Abstract
Mechanisms of brain metastatic melanoma (BMM) remain largely unknown. Understanding the modulation of signaling pathways that alter BMM cell invasion and metastasis is critical to develop new therapies for BMM. Heparanase has been widely implicated in cancer and is the dominant mammalian endoglycosidase which degrades heparan sulfate chains of proteoglycans (HSPG) including syndecans (SDCs). Recent findings also indicate that heparanase possesses non-enzymatic functions in its latent form. We hypothesized that extracellular heparanase modulates BMM cell signaling by involving SDC1/4 carboxy terminal—associated proteins and downstream targets. We digested BMM cell surface HS with human recombinant active or latent heparanase to delineate their effects on cytoskeletal dynamics and cell invasiveness. We identified the small GTPase guanine nucleotide exchange factor-H1 (GEF-H1) as a new component of a SDC signaling complex that is differentially expressed in BMM cells compared to corresponding non-metastatic counterparts. Second, knockdown of GEF-H1, SDC1, or SDC4 decreased BMM cell invasiveness and GEF-H1 modulated small GTPase activity of Rac1 and RhoA in conjunction with heparanase treatment. Third, both active and latent forms of heparanase affected Rac1 and RhoA activity; notably increasing RhoA activity. Both forms of heparanase were found to mediate the expression and subcellular localization of GEF-H1, and treatment of BMM with latent heparanase modulated SDC1/4 gene expression. Finally, treatment with exogenous heparanase downregulated BMM cell invasion. These studies indicate the relevance of heparanase signaling pathways in BMM progression, and provide insights into the molecular mechanisms regulating HSPG signaling in response to exogenous heparanase.
Keywords: BRAIN METASTATIC MELANOMA, HEPARANASE, SYNDECANS, GEF-H1, Rac1/RhoA ACTIVITY AND SIGNALING
Melanoma is the third most common cancer targeting the brain. Among patients with brain metastasis, the condition is fatal in 95% of cases [Bradley and Mehta, 2004]. In spite of this high mortality rate, molecular mechanisms regulating BMM remain largely unknown.
Heparan sulfate proteoglycan (HSPG) are ubiquitous macromolecules located in the extracellular matrix and on the cell surface. They consist of a core protein and covalently attached heparan sulfate glycosaminoglycan chains (HS) [Iozzo, 2001; Beauvais and Rapraeger, 2004; Sanderson and Yang, 2008; Mythreye and Blobe, 2009; O’Connell et al., 2009]. Cell surface HSPG, as members of the syndecan and glypican families, regulate the cross-talk between tumor and host cells by acting as co-receptors for HS-binding factors [Reiland et al., 2006]. These unique functions place cell surface HSPG at the center of cell signaling integration [Tkachenko et al., 2005]. HSPG expression characteristics are linked to tumor metastasis [Iozzo, 2001; Beauvais and Rapraeger, 2004; Sanderson and Yang, 2008; O’Connell et al., 2009]. Importantly, HSPG are targets of heparanase (HPSE), the dominant mammalian endoglycosidase (endo-β-d-glucuronidase) whose activity has been widely implicated in cancer metastasis. HPSE cleaves HS at specific intrachain sites resulting in fragments (10–20 sugar subunits) which are biologically active, for example, able to bind potent growth and angiogenic factors [Ilan et al., 2006; Reiland et al., 2006; Fux et al., 2009]. Of interest, HPSE has recently been shown also to possess non-enzymatic functions in its unprocessed form (latent HPSE, 65 kDa) independent of known endoglycosidase activity ascribed to the fully processed form (active HPSE, 58 kDa) [Zetser et al., 2003; Cohen-Kaplan et al., 2008].
