Abstract
Photosynthetic signals modulate expression of nuclear genes at the levels of mRNA transcription, mRNA stability, and translation. In transgenic tobacco (Nicotiana tabacum), the pea (Pisum sativum) Ferredoxin 1 (Fed-1) mRNA dissociates from polyribosomes and becomes destabilized when photosynthesis is inhibited by photosynthetic electron transport inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea. We used polymerase chain reaction suppressive-subtractive hybridization to identify similarly regulated endogenous tobacco genes. This screen identified 14 nuclear-encoded tobacco mRNAs whose light-induced increase in abundance is suppressed in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea. Sequence analysis of the cognate cDNAs revealed that nine of the mRNAs encode putative chloroplast-targeted proteins. We asked whether the abundance of these mRNAs was regulated transcriptionally or posttranscriptionally. Of the five mRNAs with sufficient abundance to detect using nuclear run-on assays, we observed transcriptional regulation of α-tubulin, thiazole biosynthetic enzyme, and pSKA10 (an unknown gene). Photosystem A subunit L and, to a lesser extent, α-tubulin and pSKA10 mRNAs, may also be stabilized in the light. In contrast, Rubisco small subunit mRNA abundance appears to be transcriptionally up-regulated but posttranscriptionally down-regulated in the light. To determine whether, like Fed-1 mRNA, the mRNAs identified in this screen were translationally responsive to light, we characterized the polyribosome association of these mRNAs in the light and after a 15-min dark treatment. A subset of the mRNAs showed dramatic dark-induced polyribosome dissociation, similar to Fed-1 mRNA, and all of the mRNAs showed at least slight polyribosome dissociation. Thus, both posttranscriptional and translational regulation appear to be important mechanisms regulating the expression of many nuclear-encoded mRNAs encoding proteins involved in photosynthesis.
Plant fitness is enhanced by the ability to rapidly change light-harvesting capabilities in response to fluctuating light conditions (Kulheim et al., 2002). The genes for mRNAs encoding chloroplast proteins are divided between the nuclear and the chloroplast genome and are coordinately regulated in response to light, maximizing energy storage while minimizing photo-oxidative damage (for review, see Goldschmidt-Clermont, 1998; Barkan and Goldschmidt-Clermont, 2000; Brown et al., 2001; Rochaix, 2001). Photosynthetic control of the abundance of chloroplast-encoded mRNAs occurs at the levels of transcription, mRNA processing, and mRNA stability (Kubicki et al., 1994; Nickelsen et al., 1994; Alexciev and Tullberg, 1997; Deshpande et al., 1997; Yamamoto et al., 1997; Pfannschmidt et al., 1999; Tullberg et al., 2000). In addition, in depth study of many Chlamydomonas reinhardtii chloroplast-encoded mRNAs has revealed the importance of photosynthetic regulation of translation, often via binding of the encoded protein to the 5′- or 3′-untranslated region (UTR) of the corresponding chloroplast mRNA (for review, see Bruick and Mayfield, 1999). Light-regulated binding of proteins to mRNAs may be regulated, at least in part, by changes in redox state generated by photosynthesis (Kim and Mayfield, 1997; Yohn et al., 1998; Fong et al., 2000; Shen et al., 2001). Photosynthetic inhibitors have been used widely to distinguish redox regulators of chloroplastic gene expression, implicating the ferredoxin/thioredoxin, glutathione, and PRK/chloroplast protein 12 (CP12)/GAPDH complexes in the signal transduction chain within chloroplasts (Wedel et al., 1997; Vener et al., 1998; Irihimovitch and Shapira, 2000; Trebitsh and Danon, 2001).
Regulation of gene expression between the chloroplast and nucleus includes cross-signaling between the organelles. In addition to the complex effects of photoreceptor signals on nuclear mRNA transcription (for review, see Quail, 2002), signals from the developing plastid are essential for mature chloroplast formation (Oelmuller, 1989) and for accumulation of nuclear-encoded chloroplast proteins (e.g. Zubko and Day, 2002). A series of Arabidopsis mutants have been isolated that uncouple nuclear and plastid gene expression, supporting the idea that signals between the plastid and chloroplast are essential for establishing and maintaining the appropriate balance of chloroplast- and nuclear-encoded proteins (Susek et al., 1993; Mochizuki et al., 1996; Vinti et al., 2000; Mochizuki et al., 2001). The plastid signals include, but may not be limited to, redox signaling and signaling through chlorophyll biosynthetic intermediates or carotenoids (Surpin et al., 2002; for review, see Rodermel, 2001), and they may regulate gene expression at the transcriptional, posttranscriptional, and translational levels (for review, see Petracek and Thompson, 2000).
To maintain appropriate stoichiometry between photosystems in response to light quality, promoters for a number of nuclear-encoded genes encoding chloroplast-localized proteins have differential responsiveness to oxidative and reductive signals (Pfannschmidt et al., 2001). For example, in Dunaliella tertiolecta, LHCII mRNA levels are sensitive to the redox state of the plastoquinone pool. Oxidation of plastoquinone using the photosynthetic electron transport inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) or low-light levels increases LHCII mRNA levels, whereas reduction of plastoquinone using 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone or high-light levels decreases LHCII mRNA levels (Escoubas et al., 1995). Furthermore, in Arabidopsis, blockage of plastoquinone pool reduction with DCMU dominantly suppressed Suc-induced transcriptional activation from the chlorophyll a/b-binding protein 2 and plastocyanin promoters. These results suggest a redox-regulated coordination of signals that measures sugar stores and ongoing photosynthesis (Oswald et al., 2001).
As with chloroplast-encoded mRNAs, photosynthetic signals have been shown to regulate mRNA stability of a few nuclear-encoded mRNAs. Constitutively transcribed pea (Pisum sativum) Fed-1 mRNA in transgenic tobacco (Nicotiana tabacum) accumulates in the light. However, in the dark or in the presence of DCMU in the light, Fed-1 mRNA is destabilized (Petracek et al., 1998). Mutation of a (CAUU)4 repeat element in the Fed-1 5′-UTR abolishes light-regulated Fed-1 mRNA accumulation, suggesting a role of this repeat in mRNA stability (Dickey et al., 1998). Nuclear run-on and transgene analysis in tobacco of the pea plastocyanin (PetE) gene suggest that light regulates PetE abundance at the level of mRNA stability (Helliwell et al., 1997). Like Fed-1 mRNA, PetE and protochlorophyllide oxidoreductase mRNAs are destabilized in the presence of DCMU (Eguchi et al., 2002; Sullivan and Gray, 2002). Interestingly, the DCMU-induced destabilization of PetE mRNA is counterbalanced by a DCMU-induced increase in the rate of PetE transcription (Sullivan and Gray, 2002). Opposing effects of light regulation of transcription and mRNA stability have also been observed for pea and Arabidopsis LHCB1 mRNAs. Transcription of these mRNAs increases after a pulse of blue light, but the mRNA abundance is unchanged, suggesting a blue-light induced destabilization of Lhcb1*4 mRNA (Warpeha et al., 1989; Marrs and Kaufman, 1991; Anderson et al., 1999). This destabilization is likely induced through the Lhcb1*4 5′-UTR (Anderson et al., 1999).
In a few cases, photosynthetic signals have been shown to have profound effects on the translational regulation of nuclear-encoded genes. Darkness or treatment with DCMU induces a rapid (within 20 min) and reversible dissociation of Fed-1 mRNA from polyribosomes, with a concomitant decline in the in vivo translation rates of Fed-1 mRNA (Petracek et al., 1998; Hansen et al., 2001). This regulation is conferred by sequences within the 5′-UTR and the first one-third of the Fed-1 coding sequence (Petracek et al., 1997). Similarly, in spinach (Spinacia oleracea), polyribosome associations of photosynthetic photosystem A subunit D (PsaD), photosystem A subunit F (PsaF), and photosystem A subunit L (PsaL) mRNAs all decline in response to DCMU or dark treatment (Sherameti et al., 2002), suggesting regulation of translation by light. Mutational analysis of the PsaD mRNA showed a critical role of the 5′-UTR in this response (Sherameti et al., 2002). Dark treatment of maize (Zea mays) or amaranth results in the translational inhibition of Cat2 and RbcS, respectively (Berry et al., 1986, 1988; Skadsen and Scandalios, 1987). However, unlike Fed-1, PsaD, PsaF, and PsaL mRNAs, neither Cat2 nor RbcS mRNAs dissociate from polyribosomes, suggesting an inhibition of translational elongation rather than initiation. Dark-induced expression of the ATB2 transcription factor mRNA is inhibited, probably at the level of translation, in the presence of 25 mm Suc (Rook et al., 1998). Finally, light-regulated translation has been implicated in the coordination of gene expression between mesophyll and bundle sheath cells in C4 plants (Giglioli-Guivarc'h et al., 1996; McCormac et al., 1997).
