Abstract
Plant mitochondria contain non-phosphorylating NAD(P)H dehydrogenases (DHs) that are not found in animal mitochondria. The physiological function, substrate specificity, and location of enzymes within this family have yet to be conclusively determined. We have linked genome sequence information to protein and biochemical data to identify that At1g07180 (SwissProt Q8GWA1) from the Arabidopsis Genome Initiative database encodes AtNDI1, an internal NAD(P)H DH in Arabidopsis mitochondria. Three lines of evidence are presented: (a) The predicted protein sequence of AtNDI1 has high homology with other designated NAD(P)H DHs from microorganisms, (b) the capacity for matrix NAD(P)H oxidation via the rotenone-insensitive pathway is significantly reduced in the Atndi1 mutant plant line, and (c) the in vitro translation product of AtNDI1 is imported into isolated mitochondria and located on the inside of the inner membrane.
The plant mitochondrial inner membrane contains a branched electron transport pathway that includes multiple enzymes for the oxidation of both cytosolic and matrix NAD(P)H. A feature of the plant respiratory chain is the presence of alternative enzymes that are not linked to proton translocation and, hence, are non-phosphorylating. These include the alternative oxidase (AOX), which provides an alternative route for electrons from the ubiquinone pool to oxygen, and four non-proton pumping NAD(P)H dehydrogenases (DHs), identified by their insensitivity to the complex I inhibitor rotenone (Mackenzie and McIntosh, 1999; Møller, 2001). These alternative enzymes have the potential to catalyze wasteful respiration but can also decrease the production of reactive oxygen species in the respiratory chain (Møller, 2001). The synthesis and activity of AOX are strictly controlled in most plant tissues (Vanlerberghe and McIntosh, 1997), but much less is known about the alternative NAD(P)H DHs.
Yeast (Saccharomyces cerevisiae), which lacks Complex I, has at least three non-phosphorylating NAD(P)H DHs in its respiratory chain; ScNDE1, ScNDE2 (external facing), and ScNDI1 (internal facing). These have been well characterized (Marres et al., 1991; de Vries et al., 1992; Luttik et al., 1998). Kinetic analyses have also shown these enzyme activities to be present in mitochondria from the fungus Neurospora crassa (Weiss et al., 1970). Recently, the gene encoding an external-facing NADPH-specific DH has been identified, NcNDE1 (Melo et al., 1999, 2001). In plant mitochondria, kinetic and inhibitor studies suggest the presence of five distinct NAD(P)H DHs in plant mitochondria (for review, see Møller, 2001). From studies with isolated mitochondria and submitochondrial particles, it has been shown that aside from Complex I, in the inner membrane there are two external-facing (cytosolic) NAD(P)H DHs and one or two internal-facing (matrix) NAD(P)H DHs. In addition to the inner membrane NAD(P)H DHs, there is an NAD(P)H DH located in the mitochondrial outer membrane (Moreau and Lance, 1972; Day and Wiskich, 1975). It is not clear whether all of these activities are present in the mitochondria of all tissues of all plants. Furthermore, it is not known whether all of these enzymes exist in Arabidopsis. A number of attempts at purification of these proteins have provided little consensus as to the size and number of polypeptides involved (Klein and Burke, 1984; Luethy et al., 1991; Rasmusson et al., 1993; Knudten et al., 1994; Menz and Day, 1996a, 1996b).
Recently, two putative NAD(P)H DHs have been identified from potato (Solanum tuberosum; Rasmusson et al., 1999), one internal facing and one external facing. Both were found using a single Arabidopsis expressed sequence tag to probe a cDNA library, indicating a significant level of homology between the different gene sequences. The physiological functions and different regulatory mechanisms of these enzymes are not fully understood, although induction under light stress and repression of matrix NAD(P)H DH expression under cold stress has been reported (Svensson and Rasmusson, 2001; Svensson et al., 2002).
Advances in our understanding of the significance of the non-phosphorylating NAD(P)H DH family in plant mitochondria has been limited by the lack of a clear link between genomic knowledge and biochemical data. This paper describes the identification of an NAD(P)H DH gene by characterization of the mitochondrial phenotype of a publicly available Arabidopsis T-DNA insertion mutant. For the first time to our knowledge, molecular and functional evidence for the identity and location of an Arabidopsis non-phosphorylating NAD(P)H DH is presented. Our understanding of the functions, location, and substrate specificity of other plant non-phosphorylating enzymes, based on sequence analysis is discussed, and a standard nomenclature for identified plant genes and gene products that have been identified functionally is proposed.
RESULTS
Sequence Analysis
Six putative NAD(P)H DH sequences (AT1g07180 [AtNDI1], AT2g29990, AT2g20800, AT4g05020, AT4g28220, and AT4g21490) were identified from the Arabidopsis genome database, based on sequence similarity with the yeast genes ScNDE1, ScNDE2, and ScNDI1 (Table I; Figs. 1 and 2). With the exception of AT4g21490, all of the putative translated proteins from these sequences were predicted to be mitochondrially located by at least one of the public subcellular prediction programs: Psort, Predotar, and Target P (Table II). The fact that five of the six putative NAD(P)H DHs are predicted to be mitochondrial by two of the more commonly used prediction programs suggests a mitochondrial location. The poor prediction of targeting signal length by TargetP may be because at least some of these proteins are located on the intermembrane space side of the inner membrane and, thus, are not likely to contain a typical processing site for the matrix located mitochondrial processing peptidase. Thus, the predicted length of the targeting signal may not be reliable. Even though AT4g21490 was not predicted to target to a mitochondrial location, it was retained due to its high similarity to the characterized NAD(P)H DH sequences (Table I). The highest similarity was seen between AtNDI1 and AT2g29990, with 82% amino acid residue identity. The plant protein sequences separate into two groups that either show a high similarity to the previously characterized potato StNDA or StNDB sequences (Rasmusson et al., 1999). The NDA-like sequences have an average of 74% identity (ranging from 70%-82%) compared with an average of 58% identity seen between the plant NDB-like enzymes (ranging from 47%-65%).
Table I.