The invasive cell phenotype requires cytoskeletal dynamics driven by the small GTPases Rac1 and RhoA [Ridley et al., 1992; Sanz-Moreno et al., 2008; Sanz-Moreno and Marshall, 2009; Symons and Segall, 2009]. As tumor cells invade the surrounding tissue they establish, abolish, and/or relocate transient focal adhesions [Ilina and Friedl, 2009; Sanz-Moreno and Marshall, 2009]. Focal adhesion dynamics require Rac1 and RhoA activities [Symons and Segall, 2009] and recent evidence support roles of SDC cell surface HSPG [Avalos et al., 2009].
We hypothesized that SDC downstream signaling events are important for driving BMM cell invasiveness, and that heparanase modulates HSPG-associated signaling unrelated to its endoglycosidase activity. To this end, we utilized a previously characterized human BMM cell system consisting of variants exhibiting a gradient of in vitro cell invasiveness and in vivo brain metastatic behavior: tumorigenic but non-metastatic SBCl3, and syngeneic brain metastatic SB1B cells [Verschraegen et al., 1991]. We identified GEF-H1 as a novel SDC4-associated signal transduction protein in SB1B BMM cells. Of note, GEF-H1 is a RhoA GEF with microtubule binding abilities that is able to bind and inhibit Rac1 [Birkenfeld et al., 2008]. Treatment of SBCl3 and SB1B cells with recombinant human active or latent heparanase promotes an increased RhoA activity. Heparanase altered SDC gene expression, subcellular localization, and protein expression, resulting in inhibition of BMM cell invasiveness. Furthermore, we demonstrate HPSE-induced modulation of BMM cell invasion and GTPase activity using both active and latent forms of heparanase, suggesting roles for this molecule in melanoma pathogenesis which are independent of its enzymatic activity.
MATERIALS AND METHODS
CELL CULTURE
Previously characterized SBCl3 and SB1B BMM cells [Verschraegen et al., 1991] were generously provided by Dr. Beppino Giovanella (Stehlin Foundation, Houston, TX). Cells were grown in DMEM/F-12 media with 10% (v/v) fetal bovine serum (FBS), 1% (w/v) penicillin/streptomycin and 1% (w/v) l-glutamine (growth media), then used as indicated. For recombinant human heparanase (rhHPSE) treatment, cells were plated in normal growth media for 24 h, then media was changed to contain only 5% (v/v) serum with or without (500 ng/ml) recombinant human active or latent heparanase (A-HPSE or L-HPSE, respectively) for 16 h at 37°C [Reiland et al., 2006]. Removal of HS from cell surface HSPG was accomplished by digestion with Heparitinase III (HepIII) from Flavobacterium heparinum (Sigma, St. Louis, MO). Final HepIII concentration was 0.05 U/ml culture medium [Reiland et al., 2006]. Digestions were carried out for 1 h at 37°C, 5% (v/v) CO2. Cells were then washed with PBS and used as indicated. Human brain endothelial cells (HBEC, Stehlin Foundation) were grown in RPMI 1640+10% (v/v) FBS+1% (w/v) penicillin/streptomycin and 10 µg/ml of Endothelial Cell Growth Factor (Sigma). Conditioned media was collected when cells were passaged and frozen at −80°C until use.
CELL LYSATES
RIPA buffer (Sigma) containing 150 mM NaCl, 1% (v/v) IGEPAL CA-630 (Sigma), 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 50 mM Tris (pH 8.0), was supplemented with the protease inhibitor cocktail: 1 mM PMSF (Sigma) and complete-Mini (11836153001, Roche, Mannheim, Germany) to generate whole cell lysates. Nuclear and cytoplasmic lysates were generated using NE-PER (Thermo, Waltham, MA), supplemented with a protease inhibitor cocktail provided by the kit, and 1 mM PMSF (Sigma). Lysates for syndecan core proteins were generated as previously described [Beauvais and Rapraeger, 2003]. Briefly, 100 µg of cell lysates were precipitated at −20°C overnight (16 h) with 2.5 volumes of methanol (−20°C), then precipitates were washed with 0.5 ml of acetone (−20°C) and dried. The pellets were resuspended in 50 µl of heparitinase buffer [50 mM HEPES (pH 6.5), 50 mM sodium acetate, 150 mM NaCl, 5 mM CaCl2] with 0.004 U/ml heparitinase (Sigma) and 0.1 U/ml chondroitin ABC lyase (Sigma) for 4 h at 37°C, supplementing with additional enzymes after 2 h.