Little is known about the mechanisms by which light regulates cytoplasmic mRNA translation. However, recent results suggest the striking possibility that translation of the rye (Secale cereale) Cat1 mRNA may occur via direct modification of the mRNA. Rye Cat1 mRNA isolated from light-exposed leaves was translated more efficiently in vitro than Cat1 mRNA isolated from dark-exposed leaves (Schmidt et al., 2002). The presence of the methylation inhibitors cyclo-Leu and phosphinothricin prevented the enhanced translation of Cat1 mRNA isolated from light-exposed leaves, suggesting that methylation of the Cat1 mRNA may be involved.
Here, we discuss the isolation of endogenous tobacco mRNAs whose abundance is either repressed or enhanced in the presence of DCMU. Strikingly, many of these mRNAs identified encode proteins involved in photosynthesis, suggesting that photosynthesis may regulate the mRNA levels of many genes involved in photosynthesis. In addition, we investigated the possibility that the mRNAs regulated at the level of abundance are also regulated at the level of translation. We examined the polyribosome loading profile of each of these mRNAs in the light and after a short dark treatment. A subset of the identified mRNAs showed substantial and rapid dark-induced dissociation from polyribosomes, suggesting that light has a substantial impact on the translation of many nuclear mRNAs encoding proteins involved in photosynthesis.
RESULTS
Isolation of Tobacco Leaf mRNAs Less Abundant in the Presence of Photosynthetic Electron Transport Inhibitor DCMU
Previously, we showed that the abundance of Fed-1 mRNA in transgenic tobacco is regulated by photosynthesis. Upon shift from dark to light, Fed-1 mRNA abundance increases 3- to 4-fold, and this increase is blocked by the addition of DCMU (Petracek et al., 1997). To identify similarly regulated endogenous tobacco mRNAs, we constructed a suppressive-subtractive cDNA library. For isolation of tester and driver cDNAs, we treated tobacco plants with 1% (v/v) ethanol or with DCMU in 1% (v/v) ethanol, respectively, wrapped the plant containers in foil for a 40-h dark treatment, and then re-exposed the plants to light for 6 h. PCR was used to amplify mRNAs enriched in the light versus DCMU plus light, yielding plasmids containing 146 partial cDNAs. These 146 cDNAs were transferred to a nylon membrane and hybridized with 32P-labeled light-enriched (forward-subtracted) or DCMU-enriched (reverse-subtracted) PCR-amplified cDNA. This screen identified 26 cDNAs that showed greater hybridization to the forward-subtracted than the reverse-subtracted probe, suggesting that these mRNAs display increased accumulation in light versus light plus DCMU. Sequence analysis of these 26 cDNAs identified 22 different cDNAs. Thiazole biosynthetic enzyme (THI1) was isolated three times, and α-tubulin (TUBA3) and light-harvesting complex (LHC-I) were each isolated twice. The remaining 19 mRNAs were identified as unique cDNAs, suggesting that this screen was not saturated.
We used northern-blot analysis as an independent method to determine whether photosynthesis regulates the abundance of the identified mRNAs. Total RNA was extracted from plants transferred from darkness to light in the absence or presence of DCMU, transferred to nylon membrane, and hybridized with cDNA-specific probes. Examples of these analyses are shown in Figure 1, and a summary of the quantitative analyses is presented in Table I. Three cDNAs (not listed in Table I) were identified as differentially regulated in the secondary screen but were not detectable by northern analysis and thus were not analyzed further (pSK175, a putative 9-cis-epoxycarotenoid dioxygenase 4 [NCED1], and pSKD70). Of the detectable mRNAs, five showed a 3-fold or greater accumulation, and 14 showed at least a 1.4-fold greater accumulation in light versus light plus DCMU (Table I). Thus, this screen successfully identified mRNAs whose abundance is regulated by light in the absence, but not the presence, of DCMU.
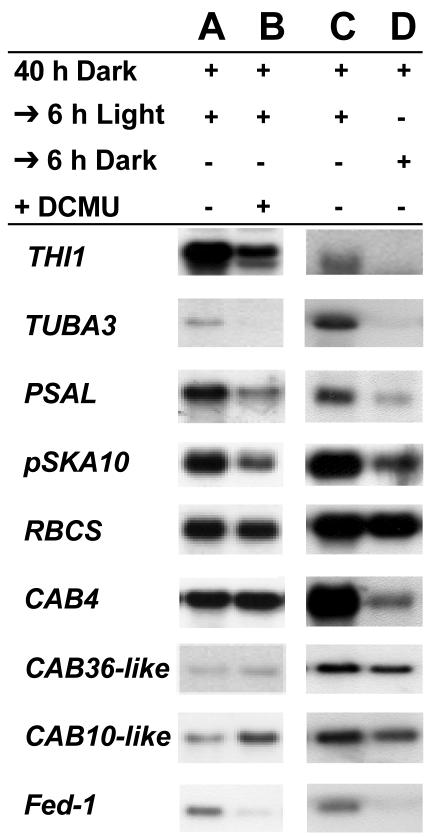
Figure 1.
mRNA abundance of differentially selected mRNAs in the presence of DCMU. Tobacco seedlings grown on plates containing 1.5% (w/v) agar were treated with 1% (v/v) ethanol (A) or DCMU/1% (v/v) ethanol (B), or not treated (C and D), placed in the dark for 40 h, and then re-exposed to the light for an additional 6 h (A-C) or kept in the dark for an additional 6 h (D). Five micrograms of total RNA were isolated from a pooled sample containing at least 10 plants, resolved by gel electrophoresis, blotted, and probed with 32P-labeled antisense probes to the indicated mRNAs. A summary of the analysis for all differentially selected mRNAs, repeated with at least three separate hybridization experiments, is presented in Table I.
Table I.
The effects of light, dark, and inhibition of photosynthesis by DCMU on tobacco mRNA accumulation
Tobacco plants were put in the dark for 40 h and then re-exposed to light for 6 h in the absence (40D→6 L) or presence (40D→6 L + DCMU) of DCMU or left in the dark for an additional 6 h (40D→6D). Ratios for mRNA abundance for 40D→6 L:40D→6 L + DCMU and 40D→6 L:40D→6D are followed by ses and were calculated by combining phosphor imager results from at least three separate experiments. The statistical significance of the differences in the ratios of 40D→6 L:40D→6 L + DCMU and 40D→6 L:40D→6D and for the differences in the ratios of 40D→6 L:40D→6D and run-on 40D→6 L:40D→6D were determined by using the two-tailed two independent samples t test (*, P < 0.05; **, P < 0.01). Chloroplast signal peptides were predicted using TargetP (http://www.cbs.dtu.dk/services/TargetP/).
