Abstract
The title compound, C15H13Br4NO4, was obtained via radical bromination reaction of ethyl 6,7-dimethoxy-2-methylquinoline-3-carboxylate and N-bromosuccinimide (NBS) in the presence of benzoyl peroxide (BPO) under photocatalytic conditions. The quinoline ring system is approximately planar with a maximum deviation from the mean plane of 0.035 (1) Å. The dihedral angle between the six-membered rings is 2.33 (2)°. The methoxy O atoms of the two neighboring methoxy groups are in-plane while their methyl C atoms are located on either side of the quinolyl ring plane at distances of −1.207 (1) and 1.223 (1) Å.
Related literature
The quinoline nucleus is widely present in numerous natural compounds, see: Michael et al. (1997 ▶, 2002 ▶). For the biological activity of quinoline derivatives, see: Heath et al. (2004 ▶); Keyaerts et al. (2004 ▶); Ko et al. (2001 ▶). For our previous work on the preparation of quinoline derivatives, see: Yang et al. (2007 ▶, 2008 ▶).
Experimental
Crystal data
C15H13Br4NO4
M r = 590.90
Triclinic,

a = 8.992 (2) Å
b = 9.632 (2) Å
c = 11.454 (3) Å
α = 84.868 (3)°
β = 71.948 (3)°
γ = 77.552 (3)°
V = 920.8 (4) Å3
Z = 2
Mo Kα radiation
μ = 8.76 mm−1
T = 298 K
0.25 × 0.20 × 0.18 mm
Data collection
Bruker APEXII CCD area detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2002 ▶) T min = 0.218, T max = 0.302
4750 measured reflections
3257 independent reflections
2373 reflections with I > 2σ(I)
R int = 0.035
Refinement
R[F 2 > 2σ(F 2)] = 0.043
wR(F 2) = 0.105
S = 0.97
3257 reflections
220 parameters
H-atom parameters constrained
Δρmax = 0.97 e Å−3
Δρmin = −0.74 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks quinoline, I. DOI: 10.1107/S1600536810029351/si2277sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810029351/si2277Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
We are grateful to the National Natural Science Foundation of China (grant No. 20802021) and the Natural Science Foundation of Guangdong Province, China (grant No. 8251063101000002).
supplementary crystallographic information
Comment
The quinoline ring system is widely present in numerous natural compounds (Michael et al., 1997, 2002). Quinoline derivatives are pharmacologically active compounds displaying a wide range of biological activity (Heath et al., 2004; Keyaerts et al., 2004; Ko et al., 2001)).
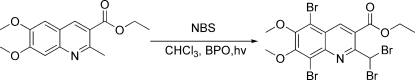
In previous works, we have reported the synthesis of some new quinoline derivatives (Yang et al., 2007, 2008). Herein, we report the synthesis and structure determination of a new Bromine-containing quinoline derivative, resulting from the radical bromination of ethyl 6,7-dimethoxy-2-methylquinoline-3-carboxylate under photocatalytic conditions. Our attempt to brominate the methyl group linked at C-2 position of quinoline ring, which has an acetal function at C-3, failed and led to 5,8-Dibromo-2-dibromomethyl-6,7- dimethoxy-quinoline-3-carboxylic acid, ethyl ester and other by-pruducts. This compound is the result of a unwanted reaction.
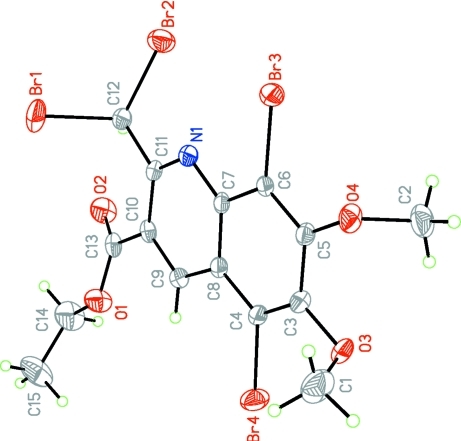
The molecular geometry of the title compound is illustrated in Fig 1. The title molecule contains an approximate planar quinolyl moiety with a maximum deviation from the mean plane of 0.035 (1)Å. The dihedral angle between the six-membered rings is 2.33 (2)°. The methoxy O atoms of the two neighboring methoxy groups are in-plane and their methyl C atoms locate on both sides of the quinolyl ring plane with maximun out-of-plane deviations of -1.207 (1) and 1.223 (1)Å, respectively.
