Abstract
The title compound, C20H26N2O5, was prepared in good yield (76%) through condensation of diethyl (4-methoxybenzyl)propanedioate with 3,5-dimethyl-1H-pyrazole. The dihedral between the benzene and pyrazole rings is 83.96 (10)°. The crystal packing is stabilized by a C—H⋯O interaction, which links the molecules into centrosymmetric dimers.
Related literature
For related compounds displaying biological activity, see: Dayam et al. (2007 ▶); Patil et al. (2007 ▶); Ramkumar et al. (2008 ▶); Sechi et al. (2009 ▶) & Zeng et al. (2008 ▶). For the synthetic procedure, see: Pommier & Neamati (2006 ▶).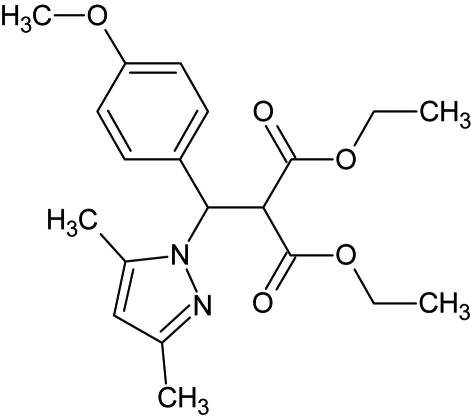
Experimental
Crystal data
C20H26N2O5
M r = 374.43
Monoclinic,
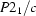
a = 11.9618 (3) Å
b = 7.9681 (2) Å
c = 21.1269 (6) Å
β = 96.504 (1)°
V = 2000.70 (9) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 296 K
0.23 × 0.17 × 0.14 mm
Data collection
Bruker X8 APEXII CCD area-detector diffractometer
18616 measured reflections
3921 independent reflections
3177 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.056
wR(F 2) = 0.154
S = 1.05
3921 reflections
249 parameters
H-atom parameters constrained
Δρmax = 0.68 e Å−3
Δρmin = −0.45 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810025572/bt5280sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810025572/bt5280Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Enhanced figure: interactive version of Fig. 3
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C12—H12⋯O2i | 0.93 | 2.51 | 3.358 (3) | 152 |
Symmetry code: (i) 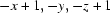 .
.
Acknowledgments
This work was supported by grants from Project PGR-UMP-BH-2005, the Centre National de Recherche Scientifique, CNRS (France), the Centre National pour la Recherche Scientifique et Technique, CNRST (Morocco), and the CURI (Morocco).
supplementary crystallographic information
Comment
For the rational design of new HIV-1 Integrase (H—I) inhibitors, one validated target for chemotherapeutic intervention (Dayam et al., 2007), is fundamentally based on intermolecular coordination between H—I / chemical inhibitor / metals (Mg+2 and Mn+2, co-factors of the enzyme), leading to the formation of bimetallic complexes (Zeng et al., 2008; Sechi et al., 2009). Thereby, several bimetallic metal complexes, in many cases exploring the known-well polydentate ligands, appear in this scenario as the most promising concept to be employed in either enzyme / drug interaction or electron transfer process, in the last case involving the biological oxygen transfer (Sechi et al., 2009; Ramkumar et al., 2008). Another exciting example of application for such polydentate ligands involves the synergic water activation, that occurs via the so-called -remote metallic atoms. Such organometallic compounds are structurally deemed to promote or block the H—I activity (Zeng et al., 2008).
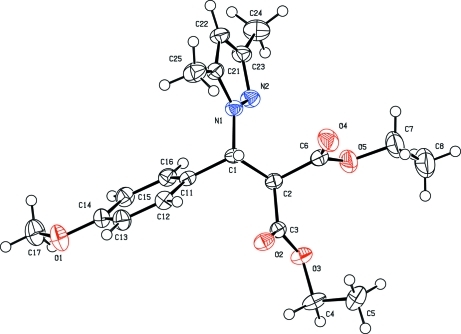
In the molecule of the title compound (Fig.1), the dihedral angle between the planes of the pheny and the pyrazol ring is 83.96 (10)°.
