Abstract
Phosphorylation of phosphoenolpyruvate carboxylase (PEPc; EC 4.1.1.31) plays an important role in the control of central metabolism in higher plants. Two PPCK (PEPc kinase) genes have been identified in tomato (Lycopersicon esculentum cv Alicante), hereafter termed LePPCK1 and LePPCK2. The function of the gene products has been confirmed by transcription of full-length cDNAs, translation, and in vitro assay of kinase activity. Previously studied PPCK genes contain a single intron. LePPCK2 also contains a novel second intron that exhibits alternative splicing. The correctly spliced transcript encodes a functional PEPc kinase, whereas unspliced or incorrectly spliced transcripts encode a truncated, inactive protein. The relative abundance of the transcripts depends on tissue and conditions. Expression of LePPCK2 was markedly increased during fruit ripening. In ripe Alicante fruit, the locule and seeds contained only the correctly spliced LePPCK2 transcripts, whereas in ripe fruit of the tomato greenflesh mutant, they contained correctly and incorrectly spliced transcripts. Potato (Solanum tuberosum) contains genes that are very similar to LePPCK1, and LePPCK2; StPPCK2 exhibits alternative splicing. Aubergine (Solanum melongena) and tobacco (Nicotiana tabacum) also contain a PPCK2 gene; the sequence of the alternatively spliced intron is highly conserved between all four species. The data suggest that the two PPCK genes have different roles in tissue-specific regulation of PEPc and that the alternative splicing of PPCK2 transcripts is functionally significant.
Phosphoenolpyruvate carboxylase (PEPc; EC 4.1.1.31) is a ubiquitous enzyme in higher plants. It catalyzes the carboxylation of phosphoenolpyruvate to form oxaloacetate and inorganic phosphate and plays a range of roles in different tissues. In C4 and Crassulacean acid metabolism (CAM) plants, a photosynthetic isoform of PEPc catalyzes the primary fixation of CO2 (O'Leary, 1982; Andreo et al., 1987). In most non-photosynthetic tissue and in C3 leaves, PEPc is the major anapleurotic enzyme. It allows the replenishment of tricarboxylic acid cycle intermediates to provide the precursors for biosynthetic pathways such as amino acid biosynthesis. It also provides malate in guard cells, legume root nodules, and developing fruit (Andreo et al., 1987; Chollet et al., 1996; Vidal and Chollet, 1997). To fulfill these different functions, PEPc is encoded by a small gene family whose members are expressed in different types of tissue (Lepiniec et al., 1994).
PEPc is strongly regulated by pH and by positive and negative effectors (Glc 6-phosphate and malate, respectively; Andreo et al., 1987). Superimposed on this, reversible phosphorylation of a strictly conserved Ser residue near the N-terminal end of the protein causes a change in the allosteric properties of PEPc. This phosphorylation reduces the sensitivity of the enzyme to its inhibitor, malate, but increases its sensitivity to its activator, Glc 6-phosphate (Chollet et al., 1996; Vidal and Chollet, 1997; Nimmo, 2000). The phosphorylation state of PEPc and, therefore, also its activity increase in response to different signals in different organs, including light in C4 leaves, a circadian oscillator in CAM leaves, and light plus nitrogen supply in C3 leaves; these signals control the activity of PEPc kinase (Chollet et al., 1996; Vidal and Chollet, 1997; Nimmo, 2000, 2003).
After the cloning of PEPc kinase genes (termed PPCK), it has become clear that PEPc kinase comprises a Ser/Thr kinase catalytic domain with essentially no extensions. It resembles the protein kinase catalytic domain of plant calcium-dependent protein kinases but lacks the C-terminal Ca2+-binding EF hands and the N-terminal extensions of these enzymes (Hartwell et al., 1999a). PEPc kinase is regulated largely at the level of transcript abundance in CAM, C4, and C3 plants (Hartwell et al., 1999a, 1999b; Nimmo, 2000, 2003; Taybi et al., 2000; Tsuchida et al., 2001; Fontaine et al., 2002; Nakagawa et al., 2003; Xu et al., 2003).
Recent work in the model C3 species Arabidopsis showed the existence of two isoforms of PEPc kinase with different expression patterns (Fontaine et al., 2002), and soybean (Glycine max) contains at least three PPCK genes expressed in different organs (Nimmo, 2003; Xu et al., 2003). This raises the possibility that distinct isoforms of the PEPc kinase play different metabolic roles. We have investigated this question in tomato (Lycopersicon esculentum) because of the importance of PEPc in the production of organic acids in developing fruit. Malic and citric acid levels increase at the end of the cell division phase of development, peak at the end of the cell expansion phase, and decline during ripening (Varga and Bruinsma, 1986; Guillet et al., 2002). Expression of one PEPc gene, termed Ppc2, correlated with high levels of PEPc activity and the accumulation of organic acids during development of tomato fruit (Guillet et al., 2002). This raises the question of whether any PPCK gene shows a similar expression pattern.