Experimental
The title compoud was syntheized by treating 1mmol of ethyl 6,7-dimethoxy-2-methylquinoline-3-carboxylate with 1.5mmol of N-bromosuccinimide (NBS) in presence of 0.5mmol of Benzoyl Peroxide (BPO) in CCl3 under photocatalytic conditions. The mixture was then cooled and filtered off and the filtrate was concentrated under reduced pressure. The crude product was purified by silica gel column chromatography with the gradient mixture of petroleum ether and ethyl acetate (v : v = 30 : 1) to afford the white product. Crystals suitable for X-ray analysis were obtained by slow evaporation of the mixted solution of petroleum ether and ethyl acetate of the title compound.
Refinement
The H atoms were positioned geometrically and allowed to ride on their parent atoms, with C—H = 0.93–0.98 Å, with Uiso(H) = 1.2 Ueq(C) or 1.5Ueq(Cmethyl).
Figures
Fig. 1.
Molecular structure of the title compound showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
Synthesis of the title compound.
Crystal data
| C15H13Br4NO4 | Z = 2 |
| Mr = 590.90 | F(000) = 564 |
| Triclinic, P1 | Dx = 2.131 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.992 (2) Å | Cell parameters from 3257 reflections |
| b = 9.632 (2) Å | θ = 1.9–25.2° |
| c = 11.454 (3) Å | µ = 8.76 mm−1 |
| α = 84.868 (3)° | T = 298 K |
| β = 71.948 (3)° | Block, colourless |
| γ = 77.552 (3)° | 0.25 × 0.20 × 0.18 mm |
| V = 920.8 (4) Å3 |
Data collection
| Bruker APEXII CCD area detector diffractometer | 3257 independent reflections |
| Radiation source: fine-focus sealed tube | 2373 reflections with I > 2σ(I) |
| graphite | Rint = 0.035 |
| phi and ω scans | θmax = 25.2°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2002) | h = −10→10 |
| Tmin = 0.218, Tmax = 0.302 | k = −8→11 |
| 4750 measured reflections | l = −13→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.105 | H-atom parameters constrained |
| S = 0.97 | w = 1/[σ2(Fo2) + (0.0511P)2] where P = (Fo2 + 2Fc2)/3 |
| 3257 reflections | (Δ/σ)max = 0.001 |
| 220 parameters | Δρmax = 0.97 e Å−3 |
| 0 restraints | Δρmin = −0.74 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.57694 (8) | 1.26070 (8) | 0.23987 (7) | 0.0629 (2) | |
| Br2 | 0.89932 (9) | 1.37644 (7) | 0.12167 (7) | 0.0596 (2) | |