RECOMBINANT HUMAN HEPARANASE
Recombinant human active heparanase was purified as previously described [McKenzie et al., 2003; Reiland et al., 2004; Murry et al., 2006]. Briefly, 500 ml of supernatant from Tni cells infected with baculovirus transfer vectors containing heparanase subunits was passed over a HiTrap heparin column (Amersham Biosciences, Piscataway, NJ). The column was then washed with wash buffer (150 mM NaCl, 25 mM Tris–HCl, pH 7.5), and eluted using a gradient of 0.15 M, 0.70 M, or 1.0 M NaCl, 25 mM Tris, pH 7.5. Collected fractions (5 ml) were concentrated using Centricon filtering devices (Millipore, Temecula, CA), and screened for heparanase activity using a heparan sulfate degrading enzyme assay kit (Takara Mirus, Madison, WI). Heparanase eluted at 0.70 M NaCl [McKenzie et al., 2003; Murry et al., 2006]. Preparations of recombinant human latent heparanase were generously provided by Dr. Neta Ilan and Dr. Israel Vlodavsky (The Bruce Rappaport Faculty of Medicine, Technion, Israel), and prepared as previously reported [Zetser et al., 2003].
RT-PCR
We performed reverse transcriptase-polymerase chain reaction (RT-PCR) using total RNA isolated from the BMM cell lines using the RNeasy Plus mini-kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. RNA yield was determined using a NanoDrop spectrophotometer (ND1000, NanoDrop products, Wilmington, DE). To ensure lack of genomic DNA contamination, 1 µg total RNA was digested with DNase I (Invitrogen, Carlsbad, CA) prior to first-strand synthesis. A first-strand synthesis kit utilizing SuperScript II reverse transcriptase (Invitrogen) was used according to manufacturer’s instructions. We utilized 2 µl from the inactivated DNase I (Invitrogen) digestion reaction. The first-strand synthesis reaction was then diluted 1:1 with H2O(DEPC) and utilized as a single-strand cDNA template. PCR amplification was performed in 20 µl reactions consisting of: 1× AmpliTaq Gold buffer (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, 300 µM dNTP mix, 400 nM primer pair, 2 µl single-strand cDNA template, and 0.1 U AmpliTaq Gold Taq polymerase (Applied Biosystems). The PCR conditions were: 94°C, 2 min; 40 cycles of 94°C; 20 s; 58°C; 15 s; 72°C, 42 s; 72°C, 30 s, performed in a Mastercycler epgradient thermocycler (Eppendorf North America, Westbury, NY). The gene accession numbers and DNA sequences for the oligonucleotide primer pairs utilized are listed in Supplemental Table III.
CLONING
For affinity chromatography experiments we used the pGEX-4T-3 vector (GE Healthcare, Piscataway, NJ), and RT-PCR to amplify the cDNA regions encoding SDC intracellular carboxy terminal (CT) domains. Oligonucleotide primer pairs were designed for insertion into the BamHI/XhoI (New England Biolabs, Ipswich, MA) restriction sites within the pGEX vector. Gene accession numbers and DNA sequences for the oligonucleotide primer pairs utilized were: SDC1, NM_001006946.1; GST-SDC1 CT Forward 5′-TTT GGA TCC CGC ATG AAG AAG AAG GAC GA-3′; GST-SDC1 CT Reverse 5′-TTT CTC GAG TCA GGC ATA GAA TTC CTC CTG-3′; SDC4, NM_002999.2; GST-SDC4 CT Forward 5′-TTT GGA TCC CGT ATG AAG AAG AAG GAT GAA G-3′; GST-SDC4 CT Reverse 5′-TTT CTC GAG TCA CGC GTA GAA CTC ATT GGT-3′. Subsequent to amplification by PCR, we cloned the PCR product into the pCRII cloning vector, and selected with ampicillin (40 µg/ml). We excised the SDC CT cDNA fragment from the cloning vector using BamHI/XhoI restriction enzymes (New England Biolabs). The pGEX-4T-3 bacterial expression vector was digested with BamHI/XhoI in preparation to accept the SDC CT cDNA fragment. Ligation of cDNA fragments into the pGEX-4T-3 was accomplished with T4 DNA ligase (Invitrogen) and selected using kanamycin (25 µg/ml). The DNA construct sequences were verified by DNA sequencing (SeqWright, Houston, TX).