| Putative Gene Name | 40D→6 L:40D→6 L + DCMU | 40D→6 L: 40D→6D | Run-On 40D→6 L:40D→6D | Chloroplast Signal Peptide |
|---|---|---|---|---|
| LHC-I | 6.8 ± 1.2 | 6.2 ± 1.3 | Yes | |
| Light harvesting complex I | ||||
| THII | 6.1 ± 0.7 | 6.2 ± 0.4 | 4.3 ± 0.8 | Yes |
| Thiazole biosynthetic enzyme | ||||
| TUBA3 | 3.5 ± 0.4 | 4.7 ± 0.9 | 2.2 ± 0.4 | No |
| Alpha tubulin | ||||
| pSKA10 | 3.1 ± 1.4 | 2.8 ± 0.3 | 1.6 ± 0.4 | ND |
| unknown | ||||
| RPL29 | 3.0 ± 0.4 | 8.0 ± 2.0 | Yes | |
| Ribosomal protein L29 | ||||
| PER | 2.6 ± 0.8 | 3.6 ± 0.2 | No | |
| Peroxidase | ||||
| CP12 | 2.4 ± 0.9 | 6.9 ± 1.0* | Yes | |
| Chloroplast Protein 12 | ||||
| PSII-X | 2.1 ± 0.3 | 6.2 ± 0.7* | Yes | |
| Photosystem II subunit X | ||||
| RBCS | 1.8 ± 0.5 | 2.0 ± 0.0 | 12.3 ± 1.7* | Yes |
| Small subunit Rubisco | ||||
| PSAL | 1.7 ± 0.4 | 2.9 ± 0.8 | 1.1 ± 0.1 | Yes |
| Photosystem A subunit L | ||||
| PSAK | 1.7 ± 0.2 | 3.0 ± 0.5 | Yes | |
| Photosystem A subunit K | ||||
| PSAF | 1.7 ± 0.3 | 2.1 ± 0.3 | Yes | |
| Photosystem A subunit F | ||||
| OEC 33 kDa | 1.4 ± 0.1 | 1.2 ± 0.1 | Yes | |
| Oxygen evolving complex 33kDa subunit | ||||
| CAT-1 | 1.4 ± 0.2 | 1.3 ± 0.1 | No | |
| Catalase I | ||||
| Annexin | 1.2 ± 0.1 | 2.5 ± 0.2* | No | |
| CAB16 | 1.1 ± 0.1 | 5.9 ± 0.2** | Yes | |
| Light harvesting complex B | ||||
| CAB4 | 0.9 ± 0.3 | 3.8 ± 1.0 | Yes | |
| Light harvesting complex B | ||||
| CAB10-like | 0.8 ± 0.1 | 1.8 ± 0.8 | Yes | |
| Light harvesting complex B | ||||
| CAB36-like | 0.7 ± 0.0 | 3.8 ± 1.0 | Yes | |
| Light harvesting complex B |
Nine of the predicted proteins (LHC-I, ribosomal protein L29 [RPL29], CP12, photosystem II subunit X [PSII-X], PSAL, photosystem A subunit K [PSAK], PSAF, and the 33-kD precursor protein of oxygen-evolving complex [OEC 33 kDa]) contain a predicted chloroplast signal peptide (Table I) and localize to the chloroplast (Palomares et al., 1991; Obokata et al., 1993; Belanger et al., 1995; Kim et al., 1996; Pohlmeyer et al., 1996; Zhao et al., 1999; Haldrup et al., 2000). NCED1 also is predicted to carry a chloroplast signal peptide, suggesting it may function in the chloroplast. Four of the identified mRNAs (TUBA3, PER, CAT-1, and Annexin) encode proteins (α tubulin, Nicotiana tabacum secretory peroxidase, SR1 catalase, and annexin) that are not implicated in functioning in the chloroplast, but both Nicotiana tabacum secretory peroxidase and SR1 catalase have roles in stress responses. The previously unknown pSKA10 cDNA is a partial cDNA, with the predicted protein displaying limited homology to two Arabidopsis proteins (At5g37540, nucleoid DNA binding protein; At1g66180, unknown protein), the first of which is predicted to localize to the chloroplast. The striking enrichment for chloroplast-localized proteins in this screen suggests that light/photosynthesis may regulate the abundance of many mRNAs encoding chloroplast-localized proteins.
Four of the identified cDNAs (CAB16, CAB4, CAB10-like, and CAB36-like) represent light-harvesting complex II (B) mRNAs and were named in relation to genes already in GenBank. Northern-blot analyses (Fig. 1; Table I) indicate that DCMU has either little or no effect on the abundance of these mRNAs, or in some cases, DCMU may even slightly increase their abundance. Although such a category of mRNAs was unexpected based on our experimental design, these results are similar to previously published reports on the lack of effect of DCMU on CAB mRNA levels (Escoubas et al., 1995; Petracek et al., 1997). Notably, all four of these CAB mRNAs show a decreased abundance in response to darkness (Table I). Together, these data suggest that the dark-induced decrease in abundance of these CAB mRNAs is in response to a signal that is not directly related to the cessation of photosynthesis, such as a photoreceptor response.
Light regulation of mRNA abundance can occur either via regulation of promoter activity (transcriptional regulation) and/or regulation of mRNA stability (posttranscriptional regulation). We used nuclear run-on assays to determine whether light altered the rates of transcription of the genes we identified in our screen. Five-week-old 35S:Fed-1 transgenic tobacco plants were transferred to the dark for 40 h and then either brought to the light or left in the dark for an additional 6 h. Nuclei were isolated, and run-on nuclear RNAs were labeled with 32P and hybridized to each of the differentially expressed cDNAs. Run-on analysis of many of these RNAs was not possible because the signals were not sufficiently above background levels of hybridization to vector only (pKS-; Fig. 2) and salmon sperm DNA (S. Bhat, data not shown). However, five endogenous tobacco mRNAs and the Fed-1 RNA expressed from the transgene were clearly detectable above background (Fig. 2). These assays indicate that Fed-1 transcription rates are the same in the light and the dark (Fig. 2), whereas Fed-1 mRNA shows a 4-fold increase in the light compared with the dark (Fig. 1), consistent with our observation that dark results in a decreased half-life of Fed-1 mRNA (Petracek et al., 1998). Of the five experimental mRNAs, TUBA3 and PSAL (and perhaps pSKA10) showed a slightly greater difference in steady-state mRNA accumulation than in transcriptional run-on assays after a 40-h dark + 6-h light treatment compared with a 46-h dark treatment (Table I; Figs. 1 and 2). Thus, the differential accumulation of these mRNAs is, at least in part, posttranscriptionally regulated.
Figure 2.
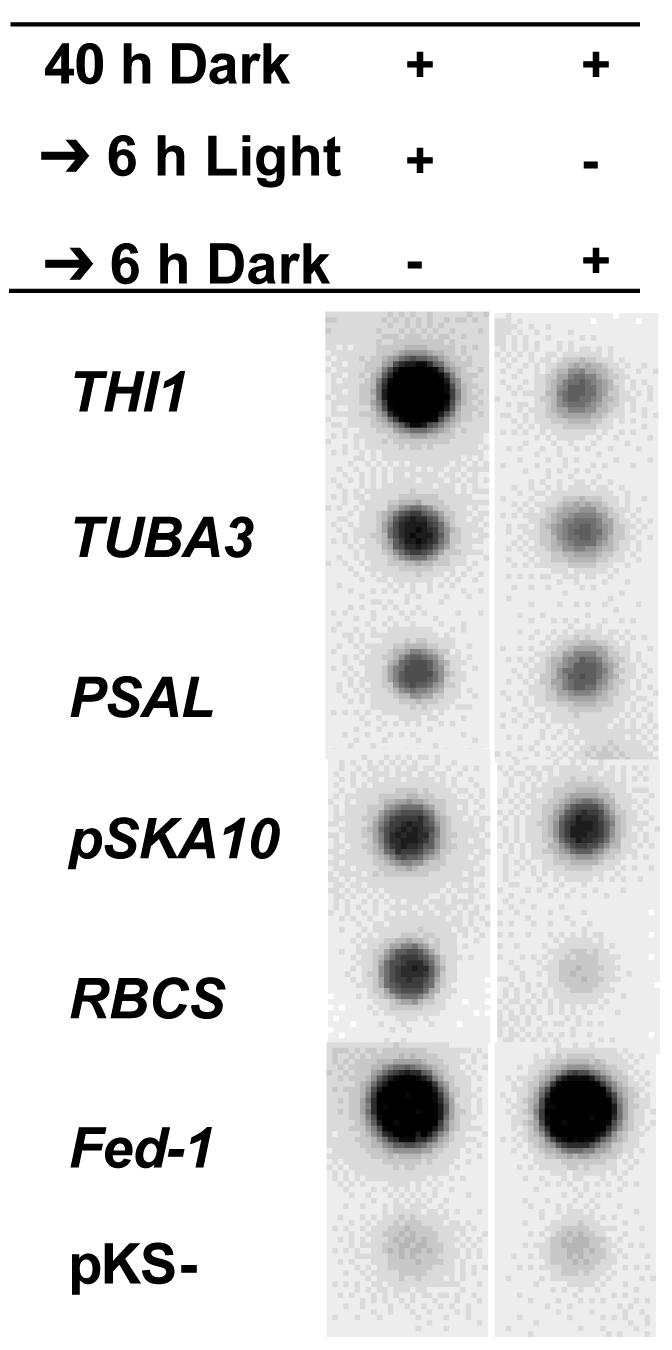
Nuclear run-on analysis of differentially selected cDNAs. Nuclei were isolated from soil-grown plants treated with 46 or 40 h of darkness followed by 6 h of light. [32P]UTP nuclear run-on RNAs were then hybridized to dot blots of plasmids carrying the indicated differentially selected cDNAs. Each hybridization was repeated at least three independent times, and the data presented are the results from one representative blot. Hybridization was quantified using phosphor imager analysis, and quantitative results are presented in Table I.
