Abstract
Excess manganese (Mn) supply causes formation of visible brown depositions in the cell walls of leaves of cowpea (Vigna unguiculata), which consist of oxidized Mn and oxidized phenols. Because oxidation of Mn and phenolic compounds in the leaf apoplast was proposed to be catalyzed by apoplastic peroxidases (PODs), induction of these enzymes by Mn excess was investigated. POD activity increased upon prolonged Mn treatment in the leaf tissue. Simultaneously, a significant increase in the concentration of soluble apoplastic proteins in “apoplastic washing fluid” was observed. The identity of the released proteins was systematically characterized by analysis of the apoplast proteome using two-dimensional gel electrophoresis and liquid chromatography-tandem mass spectrometry. Some of the identified proteins exhibit sequence identity to acidic PODs from other plants. Several other proteins show homologies to pathogenesis-related proteins, e.g. glucanase, chitinase, and thaumatin-like proteins. Because pathogenesis-related-like proteins are known to be induced by various other abiotic and biotic stresses, a specific physiological role of these proteins in response to excess Mn supply remains to be established. The specific role of apoplastic PODs in the response of plants to Mn stress is discussed.
For a wide range of plant species, formation of brown spots is part of a characteristic development of Mn toxicity symptoms in older leaves. The subsequent development of chlorosis and necrosis and finally leaf shedding occurs before a reduction in vegetative growth on the whole plant level (Horst, 1988; El-Jaoul and Cox, 1998). Analysis of the Mn-induced formation of brown spots revealed the presence of oxidized Mn and oxidized phenols, especially in the cell wall of the epidermis layer (Horiguchi, 1987; Wissemeier and Horst, 1992). The formation of visible Mn toxicity symptoms is accompanied by the spatial formation of callose in the area of brown spots (Wissemeier and Horst, 1987). The physiological role of callose formation in response to toxic Mn levels in the tissue is unknown, but its detection serves as an additional sensitive parameter for Mn-induced injury of the leaf tissue. In cowpea (Vigna unguiculata), the leaf apoplast has been proposed to be the most important compartment for the defense of Mn stress (Horst et al., 1999). The oxidation of MnII by a H2O2-consuming peroxidase (POD) has been proposed to be the key reaction leading to Mn toxicity symptoms, probably accompanied by the formation of reactive intermediate compounds like phenoxy radicals and MnIII (Horst, 1988). This hypothesis is supported by a close relationship between the Mn-induced formation of brown spots, activation of constitutive apoplastic POD, and Mn-induced release of POD into the apoplast (Fecht-Christoffers et al., 2003). Among the existing information about the physiological functions of POD in plant tissue, its activation in response to a broad range of biotic and abiotic factors plays a particularly important role (Greppin et al., 1986; Obinger et al., 1996). POD activity is often used as a physiological marker for plant stress response as part of a complex cascade of reactions with an apparent lack of specificity. However, some specific relationships between horseradish POD and the oxidation of MnII and phenolic compounds in the apoplast were reported by Kenten and Mann (1950). In particular, the stimulating effect of Mn on apoplastic H2O2-producing PODs (Halliwell, 1978) provides evidence of a specific role of Mn on the functionality of PODs in the leaf apoplast.
To get a better understanding of the specific involvement of POD and other proteins in the response of cowpea plants to Mn excess, observations on changes of POD activities and further physiological changes in the apoplast were investigated at different stages of Mn toxicity. Furthermore, two-dimensional (2D) gel electrophoresis techniques were used for the separation of apoplastic water-soluble proteins followed by a systematic identification of proteins using nanoscale capillary liquid chromatography-tandem mass spectrometry (nano LC-MS/MS). Most of the identified proteins exhibit sequence similarity to previously characterized proteins from other organisms, which partially are known to play roles in stress defense. These proteins represent new tracks for the investigation of Mn stress in higher plants.
RESULTS
Effect of Mn Treatment Duration on Mn Uptake, Activity of Apoplastic Guaiacol- and NADPH-POD, Protein Concentration in the “Apoplastic Washing Fluid” (AWF), and Callose Formation
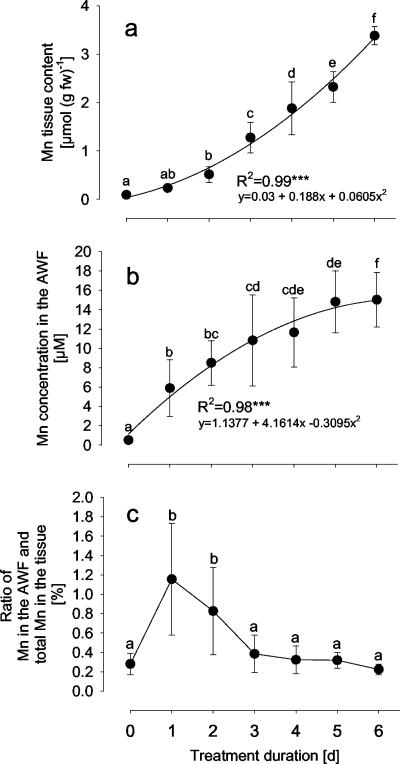
Mn is readily taken up by cowpea plants and transported to leaves. The Mn tissue content increased exponentially 40-fold during 6 d of treatment with 50 μm Mn (Fig. 1A). The related increase of the Mn concentration in the AWF followed a saturation curve (Fig. 1B) with a significant increase already after 1 d of Mn treatment. The ratio of water-soluble Mn in the AWF and total Mn of cowpea leaves was about 0.3% (Fig. 1C). After 1 d of Mn treatment, it increased significantly to 1.2%. After 3 d of Mn treatment, it decreased to the initial ratio which seems to represent an equilibrium with the total Mn in the tissue.
Figure 1.
Effect of treatment duration on the total Mn content in the leaf tissue (A), the Mn concentration in the AWF of leaves (B), and the ratio of water-soluble Mn in the apoplast to total Mn in the leaf tissue (C). Plants of cowpea were treated with 50 μm Mn, whereas control plants received 0.2 μm Mn, continuously. AWF was collected by vacuum-infiltration and centrifugation of leaves. Means of 14 replicates each day are significantly different at P < 0.05 (Tukey) as indicated by different letters. Coefficients of determination of regression analysis are significant as ***, **, and * for P < 0.001, 0.01, and 0.05, respectively.
After 2 d of Mn treatment, the first visible Mn toxicity symptoms (brown spots) were detectable (Fig. 2). The development of Mn toxicity symptoms was significantly correlated with the Mn tissue content (Fig. 2). An equally sensitive indicator of Mn toxicity appeared to be the induction of callose synthesis in the leaves (Fig. 3). After 2 d of Mn treatment, callose content started to increase and was significantly higher after 3 d. The activities of both guaiacol- and NADH-PODs (Fig. 4, A and B) in the leaf AWF increased after 2 d of exposure to elevated Mn supply. This was accompanied by the release of proteins into the apoplast (Fig. 4C). In summary, formation of brown spots, activity of enzymes, the protein concentration, and the callose induction were significantly correlated with the Mn tissue contents (Figs. 2, 3, 4).
Figure 2.
Relationship between the Mn tissue content and the number of brown spots on the trifoliate leaves of cowpea. Four leaf discs per leaf were incubated in ethanol to remove chlorophyll. The number of brown spots was then determined by direct counting. Means of 14 replicates each day are significantly different at P < 0.05 (Tukey) as indicated by different letters. Coefficients of determination of regression analysis are significant as ***, **, and * for P < 0.001, 0.01, and 0.05, respectively.
Figure 3.
Relationship between the Mn and callose contents of leaves. Means of 14 replicates each day are significantly different at P < 0.05 (Tukey) as indicated by different letters. Leaf discs, previously used for counting brown spots, were homogenized in NaOH. The extracted callose was detected by anilin blue staining. Coefficients of determination of regression analysis are significant as ***, **, and * for P < 0.001, 0.01, and 0.05, respectively.