Experimental
To a solution of the diethyl (4-methoxybenzyl)propanedioate (5 mmol) in water (20 ml) was added the 3,5-dimethyl-1H-pyrazole (6 mmol) and the mixture and the stirring was continued at room temperature until the complete consume of the starting material. After removing solvent, the crude products were dissolved in diethyl ether (2x40 ml) and washed with water until the pH became neutral. The organic solvent was dried with sodium sulfate and then evaporated to give the pure compound (I) with 76% yield.. White crystals are obtained by recrystallization in ether/hexane (2/1).
Suitable single-crystal of malonate derivative (I) was obtained by recrystallization from ethanol. A white-transparent crystal was mounted on a glass fibre.
Refinement
All H atoms attached to C atoms were fixed geometrically and treated as riding with C—H = 0.96 Å (methyl), C—H = 0.93 Å (aromatic), 0.97 Å (methylene) and 0.98 Å (methine) with Uiso(H) = 1.2Ueq or Uiso(H) = 1.5Ueq(methyl).
Figures
Fig. 1.
Molecular structure of the title compound with the atom-labelling scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are represented as small spheres of arbitrary radii.
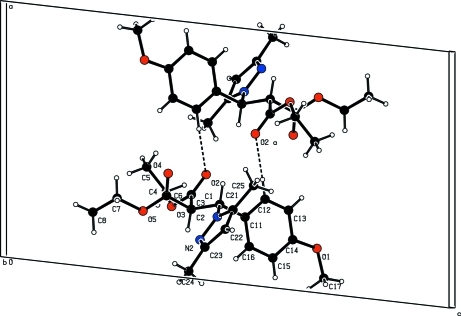
Fig. 2.
Partial packing view showing the chain generated by C—H···O hydrogen bonds shown as dashed lines. Symmetry code for generating the second molecule: 1 - x,-y,1 - z.
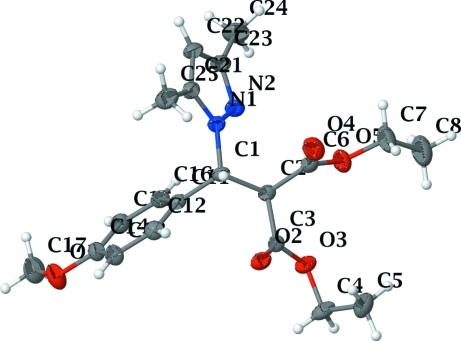
Fig. 3.
View of the title compound showing displacement ellipsoids at the 50% probability level.
Crystal data
| C20H26N2O5 | F(000) = 800 |
| Mr = 374.43 | Dx = 1.243 Mg m−3 |
| Monoclinic, P21/c | Melting point: 361 K |
| Hall symbol: -P 2ybc | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.9618 (3) Å | Cell parameters from 2174 reflections |
| b = 7.9681 (2) Å | θ = 2.3–27.1° |
| c = 21.1269 (6) Å | µ = 0.09 mm−1 |
| β = 96.504 (1)° | T = 296 K |
| V = 2000.70 (9) Å3 | Block, colourless |
| Z = 4 | 0.23 × 0.17 × 0.14 mm |
Data collection
| Bruker X8 APEXII CCD area-detector diffractometer | 3177 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.027 |
| graphite | θmax = 26.0°, θmin = 2.7° |
| φ and ω scans | h = −14→14 |
| 18616 measured reflections | k = −9→9 |
| 3921 independent reflections | l = −26→26 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.056 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.154 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0687P)2 + 1.8319P] where P = (Fo2 + 2Fc2)/3 |
| 3921 reflections | (Δ/σ)max = 0.007 |
| 249 parameters | Δρmax = 0.68 e Å−3 |
| 0 restraints | Δρmin = −0.45 e Å−3 |
Special details