| Br3 | 1.17101 (7) | 1.17543 (6) | 0.39139 (6) | 0.04485 (19) | |
| Br4 | 1.13760 (9) | 0.53496 (6) | 0.26742 (6) | 0.0561 (2) | |
| C1 | 1.2668 (10) | 0.5760 (8) | 0.5327 (7) | 0.075 (2) | |
| H18A | 1.2422 | 0.6536 | 0.5870 | 0.112* | |
| H18B | 1.3424 | 0.4998 | 0.5547 | 0.112* | |
| H18C | 1.1710 | 0.5430 | 0.5393 | 0.112* | |
| C2 | 1.5111 (8) | 0.8569 (7) | 0.4025 (6) | 0.0580 (18) | |
| H15A | 1.5351 | 0.7702 | 0.3592 | 0.087* | |
| H15B | 1.5688 | 0.8449 | 0.4618 | 0.087* | |
| H15C | 1.5419 | 0.9323 | 0.3454 | 0.087* | |
| C3 | 1.2399 (6) | 0.7379 (5) | 0.3697 (5) | 0.0342 (12) | |
| C4 | 1.1433 (6) | 0.7213 (5) | 0.3041 (5) | 0.0336 (12) | |
| C5 | 1.2496 (6) | 0.8762 (5) | 0.3958 (5) | 0.0314 (12) | |
| C6 | 1.1574 (6) | 0.9927 (5) | 0.3568 (5) | 0.0298 (11) | |
| C7 | 1.0550 (6) | 0.9772 (5) | 0.2876 (4) | 0.0283 (11) | |
| C8 | 1.0484 (6) | 0.8386 (5) | 0.2603 (5) | 0.0304 (12) | |
| C9 | 0.9471 (6) | 0.8295 (5) | 0.1901 (5) | 0.0345 (12) | |
| H19 | 0.9363 | 0.7405 | 0.1723 | 0.041* | |
| C10 | 0.8643 (6) | 0.9490 (5) | 0.1478 (5) | 0.0333 (12) | |
| C11 | 0.8822 (6) | 1.0830 (5) | 0.1777 (5) | 0.0321 (12) | |
| C12 | 0.7951 (6) | 1.2193 (5) | 0.1337 (5) | 0.0364 (13) | |
| H14 | 0.7917 | 1.2035 | 0.0513 | 0.044* | |
| C13 | 0.7654 (7) | 0.9337 (6) | 0.0670 (5) | 0.0397 (13) | |
| C14 | 0.6348 (9) | 0.7820 (7) | 0.0037 (6) | 0.072 (2) | |
| H16A | 0.7074 | 0.7516 | −0.0760 | 0.086* | |
| H16B | 0.5601 | 0.8673 | −0.0079 | 0.086* | |
| C15 | 0.5464 (10) | 0.6653 (8) | 0.0669 (7) | 0.082 (3) | |
| H17A | 0.6213 | 0.5837 | 0.0820 | 0.123* | |
| H17B | 0.4910 | 0.6396 | 0.0152 | 0.123* | |
| H17C | 0.4709 | 0.6988 | 0.1435 | 0.123* | |
| N1 | 0.9716 (5) | 1.0961 (4) | 0.2468 (4) | 0.0330 (10) | |
| O1 | 0.7234 (5) | 0.8094 (4) | 0.0843 (4) | 0.0535 (12) | |
| O2 | 0.7315 (5) | 1.0223 (5) | −0.0062 (4) | 0.0556 (12) | |
| O3 | 1.3345 (5) | 0.6235 (4) | 0.4075 (4) | 0.0436 (10) | |
| O4 | 1.3441 (5) | 0.8917 (4) | 0.4638 (3) | 0.0429 (10) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0304 (4) | 0.0793 (5) | 0.0675 (5) | 0.0064 (3) | −0.0113 (3) | 0.0034 (4) |
| Br2 | 0.0622 (5) | 0.0435 (4) | 0.0804 (5) | −0.0165 (3) | −0.0315 (4) | 0.0111 (3) |
| Br3 | 0.0409 (4) | 0.0368 (3) | 0.0633 (4) | −0.0074 (3) | −0.0231 (3) | −0.0080 (3) |