Statistical summary of alignments between characterized NAD(P)H DH and putative Arabidopsis NAD(P)H DH protein sequences
Sequence identity is shown as the no. of amino acid residues that are identical between two sequences expressed as a percentage of the total no. of self-amino acids (bold). Sequences with high similarity to each other are shaded.
| ScNDI1 | AtNDI1 | AT2g29990 | StNDA | ScNDE1 | ScNDE2 | AT2g20800 | AT4g21490 | AT4g28220 | AT4g05020 | StNDB | NcNDE1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 513 | 29% | 29% | 28% | 40% | 37% | 28% | 34% | 26 | 30 | 28 | 22% | ScNDI1 |
| - | 510 | 82% | 70% | 28% | 28% | 32% | 28 | 23 | 25 | 32 | 27% | AtNDI1 |
| - | - | 508 | 71% | 32% | 32% | 24% | 28 | 23 | 24 | 31 | 21% | AT2g29990 |
| - | - | - | - | 31% | 31% | 29% | 33 | 25 | 28 | 32 | 20% | StNDA |
| - | - | - | - | 560 | 57% | 29% | 32 | 26 | 29 | 29 | 23% | ScNDE1 |
| - | - | - | - | - | 545 | 30% | 33 | 27 | 30 | 29 | 24% | ScNDE2 |
| - | - | - | - | - | - | 582 | 55 | 54 | 65 | 57 | 26% | AT2g20800 |
| - | - | - | - | - | - | - | 478 | 47 | 69 | 57 | 29% | AT4g21490 |
| - | - | - | - | - | - | - | - | 559 | 57 | 64 | 31% | AT4g28220 |
| - | - | - | - | - | - | - | - | - | 582 | 61 | 27% | AT4g05020 |
| - | - | - | - | - | - | - | - | - | - | 577 | 26% | StNDB |
| - | - | - | - | - | - | - | - | - | - | - | 673 | NcNDE1 |
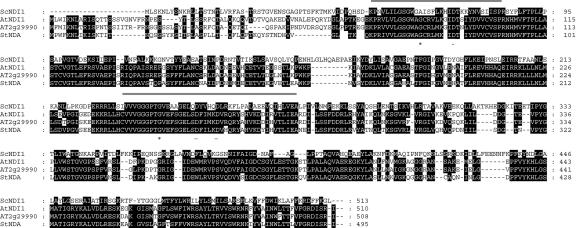
Figure 1.
Alignment of Arabidopsis NAD(P)H DH protein sequences with external-facing NADH DHs from yeast (ScNDE1, accession no. CAA87359; and ScNDE2, accession no. CAA98651) and N. crassa NcNDE1 (accession no. CAB41986) and potato StNDB (accession no. CAB52797). The Arabidopsis sequences are AT2g20800, AT4g21490, AT4g28220, and AT4g05020, with respective SwissProt or GenBank accession numbers Q9SKT7, O65414, NP_567801, and Q94BV7. Regions of high homology (80% or more) are boxed in black. Dashes represent gaps in the sequence. The dinucleotide folds (DNF) are indicated by a shaded line above the sequence, the insert region is indicated by a dashed line, and the EF hand motifs, D-x-[DNSs]-{ILVFYW}-[DENSTG]-[DNQGHRK]-{GP}-[LIVMC]-[DENQSTAGC]-x(2)-[DE]-[LIVMFYW], as defined by the PROSITE database, are indicated by a bold line below the sequence. The conserved third Gly (*) and negative charge residue (-) in both DNFs as discussed in the text are indicated below the sequence.
Figure 2.
Alignment of Arabidopsis NAD(P)H DH protein sequences with the internal-facing NADH DH from yeast (ScNDI1, accession no. S26704) and potato StNDA (accession no. CAB52796). The Arabidopsis sequences are AtNDI1(accession no. Q8GWA1) and AT2g29990 (accession no. O80874). Regions of high homology (75% or more) are boxed in black. Dashes represent gaps in the sequence. The dinucleotide folds (DNF) are indicated by a shaded line above the sequence. The conserved third Gly (*) and negative charge residue (-) in both DNFs as discussed in the text are indicated below the sequence.
Table II.
Predicted subcellular locations of the putative Arabidopsis and potato NAD(P)H DH protein sequences using Psort, Predotar, or TargetP programs
Predicted length of presequences refer to the no. of residues from the N terminus and were determined using TargetP. For some sequences, - means that the program was unable to determine the presequence cleavage site.
| Gene Product
|
Results of Predicted Subcellular Location
|
Length of Presequence.
|
||
|---|---|---|---|---|
| Psort | Predotar | TargetP | ||
| AtNDI1 | Chloroplast/mitochondria | Mitochondria | Mitochondria | 18 |
| AT2g29990 | Chloroplast/mitochondria | Mitochondria | Mitochondria | 31 |
| AT2g20800 | Mitochondria | Mitochondria | Mitochondria | 9 |
| AT4g21490 | Endoplasmic reticulum | Neither | Endoplasmic reticulum | - |
| Chloroplast/mitochondria | ||||
| AT4g28220 | Mitochondria | Mitochondria | Mitochondria | 120 |
| AT4g05020 | Mitochondria | Mitochondria | Not endoplasmic reticulum/chloroplast/mitochondria | - |
When aligned with the previously characterized proteins ScNDE1, ScNDE2, and ScNDI1 of yeast, NcNDE1 of N. crassa and with StNDB and StNDA of potato, the Arabidopsis sequences fall into two clear groups, based both on the presence of common and unique functional motifs within the ScNDE-like and ScNDI-like proteins and similarity to StNDB (Fig. 1) or StNDA (Fig. 2).
Both the NDA-like and NDB-like NAD(P)H DH sequences contain two dinucleotide fold motifs, characterized by the core GXGXXG signature (Wierenga et al., 1986). The first site is close to the N terminal with the second approximately 150 amino acids downstream. These regions of the proteins could be involved in binding the cofactor FAD and/or the substrate NAD(P)H. Their presence is consistent with observations made by Kerscher (2000) based on sequence alignments of microbial NADH DH proteins.
It is interesting to note that the predicted NDB-like enzymes are generally longer in length than the NDA-like enzymes (Table I; Figs. 1 and 2). This is due to an insertion of additional sequence that is present toward the C-terminal end of NcNDE1, three of the four Arabidopsis sequences, and the potato NDB protein (Fig. 1). This “insert” region contains two motifs that have homology to Ca2+-binding EF hand motif (Marsden et al., 1990), of which only the first is well conserved in potato NDB (Rasmusson et al., 1999) and Arabidopsis, and only the second is well conserved in NcNDE1 (Melo et al., 1999). The presence of this motif in the plant NDB-like NAD(P)H DH sequences is not unexpected because the corresponding activities are sensitive to calcium levels (Møller, 2001). This motif was not seen in ScNDE1 or ScNDE2, which is consistent with these yeast NADH DHs being calcium insensitive (Luttik et al., 1998). However, the insertion sequence and EF hand motif was also absent from the AT4g21490 sequence and the NDA-like NAD(P)H DH sequences. It has been proposed that the EF hand motif could play a role in binding the enzymes to the inner membrane (Rasmusson et al., 1999). However, this hypothesis is not consistent with the non-phosphorylating NAD(P)H DHs found in other species (specifically yeast enzymes) as suggested by Kerscher (2000).
Identification of the Atndi1 Mutant
The Arabidopsis T-DNA database of insertion mutants (Sessions et al., 2002) was searched by BLAST analysis using the six putative NAD(P)H DH gene sequences from Arabidopsis. Due to the high level of sequence identity between the two NDA-like genes, the results indicated that either or both of the genes appear to have been mutated in the same seed line (SAIL_779_G05, referred to here as the Atndi1 mutant). Molecular analysis of the Atndi1 mutant was undertaken to confirm suppression of AtNDI1 and/or AT2g29990.
