Abstract
An early auxin-induced gene was isolated from rice (Oryza sativa L. subsp. japonica cv Nihonmasari) coleoptiles by a fluorescent-labeled differential display screen. The full-length gene contains conserved domains characteristic for the cytochrome P450 superfamily. This gene, designated as CYP87A3, was weakly expressed in dark-grown coleoptiles but was up-regulated rapidly and transiently when coleoptile segments were incubated in 5 μm indole-3-acetic acid. This induction by auxin could not be suppressed by cycloheximide. Depletion of segments from endogenous auxin reduced the amount of CYP87A3 transcripts. The CYP87A3 transcript level was rapidly, although transiently, up-regulated in response to light as well. The observed pattern of gene regulation might indicate a role in the suppression of auxin-induced coleoptile growth. The role of CYP87A3 is discussed with respect to auxin signaling in the regulation of coleoptile growth.
Coleoptiles represent a classical model system to study the control of growth-related signaling, because they grow exclusively by cell elongation (Wada, 1961) and respond to environmental signals. The plant hormone auxin plays a central role in the control of coleoptile elongation. As a matter of fact, auxin was discovered as a powerful growth substance that is produced mainly in the coleoptile tip, migrates basipetally, and is crucial for the signal-dependent regulation of growth (Went, 1928).
Although the importance of basipetal auxin transport has been confirmed by a wealth of data collected over several decades (for review, see Masuda et al., 1998), the role of auxin in the signal-dependent growth response appears to be complex. As shown by recent data, auxin distribution depends on several intrinsic activities, mainly synthesis, conjugation/deconjugation, and catabolism (for review, see Normanly, 1997). Moreover, changes in auxin transport between neighboring tissues and/or local concentrations can influence growth. For instance, a correlation between growth inhibition and a decrease in the level of extractable indole-3-acetic acid (IAA) could be demonstrated in the epidermis of irradiated pea (Pisum sativum) seedlings (Behringer and Davies, 1992). In addition, sensitivity/responsiveness of the target tissue to auxin have to be considered (Trewavas, 1981). Because of this complexity, the events underlying signaling mediating auxin-triggered cell elongation are still far from being understood.
Growing evidence has appeared concerning regulation of auxin action at the level of gene expression (for example, expressional control of primary auxin-responsive genes, such as members of GH3 or Aux/IAA family; see Leyser, 2002; Hagen and Guilfoyle, 2002; Nakazawa et al., 2001). From the expression pattern of such genes, their role in signaling as well as their putative function have been inferred. We therefore ventured to isolate auxin-regulated gene(s) related to auxin-induced elongation. Using fluorescent differential display (FDD; Kuno et al., 2000), we screened for a subset of auxin-induced genes, whose expression was correlated with the auxin-induced elongation. This was accomplished by including a rice (Oryza sativa) mutant line, Yin-Yang, with an elevated auxin responsiveness of growth (Wang and Nick, 1998) as a reference system for the differential display screen. We identified several auxin-inducible cDNA fragments that were differentially regulated between wild type (WT) and mutant (Waller et al., 2002). One of these, CYP87A3, a new member of the cytochrome P450 (CYP) superfamily, was induced by auxin in the WT, but not in the mutant. In addition, this gene was light-responsive.
The CYP proteins are encoded by a highly divergent gene superfamily, present in both pro- and eukaryotes. Classic CYP proteins are heme-containing monooxygenases that catalyze numerous enzymatic reactions involved in detoxification as well as in biosynthetic pathways. In plants, they have been shown to participate in the synthesis of hormones, lignin intermediates, sterols, terpenes, and flavonoids (for review, see Schuler, 1996). Some CYP proteins have been shown to be important regulators during plant growth and development (Szekeres et al., 1996; Choe et al., 1998; Kim et al., 1998; Zhao et al., 2002).
To characterize this putative negative regulator of the auxin response, we cloned the full-length cDNA of CYP87A3 and analyzed the expression pattern in coleoptiles under various treatments. We report here that CYP87A3 is a primary auxin response gene. It is transiently up-regulated by auxin (accompanied by a stimulation of growth) but also by irradiation (accompanied by an inhibition of growth). We therefore discuss the highly dynamic CYP87A3 expression and possible mechanisms of its regulation in the context of auxin-mediated growth regulation.
RESULTS
CYP87A3 Was Isolated from a FDD Screen for Auxin-Induced Transcripts
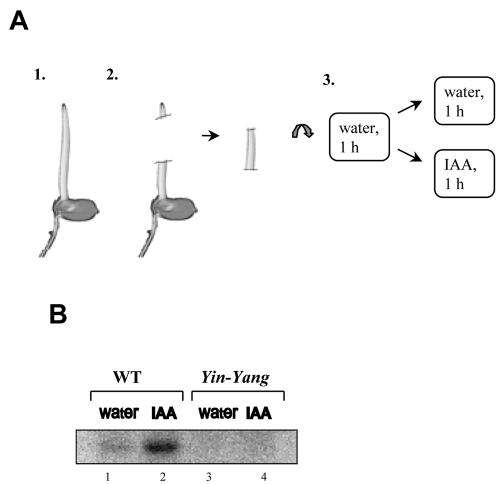
Using fluorescence-labeled differential display, auxin-induced transcripts were compared between WT rice (O. sativa L. subsp. japonica cv Nihonmasari) and the Yin-Yang mutant, where the growth response to auxin is increased by about 50% (Wang and Nick, 1998). Excised coleoptiles of 6-d-old WT and Yin-Yang plants were incubated for 1 h in water to deplete them from endogenous auxin and then incubated for 1 h in 5 μm IAA (Fig. 1A), the IAA concentration optimal for growth induction (Waller et al., 2002). As a negative control, coleoptiles were kept in water for 2 h. FDD analysis of total RNA from each treatment showed that only a few auxin-induced genes were differentially expressed in WT and Yin-Yang (Waller et al., 2002). The expression of one of them, here and further referred to as CYP87A3, was up-regulated by auxin in the WT, whereas it was virtually absent in the Yin-Yang mutant (Fig. 1B). We therefore focused on two main objectives: to clone the complete gene, and to analyze the regulation of its expression with respect to auxin signaling.