Here, we report on the cloning and expression of two isoforms of PEPc kinase from tomato. The data show one unique feature of PEPc kinase in the Solanaceae. All PEPc kinase genes reported to date contain just one intron, close to the 3′ end of the coding sequence. However, one of the PEPc kinase genes of tomato contains a novel second intron, which is subject to alternative splicing. One transcript encodes a functional PEPc kinase, but two transcripts encode a truncated, inactive protein. Our results show that the relative abundance of the transcripts is dependent on tissue and conditions. Similar genes, also subject to alternative splicing, have been detected in potato (Solanum tuberosum), tobacco (Nicotiana tabacum), and aubergine (Solanum melongena).
RESULTS
Identification of LePPCK1 and LePPCK2
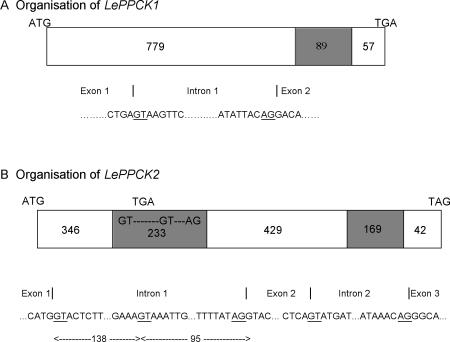
An examination of the expressed sequence tag (EST) database suggested that tomato contains two PPCK genes. One putative full-length PEPc kinase EST (GenBank accession no. AW033195), hereafter termed LePPCK1, was completely sequenced (Gen-Bank accession no. AF203481). The deduced amino acid sequence (279 residues, 31.4 kD) is aligned with several other PEPc kinase sequences in Figure 1. To examine the structure of the LePPCK1 gene, we designed PCR primers (see Table I) to amplify genomic DNA. A single band of 1148 bp was generated, cloned, and sequenced (GenBank accession no. AY190084). Inspection of this sequence showed that the coding sequence comprises two exons, with one 89-bp intron from base 808 to 896 of AY190084 inclusive (see Fig. 2).
Figure 1.
Alignment of deduced PPCK amino acid sequences. FtPPCK, Flaveria trinervia (AB065100); LePPCK1 and LePPCK2, tomato (this work); KfPPCK, Kalanchoë fedtschenkoi (AF162661); StPPCK1, potato (this work); and McPPCK, ice plant (Mesembryanthemum crystallinum, AF158091). Dots and vertical lines, Identities and similarities respectively.
Table I.
Primers used in reverse transcription (RT)-PCR analyses and cloning
| Primer Name and Specificity | Sequence |
|---|---|
| RT-PCR | |
| LePPCK1, 5′ | 5′-TCA AAT TTG CGA AGA AAT CG-3′ |
| LePPCK1, 3′ | 5′-CCT TCT CCT TCT CTC TTC CAC A-3′ |
| LePPCK2, 5′ | 5′-GAG CTA TTC GCC GTC AAG TC-3′ |
| LePPCK2, 3′ | 5′-AAA TCA GCC AAT TTC AGT TCG-3′ |
| Le Actin 52, 5′ | 5′-GAT GCC TAT GTT GGT GAC GA-3′ |
| Le Actin 52, 3′ | 5′-ATC CTC CGA TCC AGA CAC TG-3′ |
| StPPCK2, 5′ | 5′-GAG TCA TTC GCC GTC AAG TC-3′ |
| StPPCK2, 3′ | 5′-AAA TCA GCC AAT TTC AGC TCG-3′ |
| Cloning | |
| LePPCK2, 5′ FL | 5′-AGG AAT TCG GCA CGA GAA A-3′ |
| LePPCK2, 3′ FL | 5′-GAC CAG AGT ATA TTG GTG CCT GT-3′ |
| StPPCK1, 5′ | 5′-ATG TTC CAA ACC TTG AAA AA-3′ |
| StPPCK1, 3′ | 5′-TCA GTT TAG ATC AGC CAT TG-3′ |
Figure 2.
Organization of the LePPCK1 and LePPCK2 genes. White boxes, Exons; shaded boxes, introns. A, LePPCK1; B, LePPCK2. The numbers show the lengths of exons and introns in base pairs. The sequences round the splice sites are shown in full; the intron start GT and end AG sequences are underlined.
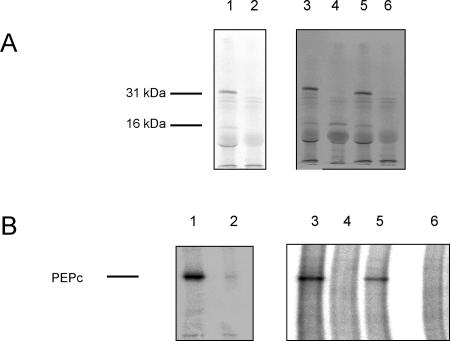
The LePPCK1 cDNA was subcloned behind a T3 promoter in pBluescript. To test whether the full-length cDNA was functional, the plasmid was linearized with NotI, transcribed, and translated. The translation product was then assayed for PEPc kinase activity. Figure 3 shows that the translation product was of the expected size (31 kD) and was able to phosphorylate PEPc.