Figure 4.
Relationships between the Mn tissue content and the activity of guaiacol-POD (A), the activity of NADH-POD (B), and the concentration of total protein in the AWF of leaves (C). Guaiacol-POD activity was measured in the presence of 20 mm guaiacol and 0.03% (w/w) H2O2 (pH 6). NADH-POD was measured in the presence of 0.6 mm NADH, 1.6 mm p-coumaric acid, and 16 mm MnCl2 (pH 5). Protein concentrations were measured according to Bradford (1976). Means of 14 replicates each day are significantly different at P < 0.05 (Tukey) as indicated by different letters. Coefficients of determination of regression analysis are significant as ***, **, and * for P < 0.001, 0.01, and 0.05, respectively.
Analysis of Water-Soluble Proteins from the Leaf Apoplast by 2D Blue-Native (BN)/SDS-PAGE and LC-MS/MS
To systematically monitor the induction of apoplastic proteins upon Mn treatment, AWF was analyzed by BN-PAGE. For this procedure, Coomassie dyes are used before electrophoresis to introduce negative charges into proteins and protein complexes without denaturing them. BN-PAGE allows proteins to be efficiently resolved under native conditions. Subsequently, proteins can be visualized by Coomassie Blue staining and PODs by in-gel activity measurements using guaiacol as a substrate, which leads to the formation of a brown-colored oxidation product (Fig. 5). Mn treatment causes an induction of several apoplastic PODs in the 30-kD range (Fig. 5A). Before treatment (d = 0), POD activity was expressed by two to three bands in the gel. These bands became stronger upon prolonged Mn treatment. Furthermore, new bands became visible after 2 d of Mn treatment (Fig. 5A). The staining of the gel with Coomassie Blue showed additional proteins, which also were induced by Mn treatment (Fig. 5B).
Figure 5.
One-dimensional (1D) resolution of water-soluble proteins from the leaf apoplast of cowpea. Separation was carried out by BN-PAGE using a 12% to 20% (w/v) gradient gel. PODs were detected by staining with 20 mm Guaiacol+0.01% (w/w) H2O2 (A) and total protein with colloidal Coomassie Blue (B). The numbers above the gels indicate duration of the Mn treatment (in days); numbers on the right indicate molecular masses of proteins.
Because the Mn toxicity-induced PODs have very similar molecular masses, BN gel electrophoresis was combined with SDS-PAGE to increase resolution capacity (Fig. 6). Most protein bands of the native gel dimension were separated into at least two protein spots in the second gel dimension. Several protein spots became specifically visible upon Mn treatment (Fig. 6B). Representative spots of the constitutive POD and Mn-induced proteins were cut out of the gel and analyzed by LC-MS/MS (Table I). The sequence of one peptide of protein number 4 exhibits significant sequence identity to previously characterized PODs from other plant species (Table I). Additional bands identified as POD by guaiacol staining showed sequence similarity to thaumatin-like proteins (spot 5). Further Mn-induced proteins were identified as chitinases (spot 1 and 7), glucanases (spot 1), fascilin-like arabinogalactan-protein (spot 7), hevein-like and wound-induced proteins (spot 8), and pathogenesis-related (PR) proteins class 1 (spot 10). Most of the proteins show similar molecular masses in the range of 20 to 30 kD, which caused overlappings of proteins on 1D and 2D gels. To further improve resolution of proteins from the AWF, a second 2D gel system was employed, which is based on isoelectric focusing (IEF) for the first gel dimension and SDS-PAGE for the second (Fig. 7).
Figure 6.
2D resolution of water-soluble proteins from the leaf apoplast of cowpea by BN/SDS-PAGE. PODs were detected by staining with 20 mm guaiacol + H2O2 (i) and total proteins with colloidal Coomassie Blue (ii and second dimension). Plants precultured for 13 d were treated with 50 μm Mn for 5 d (B), whereas control plants received 0.2 μm Mn continuously (A). Numbers on the top of the gels indicate the acrylamide concentration, and the numbers on the left indicate molecular masses of proteins. Marked spots were identified by nano LC-MS/MS.
Table I.
Identified proteins of the apoplast of cowpea after separation by BN-/SDS-PAGE
| Noa | Ionb | Sequencec | Identified Proteind | Accession No. | Matching Sequence | ple | Mre | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 992.38 | QEVVDLYK | *1,3-β-glucanase (soybean [Glycine max]) | T05957 | QEVVDLYK | 4.27 | 26,074.11 | |
| 1,074 | HFGLFNPDK | 1,3-β-glucosidase (barley [Hordeum vulgare]) | AAA32960 | HFGLFNPDK | 8.46 | 32,583.82 | Xu et al. (1992) | |
| 1,266.36 | VNYYTEYCR | Chitinase class 4 (cowpea) | S57476 | VNYYTEYCR | 4.25 | 26,876.36 | ||
| 4 | 762.32 | MGASLLR | Peroxidase (wheat [Triticum aestivum]) | AAM76682 | MGASLLR | 8.89 | 32,425.66 | |
| Peroxidase POC1 (rice [Oryza sativa]) | AAF65464 | MGASLLR | 6.40 | 32,461.60 | ||||
| 5 | 978 | GSDGSVLGCK | *Thaumatin-like protein 1 precursor (sand pear [Pyrus pyrifolia]) | O80327 | GSDGSVIGCK | 5.07 | 25,308.31 | Sassa and Hirano (1998) |
| 1,693 | SACLAMGDDQYCCK | *Thaumatin-like protein 1 precursor (sand pear) | O80327 | SACLALNQPQYCC | 5.07 | 25,308.31 | Sassa and Hirano (1998) | |
| 1,069.42 | TGCNFDGDGK | Thaumatin-like protein PR-5a (chickpea [Cicer arietinum]) | CAA09229 | TGCNFDGSG | 5.51 | 18,732.00 | ||
| Pathogenesis-related protein 5 precursor (Arabidopsis) | JQ1695 | TGCNFDASGNG | 4.75 | 25,252.31 | Uknes et al. (1992) | |||
| 898 | LPSPWSGR | Thaumatine-like protein (apple [Malus × domestica]) | AAM12886 | PSPWSGR | 4.60 | 22,107.61 | Venisse et al. (2002) | |
| 7 | 1,095 | YGGVMLWDR | *Chitinase (EC 3.2.1.14) class III, acidic (soybean) | T05187 | YGGVMLWDR | 4.96 | 37,110.79 | Yeboah et al. (1998) |
| 1,909 | CNPSLNCNVFSDLQK | *Chitinase (EC 3.2.1.14) class III, acidic (cowpea) | S57468 | CNPSINNCNVFSD | 4.81 | 26,326.12 | ||
| 966 | VGFGSAASGSK | Endosperm-specific protein-like protein (Arabidopsis) | AAM66074 | VGFGSAASGSK | 5.43 | 43,060.81 | Haas et al. (2002) | |
| Fasciclin-like arabinogalactan-protein (Arabidopsis) | NP_566043 | VGFGSAASGSK | 5.43 | 43,074.83 | ||||
| 1,434 | QQFPLAVXXXDK | Fasciclin-like arabinogalactan-protein (FLA6; Arabidopsis) | NP_565475 | QQFPLAV | 5.51 | 26,506.10 | ||
| 8 | 1,454 | AVSAYCSTYDADK | Hevein-like protein precursor (Arabidopsis) | P43082 | AVSAYCSTWDADK | 7.89 | 22,936.63 | Potter et al. (1993) |
| Wound-induced protein (clone TAB7; tomato [Lycopersicon esculentum]) | T07729 | AVSAYCSTWDANK | 8.38 | 21,607.17 | Harris et al. (1997) | |||
| *Putative pathogen-induced protein (tomato) | CAC81819 | AVSAYCSTWDANK | 6.62 | 14,960.57 | ||||
| 10 | 1,636 | YDYGSNTCVGGECR | PR-1 protein (wine grape [Vitis vinifera]) | CAA05868 | YDYNSNSCVGGEC | 6.05 | 11,078.28 | |
| 1,013 | LWVDEKP | Pathogenesis-related protein 1-1a (cucumber [Cucumis sativus]) | AAL84768 | LWVDEKP | 6.92 | 15,655.49 | ||
| 1,453 | YGENLAGSSGDLSG | *Pathogenesis-related protein 1 (European white birch [Betula pendula]) | AAF62171 | YGENLAASSGDLSG | 5.99 | 10,819.80 |
a The nos. correspond to the nos. given in Figure 6. b Ion precursor (mass/charge unit). c Amino acid sequence as determined by ESI-MS/MS. Due to similar molecular properties, Leu and Ile cannot be distinguished. L represents both. d Proteins were identified by http://www.ncbi.nlm.nih.gov/BLAST/. Stars indicate correspondences to Table II. e pl and molecular mass were calculated by http://www.expasy.org/.