As expected, we also identified genes that appear to be mainly regulated at the level of transcription by light. THI1 mRNA showed similar differences in run-on assays compared with steady-state mRNA accumulation between the light and dark, suggesting transcriptional control of THI1. In strong contrast to the other mRNAs, RBCS mRNA showed a dramatic 13-fold higher rate of transcription in the light compared with in the dark, but only a 2-fold difference in steady-state mRNA accumulation (P < 0.05; Table I; Fig. 2). These results suggest that transcription of RBCS mRNA is higher in the light but that the mRNA is destabilized in the light or stabilized in the dark, resulting in counterbalancing effects on RBCS mRNA accumulation.
Dark-Induced Polyribosome Dissociation of Differentially Regulated mRNAs
In transgenic tobacco, Fed-1 mRNA expression is responsive to the dark at the levels of both abundance and translation (Dickey et al., 1992; Petracek et al., 1997). Fed-1 mRNA dissociates from polyribosomes within 20 min of a transfer to dark and then remains dissociated for the duration of the dark treatment including 46 h of darkness (Dickey et al., 1998; Hansen et al., 2001). Furthermore, transfer of plants back to the light results in reassociation of the Fed-1 mRNA within 30 min (Hansen et al., 2001). Measurement of cellular mRNA polyribosome association by UV absorbance suggests that a dark period of 20 min or less results in a small decline in translation that may either be specific to a subset of mRNAs or may represent relatively equivalent declines in the translation of all mRNAs (Hansen et al., 2001). Dark treatment extending to 46 h results in continued decreases in overall protein synthesis (Dickey et al., 1998; Hansen et al., 2001).
To help define the mRNAs involved in the most rapid translational response to darkness, we asked whether the mRNAs we identified were represented among the mRNAs that rapidly dissociate from polyribosomes after a short dark treatment using polyribosome profile analyses. Leaves from 6-week-old tobacco grown in soil on a 12-h-light/12-h-dark cycle were harvested 6 h into the light cycle or after being transferred to the dark for 15 min. Crude extracts were resolved on Suc gradients, fractions were collected, and RNA was isolated and subjected to northern-blot analysis. During fractionation, the total UV254 absorbance profiles were recorded, allowing detection of RNA in polyribosomes (fractions approximately 8-12) and the free 80S monosome (Fig. 3A). As seen previously for a 20-min dark treatment (Hansen et al., 2001), after a 15-min dark treatment, we observed a small but reproducible decline in the overall fraction of RNA in the polyribosomes along with a concurrent increase in the 80S monosome peak (Suc gradient fraction 5) and the nontranslated mRNP fractions (Suc gradient fractions 1-4; Fig. 3A).
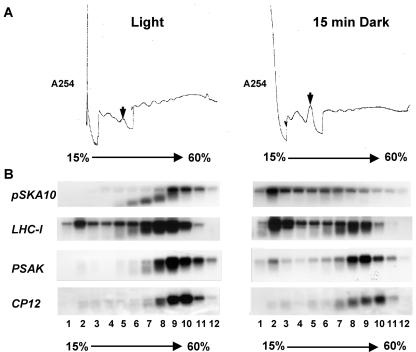
Figure 3.
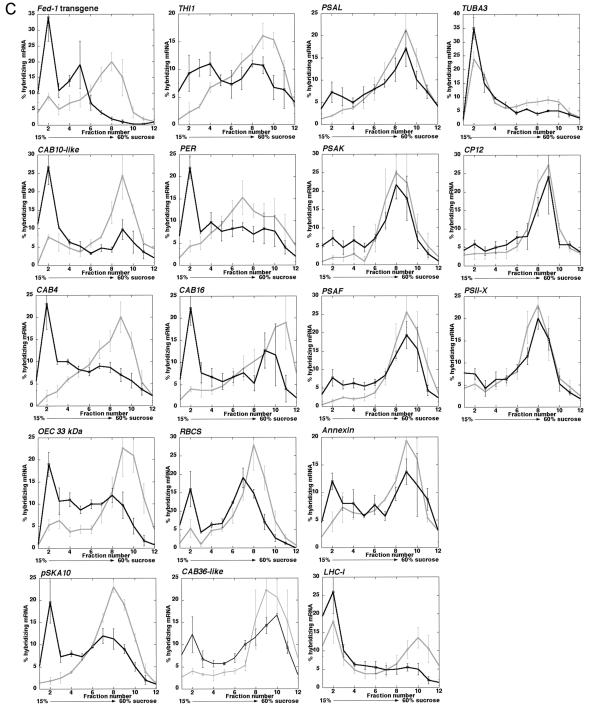
Polyribosome analysis of differentially selected endogenous tobacco mRNAs. Tobacco plants were grown on a 12-h-light/12-h-dark cycle in soil. Plants were transferred from the light to darkness for 15 min and harvested into liquid N2. Extracts were resolved on a 15% to 60% (w/v) Suc gradient, and RNA from each fraction were resolved by gel electrophoresis, blotted, and probed with antisense 32P-labeled RNA to the indicated cDNAs. A, Absorbance UV254 (A254) profile of tobacco Suc gradients. Extracts of tobacco plants harvested in the light or after 15 min of darkness were resolved on Suc gradients and fractionated. UV254 profiles of the gradient from top to bottom are presented from left to right and are aligned over the gradient fractions to indicate the relative position in the gradient for hybridizing mRNA. B, Examples of northern-blot analyses showing Suc gradient fractions from light- and 15-min-dark-treated plants hybridized with mRNAs showing varying degrees of dark-induced polyribosome dissociation. C, Graphical analysis of polyribosomal RNA hybridization results. Hybridizing mRNA levels were quantified by phosphor imager analysis. Each graph represents the results from three separate experiments, with se bars shown. The dark line represents RNA levels in the fraction from dark-treated plants, and the light-gray line represents RNA levels from light-grown plants. Fractions from left to right are taken from the top to the bottom of the gradient with fraction 5 being the approximate location of the monosome peak and polyribosomes sedimenting in fractions 6 to 12.
The mRNAs we identified have different levels of reduced polyribosome dissociation after a 15-min dark treatment (Fig. 3, B and C; Table II). Figure 3B shows northern-blot hybridization of four representative mRNAs covering the range of dark-induced polyribosome dissociation, from the greatest to the least dissociation response. Figure 3C shows the quantitation of specific RNAs in the Suc gradient fractions. The gene-specific mRNAs showing the greatest to the least degree of polyribosome dissociation are arranged from the left to the right. All mRNAs examined after a short 15-min dark treatment show at least a slight dissociation from polyribosomes, with a shift primarily to fraction 2 (Fig. 3C; Table II). Furthermore, the degree of polyribosome dissociation in the dark seems to be independent of the level of initial polyribosome association in the light (e.g. compare PSAK and LHCI in Fig. 3B; Table II). By comparison with the overall UV profile (Fig. 3A), fraction 2 is likely to represent non-translated mRNP complexes, and an increase of many mRNAs in these fractions with a concomitant decrease in the polyribosome fractions suggests that there is a rapid but modest overall decrease of total translation in response to dark. Despite this general trend, the mRNAs show strikingly different degrees of response. Using a one-tailed two independent sample t test, we asked whether the observed increases in fraction 2 hybridizing mRNA in the dark or the decreases in polyribosomal hybridizing mRNA were statistically significant. In the dark, 15 of the mRNAs showed a statistically significant decrease in polyribosome association and eight of the mRNAs show statistically significant increases in fraction 2 hybridizing mRNA (Table II). Some mRNAs respond similarly to the transgenic Fed-1 mRNA (e.g. CAB10-like, CAB4, OEC 33 kDa, and pSKA10). These mRNAs exhibit a ≥2-fold dissociation from polyribosomes (P < 0.01; Table II) with a concomitant ≥3.5-fold increase in fraction 2 (P < 0.05; Table II), whereas others exhibit only a minimal increase in fraction 2 hybridizing mRNA in the dark (e.g. TUBA3, CP12, and PSII-X). These results indicate that, like Fed-1, a subset of endogenous tobacco mRNAs shows a rapid and dramatic decrease in translation in response to dark.