Figure 3.
Functional analysis of LePPCK cDNAs. A, Phosphor images of [35S]Met-labeled products from in vitro translation of RNA samples, separated on a 12.5% (w/v) SDS polyacrylamide gel. B, Phosphor images of immunoprecipitated, 32P-labeled PEPc from assays of the PEPc kinase activity of translation products, separated on an 8% (w/v) SDS polyacrylamide gel. The bar indicates PEPc. In each panel, the lanes are: 1, RNA transcribed from the cDNA clone of LePPCK1; 2, no RNA control; 3, RNA transcribed from the cDNA clone of LePPCK2 transcript 1; 4, RNA transcribed from the cDNA clone of LePPCK2 transcript 2; 5, RNA transcribed from the cDNA clone of K. fedtschenkoi PPCK as a positive control; and 6, no RNA control.
A second putative full-length tomato PEPc kinase EST (GenBank accession no. AW223421), hereafter termed LePPCK2, was also completely sequenced (data not shown; see below). Inspection of this sequence showed that the cDNA contains an insertion of 138 bp near the middle of the coding region relative to other PEPc kinase cDNAs. The insertion also included an in-frame stop codon. To examine the structure of the gene, PCR primers were designed to amplify cDNA (from tomato fruit) and genomic DNA, and the products were sequenced. Analysis of these sequences revealed that the LePPCK2 gene (GenBank accession no. AY188444) contains two introns (Fig. 2). Like other PEPc kinase genes, one is located close to the 3′ end of the coding sequence (bases 1,044-1,212 of AY188444). The second intron (bases 372-604 of AY188444) is located close to the middle of the coding region and possesses two alternative 5′ splice start sites (bases 372 and 510, see Fig. 2). The in-frame stop codon is between these sites (bases 450-452 of AY188444). Hence, splicing can give rise to a transcript that encodes a functional PEPc kinase (transcript 1, the full sequence of which is GenBank accession no. AY187634), an incorrectly spliced transcript with a premature stop codon (transcript 2), and an unspliced transcript with a premature stop codon (transcript 3). Transcripts 2 and 3 both encode a truncated, nonfunctional PEPc kinase. As shown in Table II, ESTs corresponding to each transcript have been detected. The deduced amino acid sequence encoded by transcript 1 (278 residues, 31.1 kD) is shown in Figure 1. There is 61% sequence identity between the two tomato PPCK proteins.
Table II.
Origin of PPCK ESTs
The Institute for Genomic Research tentative consensus sequences TC125298, TC117287, TC117288, and TC117289 refer to LePPCK1, LePPCK2 transcript 1, LePPCK2 transcript 2, and LePPCK2 transcript 3, respectively (see http://www.tigr.org/tdb/tgi/lgi/), whereas TC67959, TC59893, and TC59894 refer to StPPCK1, StPPCK2 transcript 2, and StPPCK2 transcript 3, respectively (see http://www.tigr.org/tdb/tgi/stgi/).
| GenBank Accession No., Source of Clone |
|---|
| LePPCK1 |
| AI774158, leaf, Pseudomonas syringae pv. tomato resistant |
| AW033195, callus |
| AW933544, green fruit |
| BE431605, breaker fruit |
| BE459112, developing immature green fruit |
| BF096819, nutrient-deficient roots |
| BG129025, shoot meristem |
| StPPCK1 |
| BG887336, dormant tuber |
| BG887353, dormant tuber |
| BG888336, dormant tuber |
| BQ045511, leaves challenged with Phytophthora infestans, incompatible interaction |
| LePPCK2 |
| AW222608, red ripe fruit |
| AW223421, red ripe fruit, transcript 2 |
| AW441584, red ripe fruit |
| AW442172, red ripe fruit |
| AW738217, flower buds, anthesis |
| BE462009, breaker fruit, transcript 2 |
| BF112946, breaker fruit, transcript 3 |
| BG127024, shoot meristem, transcript 3 |
| BI205375, suspension culture |
| BI934398, flower, anthesis, transcript 2 |
| BM409042, breaker fruit, transcript 2 |
| BM409651, breaker fruit, transcript 1 |
| BM411455, breaker fruit |
| BM536300, breaker fruit, transcript 2 |
| StPPCK2 |
| BG594065, sprouting eyes, transcript 3 |
| BG594668, sprouting eyes, transcript 2 |
| BQ505603, mixed tissues, transcript 3 |
| BQ505604, mixed tissues |
| BQ505884, mixed tissues, transcript 2 |
| BQ505885, mixed tissues, transcript 2 |
LePPCK2 transcript 1 was positioned behind a T7 promoter in pCR4-TOPO, and LePPCK2 transcript 2 was positioned behind a T3 promoter in pBluescript. The clones were linearized, transcribed, and translated. The translation products were then assayed for PEPc kinase activity. Figure 3 (lane 3) shows that the product from transcript 1 was of the expected size (31 kD) and was able to phosphorylate PEPc as effectively as did K. fedtschenkoi PEPc kinase generated in a similar way (lane 5). Transcript 2 consistently directed the synthesis of a 16-kD protein (compare Fig. 3, lane 4 with the no RNA control lane 6 in A) that did not phosphorylate PEPc. The size of this protein was as expected from the position of the in-frame stop codon in LePPCK2 transcript 2. A slightly smaller band can be seen in the translation products from the full-length PEPc kinase clones (lanes 1, 3 and 5). This presumably results from either breakdown of the major product or premature termination of translation.