Figure 7.
2D resolution of water-soluble proteins from the leaf apoplast of cowpea by IEF/SDS-PAGE. Plants precultured for 13 d were treated with 50 μm Mn for 5 d (B), whereas control plants received 0.2 μm Mn continuously (A). Numbers on the top of the gels indicate the pH gradient, and the numbers on the left indicate molecular masses of proteins. Marked spots were identified by nano LC-MS/MS.
Analysis of Water-Soluble Proteins from the Leaf Apoplast by IEF/SDS-PAGE and LC-MS/MS
In agreement with the results obtained by 2D BN-PAGE, 2D IEF/SDS-PAGE allowed visualization of several proteins that are specifically induced upon Mn treatment, including a group of proteins in the 25- to 30-kD range (protein nos. 2, 3, 8, 9, and 10 on Fig. 7B). These proteins are clustered at pH 6 to 9. Another group of proteins can be found in the 40-kD range at acidic pH (protein nos. 13, 14, 16, 18, and 20). Analysis of selected protein spots by nano LCMS/MS allowed determination of peptide sequences (Table II). On the basis of sequence similarity to characterized proteins from other organisms, the analyzed proteins represent PR proteins (spots 2, 8, 9, and 10), chitinases (spots 13, 14, and 16), 1,3-β-glucanases (spots 18 and 20), and PODs (spots 16 and 18).
Table II.
Identified proteins of the apoplast of cowpea separated by IEF/SDS-PAGE
| No.a | Ionb | Sequencec | Identified Proteind | Accession No. | Matching Sequence | ple | Mre | Reference |
|---|---|---|---|---|---|---|---|---|
| 2 | 1,449.48 | SAYCSTYDA | *Putative pathogen-induced protein (tomato) | CAC81819 | SAYCSTWDA | 6.62 | 14,960.57 | |
| Wound-induced protein 2 percursor (potato [Solanum tuberosum]) | S04927 | SAYCSTWDA | 7.37 | 22,498.23 | Stanford et al. (1989) | |||
| Hevein-like protein precursor (Arabidopsis) | P43082 | SAYCSTWDA | 7.89 | 22,936.63 | Potter et al. (1993) | |||
| 3 | 1,449.57 | GLSAYCRQ | ? | |||||
| 8 | 1,652.52 | NRDYGSNTCVG | Pathogenesis-related protein (barley) | CAA52893 | DYGSNTCAG | 8.19 | 17,439.62 | Bryngelson et al. (1994) |
| 2,794.21 | VAA(M/F)A(Q/K)NYANQR | Pathogenesis-related protein (Tobacco [Nicotiana tabacum]) | S22531 | AAFAQNYANQR | 7.62 | 20,120.38 | Eyal et al. (1992) | |
| 1,714.56 | ESPEADYLLNEHNAAR | Pathogenesis-related protein (tobacco) | S22531 | SPQ-DYL-NPHNAAR | 7.62 | 20,120.38 | Eyal et al. (1992) | |
| 9 | 1,637.52 | YGENLAGSSGDLSGK | *Pathogenesis-related protein (birch) | AAF62171 | YGENLAASSG-DLSG | 5.99 | 10,819.8 | |
| 10 | 1,636.52 | QNYGSNTCVGWCR | Pathogenesis-related protein precursor (barley) | S52627 | NYGSNTC | 9.08 | 17,678.86 | Mouradov et al. (1994) |
| 13 | 1,722.72 | SSWNQWTS | *Chitinase class III, acidic (cowpea) | S57468 | SSWNQWTS | 4.81 | 26,326.12 | |
| YGGVMLDSDR | *Chitinase class III, acidic (soybean) | T05187 | YGGVML-WDR | 4.96 | 37,110.79 | Yeboah et al. (1998) | ||
| 14 | 1,721.74 | (LS)LSSWNQWTSSQAK | Chitinase (adzuki bean [Vigna angularis]) | P29024 | SSWNQWTSSQAK | 4.9 | 31,701.55 | Ishige et al. (1993) |
| 16 | 1,721.74 | YGGV(F/M)LWDR | Chitinase class III, acidic (soybean) | T05187 | YGGVMLWDR | 4.96 | 37,110.79 | Yeboah et al. (1998) |
| 945.46 | AL(N/D)GLSSQR | Chitinase class III (African yam-bean [Sphenostylis stenocarpa]) | AAD27874 | ALNDLSSQR | 4.08 | 31,392.93 | Colucci et al. (1999) | |
| Putative peroxidase (rice) | AAL34128 | LDGLSS | 4.65 | 33,956.4 | ||||
| 1,222.46 | EGTLAETCNTK | Chitinase class III (African yam-bean) | AAD27874 | EGTLADTCNT | 4.08 | 31,392.93 | Colucci et al. (1999) | |
| 18 | 1,060.57 | HFGLFTPDK | 1,3-β-glucanase (maize [Zea mays]) | T02088 | HFGLFNPDK | 4.56 | 34,893.92 | Wu et al. (1994) |
| 1,559.79 | TQLNAACPWLKMK | Peroxidase, cationic (Townsville stylo [Stylosanthes humilis]) | AAB67737 | TQLEAACP | 7.06 | 33,781.18 | Stines et al. (1996) | |
| 978.45 | (GS)DGSVLGCK | *Thaumatin-like protein 1 precursor (sand pear) | O80327 | DGSVIGCK | 5.07 | 25,308.31 | Sassa and Hirano (1998) | |
| 20 | 992.54 | KEVVDLYK | *1,3-β-glucanase (soybean) | T05957 | EVVDLYK | 4.27 | 26,074.11 | |
| 974.57 | AAPVVDLYK | Glucan endo-1,3-β-d-glucosidase (chickpea) | CAA10167 | VVDLYK | 5.49 | 35,595.06 | ||
| 1,385.77 | QRGLFNPDKSPK | Glucan endo-1,3-β-d-glucosidase (Chickpea) | CAA10167 | GLFNPDKSPK | 5.49 | 35,595.06 |
a The nos. correspond to the nos. given in Figure 7. b Ion precursor (mass/charge unit). c Amino acid sequence as determined by ESI-MS/MS. d Due to similar molecular properties, Leu and Ile cannot be distinguished. L represents both. e Proteins were identified by http://www.ncbi.nlm.nih.gov/BLAST/. Stars indicate correspondences to Table I. Isoelectric point and molecular mass were calculated by http://www.expasy.org/.
