacic A (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A (2002) Using genomics resources to guide research directions: the arabinogalactan-protein gene family as a test case. Plant Physiol 129: 1448-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpe MD, Nothnagel EA (1999) Arabinogalactan-proteins in the multiple domains of the plant cell surface. Adv Bot Res 30: 207-289 [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu J-K (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58: 1399-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Barbar E, Leykam JF, Kieliszewski MJ (2001) Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J Biol Chem 276: 11272-11278 [DOI] [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc Natl Acad Sci USA 96: 14736-14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752-1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer-Knudsen J, Clarke AE, Bacic A (1996) A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J 9: 71-83 [DOI] [PubMed] [Google Scholar]
- Swofford DL (1999) PAUP 4.01: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.01. Sinauer, Sunderland, MA
- Tan L, Leykam JF, Kieliszewski MJ (2003) Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol 132: 1362-1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montague M, Lijsebettens MV (1988)Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536-5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A (2001) Function and interactions of integrins. Cell Tissue Res 305: 285-298 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC (2001) N-acetylglucosamine and glucosamine-containing arabinogalactan-proteins control somatic embryogenesis. Plant Physiol 125: 1880-1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Van Kammen A, de Vries S (2002) A relationship between seed development, arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol Plant 114: 637-644 [DOI] [PubMed] [Google Scholar]
- Vartanian N, Marcotte L, Giraudat J (1994) Drought rhizogenesis in Arabidopsis thaliana. Plant Physiol 104: 761-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J, Lis H, Katchalski E (1967) Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem J 105: 1c-2c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutani I, Ozawa S, Nishida T, Sugiyama M, Komamine A (1994) Isolation of temperature-sensitive mutants of Arabidopsis thaliana that are defective in the redifferentiation of shoots. Plant Physiol 105: 815-822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Nickels R, McIntosh L (2001) A genome approach to mitochondrial-nuclear communication in Arabidopsis. Plant Physiol Biochem 39: 345-353 [Google Scholar]
- Zhong Y, Shanley J (1995) Altered nerve terminal arborization and synaptic transmission in Drosophila mutants of cell adhesion molecule fasciclin I. J Neurosci 15: 6679-6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZD, Tan L, Showalter AM, Lamport DTA, Kieliszewski MJ (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431-444 [DOI] [PubMed] [Google Scholar]