PEPc Kinase Expression in Different Tissues of Tomato
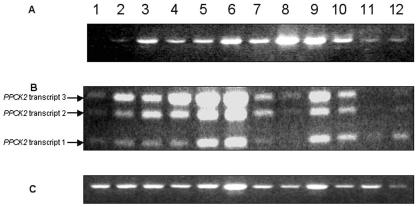
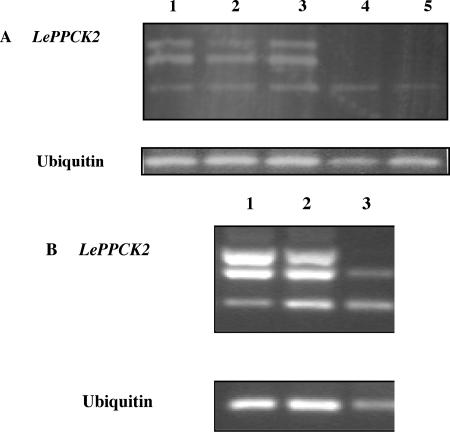
PEPc kinase is expressed at a low level in C3 plants, with the exception of legume root nodules (Nakagawa et al., 2003; Xu et al., 2003). The expression of both tomato PEPc kinases was studied, therefore, by semiquantitative RT-PCR rather than by northern blotting. The primers used for the two PPCK genes and for actin52 as a constitutive control are shown in Table I. We investigated developing fruit at several stages: young leaves, mature leaves, flowers, roots, and seedlings harvested in the light and dark. The cycle number of 35 was chosen to ensure that bands could be detected in all samples; similar band patterns were detected with lower cycle numbers (data not shown). Although there is some variation in the intensities of the actin52 bands, it is clear that LePPCK1 transcripts were present in all of the samples studied, lowest in expansion phase 1 fruit, and highest in mature leaves. Expression in seedlings was enhanced in the light (Fig. 4A, compare lane 10 with 11). The expression of LePPCK2 showed three interesting features (Fig. 4B). First, it was strongly increased during fruit ripening. Second, as for LePPCK1 expression, transcript abundance was increased in the light (Fig. 4B, compare lane 10 with 11). Third, the relative abundance of the three transcripts appeared to depend upon both tissue and conditions. In ripening fruit and illuminated seedlings, transcripts 2 and 3 were more abundant than transcript 1. In roots and in seedlings in the dark, transcript 1 was the major species. Differences in the relative abundance of the transcripts are further illustrated in Figure 5A, which shows that the skin, outer pericarp, and inner pericarp of ripe fruit contain all three transcripts, whereas in the locule and seeds, only transcript 1 is detectable.
Figure 4.
Expression pattern of LePPCK genes. This shows the products obtained from RT-PCR (35 cycles). A, LePPCK1; B, LePPCK2; C, Actin52. Lane 1, Expansion phase 1 fruit; lane 2, expansion phase 2 fruit; lane 3, green fruit; lane 4, breaker fruit; lane 5, pink fruit; lane 6, ripe red fruit; lane 7, young leaves; lane 8, mature leaves; lane 9, flowers, lane 10, seedlings in light; lane 11, seedlings in dark; lane 12, roots.
Figure 5.
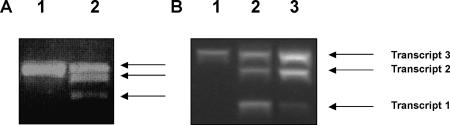
Expression pattern of LePPCK2 in fruit compartments. This shows the products obtained from RT-PCR (35 cycles) of compartments from ripe red fruit. A, LePPCK2 and ubiquitin from wild-type plants. Lane 1, Skin; lane 2, outer pericarp; lane 3, inner pericarp; lane 4, gel; lane 5, seeds. B, LePPCK2 and ubiquitin from the gf mutant. Lane 1, Skin; lane 2, pericarp; lane 3, locule plus seeds.