Figure 1.
Fluorescence differential display screen for auxin-inducible transcripts. A, Scheme of the experiment: 1, coleoptiles were grown for 6 d in darkness; 2, coleoptile segments were excised; and 3, segments were predepleted in water for 1 h and then incubated for 1 more h in IAA or in water. B, A part of an FDD gel image showing the CYP87A3 expression pattern in WT and the mutant Yin-Yang. Total RNA was isolated from coleoptile segments incubated for 1 h either in distilled water (lanes 1 and 3) or in 5 μm IAA (lanes 2 and 4).
CYP87A3 Is Coding for a Protein That Belongs to the CYP Superfamily
To obtain the full-length CYP87A3 cDNA, the RACE approach was applied. The predicted length of the mRNA (2,185 bp; accession no AJ459255) was consistent with the observed size estimated from northern-blot analysis. The TATA box (TATATAA) is located 33 bp upstream of the transcription start. By searching the Monsanto rice sequencing database (http://www.rice-research.org) we found that the CYP87A3 genomic sequence was localized in the rice clone OSM1165. The CYP87A3 gene contains nine exons and eight introns and is 4.5 kb long excluding the promoter region. The CYP87A3 genomic sequence is now available as part of the sequence of bacterial artificial chromosome clone OSJNBA0088122 (accession no. AL607001) and is located on chromosome 4.
Two reading-frame translations of the CYP87A3 cDNA yielded numerous stop codons throughout the whole length, whereas the third frame revealed an open reading frame of 1,542 bp coding for a deduced protein of 514 amino acids. The estimated molecular mass of this protein is 57.5 kD, and the predicted pI is 9.0. The putative CYP87A3 protein showed homology to members of the CYP superfamily.
By definition, CYP sequences are classified into the same family (designated by the first number) when they are >40% identical to one another at the level of the amino acid sequence, those belonging to the same subfamily (designated by the letter) are >55% identical, and those corresponding to alleles of the same locus are >97% identical (Schuler, 1996). BLAST searching indicated that CYP87A3 protein shows substantial amino acid identity (61%) with a predicted protein of Arabidopsis (chromosome 1; accession no. NP-172734) designated CYP87A2 and 55% amino acid identity with putative CYP87A1 from sunflower (Helianthus annuus; accession no. AF216313, partial mRNA sequence). Therefore, the identified gene was designated as a member of the rice CYP87A subfamily and named CYP87A3. The function of both of the closest homologs is unknown. The closest homologs of known function are constitutive photomorphogenesis and dwarfism (CPD; 39% amino acid identity; accession no. Q42569), DWF4 (steroid 22-hydroxylase; accession no. AF044216.1), and ROTUNDIFOLIA3 (34% amino acid identity; accession no. Q9M066). They belong to the A, B, and C subfamilies of the CYP90 family from Arabidopsis. Similar levels of homology were found for the tomato (Lycopersicon esculentum) DWARF protein CYP85 (38% amino acid identity; accession no. Q43147) and the maize (Zea mays) DWARF3, CYP88A1 (32% amino acid identity; accession no. Q43246). By database searching (Dr. Nelson WebPage [http://drnelson.utmem.edu/rice. html]), we identified the two closest homologs of the CYP87A3 in rice, sharing the same locus on chromosome 4 (OSJN00128): One putative protein derived from this locus was 57% identical and thus could be assigned to the same subfamily, CYP87A; a second putative protein from this locus had 48% of amino acid identity to CYP87A3.
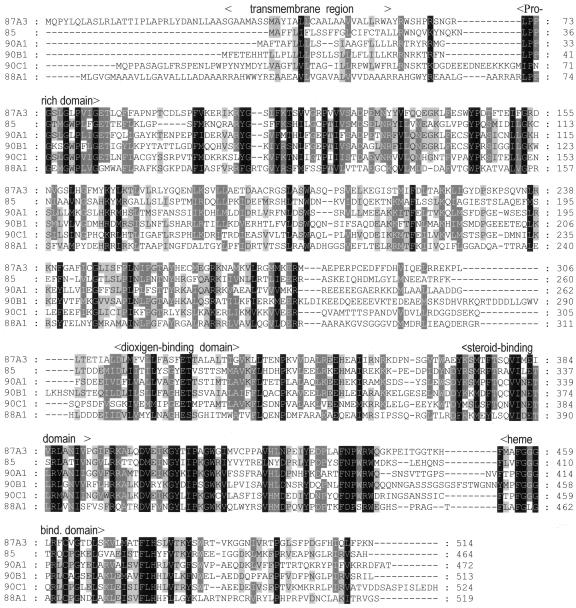
CYPs are heme-containing proteins that primarily catalyze reactions of monooxygenation/hydroxylation (Werck-Reichhart et al., 2000). The catalytic domain containing a ferriprotoporphyrin IX (heme) prostetic group covalently attached to a Cys is characterized by a highly conserved, C-terminal F-G-RC-G motif (Schuler, 1996), which is also found in CYP87A3 (Fig. 2). An amino acid alignment of two CYP proteins involved in brassinosteroid biosynthesis (22α-hydroxylase DWF4/CYP90B1 [Choe et al., 1998]; 23α-hydroxylase CPD/CYP90A1 [Szekeres et al., 1996]) and other CYP proteins that are involved in gibberellin biosynthesis and dwarfism (DWF3/CYP88A1 [Winkler and Helentjaris, 1995]; ROTUNDIFOLIA/CYP90C1 [Kim et al., 1998]; and DWARF/CYP85 [Bishop et al., 1996]) with CYP87A3 revealed highly conserved domains in these proteins in addition to the C-terminal heme-binding domain. All of these proteins exhibit a central region with steroid substrate-binding and oxygen-binding domains (Fig. 2). An N-terminal transmembrane helix can be predicted with high probability from a hydrophobicity analysis of the CYP87A3 sequence. An N-terminal endoplasmic reticulum anchor domain that is not cleaved off during posttranslational modification (Omura, 1999) is typical for most plant and many animal CYPs. A Pro-rich region (Fig. 2) downstream of the signal sequence that is highly conserved among classic CYP proteins can be identified in CYP87A3 as well (Schuler, 1996).