DISCUSSION
PODs in the Leaf Apoplast
The leaf apoplast is a compartment of storage and physiological reactions like intercellular signaling, defense against biotic and abiotic stresses, and transport of water and nutrients (Sakurai, 1998; Sattelmacher, 2001). The extracellular space of plants contains acidic (anionic) and basic (cationic) POD isoenzymes present both as cell wall-bound and soluble enzymes with differential affinities to substrates (Campa, 1991; Kärkönen et al., 2002). The acidic PODs show a high affinity to lignin precursors and H2O2 and were considered to be important for the normal functioning of the cell wall (Mäder et al., 1980; Imberty et al., 1985; Ros Barcelo et al., 1987). The expression of PODs is tissue specific and regulated by development (Mohan et al., 1993; Klotz et al., 1998). In tobacco (Nicotiana tabacum), acidic PODs were strongly expressed in trichomes and the epidermis (Klotz et al., 1998) and not expressed in tissues or regions undergoing growth, probably due to the inhibitory effect of PODs on growth and elongation (MacAdam et al., 1992a, 1992b; de Souza and MacAdam, 1998). Acidic PODs in the apoplast of tobacco were proposed to be involved in the first level of defense reactions against several stresses (Klotz et al., 1998). This was indicated by the induction of PODs and POD promoters by wounding, pathogen attack (Lagrimi and Rothstein, 1987; Mohan et al., 1993), and Cu treatment (Cuypers et al., 2002).
PODs were also proposed to be able to produce H2O2 necessary for lignification (Mäder and Amberg-Fisher, 1982). In general, basic PODs show a high affinity to NADH with subsequent formation of H2O2. The NADH oxidation by POD is strongly stimulated in the presence of Mn and monophenolic compounds (Halliwell, 1978). Therefore, NADH-POD activities were mostly measured in the presence of Mn and phenols (Otter and Polle, 1997; Chi and Kao, 2001). The H2O2 production and consumption is probably catalyzed independently by different PODs as suggested by Mäder et al. (1980). A two-step control of PODs in the apoplast was described by Gaspar et al. (1985). The rapid increase of activity of basic PODs was ascribed to demasking effects of available PODs in the apoplast or by a rapid secretion of basic PODs into the apoplast under the control of Ca2+ (Castillo et al., 1984). Basic PODs responded to several stimuli within seconds. The secretion of acidic PODs was assumed to occur at a later stage, stimulated by ethylene and Ca2+. This was proposed to be linked to the release of phenolic monomers with subsequent cross-link reactions catalyzed by acidic PODs (Castillo, 1986).
Additionally, PODs were also proposed to act as polyfunctional enzymes operating simultaneously as oxidase and POD (Pedreño et al., 1995) dependent from pH and availability of reducing compounds, e.g. NADH and IAA (Penel, 1995).
Relationship between Mn Toxicity and POD in the Leaf Apoplast of Cowpea
In cowpea, the synchronous increase in POD and the formation of brown spots suggest a close relationship between the expression of Mn toxicity symptoms and the activity of water-soluble POD extracted from the apoplast (Figs. 2 and 4). The Mn-induced increase of POD activity was also reflected by the induced presence of several proteins on BN gels (Fig. 5). Among these, one protein of approximately 32 kD was identified as POD on the basis of sequence similarity to known PODs from other organisms (no. 4 in Fig. 6 and Table I). PODs, which specifically were induced upon Mn treatment, could also be identified by 2D IEF/SDS-PAGE in the acidic pH range (Fig. 7). The estimated pI values indicate that these POD isoenzymes belong to the group of acidic PODs. These pI values differ in some cases from the pI values reported in the literature. This discrepancy might be due to the broad variance of POD isoenzymes in plants. Even within plant species, POD isoenzymes may differ by more than 50% in peptide sequence (Welinder, 1992). Therefore, identification of PODs on the basis of sequence similarities is difficult. The browning of the leaf tissue in cowpea in response to toxic Mn supply might be due to the oxidation of phenolic compounds especially by H2O2-consuming acidic PODs, which have a high affinity to monophenols in the leaf apoplast. The Mn toxicity symptoms occur in organs with reportedly high POD expression, e.g. epidermis and trichomes of older plant organs. The spatial distribution of POD expression could be the cause for the formation of Mn toxicity symptoms especially in the epidermis of cowpea. This relationship is confirmed by observations in sunflower (Helianthus annuus; Blamey et al., 1986), Arabidopsis, tobacco, and rape seed (W.J. Horst, P. Maier, and M.M. Fecht-Christoffers, unpublished data). Mn toxicity in these species leads to brown depositions in and at the base of trichomes. Additionally, leaf age plays a major role for the expression of Mn toxicity symptoms. In cowpea, older leaves are significantly more sensitive than young leaves at elevated Mn tissue contents (Horst, 1988). Also in tobacco, Mn-induced formation of brown spots was observed exclusively on older leaves (W.J. Horst, P. Maier, and M.M. Fecht-Christoffers, unpublished data).
The NADH oxidation rate showed a similar reaction pattern as the “guaiacol-PODs,” indicating that PODs in the apoplast of cowpea were able to react with guaiacol and NADH as well (Fig. 4). After the 1st d of treatment, a slight increase of NADH-POD was detectable without a marked change in POD-isoenzyme composition. This was accompanied by a slight increase in callose formation. The physiological role of callose formation in response to toxic Mn levels in the tissue is unknown, but its detection serves as an additional sensitive parameter for Mn-induced injury of the leaf tissue (Wissemeier and Horst, 1987). A Mn-enhanced release of H2O2 from cowpea-leaf segments (Horst et al., 1999) and from soybean cells grown in suspension culture (W.J. Horst, P. Maier, and M.M. Fecht-Christoffers, unpublished data) was observed, indicating that Mn stimulates the H2O2-production in the leaf apoplast. Assuming 5- to 10-fold higher apoplastic Mn concentrations in vivo than in the AWF (Lohaus et al., 2001), a direct enhancement of the NADH-POD activity by Mn (10-160 μm) cannot be excluded leading to H2O2 production and subsequent stimulation of H2O2-consuming PODs. It remains unclear whether H2O2 production or H2O2 consumption is preferred by apoplastic water-soluble PODs in vivo. Because a high spatial variability of apoplastic pH in leaves has been demonstrated (Hoffmann and Kosegarten, 1995; Sattelmacher et al., 1998; Mühling and Läuchli, 2000), a relationship with the typically uneven distribution of Mn toxicity symptoms in the leaves (Horst and Marscher, 1978; Horst, 1980) could be assumed.
Other Proteins in the Apoplast That Are Induced by Mn Stress
The release of PODs in the apoplast was accompanied by the secretion of proteins that show similarities to wound-induced proteins and PR enzymes, e.g. glucanase, chitinase, and thaumatin-like proteins and PR proteins class I. Furthermore, the gels in Figures 6 and 7 still exhibit regions with high spot density, indicating that further proteins might exist that overlap in the 2D gels. This fact is also reflected by the identification of peptides from single protein spots, which exhibit sequence identity to different proteins from databases. Despite this variability, the partial agreement between the data in Tables I and II demonstrates their reproducibility. PR-denominated proteins in plant tissues are induced by pathogens, and their expression often was confirmed independently for more than one plant-pathogen combination. Homologous proteins not induced by pathogens are denominated PR-like proteins (van Loon and van Strien, 1999). The Mn toxicity-induced proteins released into the AWF of cowpea leaves show similarities to PR-like proteins induced by heavy metals (Didierjean et al., 1996), during plant development (Malehorn et al., 1993) and senescence (Hanfrey et al., 1996), by wounding (Standford et al., 1989; Ishige et al., 1993; Harris et al., 1997), and by ripening of fruits (Fils-Lycaon et al., 1996; Ruperti et al., 2002). Their induction was related to the presence of plant signaling-molecules, e.g. ethylene (Watanabe et al., 1999), abscisic acid (Akiyama and Pillai, 2001), and salicylic acid (Uknes et al., 1992; Busam et al., 1997). In general, PR-like proteins were reported to be induced by several abiotic stresses, e.g. ozone (Schraudner et al., 1992; Yalpani et al., 1994), UV irradiation (Didierjean et al., 1996; Rakwal et al., 1999), heat stress (Margis-Pinheiro et al., 1994), wounding (Zhang and Punja, 1994; Ruperti et al., 2002), heavy metals (Edreva, 1990; Jacobsen et al., 1992; Rakwal et al., 1999), and during salt (Esaka et al., 1994) and cold adaptation (Antikainen et al., 1996; Hiilovaara-Teijo et al., 1999). Mn-induced expression of PR-like proteins was observed in tobacco (Edreva, 1990) and sunflower leaves (Jung et al., 1995), whereas several PR-proteins were also induced by a physiological, non-pathogenic disorder (Edreva, 1990), mercury, and UV light (Jung et al., 1995). These findings led to the conclusion that the Mn-induced secretion of the PR-like proteins is part of a general stress response of plants. This unspecific induction of PR-like proteins was also observed in maize plants treated with mercury chloride and gave evidence of the presence of multiple pathways of gene regulation in response to abiotic stresses (Didierjean et al., 1996). The induction of PR-like proteins during development and cold adaptation was proposed to enhance resistance against other abiotic and biotic stresses (Antikainen et al., 1996; Hiilovaara-Teijo et al., 1999; Ruperti et al., 2002). In cowpea, a potential beneficial effect of induced PR-like proteins is unknown and to this date not considered. The development of symptoms accompanied by the release of proteins into the apoplast represents a sensitive response towards increased Mn supply leading to reduced plant growth and yield production.