In view of the developmental control of expression exhibited by LePPCK2 and the positional control of splicing shown in Figures 4 and 5A, we examined the expression of this gene in the gf (greenflesh) tomato mutant, in which the breakdown of chlorophyll during fruit ripening is markedly reduced (Akhtar et al., 1999). One notable difference was observed between gf and Alicante: In the gf mutant, the splicing pattern of LePPCK2 was affected such that the seeds and locule of ripe gf fruit contained both transcripts 1 and 2 (Fig. 5B), whereas Alicante contained only transcript 1 (Fig. 5A).
Identification of Potato, Tobacco, and Aubergine PEPc Kinases
Further analysis of the EST database revealed two putative potato PEPc kinase genes. As summarized in Table II, several ESTs closely resemble LePPCK1; none of these, however, are full length. Other ESTs resemble either transcript 2 or 3 of LePPCK2. To examine the structure of the corresponding genes, hereafter termed StPPCK1 and StPPCK2, we designed PCR primers (see Table I) to amplify cDNA (from mature leaves) and genomic DNA. Using StPPCK1 primers, a partial genomic sequence of 913 bp was cloned and sequenced (GenBank accession no. AY219178); this contains only one intron as for LePPCK1 (bases 765-877 of AY219178). Comparison of the deduced amino acid sequences of StPPCK1 and LePPCK1 shows 98.5% identity over 265 amino acids (see Fig. 1). To test whether StPPCK2 exhibits alternative splicing, we designed primers to the region flanking its putative additional intron based on the sequence of the ESTs BG594065 and BG594668. Using these, we amplified three bands of the predicted mobilities from potato leaf cDNA but only one from genomic DNA (Fig. 6A), indicating that StPPCK2 does contain an additional intron that is subject to alternative splicing. The sequence of the partial genomic fragment of StPPCK2 (555 bp, GenBank accession no. AY293738) matches the sequences of the ESTs corresponding to StPPCK2 (Table II) almost exactly (not illustrated).
Figure 6.
Analysis of potato and tobacco PPCK2 transcripts. A, Potato; lane 1, products obtained from PCR of genomic DNA; lane 2, products obtained from RT-PCR of mature leaf RNA (35 cycles). B, tobacco; lane 1, products obtained from PCR of genomic DNA; lane 2, products obtained from RT-PCR of flower RNA; lane 3, products obtained from RT-PCR of mature leaf RNA (33 cycles). The arrows indicate (from top to bottom) transcripts 3, 2, and 1.
To check other members of the Solanaceae, we amplified and sequenced a 553-bp fragment of aubergine genomic DNA (GenBank accession no. AY236482) and a 538-bp fragment of tobacco genomic DNA (GenBank accession no. AY347261) using the primers designed for RT-PCR of LePPCK2 (Table I) that span the unusual intron in LePPCK2. The resulting sequences are very similar to the sequences of the LePPCK2 and StPPCK2 genes. Figure 7 shows a comparison of the intron sequences, which comprise 233, 234, 232, and 217 bp in tomato, potato, aubergine, and tobacco, respectively. The two donor splice sites and the acceptor site are conserved. All four sequences contain at least one in-frame stop codon 70 to 80 bp into the intron (bold in Fig. 7); aubergine has two closely adjacent stop codons. The tobacco intron has a deletion of 18 bp relative to the other introns just 3′ to the second (“incorrect”) donor splice site. Using the same primers, we amplified three bands of the predicted mobilities from tobacco flower and leaf cDNA but only one from genomic DNA (Fig. 6B), indicating that the tobacco PPCK2 gene does contain an additional intron that is subject to alternative splicing. As with tomato, the relative abundance of the three transcripts differs between organs (Fig. 6B). The data argue strongly that both aubergine and tobacco contain a gene equivalent to the PPCK2 genes of potato and tomato.
Figure 7.
Sequences of the unusual PPCK2 intron. Tomato, Le; potato, St; aubergine, Sm; tobacco, Nt. The in-frame stop codons are in bold. The sequences conserved around the strong internal splice site used in LePPCK2 transcript 2 are underlined, with the intron start GT in italics.
DISCUSSION
The results presented here demonstrate that tomato contains at least two genes that encode functional PEPc kinases. LePPCK1 is similar to other reported PEPc kinase genes (Hartwell et al., 1999a; Taybi et al., 2000; Tsuchida et al., 2001; Fontaine et al., 2002; Nakagawa et al., 2003; Nimmo, 2003; Xu et al., 2003) in that it encodes a protein kinase catalytic domain with minimal N- and C-terminal extensions and contains one small intron toward the 3′ end of the coding region. LePPCK2 has one major difference from other PPCK genes studied to date, namely the presence of an additional intron that exhibits alternative splicing. Our data show that three types of transcript can be detected, corresponding to correctly spliced, incorrectly spliced, and unspliced transcripts. The correctly spliced transcript encodes a functional PEPc kinase very similar to the enzyme from other species, whereas the other two encode truncated, nonfunctional proteins. It remains to be seen whether the truncated product accumulates in cells. A similar gene has been detected in potato, aubergine, and tobacco (Figs. 6 and 7). To date, this unusual PPCK gene with alternative splicing has been found only in the Solanaceae.