Figure 2.
Multiple sequence alignment of CYP87A3 with related CYP proteins. CYP87A3 exhibits homology to the 23α-hydroxylase CPD (CYP90A1), the 22α-hydroxylase DWF4 (CYP90B1), ROTUNDIFOLIA (CYP90C1), DWF3 (CYP88A1), and DWARF (CYP85). The locations of conserved domains are indicated above the sequences. Amino acid residues that are identical between the compared sequences are indicated by reverse font; 80% conserved, by white letters in gray boxes; 60% conserved, by gray boxes.
Features of Promoter and mRNA 3′-Untranslated Regions (UTRs)
Several putative auxin-responsive elements were found in the so far cloned 1-kb fragment of the CYP87A3 promoter using the Web Signal Scan Program and the PLACE database: (a) A GTTCCCAT motif, which differs in only one nucleotide from the (G/T) GTCCCAT found in promoters of several auxin-regulated genes such as the 165-bp fragment of pea PS-IAA4/5 promoter, reported to contain an auxin-responsive cis-element (Guilfoyle et al., 1998). (b) A TGACGTAA sequence, highly homologous to the TGA box of the E1 motif in the GH3 promoter (Liu et al., 1994). (c) Sequences highly homologous to the A1 module of the SAUR15A promoter (Xu et al., 1997). In addition, the CYP87A3 promoter contains numerous short sequences highly homologous to the auxin-responsive ocs-like element TGATGTAAGAGATTACGTAA of the GH2/4 promoter (Abel and Theologis, 1996). Ocs-like elements are activated not only by auxin, but also by other growth substances and stress-inducing agents (Abel and Theologis, 1996). It is interesting that in the CYP87A3 promoter, the ocs core sequences overlap with the TGA box or are adjacent to other putative auxin-responsive elements.
The cis-elements potentially involved in light-regulated transcription appeared to be most abundant in the 5′ region of CYP87A3. In addition to classic motifs for light regulation such as the GATA-box, the GT1-consensus, and the I-box (Donald and Cashmore, 1990; Gilmartin et al., 1990; Terzaghi and Cashmore, 1995; Rose et al., 1999; Zhou, 1999), further cis-elements characteristic for light-regulated genes in different plants are present in the CYP87A3 promoter. It also contained binding sites for embryo- and root-specific expression.
The Plant CARE promoter analysis program revealed additional binding sites for trans-factors involved in jasmonate (TGACG-motif), gibberellin (TATC-box and P-box, CCTT), ethylene (ERE, ATTT), and wounding (WUN-motif, (C)AATT) responsiveness. Our preliminary findings indicate that CYP87A3 expression is influenced by ethylene and jasmonic acid, whereas wounding had virtually no effect (data not shown).
A comparison of the CYP87A3 3′-UTR with the consensus sequence from the so-called downstream element (DST) of SAUR genes, which was shown to be responsible for rapid mRNA degradation (McClure and Guilfoyle, 1989; Gil and Green, 1996), revealed several highly homologous motifs. These multiple motifs corresponded to individual components of the DST element, but they were arranged in a different order. Although the 3′-UTR is AT-rich, its sequence did not show the multiple overlapping ATTTA motifs that are thought to be instability determinants in mammalian cells (Greenberg and Belasco, 1993).
Localization of CYP87A3
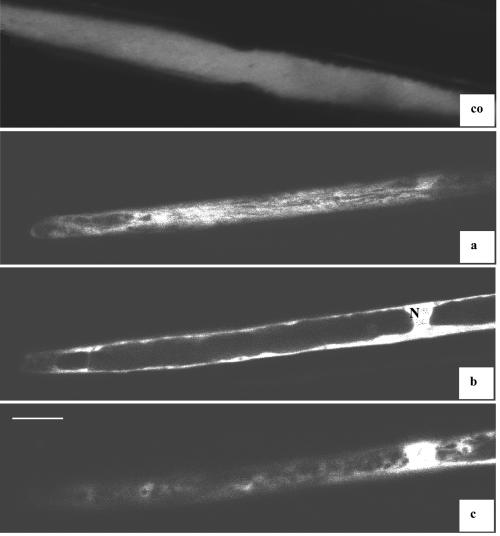
To determine the subcellular localization of the CYP87A3 protein, dark-grown rice seedlings were transiently transformed with CYP87A3:cyano-green fluorescent protein (CFP) fusion constructs using a biolistic method. Both cells with strong and weak expression were examined. Whereas in cells transformed with the control construct 35S:CFP the CFP was distributed homogenously throughout the cell (Fig. 3, co), a specific pattern of CFP distribution was observed for the CYP87A3:CFP fusion protein (Fig. 3, a-c). In addition to a signal around the nucleus, strong expression was noted in broad longitudinal strands adjacent to the outer cell surface and as a characteristic signal surrounding unstained vesicular structures (“negative staining”) in the cytoplasm. In cells with weaker expression, only these “negatively stained” vesicular structures were observed in the cell cortex. However, a strong nuclear signal (distributed throughout the nucleus) was present irrespective of the expression level.
Figure 3.
Subcellular localization of 35S:CYP87A3:CFP fusion protein. The transient expression of CYP87A3 with in-frame C-terminally fused CFP was analyzed in coleoptile epidermal cell by confocal laser scanning microscope. co, Control, 35S::CFP; a through c, sequential confocal sections through the same cell expressing CYP87A3::CFP, beginning from the outer cell surface. White bar = 20 μm; N, nucleus.