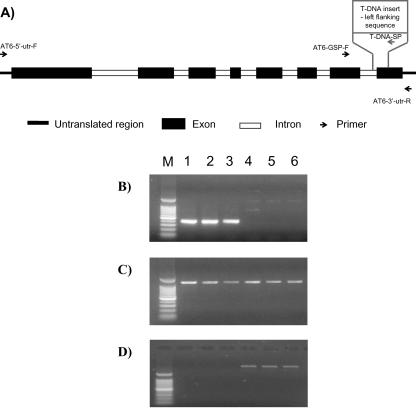
Genomic PCR was carried out using an insert-specific primer (T-DNA-SP) in combination with a gene-specific primer (ATx-GSP) to confirm the presence or absence of the insert in the genes of interest. To determine whether the NDA-like genes (AT2g29990 and AtNDI1) were being expressed, reverse transcriptase (RT)-PCR was performed using the gene-specific 5′- and 3′-untranslated region (UTR) primers (At1 and At6, Table III). All amplifications were carried out in triplicate using DNA and RNA isolated from the separate Arabidopsis shaking cultures used for mitochondrial extraction and biochemical analyses. A schematic representation of the AtNDI1 gene (including introns) indicates the location of primers used with respect to the T-DNA insertion site (Fig. 3A). In the genomic PCR, the Atndi1 shaking seedling cultures gave no amplification product using T-DNA-SP in combination with AT1-GSP (data not shown). However, products of expected size were amplified in the Atndi1 T-DNA mutant replicates using T-DNA-SP in combination with AT6-GSP (Fig. 3B). This indicates that the T-DNA insert is in the AtNDI1 gene rather than At2g29990. Figure 3C shows the amplification of AT2g29990 cDNA in each of the replicate Atndi1 mutants and in wt samples (sequence identity confirmed by sequencing). In contrast, RT-PCR using the AtNDI1 gene-specific 5′-and 3′-UTR primers failed to amplify any cDNA product from the Atndi1 mutant RNA but amplified the full-length cDNA from wt (Fig. 3D).
Table III.
Primers used for cloning the AtNDI1 gene and analysis of the Atndi1 mutant (AT1 is specific to gene AT2g29990 and AT6 to AtNDI1)
| Primer name | Specificity | Sequence |
|---|---|---|
| AT6-5′-UTR-F | AtNDI1 | CTCTCGATTGAAGAAGAGTACC |
| AT6-3′-UTR-R | AtNDI1 | ATGAGATTCGGCTAATGTCCCG |
| AT1-5′-UTR-F | AT2g29990 | AGACGAAGAAGACGAAGACG |
| AT1-3′-UTR-R | AT2g29990 | CCTTAGATACGGCTAATGTC |
| AT6-3′-UTR-R | AtNDI1 | ATGAGATTCGGCTAATGTCCCG |
| AT6-GSP-F | AtNDI1 | GGAAGTATGGCTACTATTGG |
| AT1-GSP-F | AT2g29990 | ATACAAACGCCTAGTCGACC |
| T-DNA-SP | T-DNA insert | ATTTCATAACCAATCTCGATACAC |
Figure 3.
A, Schematic representation of AtNDI1 showing exons, introns, primer locations (arrows indicating direction), and T-DNA insertion site. Molecular assays of the Atndi1 mutant compared with wild-type (wt) Arabidopsis. All reactions were carried out in triplicate using nucleic acid isolated from plant material from separate shaking cultures. Lanes: M, 100-bp marker; 1 to 3, Atndi1 T-DNA mutant shaking culture plants; 4 to 6, Arabidopsis wt shaking culture plants. B, Amplification products resulting from genomic PCR using AT6-GSP with T-DNA-SP. C, Amplification products resulting from RT-PCR using the AT2g29990 gene-specific primers, AT1-5′-UTR and At1-3′-UTR. D, Amplification products resulting from RT-PCR using the AtNDI1 gene-specific primers, AT6-5′-UTR and AT6-3′-UTR.
From these results, we concluded that the mutant plants contain an insert in AtNDI1 that has completely suppressed the transcription of this gene, whereas the highly homologous At2g29990 gene is still transcribed. As a consequence, it was assumed that changes in the biochemical properties of mutant compared with wt mitochondria are attributable to disruption of AtNDI1.
When wt and transgenic plants were grown under identical conditions, there was no observable morphological phenotype. The plants in shaking culture yielded approximately the same amount of tissue (fresh weight 20-30 g), shoot to root ratios were not significantly different, and overall appearance was similar (data not shown). This is not altogether surprising given that the transcription of only one of the two putative internal NAD(P)H DH genes is disrupted in the T-DNA insertion mutant plants.
NAD(P)H DH Capacity of the Atndi1 Mutant
Mitochondria were isolated from mutant and wt plants grown in shaking culture, and their electron transport capacity was measured. The assays were designed to measure the internal, external, and outer membrane NAD(P)H DH capacities separately in intact mitochondria. This was achieved by the selective use of FeCN as an artificial electron acceptor in conjunction with inhibitors of different parts of respiratory electron transport chain. Conditions were chosen to favor the production of NAD(P)H via malate DH and malic enzyme within the matrix (for details, see “Materials and Methods”). In all cases, enzyme activities are expressed as a ratio of the succinate DH activity for that mitochondrial preparation to compensate for any variation in the respiratory activity of individual mitochondrial extractions.
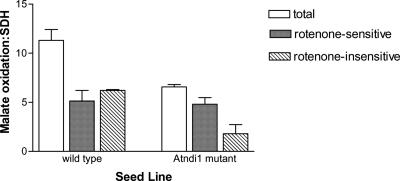
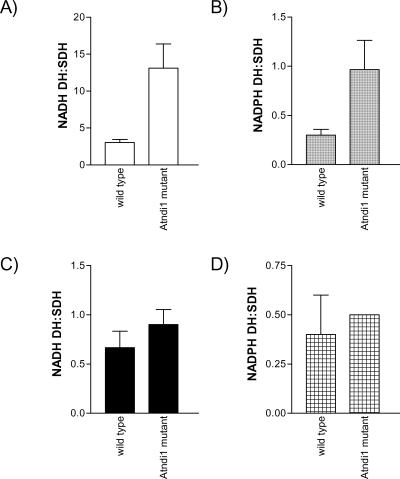
Total matrix malate-FeCN reductase activity ratio was approximate 40% lower in the mutant plants compared with wt (Fig. 4). This was due entirely to a decrease in the rotenone-insensitive pathway of approximately 65%. There was no significant change in the rate of rotenone-sensitive malate-FeCN reduction, which is catalyzed by Complex I of the respiratory chain. In addition, an immunoblot of mitochondrial proteins probed with antisera against the 51-kD subunit of bovine Complex I indicated that protein levels of this subunit were not different between wt and Atndi1 mutant mitochondria (data not shown). The activity of the external-facing inner membrane NADH DH was approximately 10-fold higher than external-facing inner membrane NADPH DH activity for both wt and Atndi1 mutant mitochondria (Fig. 5, A and B), but both of these activity ratios were approximately 4-fold greater in mutant mitochondria. No significant differences were observed in outer membrane NADH and NADPH DH activities between wt and mutant mitochondria (Fig. 5, C and D).