| Experimental. The data collection nominally covered a sphere of reciprocal space, by a combination of tree sets of exposures; each set had a different φ angle for the crystal and each exposure covered 0.5° in ω and 20 s in time. The crystal-to-detector distance was 37.5 mm. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O2 | 0.40914 (11) | −0.14261 (19) | 0.44322 (7) | 0.0301 (3) | |
| O3 | 0.26937 (12) | −0.19794 (18) | 0.36564 (7) | 0.0321 (4) | |
| O4 | 0.39768 (14) | 0.2442 (2) | 0.36381 (8) | 0.0441 (4) | |
| O5 | 0.23951 (15) | 0.1580 (2) | 0.30637 (7) | 0.0446 (4) | |
| O1 | 0.16987 (15) | −0.2065 (2) | 0.68944 (8) | 0.0468 (5) | |
| N1 | 0.25069 (14) | 0.3219 (2) | 0.47437 (8) | 0.0252 (4) | |
| N2 | 0.15274 (14) | 0.3525 (2) | 0.43620 (8) | 0.0297 (4) | |
| C1 | 0.29918 (16) | 0.1528 (2) | 0.47752 (9) | 0.0230 (4) | |
| H1 | 0.3813 | 0.1633 | 0.4845 | 0.028* | |
| C2 | 0.26772 (16) | 0.0673 (3) | 0.41296 (9) | 0.0244 (4) | |
| H2 | 0.1859 | 0.0541 | 0.4051 | 0.029* | |
| C11 | 0.26097 (16) | 0.0542 (2) | 0.53261 (9) | 0.0229 (4) | |
| C3 | 0.32439 (16) | −0.1032 (2) | 0.41081 (9) | 0.0239 (4) | |
| C12 | 0.33950 (17) | −0.0251 (3) | 0.57616 (9) | 0.0277 (4) | |
| H12 | 0.4154 | −0.0197 | 0.5706 | 0.033* | |
| C21 | 0.28468 (17) | 0.4563 (3) | 0.51094 (10) | 0.0285 (5) | |
| C16 | 0.14790 (16) | 0.0419 (3) | 0.54174 (10) | 0.0277 (4) | |
| H16 | 0.0941 | 0.0929 | 0.5127 | 0.033* | |
| C13 | 0.30680 (18) | −0.1116 (3) | 0.62736 (10) | 0.0323 (5) | |
| H13 | 0.3605 | −0.1643 | 0.6559 | 0.039* | |
| C14 | 0.19347 (18) | −0.1206 (3) | 0.63658 (10) | 0.0311 (5) | |
| C15 | 0.11364 (17) | −0.0445 (3) | 0.59307 (10) | 0.0312 (5) | |
| H15 | 0.0376 | −0.0515 | 0.5983 | 0.037* | |
| C6 | 0.30985 (19) | 0.1697 (3) | 0.35912 (10) | 0.0308 (5) | |
| C23 | 0.12710 (18) | 0.5105 (3) | 0.44907 (11) | 0.0322 (5) | |
| C4 | 0.32363 (19) | −0.3556 (3) | 0.35159 (12) | 0.0370 (5) | |
| H4A | 0.2679 | −0.4329 | 0.3314 | 0.044* | |
| H4B | 0.3575 | −0.4063 | 0.3909 | 0.044* | |
| C22 | 0.20634 (18) | 0.5797 (3) | 0.49546 (10) | 0.0330 (5) | |
| H22 | 0.2060 | 0.6874 | 0.5124 | 0.040* | |
| C5 | 0.4121 (2) | −0.3243 (4) | 0.30837 (11) | 0.0466 (6) | |
| H5A | 0.3789 | −0.2693 | 0.2704 | 0.070* | |
| H5B | 0.4442 | −0.4292 | 0.2973 | 0.070* | |
| H5C | 0.4699 | −0.2542 | 0.3296 | 0.070* | |
| C25 | 0.3888 (2) | 0.4563 (3) | 0.55689 (12) | 0.0416 (6) | |
| H25A | 0.3743 | 0.4000 | 0.5953 | 0.062* | |
| H25B | 0.4114 | 0.5699 | 0.5666 | 0.062* | |
| H25C | 0.4478 | 0.3990 | 0.5384 | 0.062* | |
| C7 | 0.2781 (3) | 0.2351 (4) | 0.24975 (12) | 0.0612 (8) | |
| H7A | 0.2695 | 0.3560 | 0.2515 | 0.073* | |