Northern-blot analyses of different organs—primary leaves, distal and basal parts of young leaves, old leaves, and roots—that had been incubated either in water or in auxin indicate that CYP87A3 is only expressed in roots and coleoptiles, but not in leaves (data not shown).
Time Course and Dose Response of Auxin Induction
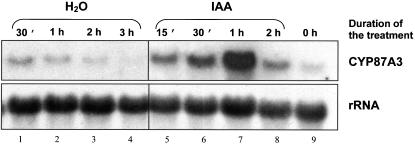
For a kinetic study, RNA was isolated from coleoptile segments of the WT that had been incubated in IAA or water for different time intervals. Expression of CYP87A3 was low in freshly excised, dark-grown coleoptiles (Fig. 4, lane 9). When endogenous auxin was depleted by incubating the segments in water, the signal decreased gradually with longer incubation periods (Fig. 4, lanes 2-4). However, for short time intervals of the incubation of segments in water (30 min), the signal was slightly stronger compared with directly harvested coleoptiles (Fig. 4, compare lanes 1 and 9). When the coleoptile segments were incubated in 5 μm IAA, the CYP87A3 was strongly but transiently induced with a lag time of less then 15 min and maximal expression at 1 h. The mRNA level declined down to the initial level within the 2nd h after the addition of auxin (lanes 5-8).
Figure 4.
Kinetics of CYP87A3 expression in response to exogenous IAA. Total RNA (15 μg per lane) was isolated from coleoptile segments that either had been excised directly from seedlings grown for 6 d in darkness (lane 9) or that after excision had been incubated in water (lanes 1-4) or in 5 μm IAA (lanes 5-8), respectively. The same membrane was hybridized with probes specific to CYP87A3 and 28S rRNA (loading control).
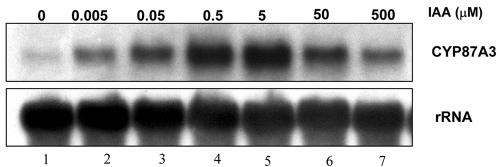
The IAA dose response curve for CYP87A3 expression revealed already for 0.005 μm IAA a slight induction as compared with the water control (Fig. 5, compare lanes 1 and 2). Between 0.005 and 5 μm IAA, the transcript was progressively enhanced with a maximal plateau at 0.5 to 5 μm (Fig. 5, lanes 4 and 5, respectively). When auxin concentration was raised further, however, the signal declined again to almost the uninduced level (Fig. 5, lanes 6 and 7).
Figure 5.
The response of CYP87A3 transcripts to different concentrations of IAA. Total RNA (15 μg per lane) was isolated from coleoptile segments that had been incubated for 1 h in IAA of different concentrations directly after excision. Top row, CYP87A3-specific probe was used; bottom row, loading control shown by hybridization with 28S rRNA-specific probe.
CYP87A3 Is Strongly Induced by Cycloheximide
Cycloheximide, an inhibitor of protein synthesis, has been often used in investigations of genes involved in early steps of auxin signaling (for review, see Abel and Theologis, 1996). A wide range of genes, termed primary auxin-responsive genes, is induced within minutes after auxin application, and this induction is not suppressed by inhibition of protein translation, indicating that all proteins necessary to initiate transcription of primary auxin-responsive genes are already present in the cell and can be activated by auxin.
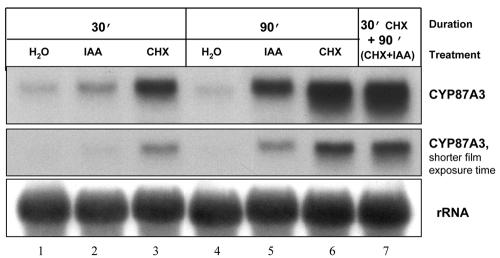
CYP87A3 transcript level was already greatly induced by 30 min of incubation in cycloheximide (Fig. 6, lane 3). During the following hour of cycloheximide treatment, the signal became even stronger (lane 6). Interestingly, cycloheximide alone stimulated the CYP87A3 transcript to a greater extent than IAA alone (Fig. 6, compare lanes 2, 3, 5, and 6). The combination of cycloheximide and IAA yielded a slight superinduction as compared with the signal obtained for cycloheximide alone (Fig. 6, lane 7; for better resolution, shorter film exposure time is shown in the middle row of the figure).
Figure 6.
CYP87A3 transcripts are elevated after cycloheximide treatment. Total RNA (15 μg per lane) was isolated from coleoptiles that have been treated with cycloheximide and/or auxin. When coleoptiles were treated by cycloheximide in combination with auxin (lane 7), cycloheximide was applied 30 min before IAA to ensure inhibition of protein synthesis before the exogenous auxin could enter the cell. The working concentrations were 5 μm for IAA and 70 μm for cycloheximide. For details, refer to the legend of Figure 4. CHX, cycloheximide.
Light Affects CYP87A3 Expression
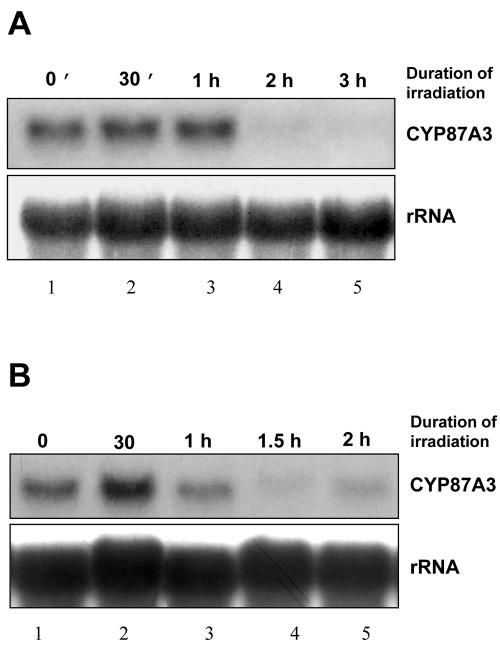
Because the FDD screen showed that light modulates the expression of CYP87A3, we followed the abundance of the transcript over time for two different intensities of red light. For the low intensity (3.8 μmol m-2 s-1), the transcript remained stable or even slightly increased up to 1 h of irradiation and then disappeared during the 2nd h of irradiation (Fig. 7A). For the high intensity (38.9 μmol m-2 s-1), the initial increase was more rapid (with a peak at 30 min) and much more pronounced in amplitude. Again, the transcript disappeared during the hour following the peak (Fig. 7B). A similar effect was observed during irradiation of seedlings with blue and far-red light (data not shown).