Figure 4.
Malate-FeCN reductase, measured in the presence and absence of rotenone, for intact, isolated mitochondria from and insertion mutation seed line, SAIL_779_G05, Arabidopsis plants. Individual mitochondrial preparation malate-FeCN reductase rates have been expressed as a ratio of succinate-phenolindo-2,6-dichlorophenol (DCPIP) reductase activity for that preparation (wt, 17 ± 7; and Atndi1, 10 ± 1 nmol DCPIP reduced mg protein-1 min-1). The means of these data are shown. Error bars = se of the mean (in all cases n = 3).
Figure 5.
NAD(P)H DH activities measured for intact, isolated mitochondria from wt and insertion mutation seed line, SAIL_779_G05, Arabidopsis plants. A, External NADH DH; B, External NADPH DH; C, Outer membrane NADH DH; D, Outer membrane NADPH DH. All rates have been expressed as a ratio of succinate-DCPIP reductase activity for the same individual mitochondrial preparation (as described in Fig. 2). The means of these data are shown. Error bars = se of the mean. A to C, n = 3; D, n = 2.
Protein Import
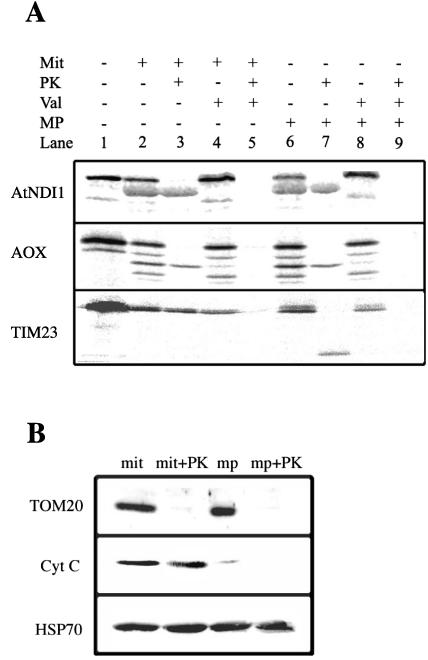
To confirm the location of the AtNDI1 protein, in vitro import assays were carried out. The full-length cDNA for AtNDI1 was cloned and sequenced using primers located in the 5′- and 3′-UTRs of the gene. Comparison of the cloned sequence with that from the database confirmed that the complete cDNA for AtNDI1 had been successfully and completely amplified (data not shown). In vitro translation of the cDNA produced a precursor protein with an apparent molecular mass of 60 kD (Fig. 6, lane 1). Upon incubation of the precursor with mitochondria, a product with an apparent molecular mass of 56 kD was produced, which was insensitive to digestion by added protein K (PK) (Fig. 6, lanes 2 and 3). The generation of this product was sensitive to valinomycin in the import assay, indicating that the protein was imported into or across the inner membrane (Fig. 6, lanes 4 and 5). A similar pattern was observed with the AOX precursor and for TIM23 except that the latter has no cleavable presequence; thus, no change in apparent mass was observed upon import.
Figure 6.
Import of AtNDI1 into isolated soybean (Glycine max) cotyledon mitochondria. A, AtNDI1 precursor protein was produced in an in vitro transcription-translation system and incubated with mitochondria to assess its ability to be imported into mitochondria. Lane 1, Translation mixture alone; the precursor protein is seen as a prominent band with an apparent molecular mass of 60 kD. Lane 2, Translation mixture incubated with mitochondria; an additional band with an apparent molecular mass of 56 kD is evident. Lane 3, As in lane 2 but treated with Proteinase K after the import assay. Lanes 4 and 5, As in lanes 2 and 3 but with valinomycin added to the import assay before the addition of precursor protein. Lanes 6 to 9, As in lanes 2 to 4 except that after the import assay, mitochondria were pelleted and their outer membranes ruptured to generate intermembrane space-depleted mitochondria by resuspending in water. B, Western-blot analysis of mitochondria (mit), mitochondria treated with PK (mit + PK) intermembrane space-depleted mitochondria (mp), and intermembrane space-depleted mitochondria treated with PK (mp + PK), using antibodies against Tom20, an outer membrane protein, cytochrome c, an intermembrane space protein, and HSP70, a matrix protein.
To determine if the imported AtNDI1 was on the outside or inside of the inner membrane, we ruptured the outer membrane and prepared intermembrane space-depleted mitochondria to allow access of added PK to the outside of the inner membrane. Under these conditions, TIM23 was partially degraded indicated by a clear inner membrane-protected fragment, showing that the PK had gained access to intermembrane space components (Fig. 6, lane 3 verse 7). In contrast, under identical conditions, neither AOX nor AtDNI1 were digested by PK, indicating complete protection by the inner membrane. To further verify the intactness of our mitochondria and the rupture of the outer membrane, we carried out western-blot analysis with an outer membrane component (TOM20), an intermembrane space component (cytochrome c), and a matrix-located component (HSP70). It was observed that no significant rupture of the inner membrane had taken place under the conditions used because the HSP70 was intact in all the fractions. Likewise, with intact mitochondria, cytochrome c was protected from PK digestion. Most of this protein was lost from salt-washed intermembrane space-depleted mitochondria, and the small amount remaining was digested by PK with treatment of intermembrane space-depleted mitochondria. Finally, it was observed that TOM20 was accessible to PK in both mitochondria and intermembrane space-depleted mitochondria. The results together indicate that the PK treatment was sufficient to digest exposed proteins in the intermembrane space-depleted mitochondria. Thus, we conclude that the AtNDI1 was located on the inside of the inner membrane.
DISCUSSION
Identification of an Internal NAD(P)H DH
In this study, evidence has been presented that directly links function with sequence information for the non-phosphorylating NAD(P)H DH from plant mitochondria. Three separate lines of evidence reported here suggest very strongly that the AtNDI1 gene encodes an NAD(P)H DH located on the inside of the mitochondrial inner membrane. First, the predicted protein sequence grouped with other suspected internal NAD(P)H DH such as ScNDI1 and potato NDA sequence. Second, the Atndi1 mutant containing an insert in the gene-encoding protein AtNDI1 has severely decreased rates of internal rotenone-insensitive NAD(P)H DH activity.