CONCLUSIONS
In this work, physiological and proteomic changes were studied at different stages of Mn toxicity to get a better understanding of the role of Mn-induced processes in the leaf apoplast in the expression of Mn toxicity. The appearance of Mn toxicity symptoms was preceded by a steep increase of the Mn concentration of the AWF indicating a particular role of free apoplastic Mn in the expression of Mn toxicity despite its low contribution to the total Mn content of the leaf. Mn excess almost simultaneously induced the formation of brown spots and callose, the activation of guaiacol- and NADH-PODs, and the release of proteins into the apoplast. The analysis of the proteome of the leaf apoplast has placed previous results and speculations about the physiology of Mn toxicity (Horst et al., 1999; Fecht-Christoffers et al., 2003) on a more solid basis. They confirm the particular role of PODs in the expression of Mn toxicity mediating H2O2 production/consumption and the oxidation of phenols in the leaf apoplast. However, on the basis of these observations, conclusions about the chronology of Mn-induced physiological changes are difficult to draw. Particularly, the enhanced release into the apoplast of PR-like proteins appears to be a late response to excess Mn. A more detailed kinetic study with emphasis on very early stages on Mn toxicity and a comparison of Mn-sensitive and -tolerant leaves (genotype, Si nutrition, and leaf age) are required.
MATERIALS AND METHODS
Plant Material
Cowpea (Vigna unguiculata [L.] Walp. cvs TVu 91 and TVu 1987) was grown hydroponically in a growth chamber under controlled environmental conditions at 30°C/25°C day/night temperature, 75% ± 5% relative humidity, and a photon flux density of 270 μmol m-1 s-1 photosynthetic active radiation at mid-plant height during a 16-h photoperiod. After germination in 1 mm CaSO4, seedlings were transferred to a constantly aerated nutrient solution. After preculture, the MnSO4 concentration in the nutrient solution was increased, whereas control plants received 0.2 μm Mn continuously. The nutrient solution was changed two to three times a week to avoid nutrient deficiencies.
Quantification of Toxicity Symptoms
For the quantification of Mn toxicity symptoms, the density of brown spots was counted on a 1 cm2 area at the base, middle, and tip on the upper side of the trifoliate leaf.
Extraction of Water-Soluble Proteins from the Leaf Apoplast
AWF was extracted by a vacuum infiltration/centrifugation technique. Leaves were infiltrated with water by reducing the pressure to 35 hPa followed by a slow relaxation for 2 min. The AWF was recovered by centrifugation at 1,324g for 5 min at room temperature.
Mineral Analysis
Mn in the bulk-leaf tissue was determined after dry ashing (480°C, 8 h) and dissolving the ash in 6 m HCl with 1.5% (w/v) hydroxylammonium chloride and diluted 1:10 with water. AWF was diluted 1:10, whereas HCl and hydroxylammonium chloride were added to give final concentration of 0.6 m HCl and 0.15% (w/v) hydroxylammoniumchloride. Measurements were carried out by optical emission spectroscopy, inductively-coupled plasma (Spectro Analytical Instruments GmbH, Kleve, Germany).
Callose Extraction and Detection
For the measurement of callose formation, four leaf discs (150 mg fresh weight) were cut out of the leaf and fixed in ethanol. After 3 d, ethanol was replaced by demineralized water and incubated over night. Leaf discs were homogenized in 1 mL of 1 m NaOH, and homogenates were incubated for 15 min at 80°C in a water bath. After centrifugation at 13,000g for 5 min, 100 μL of the supernatant was mixed with 600 μL of the anilin mix (0.59 m Gly buffer [pH 9.5], 0.21 m HCl, and 0.04% [w/v] anilin blue) and incubated in a 50°C water bath for 20 min. After cooling the samples down to room temperature, the callose concentration was measured by detecting the fluorescence at excitation wavelength/bandpath of 400 nm/30 nm and emission wavelength/bandpass of 485 nm/40 nm with a microplate reader (BioTek-Instruments, Germany). Control measurements were done with Gly-HCl-buffer solution without anilin blue. For the calculation, the molar extinction coefficient ε = 11.32 (mg L-1)-1 was used.
POD Activity in the AWF
Guaiacol-POD activities in the AWF were determined spectrophotometrically at λ = 470 nm by following the H2O2-dependent oxidation of guaiacol. Samples were mixed with guaiacol solution (20 mm guaiacol in 10 mm Na2HPO4 buffer [pH 6] and 0.03% [w/w] H2O2). NADH-POD activities in the AWF were determined spectrophotometrically at λ = 340 nm by following decline of NADH. Samples were mixed with 0.6 mm NADH, 1.6 mm p-coumaric acid, and 16 mm MnCl2 in 100 mm NaAc buffer (pH 5).
Detection of Total Protein in the AWF
The protein concentration in the AWF was measured according to Bradford (1976).
Extraction of Proteins from the AWF
For protein separation by electrophoresis under native conditions, the proteins of the AWF were purified at 4°C by using centrifugal concentrators with a molecular mass cut off at 5 kD (Vivaspin 6, Vivasience, Hannover, Germany). Run conditions were used according to the manufacturer's instructions.
For the separation by IEF, proteins were extracted by phenol and precipitated by acetate/methanol. The volume of the AWF was reduced by lyophilization. The sample was mixed with extraction buffer (700 mm saccharose, 500 mm Tris, 50 mm EDTA, 100 mm KCl, and 2% [v/v] mercaptoethanol), and after incubation for 10 min on ice, an equal volume of phenol (saturated solution, pH 6.6/7.9, [Tris], FA Amresco, Solon, OH) was added and shaken for 30 min. The aqueous and organic phases were separated by centrifugation for 10 min at 5,000g and 4°C. The phenolic phase was re-extracted with extraction buffer and centrifuged once more. The phenol phase was combined with 5 volumes of 0.1 m ammonium acetate in methanol and incubated for approximately 14 h at -20°C. After centrifugation at 20,000g for 5 min at 4°C, precipitated proteins were washed three times with ammonium acetate in methanol and finally with acetone. The protein samples were air dried and resuspended immediately before electrophoresis.
2D BN/SDS-PAGE
A detailed protocol for 2D BN/SDS-PAGE was published by Schägger (2001). Protein samples were combined with a Coomassie Blue solution (5% [w/v] Serva Blue G and 750 mm aminocaproic acid). Samples were loaded onto a native acrylamid gel with a 4% (w/v) acrylamide stacking gel and a 12% to 20% (w/v) gradient separation gel. After electrophoresis, the gel was soaked in 20 mm guaiacol and 0.03% (w/w) H2O2 to detect PODs in the gel. Gel slices without incubation in POD staining solution were used for the second electrophoresis dimension. For this application, gels were first incubated in 1% (w/v) SDS/1% (v/v) mercaptoethanol for 30 min. Separation gels for the second gel dimension had 10% or 16.5% (w/v) acrylamide, whereas the sample gels always had 10% (w/v) acrylamide. Gel electrophoresis was carried out in Protean II Xi and XL cells (gel dimension 20 × 18 cm) from Bio-Rad (Munich).