Figure 7.
CYP87A3 transcripts are transiently induced by red light. Total RNA (14 μg per lane) was isolated from coleoptiles after irradiation for different time intervals. A, Weak red light (RL), 3.8 μmol m-2 s-1. B, Strong red light, 38.9 μmol m-2 s-1. The irradiation time is indicated. For details refer to the legend of Figure 4.
DISCUSSION
CYP: From Regulation to Function
CYP87A3, a putative member of the rice CYP monooxygenase superfamily, was isolated in a FDD screen for auxin-inducible genes related to the auxin responsiveness of growth. CYPs are involved in oxidative metabolism of different endogenous and exogenous lipophilic substrates (Mizutani et al., 1998). The catalyzed reaction is usually a mono-oxygenation, with the insertion of one oxygen atom (as a hydroxyl group) into a substrate and formation of one water molecule, but additional more atypical activities, such as dimerizations, isomerizations, dehydrations, and reductions have also been reported (for review, see Halkier, 1996; Werck-Reichhart et al., 2000). In the generalized reaction scheme, electrons are transferred from NADPH to the CYP via FAD and FMN flavoproteins called CYP reductases. Plant CYPs are usually bound via their N terminus to the cytoplasmic surface of the endoplasmic reticulum. They have a highly hydrophobic N-terminal “signal anchor sequence,” consisting of about 25 amino acid residues, which targets and anchors the CYP molecules to the membrane (for review, see Omura, 1999). By hydropathy analysis, this plasma membrane anchor sequence was predicted to be present in CYP87A3 as well. The analysis of subcellular localization of CYP87A3 by means of transient expression of its fusion with a fluorescent protein revealed a highly specific distribution in the cell cortex of transformed epidermal cells consistent with a targeting to the endomembrane system in these highly vacuolated cells. In addition, a part of the signal was found within the nucleus. This might be caused by the overexpression of the fusion protein. As a consequence, the putative binding partner of the protein would be saturated so that the free fusion protein would become dislocalized.
Due to the diversity of pathways requiring monooxygenase activity, the expression of CYPs has been reported to be modulated by a wide variety of environmental factors such as light, fungal elicitors, and wounding, as well as by developmental signals (Dixon and Paiva, 1995; Mizutani et al., 1998). In all cases, the induction profile was found to be specific for a particular CYP or subset of CYPs and not the result of a global induction of CYP genes. Analyzing the response pattern of seven CYP transcripts after treatment of maize seedlings with chemical inducers, Persans et al. (2001) have found that each of the transcripts had distinct developmental, tissue-specific, and chemical cues regulating their expression even when they encode CYPs within the same biosynthetic pathway. These observations were consistent with results obtained from analyses on the transcriptional regulation of other CYPs, for example CYP72A2 (induced by wounding and cytokinin and involved in the defense responses of tobacco [Nicotiana tabacum]; Mujer and Smigocki, 2001), CPD (suppressed by brassinosteroids and participating in brassinolide biosynthesis; Mathur et al., 1998), or CYP83B1 (auxin inducible, containing AuxREs in the promoter, and involved in the biosynthesis of indole glucosinolates from Trp, a branching point between the biosynthesis of IAA and indole glucosinolates; Bak et al., 2001).
These findings demonstrate that the pattern of CYP regulation is strictly correlated to their biological function. Following this line of argument, the CYP87A3 gene product might be placed in the tuning of growth responses. This is supported by the FDD screen that led to the isolation of this gene, where CYP87A3 transcripts were almost undetectable in coleoptiles of the mutant Yin-Yang that are characterized by an elevated auxin responsiveness of growth, but were auxin-inducible in the WT. A similar reduction of the CYP87A3 transcript level was found for further mutant, hebiba, with elevated auxin responsiveness of growth (Riemann et al., 1993). The CYP87A3 protein is therefore most likely a negative regulator for the auxin responsiveness of growth. The transient up-regulation of this gene in irradiated coleoptiles is accompanied by a substantial decrease of endogenous auxin (Riemann et al., 1993), which might be responsible for the decrease in growth. Thus the transient induction of CYP87A3 by light might indicate an increased responsiveness to auxin. Whether light modifies the auxin responsiveness of CYP87A3 at the level of transcription or of RNA stability is still undetermined. Alternatively, irradiation might lead to auxin-independent gene regulation. In both cases, however, it may control the auxin response of downstream targets of the CYP87A3 protein.
In this context it is possible, for instance, that the CYP87A3 gene product might be directly involved in IAA de-activation/transport inhibition or, alternatively, in the biosynthesis of auxin antagonists. An attractive, but still speculative target might be, for example, flavonoids that have been discussed for several decades to be involved in regulation of auxin transport (Stenlid, 1976; Brown et al., 2001, and refs. therein). Certain flavonoids were shown also to play role in auxin breakdown in plants (Furuya et al., 1962; Stenlid, 1963; Rao, 1990 and refs. therein). For instance, Mathesius (2001) found that regulation of auxin breakdown during nodule organogenesis is under control of flavonoids in white clover (Trifolium repens). Taking into account that CYPs are prominent enzymes in flavonoid biosynthesis (Schuler, 1996), one could speculate about a parallel in rice coleoptiles: Differential CYP87A3 expression might contribute to local distributions of specific flavonoids. Because flavonoids seemed to participate in regulation of auxin transport/oxidation, CYP87A3 could be involved in the control of local auxin levels. Therefore, we plan to express recombinant functional CYP87A3 to test its effect on IAA oxidation and/or to screen for its potential substrate(s).