Based on the molecular data presented (Fig. 3), the Atndi1 mutant contains an insert in the gene encoding the protein AtNDI1; as a consequence, this gene was not transcribed. On the other hand, transcription of At2g29990, which encodes a second, highly homologous, putative internal NAD(P)H DH, was unaffected. Thus, the loss of matrix NAD(P)H activity in the Atndi1 mutant could be attributed to the At1g07180 gene. Activity of both inner and outer membrane external NAD(P)H DH was either stimulated or unchanged in the mutant plants. Finally, the in vitro translation product of AtNDI1 was imported into isolated mitochondria and protected from proteinase treatment in mitochondria from which the outer membrane had been removed, confirming that this gene encodes a matrix-located protein. This enzyme consequently has been designated AtNDI1, in keeping with the nomenclature assigned for NADH DHs characterized in other species (particularly yeast). We propose that this nomenclature be applied to all non-phosphorylating NAD(P)H DHs identified in plants, once biochemical evidence has been provided for function and location.
There was not a complete elimination of the rotenone-insensitive pathway in the Atndi1 mutant (Fig. 4), but it is possible that the residual rate was generated from the gene product of the other NDA-like NAD(P)H DH, AT2g29990, suggesting that it can compensate for the lack of AtNDI1. This observation perhaps is not surprising given the highly similar sequences of the two putative internal NAD(P)H DH genes. The high level of homology between AT2g29990 and AtNDI1 is thought to be the consequence of a duplication event in the Arabidopsis genome. It is possible that subsequent to this event, the two genes have maintained function and evolved independently. However, the presence of two separate DHs on the inner matrix surface has only been demonstrated in potato and pea (Pisum sativum) leaf mitochondria (Melo et al., 1996; Agius et al., 1998), and we do not know if there are two separate DHs for rotenone-insensitive NAD(P)H oxidation in Arabidopsis. The observed residual activity in the AtNDI1 mutant could also indicate that AT2g29990 and AtNDI1 encode the same activity but with regulation via different promoters. That is, it is possible that there is a level of redundancy in the genome and that these genes are interchangeable, the two genes perhaps being induced under different conditions or in different tissues. For example, in potato, the internal NAD(P)H DH (NDA) is thought to be cold and light inducible in leaves (Rasmusson et al., 1999; Svensson and Rasmusson, 2001; Svensson et al., 2002). Future work to characterize all of the enzymes involved in the oxidation of matrix NAD(P)H could include the analysis of a double mutant. For example, such a mutant could be generated by the suppression of the AT2g29990 gene in the Atndi1 mutant identified in the current paper.
Substrate Specificity of AtNDI1
Measurement of the maximum capacity of matrix NAD(P)H oxidation is difficult and complex, and anumber of different strategies have been developed to determine this activity. In this project, we used a malate-FeCN reductase assay to determine matrix NAD(P)H oxidation and optimized malate DH and malic enzyme activities to ensure generation of sufficient NAD(P)H within the mitochondria to drive the low-affinity rotenone-insensitive enzymes (Møller and Palmer, 1982). Malate added to plant mitochondria is converted to pyruvate via malic enzyme and/or oxaloacetate via malate DH, producing mainly NADH. However, a low level of NADPH may also be made because the two malate-oxidizing enzymes have a level of dual specificity in plants (Macrae, 1971; Agius et al., 1998). The NAD:NADP preference of malic enzyme varies between 1:5 and 1:20 depending on the tissue, but in all cases, the major product is NADH. Further to this, the matrix NAD concentration is substantially higher than that of NADP (Agius et al., 2001). This suggests that under our assay conditions, the predominant product of these reactions is likely to be NADH. Further, the NADH-specific internal-facing DH has been shown to be calcium independent, and because AtNDI1 lacks an EF-hand motif, it is likely that this is the NADH-specific enzyme.
The AtNDI1 sequence possesses two regions that meet the criteria for a dinucleotide-binding fold (DNF) domain, namely: (a) a Gly-rich consensus (GxGxxG), (b) a negatively charged residue (D or E) at the end of the second β-sheet, and (c) six positions typically occupied by small hydrophobic residues (Bellamacina, 1996). For all of the sequences listed, either of the two regions could form the binding site for either FAD or the substrate NAD(P)H (there is no functional evidence for their exact identity). However, homology of these domains with those in lipoamide DH suggests that the more N-terminal DNF domain may be the binding site for FAD (Rasmusson et al., 1999; Kerscher, 2000).
It has been proposed that if one of the motifs is an NADPH-specific-binding site, then the third Gly in the Gly-rich consensus motif would generally be replaced by an S, A, or P and also that the negative charge at the end of the second β-sheet would be missing. There is evidence from complementation studies in yeast and N. crassa that ScNDE1, ScNDE2, and ScNDE1 are NADH-specific enzymes (Marres et al., 1991; Luttik et al., 1998), whereas NcNDE1 is an NADPH-specific enzyme (Melo et al., 2001). From an examination of the NcNDE1 sequence, Melo et al. (2001) speculated that the more N-terminal DNF domain binds NADPH as the third Gly is replaced by Ser, and the negative charge at the end of the second β-sheet is missing. These features are absent from the second DNF in N. crassa. However, ScNDE1 and ScNDE2 also have either S or P in the third Gly position, but the negative charge at the end of the β-sheet is conserved. This would suggest that the conservation of the negative charged Asp at the end of the β-sheet is more significant in discriminating between NADH- and NADPH-specific-binding sites, in agreement with previous observations (Lesk, 1995). Although there is conservation of Gly in the GXGXXG motif in the Arabidopsis external sequences (Fig. 1), it is tempting to speculate that AT4g05020, AT4g21490, and AT2g20800 may be NADPH-specific enzymes because the conserved D has been substituted by N at the end of the β-sheet in the N-terminal DNF. Both AtNDI1 and AT2g29990 sequences have a conserved third Gly and a conserved negative charge at the end of the second β-sheet in both DNF domains, and we would speculate that they are both NADH-specific DHs. However, it should be kept in mind that a number of NADPH and NADH specific enzymes deviate from these rules (Bellamacina, 1996; Detarsio et al., 2003), and more experimental evidence is required to conclude which of these Arabidopsis sequences are NADPH and NADH specific.