| Br4 | 0.0643 (5) | 0.0319 (3) | 0.0826 (5) | −0.0068 (3) | −0.0370 (4) | −0.0071 (3) |
| C1 | 0.069 (6) | 0.070 (5) | 0.076 (6) | 0.002 (4) | −0.027 (5) | 0.025 (4) |
| C2 | 0.038 (4) | 0.073 (5) | 0.072 (5) | −0.013 (3) | −0.023 (4) | −0.016 (3) |
| C3 | 0.030 (3) | 0.031 (3) | 0.038 (3) | 0.000 (2) | −0.009 (3) | −0.003 (2) |
| C4 | 0.027 (3) | 0.028 (3) | 0.048 (3) | −0.003 (2) | −0.013 (3) | −0.005 (2) |
| C5 | 0.025 (3) | 0.033 (3) | 0.036 (3) | −0.002 (2) | −0.011 (2) | −0.003 (2) |
| C6 | 0.020 (3) | 0.029 (3) | 0.040 (3) | −0.005 (2) | −0.007 (2) | −0.006 (2) |
| C7 | 0.018 (3) | 0.033 (3) | 0.032 (3) | −0.003 (2) | −0.006 (2) | −0.003 (2) |
| C8 | 0.023 (3) | 0.032 (3) | 0.036 (3) | −0.005 (2) | −0.008 (2) | −0.003 (2) |
| C9 | 0.030 (3) | 0.033 (3) | 0.042 (3) | −0.007 (2) | −0.012 (3) | −0.001 (2) |
| C10 | 0.022 (3) | 0.044 (3) | 0.037 (3) | −0.009 (2) | −0.010 (2) | −0.002 (2) |
| C11 | 0.021 (3) | 0.032 (3) | 0.039 (3) | 0.000 (2) | −0.006 (2) | −0.002 (2) |
| C12 | 0.030 (3) | 0.037 (3) | 0.043 (3) | −0.004 (2) | −0.015 (3) | 0.001 (2) |
| C13 | 0.023 (3) | 0.053 (4) | 0.043 (3) | −0.006 (3) | −0.010 (3) | −0.005 (3) |
| C14 | 0.079 (6) | 0.084 (5) | 0.083 (5) | −0.030 (5) | −0.057 (5) | −0.003 (4) |
| C15 | 0.089 (7) | 0.104 (6) | 0.091 (6) | −0.060 (5) | −0.060 (5) | 0.021 (5) |
| N1 | 0.028 (3) | 0.033 (2) | 0.038 (3) | −0.004 (2) | −0.013 (2) | 0.0012 (19) |
| O1 | 0.064 (3) | 0.051 (2) | 0.068 (3) | −0.021 (2) | −0.045 (3) | 0.004 (2) |
| O2 | 0.060 (3) | 0.063 (3) | 0.061 (3) | −0.017 (2) | −0.042 (3) | 0.008 (2) |
| O3 | 0.033 (2) | 0.036 (2) | 0.060 (3) | 0.0060 (18) | −0.021 (2) | 0.0009 (18) |
| O4 | 0.037 (3) | 0.051 (2) | 0.049 (2) | −0.0027 (19) | −0.026 (2) | −0.0097 (18) |
Geometric parameters (Å, °)
| Br1—C12 | 1.939 (6) | C7—N1 | 1.358 (6) |
| Br2—C12 | 1.919 (5) | C7—C8 | 1.415 (6) |
| Br3—C6 | 1.876 (4) | C8—C9 | 1.409 (6) |
| Br4—C4 | 1.894 (5) | C9—C10 | 1.367 (7) |
| C1—O3 | 1.449 (8) | C9—H19 | 0.9300 |
| C1—H18A | 0.9600 | C10—C11 | 1.418 (7) |
| C1—H18B | 0.9600 | C10—C13 | 1.504 (7) |
| C1—H18C | 0.9600 | C11—N1 | 1.322 (6) |
| C2—O4 | 1.426 (7) | C11—C12 | 1.507 (7) |
| C2—H15A | 0.9600 | C12—H14 | 0.9800 |
| C2—H15B | 0.9600 | C13—O2 | 1.200 (6) |
| C2—H15C | 0.9600 | C13—O1 | 1.313 (6) |