Auxin Regulates CYP87A3 Expression
We have found that auxin application rapidly induced CYP87A3 expression. Such a rapid response that is not inhibited by cycloheximide is characteristic for early or primary auxin response genes. We observed a strong induction of CYP87A3 in coleoptile segments that were treated with cycloheximide alone. The combination of cycloheximide with IAA seemed to have very little additional effect on this induction. A similar cycloheximide effect was shown for several other early auxin response genes, including members of the SAUR and Aux/IAA families, which can be induced by cycloheximide alone (McClure et al., 1989; Abel and Theologis, 1996). On the basis of a detailed study of the effect of protein synthesis inhibition on the expression of Ps-IAA4/5 and Ps-IAA6, Koshiba et al. (1995) proposed a dual effect of cycloheximide by (a) activation of transcription and (b) stabilization of the inducible mRNAs. To explain the first effect, it has been proposed that cycloheximide prevents synthesis or activation of a short-lived transcriptional repressor whose subsequent degradation or deactivation results in the activation of auxin-inducible genes (Koshiba et al., 1995). In fact, Aux/IAA proteins have been shown to suppress the expression of reporter genes containing highly active auxin-responsive elements (Ulmasov et al., 1997) and are therefore good candidates for such a repressor. The second effect has been explained by the disruption of translation-coupled mRNA degradation or by the depletion of the pool of a labile enzyme that degrades specific transcripts (Koshiba et al., 1995). The analysis of the 3′-UTR of CYP87A3 revealed several sequences that are identical to motifs in the DST instability sequence. This means that CYP87A3 expression might be controlled at the level of sequence-specific mRNA degradation as well. To distinguish between the particular mechanisms responsible for the superinduction of CYP87A3 by cycloheximide, further functional analysis of the UTRs would be desirable.
The regulation pattern of CYP87A3 transcripts contains two remarkable features:
CYP87A3 transcripts are only transiently (peak at 1 h) increased in response to auxin. After 2 h of auxin treatment, the transcript level had almost returned to the original situation. In contrast, the transcripts of another primary auxin response gene, OsARF1, remain elevated for several hours after the addition of auxin (Waller et al., 2002), which is typical for other classical primary auxin response genes such as members of the Aux/IAA family (at least up to 5 h; Theologis et al., 1985), GH3 (Hagen and Guilfoyle, 1985), or the SAUR family (McClure and Guilfoyle, 1987; McClure et al., 1989; Gee et al., 1991). Transient induction is usually characteristic for regulatory components involved in signaling pathways. Assuming that CYP87A3 codes for such a regulatory component involved in tissue adaptation to higher auxin concentration, a rapid return to the initial lower transcript level would restore the cellular competence for subsequent responses to changing auxin concentration.
The dose response curve, particularly its response to higher auxin concentrations differs from classic primary auxin response genes such as GH3 and SAUR. Reaching a maximum plateau at concentration 0.5 to 5 μm, the induction of CYP87A3 sharply decreases in amplitude when IAA concentrations are raised above 5 μm. This bell-shaped curve parallels the dose response curve of auxin-induced growth (Waller et al., 2002). Interestingly, a similar dose response was shown for OsARF1 with a maximum response to 1 to 3 μm IAA (Waller et al., 2002) as well as for members of Arabidopsis Aux/IAA family (optimum response at 10 μm IAA; Abel et al., 1995). In contrast, the transcription of GH3 or SAUR genes was reported to persist even very high concentrations of auxin (100 μm 2,4-dichlorophenoxyacetic acid in the case of GH3 [Hagen and Guilfoyle, 1985]; up to 10 mm 2,4-dichlorophenoxyacetic acid in the case of SAUR [McClure and Guilfoyle, 1987]).
Currently, we are performing experiments to determine possible coregulators of CYP87A3 expression. Our further research will focus on a functional analysis of the promoter region as well as on getting insight into the cross-talk between light signaling and auxin-induced elongation growth.
MATERIALS AND METHODS
Plant Material
Japonica rice (Oryza sativa L. subsp. japonica cv Nihonmasari) was used for the experiments. For FDD analyses, seedlings of the mutant Yin-Yang (Wang and Nick, 1998) were used in addition to the WT. The seedlings were grown on floating meshes as described by Nick et al. (1994) in photobiological darkness (using black boxes wrapped in black cloth) and kept in dark chambers at 25°C. Six days after germination, the apical 2 to 3 mm of the coleoptile was removed, and subapical segments of 10 to 15 mm length were used for further experiments (Waller et al., 2002). The segments were incubated in water or in different concentrations of IAA (Fluka, Buchs, Switzerland) under continuous rotation in a top-over shaker. If not stated otherwise, all preparations were made in green safelight. For the light experiments, the seedlings were irradiated for different time intervals. The light sources for red light (λmax 660 nm, 3.8 and 38.9 μmol s-1 m-2), far red light (λmax 730 nm, 15.9 μmol s-1 m-2), blue light (λmax 430 nm, 22.8 μmol s-1 m-2) and green safelight (λmax 550 nm) are described in detail by Mohr et al. (1964). All light measurements were performed using a Tektronix-J16 photoradiometer (Tektronix, Beaverton, OR).
For the transient transformation assay, 4-d-old seedlings were attached to microscopic slides with their caryopses using surgical adhesive in green safelight. After transformation, slides with attached coleoptiles were returned to the dark and placed in vertical position to allow the seedlings grow further. The seedlings were analyzed the next day.