Table II.
Light/dark effects on polyribosomal association of tobacco mRNAs
Light (L) and 15-min Dark (D) plants were treated as described in `Materials and Methods.' Total hybridizing mRNA was measured by phosphor imager analysis and percentage of the total was calculated for each fraction. The percentage of total hybridizing RNA found in non-polyribosomal fraction 2 and polyribosomal fractions 8 to 11 was calculated for each mRNA by combining data from at least three experiments performed on different days, and the ratio of D:L % hybridizing mRNA for fraction 2 and L:D % hybridizing polyribosomal mRNA was calculated. We used the one-tailed two independent samples t test to determine whether hybridizing mRNA in polyribosomal fractions was significantly reduced in the dark and fraction 2 mRNA was significantly increased in the dark, (*, P < 0.05; **, P < 0.01).
| mRNA | %mRNA in Light in Fraction 2 ± se | % mRNA in 15-min Dark in Fraction 2 ± se | % mRNA in Light in Polyribosomal Fractions 8-11 ± se | % mRNA in Dark in Polyribosomal Fractions 8-11 ± se | D:L of Total Hybridizing RNA in Fraction 2 | L:D of Total Hybridizing RNA in Polyribosomal Fractions 8-11 |
|---|---|---|---|---|---|---|
| Fed-1 | 9.0 ± 1.1 | 34.0 ± 7.5* | 41.0 ± 2.8 | 3.6 ± 0.3** | 3.8 | 11.4 |
| CAB 10-like | 7.7 ± 0.7 | 26.7 ± 4.8* | 59.9 ± 3.2 | 24.0 ± 2.1** | 3.5 | 3.5 |
| CAB4 | 2.3 ± 1.3 | 10.0 ± 0.8* | 54.7 ± 2.6 | 25.1 ± 1.4** | 10.0 | 2.2 |
| OEC 33 kDa | 5.3 ± 1.2 | 19.0 ± 2.7* | 67.7 ± 4.3 | 28.4 ± 1.9** | 3.6 | 2.3 |
| pSKA10 | 1.8 ± 0.8 | 16.2 ± 4.9* | 58.3 ± 1.4 | 27.3 ± 2.0** | 8.9 | 2.1 |
| THI1 | 2.3 ± 1.7 | 9.3 ± 3.3 | 53.6 ± 1.5 | 34.7 ± 1.9** | 4.0 | 1.5 |
| PER | 4.3 ± 1.3 | 22.0 ± 2.5* | 42.3 ± 10.6 | 27.0 ± 6.5 | 5.1 | 1.6 |
| CAB16 | 4.0 ± 0.6 | 22.3 ± 3.9* | 59.3 ± 3.1 | 33.7 ± 3.9** | 5.6 | 1.8 |
| RBCS | 5.3 ± 2.4 | 16.0 ± 4.8 | 57.0 ± 3.5 | 25.7 ± 1.2** | 3.0 | 2.2 |
| CAB36-like | 4.0 ± 0.6 | 12.3 ± 4.0 | 74.0 ± 6.6 | 51.7 ± 1.3* | 3.1 | 1.4 |
| PSAL | 2.0 ± 0.0 | 7.3 ± 2.3 | 59.3 ± 2.2 | 46.6 ± 2.5** | 3.7 | 1.3 |
| PSAK | 2.0 ± 0.6 | 7.3 ± 2.0 | 61.3 ± 5.9 | 50.4 ± 4.3 | 3.7 | 1.2 |
| PSAF | 1.5 ± 0.5 | 7.8 ± 2.3* | 75.3 ± 4.9 | 52.8 ± 3.0** | 5.2 | 1.4 |
| Annexin | 4.7 ± 1.8 | 12.1 ± 1.5 | 53.1 ± 2.7 | 43.0 ± 2.8* | 2.6 | 1.2 |
| LHCI | 18.0 ± 6.0 | 26.0 ± 5.8 | 40.8 ± 2.8 | 7.5 ± 1.3** | 1.4 | 2.3 |
| TUB3A | 23.7 ± 2.7 | 35.0 ± 2.7 | 29.9 ± 1.3 | 17.7 ± 1.4** | 1.5 | 1.7 |
| CP12 | 3.3 ± 1.5 | 6.0 ± 1.2 | 64.3 ± 3.0 | 52.4 ± 3.6* | 1.8 | 1.2 |
| PSII-X | 5.3 ± 1.3 | 7.5 ± 1.2 | 49.5 ± 3.0 | 43.9 ± 1.8 | 1.4 | 1.1 |
DISCUSSION
We have identified endogenous tobacco nuclear-encoded mRNAs whose accumulation and/or translation is sensitive to photosynthesis. Photosynthesis regulates these mRNAs (to varying degrees) at the levels of transcription, mRNA stability, and translation. Surprisingly, it appears that these modes of regulation are sometimes in opposition to each other. As has been observed for the handful of previously studied nuclear-encoded mRNAs (Pfannschmidt, 2003), most of the DCMU-sensitive mRNAs we identified encode chloroplast-targeted proteins or stress-related proteins, suggesting a common signaling mechanism that regulates the production of these types of proteins in response to the quality and quantity of light (Table I).
Translational Control by Light
All of the mRNAs isolated in this screen show at least a small decrease in accumulation in polyribosomal fractions in response to a short dark treatment (Fig. 3C; Table I). These data are consistent with our previous studies in which we observed an overall decrease in polyribosomes within 15 to 20 min in the dark, with increased time in the dark resulting in further dissociation of mRNAs from polyribosomes (Hansen et al., 2001). The decreased fraction of total mRNA in polyribosomes in the dark was paralleled by a decrease in overall translation, as measured by [35S]Met/Cys incorporation into total cellular proteins (Hansen et al., 2001). For Fed-1 mRNA, we found that dark-induced decreases in polyribosome recruitment results in decreased translation (Hansen et al., 2001). Similar observations have been made correlating decreased polyribosome loading and decreased translation in vivo (e.g. GCN4 [Tzamarias et al., 1989]; for review, see Mathews et al., 2000). Thus, the simplest interpretation of the data presented in Figure 3C and Table II is that, to varying degrees, each mRNA is less efficiently recruited to polyribosomes in the dark. It is also possible that additional layers of translational regulation may occur for some of these genes. For example, mRNA that remains associated with polyribosomes in the light or in the dark could be inhibited at the level of translational elongation or termination (Vayda, 1995; Kim et al., 2003). Additional analyses of the individual genes will be needed to assess these possibilities.
Whether or not the decreased polyribosome recruitment for the individual mRNAs is via the same mechanism or different mechanisms remains to be determined. One possibility is that the activity of a general factor involved in translation may be regulated by dark. For example, perhaps dark regulates the association of eIF3e with a component of the COP9 signalosome (Yahalom et al., 2001), affecting translation of all or nearly all nuclear mRNAs. However, it seems likely that because our survey includes only mRNAs isolated in the differential screen, our results may be biased for mRNAs that exhibit light-regulated translation. Alternatively, it is possible that the observed moderate overall decline in polyribosome association could reflect the dark-regulated translation of only a subset of mRNAs (perhaps mainly mRNAs with roles in photosynthesis), if this subset includes several very highly expressed mRNAs. Consistent with this possibility, we have identified dark-induced polyribosome dissociation of members of the abundant Cab family. It will be important in the future to determine how general this dark-induced translational response is and to identify the mechanisms involved. In addition, there may be mRNAs that become associated with polyribosomes in the dark. Microarray analysis would appear to be a fruitful approach to answer these important questions.