| H7B | 0.3571 | 0.2098 | 0.2481 | 0.073* | |
| C24 | 0.0258 (2) | 0.5924 (3) | 0.41410 (14) | 0.0489 (7) | |
| H24A | 0.0493 | 0.6722 | 0.3842 | 0.073* | |
| H24B | −0.0160 | 0.6490 | 0.4439 | 0.073* | |
| H24C | −0.0209 | 0.5086 | 0.3917 | 0.073* | |
| C17 | 0.0566 (2) | −0.2040 (4) | 0.70479 (13) | 0.0545 (7) | |
| H17A | 0.0335 | −0.0900 | 0.7101 | 0.082* | |
| H17B | 0.0521 | −0.2649 | 0.7436 | 0.082* | |
| H17C | 0.0081 | −0.2554 | 0.6709 | 0.082* | |
| C8 | 0.2127 (3) | 0.1700 (5) | 0.19471 (13) | 0.0731 (10) | |
| H8A | 0.2245 | 0.0511 | 0.1922 | 0.110* | |
| H8B | 0.2351 | 0.2230 | 0.1573 | 0.110* | |
| H8C | 0.1345 | 0.1919 | 0.1975 | 0.110* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O2 | 0.0249 (7) | 0.0292 (8) | 0.0357 (8) | 0.0049 (6) | 0.0007 (6) | −0.0048 (6) |
| O3 | 0.0317 (8) | 0.0265 (8) | 0.0371 (8) | 0.0009 (6) | −0.0005 (6) | −0.0089 (6) |
| O4 | 0.0466 (10) | 0.0436 (10) | 0.0435 (9) | −0.0125 (8) | 0.0117 (8) | 0.0046 (8) |
| O5 | 0.0624 (11) | 0.0441 (10) | 0.0264 (8) | −0.0085 (8) | 0.0010 (7) | 0.0036 (7) |
| O1 | 0.0475 (10) | 0.0586 (12) | 0.0360 (9) | −0.0031 (9) | 0.0117 (7) | 0.0143 (8) |
| N1 | 0.0258 (8) | 0.0229 (9) | 0.0265 (8) | 0.0027 (7) | 0.0013 (7) | −0.0005 (7) |
| N2 | 0.0280 (9) | 0.0278 (9) | 0.0330 (9) | 0.0061 (7) | 0.0015 (7) | 0.0008 (7) |
| C1 | 0.0216 (9) | 0.0196 (9) | 0.0277 (10) | 0.0025 (7) | 0.0022 (7) | −0.0011 (8) |
| C2 | 0.0221 (9) | 0.0238 (10) | 0.0273 (10) | 0.0019 (8) | 0.0031 (8) | −0.0012 (8) |
| C11 | 0.0242 (9) | 0.0203 (10) | 0.0246 (9) | 0.0011 (8) | 0.0036 (7) | −0.0037 (7) |
| C3 | 0.0230 (10) | 0.0237 (10) | 0.0257 (9) | −0.0028 (8) | 0.0067 (8) | −0.0018 (8) |
| C12 | 0.0228 (10) | 0.0311 (11) | 0.0287 (10) | 0.0005 (8) | 0.0009 (8) | −0.0014 (8) |
| C21 | 0.0320 (11) | 0.0253 (11) | 0.0294 (10) | −0.0012 (8) | 0.0088 (8) | −0.0025 (8) |
| C16 | 0.0242 (10) | 0.0293 (11) | 0.0292 (10) | 0.0060 (8) | 0.0014 (8) | −0.0008 (8) |
| C13 | 0.0315 (11) | 0.0372 (13) | 0.0268 (10) | 0.0019 (9) | −0.0022 (8) | 0.0033 (9) |
| C14 | 0.0379 (12) | 0.0310 (12) | 0.0253 (10) | −0.0017 (9) | 0.0073 (9) | 0.0011 (8) |
| C15 | 0.0255 (10) | 0.0350 (12) | 0.0340 (11) | 0.0006 (9) | 0.0078 (8) | −0.0019 (9) |
| C6 | 0.0405 (12) | 0.0244 (11) | 0.0277 (10) | 0.0041 (9) | 0.0041 (9) | −0.0028 (8) |
| C23 | 0.0340 (11) | 0.0253 (11) | 0.0385 (11) | 0.0075 (9) | 0.0100 (9) | 0.0030 (9) |
| C4 | 0.0369 (12) | 0.0279 (11) | 0.0454 (13) | 0.0010 (9) | 0.0015 (10) | −0.0159 (10) |
| C22 | 0.0401 (12) | 0.0222 (10) | 0.0384 (12) | 0.0037 (9) | 0.0122 (10) | −0.0033 (9) |
| C5 | 0.0504 (15) | 0.0548 (16) | 0.0350 (12) | 0.0052 (12) | 0.0069 (11) | −0.0127 (12) |