FDD
FDD was performed as described by Waller et al. (2002). First-strand synthesis was performed with 2.5 μg of total RNA using a Texas Red-labeled 3′-anchored oligo(dT) primer (5′-Texas Red-T14G-3, Youkigouseikagaku, Tokyo) and the Superscript Preamplification System (Invitrogen, Carlsbad, CA). CDNAs were amplified by PCR using combinations of the Texas Red-labeled anchor primer and arbitrary 10-mer primers (Kit B, D, F, and X, Operon Technologies, Alameda, CA). Electrophoresis and detection of the PCR products were performed with an automated DNA sequencer (SQ5500, Hitachi).
Cloning of cDNA
The cDNA of interest was isolated by preparative gel electrophoresis and excised from the gel as described by Kuno et al. (2000). The sequence of cDNA 3′ end was obtained as described by Waller et al. (2002). For the isolation of the complete cDNA sequence of CYP87A3, reverse transcription PCR using gene-specific primers and the 5′-RACE System for Rapid Amplification of cDNA Ends (Invitrogen), FirstChoice RLM-RACE kit (Ambion, Austin, TX), and GeneRacer kit (Invitrogen) were used according to the manufacturers recommendations.
RNA Isolation and Northern-Blot Analysis
Total RNA was isolated from rice coleoptiles either by phenol/chlorophorm extraction according to Ehmann et al. (1991) or by using the Rneasy Plant Mini kit (Qiagen, Hilden, Germany). Both methods resulted in comparable quality and quantity of isolated RNA. For all samples of a given northern blot, only one of the two methods was used.
Ten to 15 μg of total RNA were separated by electrophoresis in 1% agarose-formaldehyde gels according to Davis et al. (1986) and transferred to Duralon UV membrane (Stratagene, La Jolla, CA). A [α-32P]dCTP random-labeled probe corresponding to CYP87A3 3′-UTR was used. The DNA template for the probe was obtained with the following primer pair: 5′-GTGCAAAGATGTCGTTGGGTTTTT-3′ and 5′-CACGGACTAGAGGTAGCACTCGGT-3′.
The hybridization procedure and the probes for OsARF1 detection were performed according to Waller et al. (2002). After washing with 2× SSC, 1× SSC, 0.5× SSC, and 0.1× SSC washing solutions (1× SSC is 150 mm NaCl and 15 mm sodium citrate) containing 0.1% SDS at 63°C, membranes were autoradiographed for 1 to 4 d using an intensifying screen (Biomax, Eastman Kodak, Rochester, NY). To confirm equal transfer of the RNA to the membrane, the CYP87A3/OsARF1 probe was removed after autoradiography, and the membrane was rehybridized with a probe corresponding to rice 28S rRNA.
Quantitative Analysis of Northern-Blot Signals
For quantitative analyses, films were scanned and analyzed by ImageJ algorithm (http://rsb.info.nih.gov/ij/index.html). The actual RNA loading was calculated from analysis of 28sRNA reprobed membranes. For every treatment, three to seven repetitions were analyzed. All data were statistically processed.
Transient Transformation of Rice Coleoptiles with CFP Constructs
The full-length CYP87A3 cDNA was obtained with the primer pair 5′-GTGATCTAGACATGCAGCCATATCTTCAGC-3′ and 5′-GTTCTTAGGGAAGAGCTGGATATGAAAACC-3′ and inserted in frame with a CFP using the vector (35S)2x-CFP/pUC kindly provided by K. Harter and F. Nagy (unpublished data). The CYP87A3 was C-terminally linked with CFP. The expression of the CFP fusion construct was driven by two copies of the 35S promoter. Transformation of bacteria and isolation of the plasmid DNA was performed using TOPO TA-Cloning kit (Invitrogen) and Qiaprep Spin Miniprep kit (Qiagen) according to the instructions of respective providers.
Gold particles (Sigma-Aldrich, St. Louis) were coated with the isolated plasmid DNA according to the following protocol: 25 μL of a gold stock solution (60 mg mL-1 in glycerol 50%), 5 μL of plasmid DNA, 25 μL of 2.5 m calcium chloride, and 10 μL of 0.1 m spermidine solution were mixed by continuous vortexing and centrifuged for 1 min. The gold pellet was washed three times with 1 mL of 100%, 70%, and 100% ethanol. After a final centrifugation (1 min), the pellet was resuspended in 40 μL of 100% ethanol. The whole resuspension was loaded onto particle bombardment grids. The bombardment was performed in a custom-made particle gun with a Helium pressure of 2 to 3 bar per shot and under vacuum conditions (≤0.8 bar). For transformation, etiolated 4-d-old rice seedlings glued onto a slide were used. The coleoptiles were analyzed by confocal laser scanning microscopy as described by Wang and Nick (2001) 24 h after transformation.
Acknowledgments
We thank Dr. Christina Suesslin for preparation of the particle bombardment assay and Edith Fitzke for the excellent technical assistance. We also would like to acknowledge the Pharmacia rice-research.org program for providing data on the rice genome sequence.
This work has been supported by a fellowship from the Graduiertenkolleg “Molekulare Mechanismen der pflanzlichen Differenzierung” by the Deutsch Forschungsgemeinschaft (to C.C.), by HARL(B2023) and the Program for Promotion of Basic Research Activities for Innovative Biosciences, Japan (to M.F.), and by funds from the Volkswagen Foundation (to F.W. and P.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022202.