Our data indicate that some mRNAs show a much more dramatic decline than other mRNAs in mRNA levels and/or shift off polyribosomes in response to a 15-min dark treatment (`Fig. 3, B and C). Thus, it is likely that there are also mRNA-specific dark-induced translational and mRNA abundance responses. Most of the nuclear mRNAs we identified encode proteins targeted to the chloroplast, strongly suggesting that many mRNAs of this category are dramatically regulated by dark and photosynthesis at the levels of mRNA abundance and, often, translation. The functional significance of this observation will require identification of the signaling pathways involved and the mechanisms of regulation of mRNA abundance and translation by light.
Related genes from different plant species may have different responses to dark or decreased photosynthesis. In transgenic tobacco, both the pea and tobacco ferredoxin genes show light-regulated translation (Hansen et al., 2001). Recent data from our lab suggest that in transgenic Arabidopsis, the pea ferredoxin (Fed-1) mRNA displays light-regulated translation (M.E. Petracek, unpublished data), whereas the endogenous ferredoxin (FEDA) mRNA does not (Hansen et al., 2001; M.E. Petracek, unpublished data]. Thus, it seems clear that although many mRNAs encoding proteins that localize to the chloroplast are regulated by light at the levels of mRNA abundance and at the level of translation, the response of any individual mRNA must be determined on a case-by-case basis.
That a number of mRNAs are considerably more translationally sensitive to dark than others suggests the presence of a common mechanism and, likely, common sequence elements mediate the conserved translational response. However, analysis of 5′-UTRs of this category of mRNAs does not reveal any clear conserved elements or structural patterns (data not shown). This suggests that the regulatory motifs may be complex and bipartite and/or involve sequences in addition to, or other than, those within the 5′-UTR.
Possible Signaling Mechanisms from the Chloroplast to the Cytoplasm
The pea Fed-1 mRNA in transgenic tobacco shows similar posttranscriptional regulation both by dark and by DCMU in the light (Petracek et al., 1997). However, the abundance of some mRNAs identified here are regulated by dark but not DCMU (Table I), suggesting the existence of both photosynthesis and non-photosynthesis pathways of regulation of mRNA abundance. Four of the mRNAs (CP12, P < 0.05; PSII-X, P < 0.05; Annexin, P < 0.05; and CAB16, P < 0.01) show significant differences in the light to dark and light to DCMU abundance ratios (Table I). It is notable, however, that all of the DCMU-responsive mRNAs are also regulated by dark. This suggests the DCMU response pathway is a subset of the dark-response pathway. However, because many of the mRNAs do not show significant differences in light to dark and light to DCMU abundance ratios (Table I), it is possible that in green plants, the primary control of gene regulation may be through photosynthetic activity. However, whether these accumulation patterns occur via different or overlapping signaling pathways remains to be determined.
The mechanism(s) for photosynthetic regulation of nuclear gene expression is yet unclear, but it likely involves signals from the chloroplast. A number of molecules have been previously implicated in chloroplast-mediated signaling and are potential candidates for signaling pathways. First, a small redox molecule, such as oxygen, could diffuse across the chloroplast membranes and alter the redox state of regulatory proteins (Danon, 2002). Also, calcium fluxes have been shown to occur during light-to-dark transitions, and following DCMU treatment in the light, which may have a role in signal transduction (Sai and Johnson, 2002). Similarly, an NAD(P) H/NAD(P)-signaling pathway has been identified which has a role in import of chloroplast-localized proteins (Kuchler et al., 2002). Last, tetrapyrroles synthesized within the chloroplast may be specifically transported out of the chloroplast in response to light (for review, see Surpin et al., 2002). It is notable that tetrapyrroles are the precursors to heme molecules, which are required for in vitro translation of hemin-binding protein Cat-1 mRNA in rye leaves (Schmidt et al., 2002). It is possible that one or more of these pathways are involved in light-regulated translation and abundance of the mRNAs we have identified and (likely) other mRNAs.
How could chloroplastic signals alter mRNA stability or translation? First, mRNA binding proteins recognize a subset of mRNAs, and binding of these proteins to their target mRNAs may regulate their stability and translation. If the interaction of these mRNA-binding proteins is regulated by a photosynthesis-responsive, posttranslational modification (e.g. redox changes, phosphorylation, or methylation), then photosynthesis could affect mRNA abundance and/or translation. Alternatively, mRNA-specific binding proteins may not mediate regulation of mRNA abundance. Rather, perhaps photosynthetic metabolites, such as NADPH or DHAP, directly bind target mRNAs in a light-responsive manner, resulting in an altered structure of the mRNA resulting in regulation of mRNA abundance and/or translation. Consistent with this idea, in bacteria, signaling molecules and metabolites can bind directly and specifically to target mRNAs, resulting in regulation of gene expression (Mironov et al., 2002; Nahvi et al., 2002; Winkler et al., 2002). Similarly, in plants, thiamin biosynthetic enzyme and thiazole biosynthetic protein have been identified as targets for thioredoxin in the chloroplast (Balmer et al., 2003). This is especially interesting in light of the clear regulation of the Thi1 mRNA by DCMU and dark (Table I; Fig. 3C) and the ability of thiamin to bind mRNA in bacteria. It is possible that in plants, redox sensing in the chloroplast is signaled through thiamin interactions with chloroplast and/or cytosolic mRNAs, although export of thiamin from the chloroplast has not been demonstrated. Finally, as for Cat-1 mRNA whose translational response to light may be mediated by methylation of the Cat-1 mRNA (Schmidt et al., 2002), it is possible that the regulation of abundance or translation of the mRNAs we identified may be regulated by covalent modification of the mRNAs.
MATERIALS AND METHODS
Plant Growth
Tobacco seedlings were germinated in sterile Magenta boxes with membrane rafts (V8380 and M1917, Sigma-Aldrich, St. Louis) in sterile Murashige and Skoog medium (Invitrogen, Grand Island, NY) for the PCR-select library or on 0.5× Murashige and Skoog/1.5% (w/v) agarose petri plates for subsequent DCMU or light/dark mRNA abundance analysis, as described (Petracek et al., 1997, 1998). Seedlings were grown for 3 weeks in a growth chamber at 22°C using a 12-h-light/12-h-dark cycle with 12 fluorescent and six incandescent lamps to give a light fluence of approximately 240 μmol m-2 s-1 between 380 and 780 nm. For dark treatments, plants grown in Magenta boxes or petri plates were wrapped in foil for the indicated length of time before harvest or after being unwrapped and re-exposed to the light. For polyribosome analysis, plants were grown in soil or on petri plates as described (Petracek et al., 1997) and either harvested during the light cycle or transferred to a dark room for 15 min before harvesting under a green safelight. Control experiments in which the light plant was shaken for the same length of time as the plant being transferred to the dark room indicated shaking caused no change in the hybridization pattern of the plant exposed to light (M.E. Petracek, unpublished data).
DCMU Treatment
Plants were treated with DCMU as described (Petracek et al., 1997). In brief, a 100 mm DCMU stock solution in 100% (v/v) ethanol was diluted to 1 mm in sterile water, and 3 mL was pipetted onto the roots of tobacco seedlings on membrane rafts or on 1.5% (w/v) agar plates. Control plants were treated with 3 mL of a 1% (v/v) ethanol solution. Uptake was allowed with the covers propped open for 1 h before they were wrapped in foil for a 40-h dark treatment. Plants were then brought to the light for 6 h before harvesting the leaves into liquid nitrogen and storage at -80°C.
RNA Isolation and cDNA Subtraction Library Generation
Total RNA was prepared as described (Thompson et al., 1983) with the following modifications. In brief, plant samples were homogenized with a polytron in phenol:chloroform:isoamyl alcohol (25:24:1; U.S. Biochemical, Cleveland) and RNA extraction buffer (1% [w/v] SDS [Sigma-Aldrich], 1% [w/v] tri-isopropylnaphthalene-sulfonic acid [Eastman Kodak, Rochester, NY; this chemical is no longer commercially available; we now omit this without problem], 4% [w/v] p-aminosalicyclic acid [Sigma-Aldrich], 10 mm Tris, pH 7.5, 1 mm EDTA, 2% [v/v] β-mercaptoethanol, and 1 mm aurin tricarboxylic acid [ATA; Sigma-Aldrich]). ATA was omitted from samples used for cDNA preparation. Samples were centrifuged at 11,000g for 30 min at 4°C, the supernatant was removed, and the pellet resuspended in 100 μm ATA in water or, in samples used to produce cDNA, in water alone. The resuspended RNA was then precipitated with ammonium acetate and ethanol at -80°C overnight and collected at 11,000g for 30 min at 4°C. The resulting pellet was resuspended in 100 μm ATA water or water alone. All water used was treated with DEPC.