2D IEF/SDS-PAGE
For separation of proteins by their pI, the IPGphor system (Amersham Biosciences, Uppsala) availing Immobiline DryStrip gels (18 cm) with a nonlinear pH gradient (pH 3-10) was used. Proteins were resuspended in demineralized water and supplemented with a rehydration solution (8 m urea, 2% [w/v] CHAPS, 0.5% [v/v] carrier ampholyte mixture [IPG buffer, Amersham Biosciences], and a trace of bromphenol blue). Focusing conditions were used according to Werhahn and Braun (2002). Afterward, the Immobiline DryStrip gels were incubated with equilibration solution (50 mm Tris-Cl [pH 8.8], 6 m urea, 30% [v/v] glycerin, 2% [w/v] SDS, and bromphenol blue) supplemented with (a) 1% (w/v) dithiothreitol and (b) 2.5% (w/v) iodoacetamide each for 15 min, respectively. DryStrip gels were placed horizontally on a Tricine SDS-PAGE. The second-dimension electrophoresis was carried out according to Schägger and von Jagow (1987).
Staining
2D gels and the 1D BN gels were stained with colloidal Coomassie Blue according to Neuhoff et al. (1985, 1990).
Protein Preparation and Identification by Nano LC-MS/MS
After staining with colloidal Coomassie Blue, single proteins were cut out and transferred in Eppendorf vessel. Exised slices rinsing and reduction/alkylation steps were performed by the Massprep robot (Micromass, Manchester, UK). Each piece of gel was washed with 100 μL of 25 mm NH4HCO3 and dehydrated with 100 μL of acetonitrile (ACN). This operation was repeated twice. Reduction was achieved by a 1-h treatment with 10 mm dithiothreitol at room temperature. Alkylation reaction was performed by 25 mm iodacetamide for 45 min at room temperature and protected from light. Finally, gel spots were washed three times for 5 min again alternately with 25 mm ammonium carbonate and ACN. Gel pieces were completely dried before tryptic digestion and rehydrated by trypsin addition. About 3 volumes of trypsin freshly diluted (12.5 ng μL-1 in 25 mm NH4HCO3 buffer) was added to the sample. Digestion was performed overnight. The gel pieces were centrifuged, and 60 μL of 35% (v/v) H2O/60% (v/v) ACN/5% (v/v) formic acid (HCOOH) was added to extract peptides. The mixture was sonicated for 30 min. The supernatant was recovered, and the operation was repeated once. For nano LC-MS/MS, the supernatants were transferred into a 96-well plate, and the peptide extraction volume was reduced to 10 μL by evaporation to concentrate the peptides and to remove the ACN from the sample before being injected in the HPLC system. Nano LC-MS/MS analysis of the digested proteins were performed using a CapLC capillary LC system (Micromass) coupled to a hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Q-TOF II, Micromass). Sample (6.4 μL) was loaded and concentrated onto a C18 PepMap precolumn (LC Packing) under a 30 μL min-1 flow rate and flushed for 3 min with 0.1% (v/v) ACN before gradient started to elute the peptides straight to the following separative column. Chromatographic separations were then performed on a reversed-phase capillary column (Pepmap C18, 75 μm i.d., 15 cm length; LC Packings) under a 200 μL min-1 flow rate generated by the CapLC delivering a flow rate of 4.5 μL min-1 split right after the precolumn. The gradient profile used consisted of a linear gradient from 95% A (H2O/0.1% [v/v] HCOOH) to 60% B (ACN/0.1% [v/v] HCOOH) in 35 min followed by a linear gradient to 95% B in 1 min. Mass data acquisitions were piloted by MassLynx software (Micromass).
When MS/MS required, automatic switching between MS and MS/MS modes was used, and the internal parameters of Q-TOF II were set as follows. The electrospray capillary voltage was set to 3.5 kV, the cone voltage was set to 40 V, and the source temperature was set to 120°C. The MS survey scan was m/z 300 to 1,500 with a scan time of 1 s and a interscan time of 0.1 s. When the intensity of a peak rose above a threshold of 10 counts, tandem mass spectra were acquired. Normalized collision energies for peptide fragmentation was set using the charge-state recognition files for +2 and +3 of the three more intense ion parents. The scan range for MS/MS acquisition was from m/z 50 to 1,500 with a scan time of 1 s and an interscan time of 0.1 s. MS/MS acquisition switched back to MS when threshold reached 2 or after 10 s of acquisition duration. Fragmentation was performed using argon as the collision gas and with a collision energy profile optimized for various mass ranges of ion precursors. Mass data collected during a LC-MS/MS analysis were processed and converted into a PKL file to be submitted to the Global server 1.1 and Mascot search engines. Global server 1.1 and Mascot searches were first performed against SwissProt Data Bank with a tolerance on mass measurement of 0.25 D in MS mode and 0.5 D in MS/MS mode without any pI and molecular mass restrictions, but variable modifications were taken into account, like Met oxidation. The peptide mass error was limited to 50 ppm. For confirmation, each spectra was loaded onto the Peptide Sequencing software (BioLynx, Micromass), and the sequences were reprocessed manually before being submitted to a Blast search (National Center for Biotechnology Information) without any taxonomy restriction. Most obtained sequences represent complete tryptic peptides. However, the sequences of some other peptides could only be partially determined by MS/MS.
Statistical Analysis
Statistical analysis was carried out using SAS Release v8.0 (SAS Institute, Cary, NC). Coefficients of determination from regression analysis and results from analysis of variance are given according to their level of significance as ***, **, *, and + for P < 0.001, 0.01, 0.05, and 0.1, respectively. Different letters are significantly different at P < 0.05 (Tukey).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a time manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Holger Eubel for support and introduction to 2D electrophoresis techniques.
This work was supported by the Deutsche Forschungsgemeinschaft (Special Research Programme 717 “The apoplast of higher plants”).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.029215.