Several lines of evidence suggest that this alternative splicing of PPCK2 transcripts may be functionally significant. First, the nucleotide sequence of the intron is highly conserved between four members of the Solanaceae (see Fig. 7); moreover, the intron and exon sequences are equally similar. For example, the aubergine and tomato sequences are 94% identical over 321 bases of exon sequence (not shown) and 92% identical over 233 intron bases (Fig. 7). The conservation of intron sequence in the PPCK2 genes of the four species includes the splice sites as shown both directly (Fig. 7) and functionally (Fig. 6); it also covers the presence of in-frame stop codons that would lead to premature truncation of the gene product (although in aubergine there are actually two in-frame stop codons as shown in Fig. 7). The major difference between the intron sequences is an 18-bp deletion in the tobacco sequence that is downstream of both the internal stop codon and the “incorrect” donor splice site. Second, as noted below, the relative abundance of the three transcripts in tomato depends on tissue and conditions. Third, transcripts 2 and 3 actually predominate in some samples even though they contain a premature in-frame stop codon, which often leads to instability of the mRNA (Abler and Green, 1996; Brown and Simpson, 1998). Taken separately, these arguments are not conclusive; together, they suggest strongly that the alternative splicing is significant.
Various possible roles for this alternative splicing can be envisaged. First, it could allow control of the abundance of the functional transcript 1, either in the context of tissue specificity or in response to a signal; a complex example of such behavior is found in the alternative processing of the Arabidopsis FCA gene (Macknight et al., 2002; Quesada et al., 2003). For example, the relative abundances of the three PPCK2 transcripts are very different in tomato roots, seeds, and locule (which contain mainly transcript 1) compared with leaves (which contain mainly transcripts 2 and 3; Figs. 4 and 5). In seedlings, light not only increases the expression of LePPCK2 overall but also reduces the relative abundance of transcript 1 (Fig. 4). One of the major metabolic changes that occur during fruit ripening is the breakdown of chlorophyll. Chlorophyll breakdown is reduced in the gf mutant, whereas several other parameters, such as accumulation of carotenoids, are not affected (Akhtar et al., 1999). The relative abundance of LePPCK2 transcripts is altered in the gf mutant (Fig. 5), emphasizing that the alternative splicing is dependent on metabolic context.
It is possible that cells contain limiting amounts of a splicing factor that is essential for the production of transcript 1; if so, roots must contain more of this factor than, for example, leaves. Requirement for a specific factor to give correct splicing would be consistent with the sequence of the intron. Plant introns are AU and particularly U rich (Brown and Simpson, 1998). For example, the 89-bp intron in LePPCK1 mRNA contains 15 A and 47 U residues (70% AU). Incorrect processing of the unusual intron in LePPCK2 mRNA removes a sequence of 95 bases that is 75% AU. In contrast, correct processing removes an additional 138 bases that is only 61% AU. Thus, the intron is more AU rich than the open reading frame (52% AU), but selection of the correct 5′ splice site must be made in the context of an uncommonly low AU content downstream of the site.
In a second potential role of the alternative splicing, it is possible that the truncated protein expressed in vitro from LePPCK2 transcripts 2 and 3 (Fig. 3) may accumulate in vivo and play a functional role. This truncated protein would extend just beyond the N-terminal ATP-binding domain of PEPc kinase; it may be able to fold stably, to bind ATP, and/or to interact with other proteins such as PEPc. Hence, this truncated protein could play an unsuspected role in the control of the phosphorylation state of PEPc. It clearly will be important to assess whether the truncated protein does accumulate in cells. However, it must be pointed out that PEPc kinase is a very low-abundance protein. For example, even in highly illuminated maize leaves, a rich source, it comprises less than 1 in 106 of soluble protein (Saze et al., 2001); our attempts to detect native PEPc kinase immunochemically in tomato tissue extracts have proved negative (data not shown).
To gain information about the possible functions of the two tomato PPCK genes, we examined their tissue expression patterns. Our RT-PCR data agree with the distribution of ESTs between tissues (Table II); both approaches show that LePPCK1 is expressed in many organs, whereas LePPCK2 is predominantly expressed in ripening fruit. This would suggest that LePPCK1 encodes a housekeeping kinase, mainly involved in ensuring the replenishment of the TCA cycle, whereas the main role of the kinase encoded by LePPCK2 is in late ripening. ESTs corresponding to the two potato PPCK genes are found in different libraries (Table II). Following the arguments of Ronning et al. (2003), this implies that the two genes have different functions; for example, StPPCK1 seems to be expressed in dormant tubers, whereas StPPCK2 is expressed in sprouting eyes.