| C25 | 0.0425 (13) | 0.0381 (13) | 0.0423 (13) | −0.0001 (11) | −0.0034 (10) | −0.0091 (11) |
| C7 | 0.097 (2) | 0.0572 (18) | 0.0293 (13) | −0.0235 (17) | 0.0078 (14) | 0.0054 (12) |
| C24 | 0.0432 (14) | 0.0413 (15) | 0.0613 (16) | 0.0180 (11) | 0.0019 (12) | 0.0046 (13) |
| C17 | 0.0596 (17) | 0.0595 (18) | 0.0492 (15) | −0.0086 (14) | 0.0274 (13) | 0.0059 (13) |
| C8 | 0.113 (3) | 0.072 (2) | 0.0346 (14) | −0.033 (2) | 0.0104 (16) | −0.0004 (14) |
Geometric parameters (Å, °)
| O2—C3 | 1.199 (2) | C13—H13 | 0.9300 |
| O3—C3 | 1.331 (2) | C14—C15 | 1.388 (3) |
| O3—C4 | 1.460 (3) | C15—H15 | 0.9300 |
| O4—C6 | 1.201 (3) | C23—C22 | 1.397 (3) |
| O5—C6 | 1.322 (3) | C23—C24 | 1.496 (3) |
| O5—C7 | 1.465 (3) | C4—C5 | 1.495 (3) |
| O1—C14 | 1.367 (3) | C4—H4A | 0.9700 |
| O1—C17 | 1.428 (3) | C4—H4B | 0.9700 |
| N1—C21 | 1.355 (3) | C22—H22 | 0.9300 |
| N1—N2 | 1.367 (2) | C5—H5A | 0.9600 |
| N1—C1 | 1.466 (2) | C5—H5B | 0.9600 |
| N2—C23 | 1.331 (3) | C5—H5C | 0.9600 |
| C1—C11 | 1.517 (3) | C25—H25A | 0.9600 |
| C1—C2 | 1.533 (3) | C25—H25B | 0.9600 |
| C1—H1 | 0.9800 | C25—H25C | 0.9600 |
| C2—C3 | 1.521 (3) | C7—C8 | 1.424 (4) |
| C2—C6 | 1.531 (3) | C7—H7A | 0.9700 |
| C2—H2 | 0.9800 | C7—H7B | 0.9700 |
| C11—C12 | 1.390 (3) | C24—H24A | 0.9600 |
| C11—C16 | 1.391 (3) | C24—H24B | 0.9600 |
| C12—C13 | 1.376 (3) | C24—H24C | 0.9600 |
| C12—H12 | 0.9300 | C17—H17A | 0.9600 |
| C21—C22 | 1.372 (3) | C17—H17B | 0.9600 |
| C21—C25 | 1.490 (3) | C17—H17C | 0.9600 |
| C16—C15 | 1.385 (3) | C8—H8A | 0.9600 |
| C16—H16 | 0.9300 | C8—H8B | 0.9600 |
| C13—C14 | 1.393 (3) | C8—H8C | 0.9600 |
| C3—O3—C4 | 116.03 (16) | N2—C23—C24 | 120.3 (2) |
| C6—O5—C7 | 115.4 (2) | C22—C23—C24 | 128.4 (2) |
| C14—O1—C17 | 117.87 (19) | O3—C4—C5 | 110.0 (2) |
| C21—N1—N2 | 112.18 (17) | O3—C4—H4A | 109.7 |
| C21—N1—C1 | 127.50 (16) | C5—C4—H4A | 109.7 |
| N2—N1—C1 | 119.92 (16) | O3—C4—H4B | 109.7 |
| C23—N2—N1 | 104.46 (17) | C5—C4—H4B | 109.7 |
| N1—C1—C11 | 111.05 (15) | H4A—C4—H4B | 108.2 |
| N1—C1—C2 | 108.18 (15) | C21—C22—C23 | 105.97 (19) |
| C11—C1—C2 | 112.80 (16) | C21—C22—H22 | 127.0 |
| N1—C1—H1 | 108.2 | C23—C22—H22 | 127.0 |
| C11—C1—H1 | 108.2 | C4—C5—H5A | 109.5 |
| C2—C1—H1 | 108.2 | C4—C5—H5B | 109.5 |
| C3—C2—C6 | 105.57 (15) | H5A—C5—H5B | 109.5 |
| C3—C2—C1 | 110.98 (16) | C4—C5—H5C | 109.5 |
| C6—C2—C1 | 110.86 (16) | H5A—C5—H5C | 109.5 |
| C3—C2—H2 | 109.8 | H5B—C5—H5C | 109.5 |
| C6—C2—H2 | 109.8 | C21—C25—H25A | 109.5 |
| C1—C2—H2 | 109.8 | C21—C25—H25B | 109.5 |
| C12—C11—C16 | 118.10 (18) | H25A—C25—H25B | 109.5 |