Interaction between Alternative NAD(P)H DHs
The interaction between the non-phosphorylating enzymes in plant mitochondria appears to be more complex than previously anticipated. Both the residual internal NAD(P)H oxidation rate and the increase in external NAD(P)H DH activities in the Atndi1 mutant illustrate this. Rotenone-sensitive Complex I activity was not affected in mitochondria from the mutant, yet overall matrix NAD(P)H oxidation decreased. This suggests that the rotenone-insensitive bypass contributes to electron flow in the absence of the inhibitor (Bryce et al., 1990). Further, loss of AtNDI1 transcripts resulted in an increase in external NAD(P)H DH activity in the mutant plants. The reason for this increase is not clear, but a similar increase in total capacity to oxidize external NAD(P)H has been observed when Complex I in plant mitochondria has been compromised by either mitochondrial protein synthesis inhibitors (Zhang et al., 1996) or in mitochondrial deletions mutants that exhibit cytoplasmic male sterility (Gutierres et al., 1997). One possible explanation is that the decrease in matrix NAD(P)H oxidation would result in fewer electrons being donated to the ubiquinone pool and an increase in matrix NAD(P)H. Thus, the matrix becomes over-reduced. This, in turn, could stimulate transfer of reducing equivalents from the mitochondrial matrix to the cytosol, as proposed in Yarrowia lipolytica mitochondria (Kerscher et al., 2001). The increase in external NADH DH activity presented in this study could be due, therefore, to the action of a substrate shuttle across the inner mitochondrial membrane, such as the malate/oxaloacetate shuttle described in pea leaf mitochondria (Day and Wiskich, 1981).
MATERIALS AND METHODS
Sequence Analysis
BLAST (Altschul et al., 1990) services were used to compare yeast (Saccharomyces cerevisiae) NAD(P)H DH sequences (ScNDE1, accession no. Z47071; ScNDE2, accession no. Z74133; and ScNDI1, accession no. X61590) against the Arabidopsis Genome Initiative Munich Information Center for Protein Sequences database (MAtDB; http://www.mips.gsf.d/proj/thal/). All Arabidopsis proteins described are named according to their Arabidopsis Genome Initiative numbers, and the corresponding SwissProt or GenBank protein accession numbers are: Q8GWA1, AT1g07180; O80874, AT2g29990; Q9SKT7, AT2g20800; Q94BV7, AT4g05020; NP_567801, AT4g28220; and O65414, AT4g21490. Searches conducted were either BLASTN, comparing nucleotide sequences against nucleotide sequence databases, or BLASTX comparing nucleotide sequences translated in all reading frames against protein databases.
Computer programs used to predict cellular location of proteins were: TargetP (Emanuelsson et al., 2000), Psort (Nakai and Kanehisa, 1992), and Predotar (http://www.inra.fr/predotar). The length of putative presequences was determined using TargetP (Nielsen et al., 1997). For Clustal multiple protein sequence alignments, the parameters were: gap penalty 10 and gap length penalty 10, using a BLOSUM62 similarity matrix. All sequence manipulations were performed using GeneDoc software package (Nicholas and Nicholas, 1997).
Plant Material
Arabidopsis ecotype Columbia (wt) was used for all experiments. BLAST analysis of the Syngenta (San Diego) T-DNA insertion mutation database indicated that seed line SAIL_779_G05 contained an insert in either At1g07180 and/or At2g29990 (Sessions et al., 2002). This seed line (SAIL_779_G05) was provided by Syngenta. wt and mutant plants were grown in a greenhouse, in soil, at 19°C, with 16 h of light/8 h of dark. Individual plants were isolated to ensure self-pollination. Seeds were harvested at 6 to 8 weeks.
Arabidopsis Seedling Shaking Cultures
Arabidopsis seeds were surface sterilized in 70% (v/v) ethanol for 2 min, soaked in 15% (v/v) bleach for 15 min, and then rinsed four to five times in sterile water. Shaking culture growth medium (40 mL; one-half-strength Murashige and Skoog basal salt mixture salts [Sigma, Australia], B5 vitamins, 2% [w/v] Suc, and 2 mm MES [pH 5.8]) was inoculated with approximately 50 sterile seeds. Cultures were grown for 14 d, with 18 h of light/6 h of dark at 23°C, with shaking at 80 rpm.
RNA/DNA Extractions
RNeasy and DNeasy kits (Qiagen USA, Valencia, CA) were used according to the manufacturer's protocols. DNA and RNA were run on a 1% (w/v) agarose gel to check quality and quantified spectrophotometrically.
RT-PCR/PCR
Amplification reactions were carried out using primers described in Table II. Total RNA (3 μg) was used to synthesize cDNA using Invitrogen Superscript II Reverse Transcriptase and Oligo(dT)12-18 according to the manufacturer's protocols. PCR was carried out in the presence of 1.5 mm MgCl2, 0.2 mm of each dNTP, 100 ng of forward primer and reverse primer, 0.5 units of Taq DNA polymerase (Qiagen USA), and 20 ng of cDNA, in a final volume of 20 μL. Amplifications were performed on a Cetus 2400 thermal cycler (Perkin-Elmer Applied Biosystems, Foster City, CA) with the following cycling parameters: prestep at 95°C for 5 min; 20 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s; and an extension step of 72°C for 10 min. Amplification products (5-μL aliquots) were separated on a 1.5% (w/v) agarose gel. DNA size markers were run alongside the amplification products to determine fragment size.
Cloning of AtNDI1 cDNA
The full-length cDNA (including start and stop codons) was amplified using the AT6-5′-UTR-F and AT6-3′-UTR-R primers with Arabidopsis leaf RNA as the starting template (RT-PCR carried out as described above). The amplification product was cloned into pGEMT-Easy (Promega, Madison, WI) for sequencing and pGEM3Zf(-) for translation and import experiments, using standard protocols (Sambrook et al., 1989).
In Vitro Protein Import
Mitochondria from 7-d-old soybean (Glycine max) cotyledons were isolated according to Day et al. (1985). Precursor protein was expressed in an in vitro transcription-translation system using T7 RNA polymerase using standard procedures (Sambrook et al., 1989). Mitochondrial import assays were carried out as previously described (Daley et al., 2002). In brief, 5 μL of precursor protein was added to 100 μL of purified mitochondria in a final volume of 200 μL under conditions that support import. After 20 min, the reaction was placed on ice and divided into two equal aliquots. One-half was treated with 50 μL of Proteinase K on ice for 30 min, after which 2 mm phenylmethylsulfonyl fluoride was added to inhibit further PK activity, and the mitochondria were pelleted by centrifugation in a microfuge for 3 min at 20,000g at 4°C. The mitochondrial pellets were resuspended in SDS gel sample buffer, proteins were separated by SDS-PAGE, and gels were dried and exposed to a BASTR2040 plate for 24 h and imaged according to the manufacturer's instructions using a BAS 2500 instrument (Fuji, Tokyo). A number of similar reactions were carried out at the same time to investigate the pattern of import observed: (a) Valinomycin was added to a final concentration of 1 μm before the commencement of the import assay, and (b) intermembrane space-depleted mitochondria were prepared from mitochondria after the import assay to test for the location of the imported protein. The addition of valinomycin tests for insertion into or across the inner membrane because it abolishes the membrane potential required for all proteins imported into or across the inner membrane (Neupert, 1997; Glaser et al., 1998). Intermembrane space-depleted mitochondria were prepared (i.e. the outer membrane was removed) to test if the imported protein was exposed on the outside of the inner membrane or the inside of the inner membrane. Two other proteins whose location in the inner membrane have been determined previously were used as controls. The translocase of the inner membrane subunit 23 (TIM23) is a four membrane-spanning inner membrane protein with N and C termini exposed in the intermembrane space (Murcha et al., 2003). AOX in an inner membrane protein is not accessible to protease as long as the inner membrane is intact (Day et al., 1995).