For cDNA library preparation, mRNA was isolated from 400 μg of total tobacco leaf RNA using the MicroPoly(A) Pure kit (Ambion, Austin, TX) following the manufacturer's instructions. Approximately 2 μg of the resulting poly(A) mRNA was then used to create a PCR-select cDNA library (BD Biosciences Clontech, Palo Alto, CA) following the manufacturer's instructions, using light-treated samples as the tester cDNA and DCMU/light-treated samples as the driver cDNA. The resulting PCR samples were cloned into pGEM-T (Promega Biotech, Madison, WI) or PCR-Script Amp (Stratagene, La Jolla, CA). The resulting cDNAs were screened with the reverse-subtracted cDNAs using the PCR-Select Differential Screening kit (BD Biosciences Clontech) to identify cDNAs likely to represent mRNAs differentially expressed in light-treated versus DCMU/light-treated plants. Positive candidate cDNAs were then used as probes in northern-blot hybridization analysis (Dickey et al., 1992), and cDNAs were sequenced by the North Carolina State University sequencing facility and the Oklahoma State University DNA/Protein Core Facility.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Polyribosome Analysis
Polyribosome analyses were performed as described (Petracek et al., 1997). mRNA hybridization after northern-blot hybridization was quantified using a phosphor imager (Molecular Dynamics, Sunnyvale, CA; Bio-Rad, Hercules, CA).
Nuclei Isolation and RNA Run-On Assays
Nuclei isolation was as described by Folta and Kaufman (2000) except that nylon meshes of sizes 500, 300, 100, 50, and 20 μm were used for filtering the homogenate. Run-on transcription assays used 60 μg of the isolated nuclei incubated in 50 μL of buffer containing 50 mm Tris, pH 7.9, 10 mm MgCl2, 10% (v/v) glycerol, 500 μm each ATP, CTP, and GTP, 75 mm NH4Cl, 0.8 mm dithiothreitol, 125 μCi of [α-32P]UTP (800 Ci mmol-1) and 37.5 units of RNasin (Promega). Transcription reactions were incubated for 15 min at 27°C, and the reaction was stopped with 3 μL of a solution containing 3.3 μg μL-1 Escherichia coli tRNA and 17 mm UTP. After 2 min, 1 μL of RNase-free DNase Q (Promega) was added, and the mixture was incubated an additional 5 min. One hundred microliters of 7.5 m urea, 5% (w/v) SDS, 20 mm EDTA, 100 mm LiCl2, and 10 mm ATA were added, and the mixture extracted with an equal volume (150 μL) of phenol:chloroform: isoamyl alcohol (25:24:1). The 32P-labeled RNA was centrifuged through a Sephadex G-50 column equilibrated with 10 mm Tris, pH 8.0, 1 mm EDTA, and 100 μm ATA and added to 900 μL of 1.1× hybridization buffer (1× = 50% [v/v] formamide, 50 mm NaPO4, pH 7.0, 5× SSC, 0.2% [w/v] SDS, 5× Denhardt's solution [100× Denhardt's is 2% (w/v) each of bovine serum albumin, Ficoll, and polyvinyl pyrrolidone], 100 μg mL-1 E. coli tRNA, and 100 μg mL-1 poly(A)).
Two micrograms of each denatured cDNA-containing plasmid was dot blotted onto Gene Screen nylon membranes using a Hybri-Dot Manifold (Invitrogen). The membranes were prehybridized overnight at 42°C in 1 mL of 1.1× hybridization buffer and then probed with 32P-labeled run-on RNA for 72 h at 42°C. Membranes were washed twice in 2× SSC and 0.1% (w/v) SDS at 42°C for 15 min each, followed by three washes in 0.5× SSC and 0.1% (w/v) SDS at 68°C for 30 min each. Blots were exposed to Kodak XB-1 film with intensifying screens for 24 to 48 h at -70°C and to phosphor imager screens for quantitation.
Acknowledgments
We thank Tamyra Ravenel, Darnell Graham, Jennifer Barker, Xiao-Ping Guo, Angie Phillips, Claudia Dollins, and Sabith Erra for excellent technical assistance. We are appreciative of the help with the statistical analyses provided by Larry Claypool. Finally, we are grateful to Lynn Dickey for helpful discussions and to Mark Longtine for useful editorial comments.
This work was supported by the U.S. Department of Agriculture (grant no. 98-35301-7012 to M.E.P.) and by the Oklahoma Agricultural Experiment Station (project no. H-2427).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.029686.
References
- Alexciev K, Tullberg A (1997) Regulation of petB mRNA stability in pea chloroplasts by redox poise. Physiol Plant 99: 477-485 [Google Scholar]
- Anderson M, Folta K, Warpeha K, Gibbons J, Gao J, Kaufman L (1999) Blue light-directed destabilization of the pea Lhcb1*4 transcript depends on sequences within the 5′ untranslated region. Plant Cell 11: 1579-1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100: 370-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559-572 [DOI] [PubMed] [Google Scholar]
- Belanger FC, Leustek T, Chu B, Kriz AL (1995) Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Mol Biol 29: 809-821 [DOI] [PubMed] [Google Scholar]
- Berry JO, Carr JP, Klessig DF (1988) mRNAs encoding ribulose 1,5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedlings transferred to darkness. Proc Natl Acad Sci USA 85: 4190-4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JO, Nikolau BJ, Carr JP, Klessig DF (1986) Translational regulation of light-induced ribulose 1,5-bisphosphate carboxylase gene expression in amaranth. Mol Cell Biol 6: 2347-2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EC, Somanchi A, Mayfield SP (2001) Interorganellar crosstalk: new perspectives on signaling from the chloroplast to the nucleus. Genome Biol 2: 1021.1-1021.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, Mayfield SP (1999) Light-activated translation of chloroplast mRNAs. Trends Plant Sci 4: 190-195 [DOI] [PubMed] [Google Scholar]
- Danon A (2002) Redox reactions of regulatory proteins: do kinetics promote specificity? Trends Biochem Sci 27: 197-203 [DOI] [PubMed] [Google Scholar]
- Deshpande NN, Bao Y, Herrin DL (1997) Evidence for light/redox-regulated splicing of psbA pre-RNAs in Chlamydomonas chloroplasts. RNA 3: 37-48 [PMC free article] [PubMed] [Google Scholar]
- Dickey L, Petracek M, Nguyen T, Hansen E, Thompson W (1998) Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell 10: 475-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Gallo-Meagher M, Thompson WF (1992) Light regulatory sequences are located within the 5′ portion of the Fed-1 message sequence. EMBO J 11: 2311-2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi S, Takano H, Ono K, Takio S (2002) Photosynthetic electron transport regulates the stability of the transcript for the protochlorophyllide oxidoreductase gene in the liverwort, Marchantia paleacea var. diptera. Plant Cell Physiol 43: 573-577 [DOI] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, Laroche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92: 10237-10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Kaufman LS (2000) Preparation of transcriptionally active nuclei from etiolated Arabidopsis thaliana. Plant Cell Rep 19: 504-510 [DOI] [PubMed] [Google Scholar]
- Fong CL, Lentz A, Mayfield SP (2000) Disulfide bond formation between RNA binding domains is used to regulate mRNA binding activity of the chloroplast poly(A)-binding protein. J Biol Chem 275: 8275-8278 [DOI] [PubMed] [Google Scholar]
- Giglioli-Guivarc'h N, Pierre J-N, Brown S, Chollet R, Vidal J, Gadal P (1996) The light-dependent transduction pathway controlling the regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase in protoplasts from Digitaria sanguinalis. Plant Cell 8: 573-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177: 115-180 [DOI] [PubMed] [Google Scholar]
- Haldrup A, Simpson DJ, Scheller HV (2000) Down-regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana: the PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J Biol Chem 275: 31211-31218 [DOI] [PubMed] [Google Scholar]
- Hansen ER, Petracek ME, Dickey LF, Thompson WF (2001) The 5′ end of the pea ferredoxin-1 mRNA mediates rapid and reversible light-directed changes in translation in tobacco. Plant Physiol 125: 770-778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Webster CI, Gray JC (1997) Light-regulated expression of the pea plastocyanin gene is mediated by elements within the transcribed region of the gene. Plant J 12: 499-506 [DOI] [PubMed] [Google Scholar]
- Irihimovitch V, Shapira M (2000) Glutathione redox potential modulated by reactive oxygen species regulates translation of Rubisco large subunit in the chloroplast. J Biol Chem 275: 16289-16295 [DOI] [PubMed] [Google Scholar]
- Kim J, Mayfield SP (1997) Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278: 1954-1957 [DOI] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carre IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22: 935-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Robinson D, Robinson C (1996) An Arabidopsis thaliana cDNA encoding PS II-X, a 4.1 kDa component of photosystem II: a bipartite presequence mediates SecA/delta pH-independent targeting into thylakoids. FEBS Lett 390: 175-178 [DOI] [PubMed] [Google Scholar]