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533-549 [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer FJ, Davies PJ (1992) Indol-3-acetic acid levels after phytochrome-mediated changes in stem elongation rate of dark- and light-grown Pisum seedlings. Planta 188: 85-92 [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones JD (1996) The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8: 959-969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes B, Fujioka S, Takatsuto S, Sakurai A, Feldmann K (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LG, Dibner MD, Battey JF (1986) Basic Methods in Molecular Biology. Elsevier, New York
- Dixon RA, Paiva N (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RGK, Cashmore AR (1990) Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J 9: 1717-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann B, Ocker B, Schaefer E (1991) Development- and light-dependent regulation of the expression of two different chalcone synthase transcripts in mustard seedlings. Planta 183: 416-422 [DOI] [PubMed] [Google Scholar]
- Furuya M, Galston AW, Stow BB (1962) Isolation from peas of co-factors and inhibitors of indolyl-3-acetic acid oxidase. Nature 193: 456-457 [DOI] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ (1991) Tissue-specific and organ-specific expression of the auxin-responsive transcripts, SAURs and GH3, in soybean. Plant Cell 3: 419-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Green PJ (1996) Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J 15: 1678-1686 [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua N-H (1990) Molecular light switches for plant genes. Plant Cell 2: 369-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Belasco JG (1993) Control of the decay of labile protooncogene and cytokine mRNAs. In J Belasco, G Brawerman, eds, Control of Messenger RNA Stability. Academic Press, San Diego, pp 199-218
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J (1998) How does auxin turn on genes? Plant Physiol 115: 397-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373-385 [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ (1985) Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197-1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA (1996) Catalytic reactivities and structure/function relationships of cytochrome P450 enzymes. Phytochemistry 43: 1-21 [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12: 2381-2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Kamiya Y, Iino M (1995) Biosynthesis of indole-3-acetic acid from l-tryptophan in coleoptile tips of maize (Zea mays L.). Plant Cell Physiol 36: 1503-1510 [Google Scholar]
- Kuno N, Muramatsu T, Hamazato F, Furuya M (2000) Identification by large-scale screening of phytochrome-regulated genes in etiolated seedlings of Arabidopsis using a fluorescent differential display technique. Plant Physiol 122: 15-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53: 377-398 [DOI] [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645-657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Kamisaka S, Hoson T (1998) Growth behaviour of rice coleoptiles. J Plant Physiol 152: 180-188 [Google Scholar]
- Mathesius U (2001) Flavonoids induced in cells undergoing nodule organogenesis in white clover are regulators of auxin breakdown by peroxidase. J Exp Bot 52: 419-426 [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C et al. (1998) Transcription of the arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593-602 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle TJ (1989) Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243: 91-93 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle TJ (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611-623 [DOI] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ (1989) Transcription, organization and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1: 229-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ward E, Ohta D (1998) Cytochrome P450 superfamily in Arabidopsis thaliana: isolation of cDNAs, differential expression, and RFLP mapping of multiple cytochromes P450. Plant Mol Biol 37: 39-52 [DOI] [PubMed] [Google Scholar]
- Mohr H, Meyer U, Hartmann K (1964) Die Beeinflussung der Farnsporenkeimung (Osmunda cinnamomea und O. claytoniana L.) über das Phytochromsystem und die Photosynthese. Planta 60: 483-496 [Google Scholar]
- Mujer CV, Smigocki AC (2001) Cytokinin- and wound-inducible cytochrome P450 from Nicotiana plumbaginifolia. Physiol Plant 111: 172-181 [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyls length. Plant J 25: 213-221 [DOI] [PubMed] [Google Scholar]
- Nick P, Yatou O, Furuya M, Lambert AM (1994) Auxin-dependent micro-tubule responses and seedling development are affected in a rice mutant resistant to EPC. Plant J 6: 651-663 [Google Scholar]
- Normanly J (1997) Auxin metabolism. Physiol Plant 100: 431-442 [Google Scholar]
- Omura T (1999) Forty years of cytochrome P450. Biochem Biophys Res Commun 266: 690-698 [DOI] [PubMed] [Google Scholar]
- Persans MW, Wang J, Schuler M (2001) Characterization of maize cytochrome P450 mono-oxygenases induced in response to safeners and bacterial pathogens. Plant Physiol 125: 1126-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AS (1990) Root flavonoids. Bot Rev 56: 1-84 [Google Scholar]
- Riemann A, Mueller A, Korte A, Furuya M, Weiter E, Nick P (2003) Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol 133: 1820-1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYB1 is a member of a novel class of Myb-like proteins. Plant J 20: 641-652 [DOI] [PubMed] [Google Scholar]
- Schuler MA (1996) Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci 15: 235-284 [Google Scholar]
- Stenlid G (1976) The effect of flavonoids on polar auxin transport. Physiol Plant 38: 262-266 [Google Scholar]
- Stenlid G (1963) The effects of flavonoid compounds on oxidative phosphorylation and on the enzymatic destruction of indoleacetic acid. Physiol Plant 16: 110-121 [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids resque the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171-182 [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445-474 [Google Scholar]
- Theologis A, Huynh TV, Davis RW (1985) Rapid induction of specific mRNA by auxin in pea epicotyl tissue. J Mol Biol 183: 53-68 [DOI] [PubMed] [Google Scholar]
- Trewavas A (1981) How do plant growth substances act? Plant Cell Environ 4: 203-228 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963-1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S (1961) Some characteristics of indolacetic acid oxidase in rice coleoptiles. Sci Rep Tohoku Univ 4th Ser (Biol) 29: 223-235 [Google Scholar]
- Waller F, Furuya M, Nick P (2002) OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene. Plant Mol Biol 50: 415-425 [DOI] [PubMed] [Google Scholar]
- Wang Q-Y, Nick P (1998) The auxin response of actin is altered in the rice mutant Yin-Yang. Protoplasma 204: 22-33 [DOI] [PubMed] [Google Scholar]
- Wang Q-Y, Nick P (2001) Cold acclimation can induce microtubular cold stability in a manner distinct from abscisic acid. Plant Cell Physiol 42: 999-1005 [DOI] [PubMed] [Google Scholar]
- Went FW (1928) Wuchsstoff und Wachstum. Recl Trav Bot Neerl 25: 1-116 [Google Scholar]
- Werck-Reichhart D, Hehn A, Didierjean L (2000) Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci 5: 116-123 [DOI] [PubMed] [Google Scholar]
- Winkler RG, Helentjaris T (1995) The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell 7: 1307-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Hagen G, Guilfoyle T (1997) Multiple auxin response modules in the soybean SAUR 15A promoter. Plant Sci 126: 193-201 [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100-3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DX (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4: 210-214 [DOI] [PubMed] [Google Scholar]