Salt-washed intermembrane space-depleted mitochondria were prepared based on a procedure that removes most intermembrane space components in potato (Solanum tuberosum; Lister et al., 2002). After the import assay, mitochondria were placed on ice and pelleted as outlined above. The mitochondrial pellet was resuspended in 10 μL of SEH buffer (250 mm Suc, 1 mm EDTA, and 10 mm HEPES-KOH [pH 7.4]), and 150 μL of 20 mm HEPES-KOH (pH 7.4) was added and mixed gently at 4°C for 15 min. Restoration of osmotic conditions was achieved by adding 25 μL of 2 m Suc, and 10 μL of 3 m KCL was added to promote release of intermembrane space protein from the inner membrane. The sample was then divided into two, and one lot was treated with PK and analyzed as outlined above for mitochondria. Western-blot analysis was carried out with antibodies to the outer membrane protein (TOM20, Dr. Hans-Peter Braun, University of Hannover, Germany), the intermembrane space protein cytochrome c (BD Biosciences, San Diego), and the matrix protein HSP70 (Stressgene, Victoria, BC, Canada).
Mitochondrial Isolation
Mitochondria were extracted from approximately 20 to 30 g of cultured Arabidopsis seedlings (root and shoot), washed, and purified on discontinuous Percoll gradients essentially as described by Soole et al. (1990) but using 45%, 33%, 24%, and 13.5% (v/v) Percoll steps. Protein estimates were made according to Lowry et al. (1951).
Enzyme Assays
All assays were carried out spectrophotometrically using isolated, intact mitochondria in a standard reaction medium (0.25 m Suc, 0.01 m KH2PO4, 0.1 m TES, and 5 mm MgCl2 [pH 7.2]), unless otherwise stated.
Succinate-DCPIP reductase was measured at 600 nm. Mitochondria were incubated on ice for 10 min with 20 mm succinate and 1 mm ATP. The rate was determined after the addition of 70 μm DCPIP, 0.3% (w/v) phenazine methosulphate, and azide. Rates were calculated using an extinction coefficient of 19.1 mm-1 cm-1.
Malate-FeCN reductase was measured at 420 nm. Reactions were carried out at pH 6.8 in the presence of 1 mm NAD+, 10 mm malate, 10 mm pyruvate, 10 mm acetyl-CoA, thiamine pyrophosphate [to generate internal NAD(P)H via malate DH and malic enzyme], 1 mm KCN, 10 μm octyl gallate, and 0.9 mm FeCN. Rates were measured in the presence and absence of 25 μm rotenone. Butylmalonate (10 mm) was added at the end of the assay to inhibit malate transport across the membrane. Any residual rate was subtracted from the rate in the absence of the inhibitor to ensure that the NAD(P)H: FeCN reductase rate measured was not being generated by outer membrane or external-facing inner membrane NAD(P)H DHs. Rates were calculated using an extinction coefficient of 1.05 mm-1 cm-1. Rotenone-sensitive malate-FeCN reductase activity was determined by subtracting the rate in the presence of rotenone from the rate in the absence of rotenone.
NAD(P)H-O2 was measured following NAD(P)H oxidation at 340 nm after the addition of 0.1 mm NADH or NADPH, 0.1 mm ADP, and 1 mm CaCl2. Rates were calculated using an extinction coefficient of 6.22 mm-1 cm-1 (for NADH and NADPH).
NAD(P)H-cytochrome c reductase of the outer membrane was measured in the presence of 70 μm cytochrome c, 6 μm myxothiazol, and 1 mm KCN, following the addition of 1 mm NADH or NADPH, by monitoring the reduction of cytochrome c at 550 nm. Rates were calculated using an extinction coefficient of 19.8 mm-1 cm-1.
All mitochondria preparations and assays were performed in triplicate. To normalize the data between mitochondrial preparations, the results for all assays are expressed as a ratio of the state 3 (+ADP) succinate:DCPIP reductase rate for the same mitochondrial preparation. The rates of succinate:DCPIP reductase activities were not significantly different between the wt and mutant mitochondrial preparations (P < 0.05, Mann Whitney t test).
Acknowledgments
The authors wish to acknowledge Syngenta (San Diego) for providing the T-DNA insert mutant seed. We would particularly like to thank David Oliver for his advice, especially with the seedling shaking cultures, and helpful feedback on the manuscript.
This work was fully funded by the Australian Research Council. Experiments were performed at Flinders University, with the exception of the in vitro translation and protein import studies, which were conducted at the University of Western Australia, Perth.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.029363.