- Kubicki A, Steinmuller K, Westhoff P (1994) Differential transcription of plastome-encoded genes in the mesophyll and bundle-sheath chloroplasts of the monocotyledonous NADP-malic enzyme-type C4 plants maize and sorghum. Plant Mol Biol 25: 669-679 [DOI] [PubMed] [Google Scholar]
- Kuchler M, Decker S, Hormann F, Soll J, Heins L (2002) Protein import into chloroplasts involves redox-regulated proteins. EMBO J 21: 6136-6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulheim C, Agren J, Jansson S (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91-93 [DOI] [PubMed] [Google Scholar]
- Marrs KA, Kaufman LS (1991) Rapid transcriptional regulation of the Cab and Pea207 gene families in peas by blue-light in the absence of cytoplasmic protein-synthesis. Planta 183: 327-333 [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB (2000) Origins and principles of translational control. In JWB Hershey, ed, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 1-33
- McCormac D, Boinski JJ, Ramsperger VC, Berry JO (1997) C4 gene expression in photosynthetic and nonphotosynthetic leaf regions of Amaranthus tricolor. Plant Physiol 114: 801-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudle E (2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111: 747-756 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053-2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Susek R, Chory J (1996) An intracellular signal transduction pathway between the chloroplast and nucleus is involved in deetiolation. Plant Physiol 112: 1465-1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR (2002) Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043-1049 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, van Dillewijn J, Rahire M, Rochaix JD (1994) Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J 13: 3182-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obokata J, Mikami K, Hayashida N, Nakamura M, Sugiura M (1993) Molecular heterogeneity of photosystem I: psaD, psaE, psaF, psaH, and psaL are all present in isoforms in Nicotiana spp. Plant Physiol 102: 1259-1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmuller R (1989) Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastic enzyme levels. Photochem Photobiol 49: 229-239 [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA (2001) Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA 98: 2047-2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares R, Herrmann RG, Oelmuller R (1991) Different blue-light requirement for the accumulation of transcripts from nuclear genes for thylakoid proteins in Nicotiana tabacum and Lycopersicon esculentum. J Photochem Photobiol B Biol 11: 151-162 [DOI] [PubMed] [Google Scholar]
- Petracek M, Dickey L, Nguyen T, Gatz C, Sowinski D, Allen G, Thompson W (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci USA 95: 9009-9013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Huber SC, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9: 2291-2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Thompson WF (2000) Posttranscriptional light regulation of nuclear-encoded genes. In JK Setlow, ed, Genetic Engineering, Vol 22. Plenum Press, New York, pp 1-10 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T (2003) Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci 8: 33-41 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Tullberg A, Link G, Allen JF (1999) Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life 48: 271-276 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Schutze K, Brost M, Oelmuller R (2001) A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J Biol Chem 276: 36125-36130 [DOI] [PubMed] [Google Scholar]
- Pohlmeyer K, Paap BK, Soll J, Wedel N (1996) CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol Biol 32: 969-978 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180-188 [DOI] [PubMed] [Google Scholar]
- Rochaix JD (2001) Posttranscriptional control of chloroplast gene expression: from RNA to photosynthetic complex. Plant Physiol 125: 142-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S (2001) Pathways of plastid-to-nucleus signaling. Trends Plant Sci 6: 471-478 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253-263 [DOI] [PubMed] [Google Scholar]
- Sai J, Johnson CH (2002) Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. Plant Cell 14: 1279-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Dehne S, Feierabend J (2002) Post-transcriptional mechanisms control catalase synthesis during its light-induced turnover in rye leaves through the availability of the hemin cofactor and reversible changes of the translation efficiency of mRNA. Plant J 31: 601-613 [DOI] [PubMed] [Google Scholar]
- Shen Y, Danon A, Christopher DA (2001) RNA binding-proteins interact specifically with the Arabidopsis chloroplast psbA mRNA 5′ untranslated region in a redox-dependent manner. Plant Cell Physiol 42: 1071-1078 [DOI] [PubMed] [Google Scholar]
- Sherameti I, Nakamura M, Yamamoto YY, Pfannschmidt T, Obokata J, Oelmüller R (2002) Polyribosome loading of spinach mRNAs for photosystem I subunits is controlled by photosynthetic electron transportA crucial cis element in the spinach PsaD gene is located in the 5′-untranslated region. Plant J 32: 631-639 [DOI] [PubMed] [Google Scholar]
- Skadsen RW, Scandalios JG (1987) Translational control of photo-induced expression of the Cat2 catalase gene during leaf development in maize. Proc Natl Acad Sci USA 84: 2785-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Gray JC (2002) Multiple plastid signals regulate the expression of the pea plastocyanin gene in pea and transgenic tobacco plants. Plant J 32: 763-774 [DOI] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell 14: S327-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787-799 [DOI] [PubMed] [Google Scholar]
- Thompson WF, Everett M, Polans NO, Jorgensen RA, Palmer JD (1983) Phytochrome control of RNA levels in developing pea and mung bean leaves. Planta 158: 487-500 [DOI] [PubMed] [Google Scholar]
- Trebitsh T, Danon A (2001) Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc Natl Acad Sci USA 98: 12289-12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullberg A, Alexciev K, Pfannschmidt T, Allen JF (2000) Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol 41: 1045-1054 [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Roussou I, Thireos G (1989) Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell 57: 947-954 [DOI] [PubMed] [Google Scholar]
- Vayda ME (1995) Assessment of translational regulation by run-off translation of polysomes in vitro. In HJ Bohnert, ed, Methods in Cell Biology, Vol 50. Academic Press, San Diego, pp 349-359 [DOI] [PubMed] [Google Scholar]
- Vener AV, Ohad I, Andersson B (1998) Protein phosphorylation and redox sensing in chloroplast thylakoids. Curr Opin Plant Biol 1: 217-223 [DOI] [PubMed] [Google Scholar]
- Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, Lopez-Juez E (2000) Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J 24: 883-894 [DOI] [PubMed] [Google Scholar]
- Warpeha KMF, Marrs KA, Kaufman LS (1989) Blue-light regulation of specific transcript levels in Pisum sativum: Fluence-response, time-course, and reciprocity characteristics. Plant Physiol 91: 1030-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK (1997) CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci USA 94: 10479-10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952-956 [DOI] [PubMed] [Google Scholar]
- Yahalom A, Kim TH, Winter E, Karniol B, von Arnim AG, Chamovitz DA (2001) Arabidopsis eIF3e (INT-6) associates with both eIF3c and the COP9 signalosome subunit CSN7. J Biol Chem 276: 334-340 [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Kondo Y, Kato A, Tsuji H, Obokata J (1997) Light-responsive elements of the tobacco PSI-D gene are located both upstream and within the transcribed region. Plant J 12: 255-265 [DOI] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP (1998) A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc Natl Acad Sci USA 95: 2238-2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Xu T, Zucchi P, Bogorad L (1999) Subpopulations of chloroplast ribosomes change during photoregulated development of Zea mays leaves: ribosomal proteins L2, L21, and L29. Proc Natl Acad Sci USA 96: 8997-9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Day A (2002) Differential regulation of genes transcribed by nucleus-encoded plastid RNA polymerase, and DNA amplification, within ribosome-deficient plastids in stable phenocopies of cereal albino mutants. Mol Genet Genomics 267: 27-37 [DOI] [PubMed] [Google Scholar]