| C3—C4 | 1.353 (7) | C14—O1 | 1.465 (6) |
| C3—O3 | 1.365 (6) | C14—C15 | 1.517 (11) |
| C3—C5 | 1.417 (7) | C14—H16A | 0.9700 |
| C4—C8 | 1.415 (7) | C14—H16B | 0.9700 |
| C5—O4 | 1.355 (5) | C15—H17A | 0.9600 |
| C5—C6 | 1.373 (7) | C15—H17B | 0.9600 |
| C6—C7 | 1.425 (6) | C15—H17C | 0.9600 |
| O3—C1—H18A | 109.5 | C8—C9—H19 | 119.4 |
| O3—C1—H18B | 109.5 | C9—C10—C11 | 118.0 (4) |
| H18A—C1—H18B | 109.5 | C9—C10—C13 | 119.2 (5) |
| O3—C1—H18C | 109.5 | C11—C10—C13 | 122.8 (5) |
| H18A—C1—H18C | 109.5 | N1—C11—C10 | 122.7 (5) |
| H18B—C1—H18C | 109.5 | N1—C11—C12 | 116.4 (4) |
| O4—C2—H15A | 109.5 | C10—C11—C12 | 120.9 (4) |
| O4—C2—H15B | 109.5 | C11—C12—Br2 | 113.1 (3) |
| H15A—C2—H15B | 109.5 | C11—C12—Br1 | 109.2 (4) |
| O4—C2—H15C | 109.5 | Br2—C12—Br1 | 111.5 (3) |
| H15A—C2—H15C | 109.5 | C11—C12—H14 | 107.6 |
| H15B—C2—H15C | 109.5 | Br2—C12—H14 | 107.6 |
| C4—C3—O3 | 121.2 (4) | Br1—C12—H14 | 107.6 |
| C4—C3—C5 | 120.0 (5) | O2—C13—O1 | 123.9 (5) |
| O3—C3—C5 | 118.7 (4) | O2—C13—C10 | 124.7 (5) |
| C3—C4—C8 | 122.2 (4) | O1—C13—C10 | 111.4 (5) |
| C3—C4—Br4 | 118.8 (4) | O1—C14—C15 | 106.3 (4) |
| C8—C4—Br4 | 119.0 (3) | O1—C14—H16A | 110.5 |
| O4—C5—C6 | 120.8 (4) | C15—C14—H16A | 110.5 |
| O4—C5—C3 | 119.6 (4) | O1—C14—H16B | 110.5 |
| C6—C5—C3 | 119.6 (4) | C15—C14—H16B | 110.5 |
| C5—C6—C7 | 121.1 (4) | H16A—C14—H16B | 108.7 |
| C5—C6—Br3 | 119.4 (3) | C14—C15—H17A | 109.5 |
| C7—C6—Br3 | 119.5 (4) | C14—C15—H17B | 109.5 |
| N1—C7—C8 | 122.5 (4) | H17A—C15—H17B | 109.5 |
| N1—C7—C6 | 118.7 (4) | C14—C15—H17C | 109.5 |
| C8—C7—C6 | 118.7 (4) | H17A—C15—H17C | 109.5 |
| C9—C8—C4 | 125.3 (4) | H17B—C15—H17C | 109.5 |
| C9—C8—C7 | 116.4 (4) | C11—N1—C7 | 119.2 (4) |
| C4—C8—C7 | 118.3 (4) | C13—O1—C14 | 115.0 (4) |
| C10—C9—C8 | 121.2 (5) | C3—O3—C1 | 114.0 (5) |
| C10—C9—H19 | 119.4 | C5—O4—C2 | 114.8 (4) |
| O3—C3—C4—C8 | −177.7 (5) | C8—C9—C10—C11 | −0.6 (8) |
| C5—C3—C4—C8 | −0.5 (8) | C8—C9—C10—C13 | 176.4 (5) |
| O3—C3—C4—Br4 | 1.2 (7) | C9—C10—C11—N1 | −1.7 (8) |
| C5—C3—C4—Br4 | 178.4 (4) | C13—C10—C11—N1 | −178.7 (5) |
| C4—C3—C5—O4 | 178.6 (5) | C9—C10—C11—C12 | 179.8 (5) |
| O3—C3—C5—O4 | −4.1 (7) | C13—C10—C11—C12 | 2.8 (8) |
| C4—C3—C5—C6 | 1.6 (8) | N1—C11—C12—Br2 | 26.6 (6) |
| O3—C3—C5—C6 | 178.9 (5) | C10—C11—C12—Br2 | −154.9 (4) |