| C12—C11—C1 | 120.23 (17) | C21—C25—H25C | 109.5 |
| C16—C11—C1 | 121.66 (17) | H25A—C25—H25C | 109.5 |
| O2—C3—O3 | 125.32 (19) | H25B—C25—H25C | 109.5 |
| O2—C3—C2 | 124.59 (18) | C8—C7—O5 | 108.6 (2) |
| O3—C3—C2 | 110.01 (16) | C8—C7—H7A | 110.0 |
| C13—C12—C11 | 121.13 (19) | O5—C7—H7A | 110.0 |
| C13—C12—H12 | 119.4 | C8—C7—H7B | 110.0 |
| C11—C12—H12 | 119.4 | O5—C7—H7B | 110.0 |
| N1—C21—C22 | 106.12 (18) | H7A—C7—H7B | 108.3 |
| N1—C21—C25 | 123.09 (19) | C23—C24—H24A | 109.5 |
| C22—C21—C25 | 130.8 (2) | C23—C24—H24B | 109.5 |
| C15—C16—C11 | 121.46 (19) | H24A—C24—H24B | 109.5 |
| C15—C16—H16 | 119.3 | C23—C24—H24C | 109.5 |
| C11—C16—H16 | 119.3 | H24A—C24—H24C | 109.5 |
| C12—C13—C14 | 120.21 (19) | H24B—C24—H24C | 109.5 |
| C12—C13—H13 | 119.9 | O1—C17—H17A | 109.5 |
| C14—C13—H13 | 119.9 | O1—C17—H17B | 109.5 |
| O1—C14—C15 | 124.7 (2) | H17A—C17—H17B | 109.5 |
| O1—C14—C13 | 115.74 (19) | O1—C17—H17C | 109.5 |
| C15—C14—C13 | 119.52 (19) | H17A—C17—H17C | 109.5 |
| C16—C15—C14 | 119.56 (19) | H17B—C17—H17C | 109.5 |
| C16—C15—H15 | 120.2 | C7—C8—H8A | 109.5 |
| C14—C15—H15 | 120.2 | C7—C8—H8B | 109.5 |
| O4—C6—O5 | 124.9 (2) | H8A—C8—H8B | 109.5 |
| O4—C6—C2 | 124.10 (19) | C7—C8—H8C | 109.5 |
| O5—C6—C2 | 110.92 (18) | H8A—C8—H8C | 109.5 |
| N2—C23—C22 | 111.27 (19) | H8B—C8—H8C | 109.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···O2i | 0.93 | 2.51 | 3.358 (3) | 152 |
Symmetry codes: (i) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5280).
References
- Bruker (2005). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Dayam, R., Al-Mawsawi, L. Q. & Neamati, N. (2007). Bioorg. Med. Chem. Lett.17, 6155–6159. [DOI] [PubMed]
- Patil, S., Kamath, S., Sanchez, T., Neamati, N., Schinazi, R. F. & Buolamwini, J. K. (2007). Bioorg. Med. Chem.15, 1212–1228. [DOI] [PubMed]
- Pommier, Y. & Neamati, N. (2006). Bioorg. Med. Chem.14, 3785–3792. [DOI] [PubMed]
- Ramkumar, K., Tambov, K. V., Gundla, R., Manaev, A. V., Yarovenko, V., Traven, V. F. & Neamati, N. (2008). Bioorg. Med. Chem.16, 8988–8998. [DOI] [PubMed]
- Sechi, M., Carta, F., Sannia, L., Dallocchio, R., Dessì, A., Al-Safi, R. I. & Neamati, N. (2009). Antiviral Res.81, 267–276. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst.43, 920-925.
- Zeng, L. F., Zhang, H.-S. W., ang, Y. H., Sanchez, T., Zheng, Y. T., Neamati, N. & Long, Y. Q. (2008). Bioorg. Med. Chem. Lett.18, 4521–4524. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810025572/bt5280sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810025572/bt5280Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Enhanced figure: interactive version of Fig. 3