References
- Akiyama T, Pillai MA (2001) Molecular cloning, characterization and in vitro expression of a novel endo-1,3-β-glucanase up-regulated by ABA and drought stress in rice (Oryza sativa L.). Plant Sci 161: 1089-1098 [Google Scholar]
- Antikainen M, Griffith M, Zhang J, Hon WC, Yang DSC, Pihakaski-Maunsbach K (1996) Immunolocalization of antifreeze proteins in winter rye leaves, crowns, and roots by tissue printing. Plant Physiol 110: 845-857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blamey FPC, Joyce DC, Edwards DG, Asher CJ (1986) Role of trichomes in sunflower tolerance to manganese toxicity. Plant Soil 91: 171-180 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Bryngelsson T, Sommer-Knudsen J, Gregersen PL, Collinge DB, Ek B, Thordal-Christensen H (1994) Purification, characterization, and molecular cloning of basic PR-1-type pathogenesis-related proteins from barley. Mol Plant-Microbe Interact 7: 268-275 [DOI] [PubMed] [Google Scholar]
- Busam G, Kassemeyer HH, Matern U (1997) Differential expression of chitinase in Vitis vinifera L. responding to systemic acquired resistance activators or fungal challenge. Plant Physiol 115: 1029-1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa A (1991) Biological roles of plant peroxidases: known and potential functions. In J Evers, K Evers, MB Grisham, eds, Peroxidase in Chemistry and Biology, Vol II. CRS Press, Boca Raton, FL, pp 26-49 [Google Scholar]
- Castillo FJ (1986) Extracellular peroxidases as markers of stress? In H Greppin, C Penel, T Gaspar, eds, Molecular and Physiological Aspects of Plant Peroxidases. University of Geneva, Switzerland, pp 419-426
- Castillo FJ, Penel C, Greppin H (1984) Peroxidase release induced by ozone in Sedum album leaves involvement of Ca2+. Plant Physiol 74: 846-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi LC, Kao CH (2001) Abscisic acid induced changes in cell wall peroxidase activity and hydrogen peroxide level in roots of rice seedlings. Plant Sci 160: 323-329 [DOI] [PubMed] [Google Scholar]
- Colucci G, Machuka J, Chrispeels MJ (1999) cDNA cloning of a class III acid chitinase from the African yam bean (Sphenostylis stenocarpa) (accession no. AF137070) (PGR 99-075). Plant Physiol 120: 63310447467 [Google Scholar]
- Cuypers A, Vangronsveld J, Clijsters H (2002) Peroxidases in roots and primary leaves of Phaseolus vulgaris copper and zinc phytotoxicity: a comparison. J Plant Physiol 159: 869-876 [Google Scholar]
- de Souza IRP, MacAdam JW (1998) A transient increase in apoplastic peroxidase activity precedes decrease in elongation rate of B73 maize (Zea mays) leaf blades. Physiol Plant 104: 556-562 [DOI] [PubMed] [Google Scholar]
- Didierjean L, Frendo P, Nasser W, Genot G, Marivet J, Burkard G (1996) Heavy-metal-responsive genes in maize: identification and comparison of their expression upon various forms of abiotic stress. Planta 199: 1-8 [DOI] [PubMed] [Google Scholar]
- Edreva AM (1990) Induction of “pathogenesis-related” proteins in tobacco leaves by physiological (non-pathogenic) disorders. J Exp Bot 41: 701-703 [Google Scholar]
- El-Jaoul T, Cox DA (1998) Manganese toxicity in plants. J Plant Nutr 21: 353-386 [Google Scholar]
- Esaka M, Toyota A, Hayakawa H (1994) Secretion of basic and acidic chitinase from salt-adapted and -unadapted winged bean cells. Physiol Plant 92: 90-96 [Google Scholar]
- Eyal Y, Sagee O, Fluhr R (1992) Dark-induced accumulation of a basic pathogenesis-related (PR-1) transcript and a light requirement for its induction by ethylene. Plant Mol Biol 19: 589-599 [DOI] [PubMed] [Google Scholar]
- Fecht-Christoffers MM, Maier P, Horst WJ (2003) Apoplastic peroxidase and ascorbate are involved in manganese toxicity and tolerance of Vigna unguiculata. Physiol Plant 117: 237-244 [Google Scholar]
- Fils-Lycaon BR, Wiersma PA, Eastwell KC, Sautiere P (1996) A cherry protein and its gene, abundantly expressed in ripening fruit, have been identified as thaumatin-like. Plant Physiol 111: 269-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar T, Penel C, Castillo FJ, Greppin H (1985) A two-step control of basic and acidic peroxidases and its significance for growth and development. Physiol Plant 54: 418-423 [Google Scholar]
- Greppin H, Penel C, Gaspar T, editors (1986) Molecular and Physiological Aspects of Plant Peroxidases. University of Geneva, Switzerland
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL (2002) Full-length messenger RNA sequence greatly improve genome annotation. http://genomebiology.com/2002/3/6/Research/0029 [DOI] [PMC free article] [PubMed]
- Halliwell B (1978) Lignin synthesis: the generation of hydrogen peroxide and superoxide by horseradish peroxidase and its stimulation by manganese (II) and phenols. Planta 140: 81-88 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Fife M, Buchanan-Wollaston V (1996) Leaf senescence in Brassica napus: expression of genes encoding pathogenesis-related proteins. Plant Mol Biol 30: 597-609 [DOI] [PubMed] [Google Scholar]
- Harris N, Taylor JE, Roberts JA (1997) Characterization and expression of an mRNA encoding a wound-induced (Win) protein from ethylene-treated tomato leaf abscission zone tissue. J Exp Bot 48: 1223-1227 [Google Scholar]
- Hiilovaara-Teijo M, Hannukkala A, Griffith M, Yu XM, Pihakaski-Maunsbach K (1999) Snow-mold-induced apoplastic proteins in winter rye leaves lack antifreeze activity. Plant Physiol 121: 665-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Kosegarten H (1995) FITV-dextran for measuring apoplast pH and apoplastic pH gradients between various cell types in sunflower leaves. Physiol Plant 95: 327-335 [Google Scholar]
- Horiguchi T (1987) Mechanism of manganese toxicity and tolerance of plants: II. Deposition f oxidized manganese and plant tissue. Soil Sci Plan Nutr 33: 595-606 [Google Scholar]
- Horst WJ (1980) Genotypische Unterschiede in der Mangan-Toleranz von Cowpea (Vigna unguiculata). Angew Bot 54: 377-392 [Google Scholar]
- Horst WJ (1988) The physiology of Mn toxicity. In MJ Webb, RO Nable, RD Graham, RJ Hannam, eds, Manganese in Soil and Plants. Kluwer Academic Publishers, Dodrecht, The Netherlands, pp 175-188
- Horst WJ, Fecht M, Naumann, A Wissemeier AH, Maier P (1999) Physiology of manganese toxicity and tolerance in Vigna unguiculata (L.) Walp. J Plant Nutr Soil Sci 162: 263-274 [Google Scholar]
- Horst WJ, Marscher H (1978) Symptome von Mangan-Überschuβ bei Bohnen. Z Pflanzenernähr Bodenkd 141: 129-142 [Google Scholar]
- Imberty A, Goldberg R, Catesson AM (1985) Isolation and characterization of Populus isoperoxidases involved in the last step of lignin formation. Planta 164: 221-226 [DOI] [PubMed] [Google Scholar]
- Ishige F, Mori H, Ymazaki K, Imaseki H (1993) Cloning of a complementary DNA that encodes an acidic chitinase which is induced by ethylene and expression of the corresponding gene. Plant Cell Physiol 34: 103-111 [PubMed] [Google Scholar]
- Jacobsen S, Hauschild MZ, Rasmussen U (1992) Induction by chromium ions of chitinases and polyamines in barley (Hordeum vulgare L.) and rape (Brassica napus L. ssp. oleifera). Plant Sci 84: 119-128 [Google Scholar]
- Jung JL, Maurel S, Fritig B, Guenther H (1995) Different pathogenesis-related proteins are expressed in sunflower (Helianthus annuus L.) in response to physical, chemical and stress factors. J Plant Physiol 145: 153-160 [Google Scholar]
- Kärkönen A, Koutaniemi S, Mustonen M, Syrjänen K, Brunow G, Kilpeläinen I, Teeri TH, Simola LK (2002) Lignification related enzymes in Picea abies suspension cultures. Physiol Plant 114: 343-353 [DOI] [PubMed] [Google Scholar]
- Kenten RH, Mann PJG (1950) The oxidation of manganese by peroxidase systems. Biochem J 46: 67-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz KL, Liu TTY, Liu L, Lagrimini LM (1998) Expression of the tobacco anionic peroxidase gene is tissue-specific and developmentally regulated. Plant Mol Biol 36: 509-520 [DOI] [PubMed] [Google Scholar]