References
- Agius SC, Bykova NV, Igamberdiev AU, Møller IM (1998) The internal rotenone-insensitive NADPH dehydrogenase contributes to malate oxidation by potato tuber and pea leaf mitochondria. Physiol Plant 104: 329-336 [Google Scholar]
- Agius SC, Ramusson AG, Møller IM (2001) NAD(P) turnover in plant mitochondria. Aust J Plant Physiol 28: 461-470 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410 [DOI] [PubMed] [Google Scholar]
- Bellamacina CR (1996) The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J 10: 1257-1269 [DOI] [PubMed] [Google Scholar]
- Bryce JH, Azcon-Bieto J, Wiskich JT, Day DA (1990) Adenylate control of respiration in plants: the contribution of rotenone-insensitive electron transport to ADP-limited oxygen consumption by soybean mitochondria. Physiol Plant 78: 105-111 [Google Scholar]
- Daley D, Adams KL, Clifton R, Qualmann S, Millar AH, Palmer JD, Pratje E, Whelan J (2002) Gene transfer from mitochondrion to nucleus: novel mechanisms for gene activation from Cox2. Plant J 30: 11-21 [DOI] [PubMed] [Google Scholar]
- Day DA, Neuberger M, Douce R (1985) Interactions between glycine decarboxylase, the tricarboxylic acid cycle and the respiratory chain in pea leaf mitochondria. Aust J Plant Physiol 12: 119-130 [Google Scholar]
- Day DA, Wiskich JT (1975) Isolation and properties of the outer membrane of plant mitochondria. Arch Biochem Biophys 171: 117-123 [DOI] [PubMed] [Google Scholar]
- Day DA, Wiskich JT (1981) Glycine metabolism and oxaloacetate transport by pea leaf mitochondria. Plant Physiol 68: 425-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT (1995) Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol 22: 497-509 [Google Scholar]
- Detarsio E, Wheeler MCG, Campos Bermudez VA, Andreo CS, Drincovich MF (2003) Maize C4 NADP-malic enzyme: expression in E. coli and characterisation of site-directed mutants at the putative nucleotide-binding sites. J Biol Chem 278: 13757-13764 [DOI] [PubMed] [Google Scholar]
- de Vries S, van Witzenburg R, Grivell LA, Marres CAM (1992) Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem 203: 587-592 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005-1016 [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjoling S, Tanudji M, Whelan J (1998) Mitochondrial protein import in plants: signals, sorting, targeting, processing and regulation. Plant Mol Biol 38: 311-338 [DOI] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Dioliez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94: 3436-3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher SJ (2000) Diversity and origin of the alternative NADH:ubiquinone oxidoreductases. Biochim Biophys Acta 1459: 274-283 [DOI] [PubMed] [Google Scholar]
- Kerscher SJ, Eschemann A, Okun PM, Brandt U (2001) External alternative NADH:ubiquinone oxidoreductase redirected to the internal face of the mitochondrial inner membrane rescues Complex I deficiency. J Cell Sci 114: 3915-3921 [DOI] [PubMed] [Google Scholar]
- Klein RR, Burke JJ (1984) Separation procedure and partial characterisation of two NAD(P)H dehydrogenases from cauliflower mitochondria. Plant Physiol 76: 436-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudten AF, Thelen JJ, Luethy MH, Elthon TE (1994) Purification, characterisation, and submitochondrial localisation of the 32kDa NADH dehydrogenase from maize. Plant Physiol 106: 1115-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk AM (1995) NAD-binding domains of dehydrogenases. Curr Opin Struct Biol 5: 775-783 [DOI] [PubMed] [Google Scholar]
- Lister R, Mowday B, Whelan J, Millar AH (2002) Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J 30: 555-566 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Roesbrough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin phenol reagent. J Biol Chem 193: 265-275 [PubMed] [Google Scholar]
- Luethy MH, Hayes MK, Elthon TE (1991) Partial purification and characterisation of three NAD(P)H dehydrogenases from Beta vulgaris mitochondria. Plant Physiol 97: 1317-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik MAH, Overkamp KM, Kötter P, de Vries S, van Dijken P, Pronk JT (1998) The Saccharomyces cerevisiae nde1 and nde2 genes encode separate mitochondrial NADH dehydrogenases catalysing the oxidation of cytosolic NADH. J Biol Chem 273: 24529-24534 [DOI] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11: 571-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae AR (1971) Malic enzyme activity of plant mitochondria. Phytochem 10: 2343-2347 [Google Scholar]
- Marres CAM, de Vries S, Grivell L (1991) Isolation and inactivation of the nuclear gene encoding the rotenone-insensitive internal NADH:ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem 195: 857-862 [DOI] [PubMed] [Google Scholar]
- Marsden BJ, Shaw GS, Sykes BD (1990) Calcium binding proteins: elucidation of the contributions to calcium affinity from an analysis of species variants and peptide fragments. Biochem Cell Biol 68: 587-601 [DOI] [PubMed] [Google Scholar]
- Melo AMP, Duarte M, Møller IM, Prokisch H, Dolan PL, Pinto L, Nelson MA, Videira A (2001) The external calcium-dependent NADPH dehydrogenase from Neurospora crassa mitochondria. J Biol Chem 276: 3947-3951 [DOI] [PubMed] [Google Scholar]
- Melo AMP, Duarte M, Videira A (1999) Primary structure and characterisation of a 64kDa NADH dehydrogenase from the inner membrane of Neurospora crassa mitochondria. Biochim Biophys Acta 1412: 282-287 [DOI] [PubMed] [Google Scholar]
- Melo AMP, Roberts TH, Møller IM (1996) Evidence for the presence of two rotenone-insensitive NAD(P)H dehydrogenases on the inner surface of the inner membrane of potato tuber mitochondria. Biochim Biophys Acta 1276: 133-139 [Google Scholar]
- Menz RI, Day DA (1996a) Identification and characterisation of an inducible NAD(P)H dehydrogenase from red beetroot mitochondria. Plant Physiol 112: 607-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz RI, Day DA (1996b) Purification and characterisation of a 43kDa rotenone-insensitive NADH dehydrogenase from plant mitochondria. J Biol Biochem 271: 23117-23120 [DOI] [PubMed] [Google Scholar]
- Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561-591 [DOI] [PubMed] [Google Scholar]
- Møller IM, Palmer JM (1982) Direct evidence for the presence of a rotenone-resistant NADH dehydrogenase on the inner surface of the inner membrane of plant mitochondria. Physiol Plant 54: 267-274 [Google Scholar]
- Moreau, Lance C (1972) Isoelement et propriétés des membranes externes et internes de mitochondries végétales. Biochimie 54: 1335-1348 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Lister R, Ho AYY, Whelan J (2003) Identification, expression and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol 131: 1737-1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14: 897-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: Analysis and Visualization of Genetic Variation, EMBNEW.NEVS 4:14. http://www.psc.edu/biomed/genedoc
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng 10: 1-6 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863-917 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Fredlund KM, Møller IM (1993) Purification of a rotenone-insensitive NAD(P)H dehydrogenase from the inner surface of the inner membrane of red beetroot mitochondria. Biochim Biophys Acta 1141: 107-110 [Google Scholar]
- Rasmusson AG, Svensson AS, Knoop V, Grohman L, Brennicke A (1999) Homologues of yeast and bacterial rotenone-insensitive NADH dehydrogenases in higher eukaryotes: two enzymes are present in potato mitochondria. Plant J 20: 79-87 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sessions A, Burke E, Presting G, Aux G, McElver, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985-2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soole KL, Dry IB, Wiskich JT (1990) Oxidation of NADH by plant mitochondria: kinetics and effects of calcium ions. Physiol Plant 78: 205-210 [Google Scholar]
- Svensson AS, Johansson FI, Moller IM, Rasmusson AG (2002) Cold stress decreases the capacity for respiratory NADH oxidation in potato leaves. FEBS Lett 517: 79-82 [DOI] [PubMed] [Google Scholar]
- Svensson AS, Rasmusson AG (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28: 73-82 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703-734 [DOI] [PubMed] [Google Scholar]
- Weiss H, von Jagow G, Klingerberg M, Butcher T (1970) Characterisation of Neurospora crassa mitochondria prepared with a grind mill. Eur J Biochem 14: 75-82 [DOI] [PubMed] [Google Scholar]
- Wierenga RK, Terpstra P, Hol WGJ (1986) Prediction of the occurrence of the ADP-binding βαβ-fold in proteins using an amino acid fingerprint. J Mol Biol 187: 101-107 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Mischis L, Wiskich JT (1996) Respiratory responses of pea and wheat seedlings to chloramphenicol treatment. Aust J Plant Physiol 23: 583-592 [Google Scholar]