| O4—C5—C6—C7 | −178.5 (5) | N1—C11—C12—Br1 | −98.2 (5) |
| C3—C5—C6—C7 | −1.6 (8) | C10—C11—C12—Br1 | 80.3 (5) |
| O4—C5—C6—Br3 | 3.2 (7) | C9—C10—C13—O2 | −154.5 (6) |
| C3—C5—C6—Br3 | −179.9 (4) | C11—C10—C13—O2 | 22.4 (9) |
| C5—C6—C7—N1 | −177.2 (5) | C9—C10—C13—O1 | 24.0 (7) |
| Br3—C6—C7—N1 | 1.1 (7) | C11—C10—C13—O1 | −159.1 (5) |
| C5—C6—C7—C8 | 0.5 (7) | C10—C11—N1—C7 | 2.5 (8) |
| Br3—C6—C7—C8 | 178.8 (4) | C12—C11—N1—C7 | −179.0 (5) |
| C3—C4—C8—C9 | 179.0 (5) | C8—C7—N1—C11 | −0.8 (8) |
| Br4—C4—C8—C9 | 0.1 (7) | C6—C7—N1—C11 | 176.8 (5) |
| C3—C4—C8—C7 | −0.6 (8) | O2—C13—O1—C14 | 2.1 (9) |
| Br4—C4—C8—C7 | −179.5 (4) | C10—C13—O1—C14 | −176.4 (5) |
| N1—C7—C8—C9 | −1.4 (7) | C15—C14—O1—C13 | −159.2 (6) |
| C6—C7—C8—C9 | −179.0 (5) | C4—C3—O3—C1 | −97.1 (6) |
| N1—C7—C8—C4 | 178.2 (5) | C5—C3—O3—C1 | 85.7 (6) |
| C6—C7—C8—C4 | 0.5 (7) | C6—C5—O4—C2 | −111.0 (6) |
| C4—C8—C9—C10 | −177.5 (5) | C3—C5—O4—C2 | 72.0 (6) |
| C7—C8—C9—C10 | 2.1 (8) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SI2277).
References
- Bruker (2002). SADABS Bruker AXS Inc., Madison. Wisconsin, USA.
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison. Wisconsin, USA.
- Heath, J. A., Mehrotra, M. M., Chi, S., Yu, J. C., Hutchaleelaha, A., Hollenbach, S. J., Giese, N. A., Scarborough, R. M. & Pandey, A. (2004). Bioorg. Med. Chem. Lett.14, 4867–4872. [DOI] [PubMed]
- Keyaerts, E., Vijgen, L., Mae, P., Neyts, J. & Ranst, M. V. (2004). Biochem. Biophys. Res. Commun.323, 264–268. [DOI] [PMC free article] [PubMed]
- Ko, T. C., Hour, M. J., Lien, J. C., Teng, C. M., Lee, K. H., Kuo, S. C., Huang, L. J. (2001). Bioorg. Med. Chem. Lett.11, 279–282. [DOI] [PubMed]
- Michael, J. P. (1997). Nat. Prod. Rep.14, 605–618.
- Michael, J. P. (2002). Nat. Prod. Rep.19, 742–760. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Yang, D. Q., Guo, W., Cai, Y. P., Jiang, L. S., Jiang, K. L. & Wu, X. B. (2008). Heteroat. Chem.19, 229–233.
- Yang, D. Q., Jiang, K. L., Li, J. N. & Xu, F. (2007). Tetrahedron, 63, 7654–7658.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks quinoline, I. DOI: 10.1107/S1600536810029351/si2277sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810029351/si2277Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report