Recently, Guillet et al. (2002) have reported the cloning and expression analysis of two PEPc genes from tomato (Ppc1 and Ppc2). The Ppc1 gene product is thought to be anapleurotic, whereas expression of Ppc2 is increased during the cell expansion phase of fruit development, before breaker phase. Ppc2 transcripts in unripe fruit were detected in the vacuolated cells in the pericarp, enlarging cells near the vicinity of the seeds and in the periphery of the vascular bundles. In ripening fruit, both expression of Ppc2 and PEPc activity drop considerably. Guillet et al. (2002) suggested that the Ppc2 gene product is probably involved in accumulation of malate, aiding cell expansion. Hence, the observation that LePPCK2 is expressed during ripening is surprising because it occurs at a time when the PEPc activity and malate content of the fruit are falling. It is possible that the LePPCK2 gene product either phosphorylates another PEPc or phosphorylates a different protein. In any event, it will be important to determine the precise spatial distribution of the functional and nonfunctional LePPCK2 transcripts and compare these with the distribution of Ppc2 transcripts.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum cv Alicante) plants were grown from seed in bedding compost (William Sinclair Horticulture Ltd., Lincoln, UK) in a greenhouse under a 16-h photoperiod supplemented with mercury vapor lamps (300 μmol m-2 s-1). Plants were repotted at 4 weeks and were watered every 2 d. Tomato fruits were allowed to ripen on the vine. RNA was isolated from quartered segments. For localization studies, fruit were collected at the ripe red stage and separated into skin, outer pericarp, inner pericarp, locule, and seeds. Young leaves were harvested as the primary leaf 14 d after sowing, whereas mature leaves were harvested after 6 weeks. Seedlings (8 d old) were harvested after either 10 h of darkness or 3 h of light.
Isolation of RNA and DNA
Plant material was frozen under liquid nitrogen and stored at -70°C. Frozen plant tissue (2-3 g) was ground to a fine powder using an autoclaved mortar and pestle. RNA was then isolated according to the protocol of Chang et al. (1993), with the following modifications. Ten milliliters of extraction buffer was used for 2 to 3 g of tissue. Chloroform was used rather than chloroform:isoamyl alcohol. After precipitation with LiCl, the pellet was resuspended in 500 μL of 10 mm Tris-HCl (pH 7.5) and 1 mm EDTA and extracted once with an equal volume of chloroform. Sodium acetate (50 μL of a 3 m solution [pH 5.2]) was added to the aqueous layer, followed by 2.5 volumes of ice-cold ethanol. RNA was allowed to precipitate overnight at 4°C. The quantity and purity of the RNA was determined spectrophotometrically according to the method described by Sambrook et al. (1989). Intactness of the RNA was determined by detection of ribosomal RNA bands after agarose gel electrophoresis. A DNase treatment (DNA-free, Ambion, Huntingdon, UK) was used to ensure elimination of contaminating DNA from RNA preparations.
Genomic DNA was extracted using a DNA isolation kit (PUREgene DNA isolation kit, Gentra Systems, Gentra Systems, Minneapolis). The quantity and purity of the DNA was determined spectrophotometrically according to the method described by Sambrook et al. (1989).
RT-PCR and PCR
The RNA samples (100 ng) were mixed with 0.25 μm oligo(dT) for 5 min at 70°C and cooled at 4°C for 5 min. RT was carried out in a reaction mixture (50 μL) containing avian myeloblastosis virus reverse transcriptase buffer, 1 mm dNTPs, 1 unit μL-1 RNase inhibitor, and 0.4 units μl-1 avian myeloblastosis virus reverse transcriptase (all from Promega, Madison, WI). The reaction was performed at 48°C for 45 min. The enzyme was then heat inactivated at 95°C for 5 min, and the samples were used directly for PCR.
PCR reactions were performed using 2.5 μL of each cDNA sample in a reaction mixture (25 μL) containing 12.5 μL of 2× Reddy Mix (Abgene, Epsom, UK) and 0.5 μm of the 5′ and 3′ primers. The primer sequences are shown in Table I. The PCR reactions were conducted in a programmable thermocycler (PCR Sprint, Hybaid, Ashford, UK). The reaction conditions for the amplification of LePPCK1 were an initial denaturation step of 94°C for 5 min, 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 60 s, and a final extension step of 72°C for 5 min. The reaction conditions for the amplification of LePPCK2 were an initial denaturation step of 94°C for 5 min, 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s, and a final extension step of 72°C for 5 min. Primers for either Actin52 or ubiquitin were used as a constitutive control in both conditions. After amplification, reactions were resolved by electrophoresis on a 1% (w/v) agarose gel and stained with ethidium bromide.
Cloning
PCR fragments were extracted from an agarose gel by purifying with QIAquik gel extraction kit (Qiagen USA, Valencia, CA) and cloned directly into pCR4-TOPO vectors using the TOPO TA cloning kit with one shot cells (Invitrogen, Carlsbad, CA). All clones were sequenced using universal primers (MWG-Biotech, Ebersberg, Germany).