- Lin CD, Kao CH (2001) Abscisic acid induced changes in cell wall peroxidase activity and hydrogen peroxide level in roots of rice seedlings. Plant Sci 160: 323-329 [DOI] [PubMed] [Google Scholar]
- Lohaus G, Pennewies K, Sattelmacher B, Hussmann M, Muehling KH (2001) Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol Plant 111: 457-465 [DOI] [PubMed] [Google Scholar]
- MacAdam JW, Nelson CJ, Sharp RE (1992a) Peroxidase activity in the leaf elongation zone of tall fescue: I. Spatial distribution of ionically bound peroxidase activity in genotypes differing in length of the elongation zone. Plant Physiol 99: 872-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAdam JW, Sharp RE, Nelson CJ (1992b) Peroxidase activity in the leaf elongation zone of tall fescue: II. Spatial distribution of apoplastic peroxidase activity in genotypes differing in length of the elongation zone. Plant Physiol 99: 879-885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäder M, Amberg-Fisher V (1982) Role of peroxidase in lignification of tobacco cells: I. Oxidation of nicotinamide adenine dinucleotide and formation of hydrogen peroxide by cell wall peroxidase. Plant Physiol 70: 1128-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäder M, Ungemach J, Schloss P (1980) The role of peroxidase isoenzyme groups of Nicotiana tabacum in hydrogen peroxide formation. Planta 147: 467-470 [DOI] [PubMed] [Google Scholar]
- Malehorn DE, Scott KJ, Shah DM (1993) Structure and expression of a barley acidic β-glucanase gene. Plant Mol Biol 22: 347-360 [DOI] [PubMed] [Google Scholar]
- Margis-Pinheiro M, Marivet J, Burkard G (1994) Bean class IV chitinase gene: structure, developmental expression and induction by heat stress. Plant Sci 98: 163-173 [Google Scholar]
- Mohan R, Vijayan P, Kolattukudy PE (1993) Developmental and tissue-specific expression of a tomato anionic peroxidase (tap1) gene by a minimal promotor, with wound and pathogen induction by an additional 5′-flanking region. Plant Mol Biol 22: 475-490 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Mouradova E, Scott KJ (1994) Gene family encoding basic pathogenesis-related 1 protein in barley. Plant Mol Biol 26: 503-507 [DOI] [PubMed] [Google Scholar]
- Mühling KH, Läuchli A (2000) Light-induced pH and K+ changes in the apoplast of intact leaves. Planta 212: 9-15 [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Stamm R, Eibl H (1985) Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6: 427-448 [Google Scholar]
- Neuhoff V, Stamm R, Pardowitz I, Arold N, Ehrhardt W, Taube D (1990) Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11: 101-117 [DOI] [PubMed] [Google Scholar]
- Obinger C, Burner U, Ebermann R, Pene C, Greppin H (1996) Plant peroxidases: biochemistry and physiology. IV International Symposium 1996. Proceedings University of Agriculture, Vienna and University of Geneva, Switzerland
- Otter T, Polle A (1997) Characterisation of acidic and basic apoplastic peroxidases from needles of Norway spruce (Picea abies, L., Karsten) with respect to lignifying substrates. Plant Cell Physiol 38: 595-602 [Google Scholar]
- Pedreño MA, Ferrer MA, Gaspar T, Munoz R, Ros Barcelo A (1995) The polyfunctionality of cell wall peroxidases avoids the necessity of an independent H2O2-generating system for phenolic coupling in the cell wall. Plant Perox Newslett 5. http://www.unige.ch/LABPV/perox.html
- Penel C (1995) Does oxyferroperoxidase exist in vivo? Plant Perox Newslett 5. http://www.unige.ch/LABPV/perox.html
- Potter S, Uknes S, Lawton K, Winter AM, Chandler D, DiMaio J, Novitzky R, Ward E, Ryals J (1993) Regulation of hevein-like gene in Arabidopsis. Mol Plant-Microbe Interact 6: 680-685 [DOI] [PubMed] [Google Scholar]
- Rakwal R, Agrawal GK, Yonekura M (1999) Separation of proteins from stressed rice (Oryza sativa L.) leaf tissues by two-dimensional polyacrylamide gel electrophoresis: induction of pathogenesis-related and cellular protectant proteins by jasmonic acid, UV irradiation and copper chloride. Electrophoresis 20: 3472-3478 [DOI] [PubMed] [Google Scholar]
- Ros Barcelo A, Muñoz R, Sabater F (1987) Lupin peroxidases: I. Isolation and characterization of cell wall-bound isoperoxidase activity. Physiol Plant 71: 448-454 [Google Scholar]
- Ruperti B, Cattivelli L, Pagni S, Ramina A (2002) Ethylene-responsive genes are differentially regulated during abscission, organ senescence and wounding in peach (Prunus persica). J Exp Bot 53: 429-437 [DOI] [PubMed] [Google Scholar]
- Sakurai N (1998) Dynamic function and regulation of apoplast in the plant body. J Plant Res 111: 133-148 [Google Scholar]
- Sassa H, Hirano H (1998) Style-specific and developmentally regulated accumulation of a glycosylated thaumatin/PR5-like protein in Japanese pear (Pyrus serotina Rehd.). Planta 205: 514-521 [DOI] [PubMed] [Google Scholar]
- Sattelmacher B (2001) The apoplast and its significance for plant mineral nutrition. New Phytol 149: 167-192 [DOI] [PubMed] [Google Scholar]
- Sattelmacher B, Mühling KH, Pennewiss K (1998) The apoplast: its significance for the nutrition of higher plants. Z. Pflanzenernähr Bodenk 161: 485-498 [Google Scholar]
- Schägger H (2001) Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol 65: 231-244 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368-379 [DOI] [PubMed] [Google Scholar]
- Schraudner M, Ernst D, Langebartels C, Sandermann H (1992) Biochemical plant responses to ozone: III. Activation of the defense-related proteins β-1,3-glucanase and chitinase in tobacco leaves. Plant Physiol 99: 1321-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standford A, Bevan M, Northcote D (1989) Differential expression within a family of novel wound-induced genes in potato. Mol Gen Genet 215: 200-208 [DOI] [PubMed] [Google Scholar]
- Stines AP, Curtis MD, Aitken EAB, Manners JM (1996) Nucleotide sequence of a pathogen-inducible cationic peroxidase gene (accession no. L77080) from Stylosanthes humilis (PGR96-029). Plant Physiol 111: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85-97 [Google Scholar]
- Venisse JS, Malnoy M, Faize M, Paulin JP, Brisset MN (2002) Moldulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol Plant-Microbe Interact 15: 1204-1212 [DOI] [PubMed] [Google Scholar]
- Watanabe A, Nong VH, Zhang D, Arahira M, Yeboah NA, Udaka K, Fukazawa C (1999) Molecular cloning and ethylene-inducible expression of Chib1 chitinase from soybean (Glycine max (L.) Merr.). Biosci Biotechnol Biochem 63: 251-260 [DOI] [PubMed] [Google Scholar]
- Welinder KG (1992) Superfamiliy of plant, fungal, and bacterial peroxidases. Curr Opin Struct Biol 2: 388-393 [Google Scholar]
- Werhahn W, Braun HP (2002) Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis 23: 640-646 [DOI] [PubMed] [Google Scholar]
- Wissemeier AH, Horst WJ (1987) Callose deposition in leaves of cowpea (Vigna unguiculata (L.) Walp.) as a sensitive response to high Mn supply. Plant Soil 102: 283-286 [Google Scholar]
- Wissemeier AH, Horst WJ (1992) Effect of light intensity on manganese toxicity symptoms and callose formation in cowpea (Vigna unguiculata (L.) Walp.). Plant Soil 143: 299-309 [Google Scholar]
- Wu S, Kriz AL, Widholm JM (1994) Nucleotide sequence of a maize cDNA for a class II, acidic β-1,3-glucanase. Plant Physiol 106: 1709-1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Wang J, Fincher GB (1992) Evolution and differential expression of the (1→3)-beta-glucan endohydrolase-encoding gene family in barley, Hordeum vulgare. Gene 120: 157-165 [DOI] [PubMed] [Google Scholar]
- Yalpani N, Enyedi AJ, León J, Raskin I (1994) Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta 193: 372-376 [Google Scholar]
- Yeboah NA, Arahira M, Nong VH, Zhang D, Kadokura K, Watanabe A, Fukazawa C (1998) A class III acidic endochitinase s specifically expressed in the developing seeds of soybean (Glycine max (L.) Merr.). Plant Mol Biol 36: 407-415 [DOI] [PubMed] [Google Scholar]
- Zhang YY, Punja ZK (1994) Induction and characterization of chitinase isoforms in cucumber (Cucumis sativus L.): effect of elicitors, wounding and pathogen inoculation. Plant Sci 99: 141-150 [Google Scholar]