Assessment of PEPc Kinase Activity Encoded by cDNAs
This was carried out as described previously (Fontaine et al., 2002). In outline, plasmids were linearized and transcribed from either a T3 or T7 promoter as appropriate. The transcripts were translated in a rabbit reticulocyte lysate system using [35S]Met, and the PEPc kinase activity of the translation products was assayed (Hartwell et al., 1996). Figure 3 shows phosphor images of 35S incorporated into protein and 32P incorporated into PEPc after SDS-PAGE.
Acknowledgments
We thank Prof. Don Grierson for the gift of gf tomato seeds.
This work was supported by the Biotechnology and Biological Sciences Research Council (PhD studentships to J.T.M. and S.S. and research support to H.G.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.030775.
References
- Abler ML, Green PJ (1996) Control of mRNA stability in higher plants. Plant Mol Biol 32: 63-78 [DOI] [PubMed] [Google Scholar]
- Akhtar MS, Goldschmidt EE, John I, Rodonio S, Matile P, Grierson D (1999) Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato (Lycopersicon esculentum Mill.). J Exp Bot 50: 1115-1122 [Google Scholar]
- Andreo CS, Gonzalez DH, Iglesias AA (1987) Higher plant phosphoenol- pyruvate carboxylase: structure and regulation. FEBS Lett 213: 1-8 [Google Scholar]
- Brown JWS, Simpson CG (1998) Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol 49: 77-95 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113-116 [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Phys Plant Mol Biol 47: 273-298 [DOI] [PubMed] [Google Scholar]
- Fontaine V, Hartwell J, Jenkins GI, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25: 115-122 [Google Scholar]
- Guillet C, Just D, Bénard N, Destrac-Irvine A, Baldet P, Hernould M, Causse M, Raymond P, Rothan C (2002) A fruit-specific phosphoenol- pyruvate carboxylase is related to rapid growth of tomato fruit. Planta 214: 717-726 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999a) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20: 333-342 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Jenkins GI, Wilkins MB, Nimmo HG (1999b) The light induction of maize phosphoenolpyruvate carboxylase kinase translatable mRNA requires transcription but not translation. Plant Cell Environ 22: 883-889 [Google Scholar]
- Hartwell J, Smith LH, Wilkins MB, Jenkins GI, Nimmo HG (1996) Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J 10: 1071-1078 [Google Scholar]
- Lepiniec L, Vidal J, Chollet R, Gadal P, Cretin C (1994) Phosphoenolpyruvate carboxylase:structure, regulation and evolution. Plant Sci 99: 111-124 [Google Scholar]
- Macknight R, Duroux M, Laurie C, Dijkwel P, Simpson G, Dean C (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14: 877-888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Izumi T, Banba M, Umehara Y, Kouchi H, Izui K, Hata S (2003) Characterization and expression analysis of genes encoding phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase of Lotus japonicus, a model legume. Mol Plant Microbe Interact 16: 281-288 [DOI] [PubMed] [Google Scholar]
- Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5: 75-80 [DOI] [PubMed] [Google Scholar]
- Nimmo HG (2003) Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch. Biochem Biophys 414: 189-196 [DOI] [PubMed] [Google Scholar]
- O'Leary MH (1982) Phosphoenolpyruvate carboxylase: an enzymologist's view. Annu Rev Plant Phys 33: 297-315 [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson G (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J 22: 3142-3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronning CM, Stegalkina SS, Ascenzi RA, Bougri O, Hart AL, Utterbach TR, Vanaken SE, Riedmuller SB, White JA, Cho J et al. (2003) Comparative analyses of potato expressed sequence tag libraries. Plant Physiol 131: 419-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Saze H, Ueno Y, Hisabori T, Hayashi H, Izui K (2001) Thioredoxin-mediated reductive activation of a protein kinase for the regulatory phosphorylation of C4-form phosphoenolpyruvate carboxylase from maize. Plant Cell Physiol 42: 1295-1302 [DOI] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in Crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471-1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida Y, Furumato T, Izumida A, Hata S, Izui K (2001) Phosphoenol- pyruvate carboxylase kinase involved in Flaveria trinervia: cDNA cloning and characterization. FEBS Lett 507: 318-322 [DOI] [PubMed] [Google Scholar]
- Varga A, Bruinsma J (1986) Tomato. In SP Monselise, ed, CRC Handbook of Fruit Set and Development. CRC Press, Boca Raton, FL, pp 461-491
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2: 230-237 [Google Scholar]
- Xu W, Zhou Y, Chollet R (2003) Identification and expression of a soybean nodule-enhanced PEP-carboxylase kinase gene (NE-PpcK) that shows striking up-/down-regulation in vivo. Plant J 34: 441-452 [DOI] [PubMed] [Google Scholar]