GST-PD
Affinity chromatography glutathione-S-transferase pulldowns (GST-PD) were performed as previously described [Ridgway et al., 2009]. Briefly, GST-SDC1 or GST-SDC4 fusion constructs were transformed into the Escherichia coli strain BL21. Expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to the LB culture media. Induced fusion proteins from bacteria were purified by lysing using the B-PER reagent (Thermo), and by passing cell lysates over columns containing glutathione beads (Thermo). GST fusion proteins were eluted from the columns with reduced glutathione. Thirty micrograms of purified GST fusion proteins were bound to glutathione bead columns and whole cell lysates (500 µg) from BMM cell lines were passed over the columns. These were then washed thrice and bound proteins eluted with 2× Laemmli buffer with subsequent heating to 95°C for 5 min. Samples were separated by SDS–PAGE and proteins were identified by Western blotting.
SIRNA
Cells were treated with 100 nmol siRNA using FuGene 6 reagent (Roche) at a 3:2 ratio for 36 h before further use. Sequences for SMART siRNA pools for GEF-H1 (L-009883, Dharmacon, Denver, CO) were: GEF-H1 5′-GAA UUA AGA UGG AGU UGC A-3′; 5′-GUG CGG AGC AGA UGU GUA A-3′; 5′-GAA GGU AGC AGC CGU CUG U-3′; 5′-CCA CGG AAC UGG CAU UAC U-3′. SDC1 (L-020621-10) sequences were: 5′-ACG CAG CUC CUG ACG GCU A-3′; 5′-GUA ACG ACA AUA AAC GGU A-3′; 5′-CAU CAG GCC UCA ACG ACA A-3′; 5′-UGG GSS SCU UGG CUC GAA U-3′. SDC4 (L-′03706-00) sequences were 5′-GAU CGG CCC UGA AGU UGU C-3′; 5′-GUG AGG AUG UGU GUC CCA ACA A-3′; 5′-GAA UCU CAC CGU UGA AGA-3′; 5′-UAG AGG AGA AUG AGG UUA U-3′. We also used 100 nM ON-TARGETplus SMART siRNA Non-targeting Pool (D-001810-10-05, Dharmacon) as negative control. The non-overlapping siRNA sequences used were: 5′-UGG UUU ACA UGC GAC UAA-3′; 5′-UGG UUU ACA UGU UGU GUG A-3′; 5′-UGG UUU ACA UGU UUC UGA-3′; and 5′-UGG UUU ACA UGU UUU CCU A-3′.
WESTERN BLOTTING
Proteins were resolved using SDS–PAGE, transferred to PVDF membranes (Bio-Rad, Hercules, CA), and blocked with 5% (w/v) milk (for most antibodies), or 3% (w/v) bovine serum albumin (for anti-Rac1 or anti-RhoA antibodies) in TBS with 0.5% (v/v) Tween®-20 before being probed with appropriate antibodies. Those used in experiments were: SDC1 (clone B-A38, 1:1,000 dilution) purchased from Cell Sciences (Canton, MA), SDC4 (ab24511, 1:1,000 dilution) was from Abcam (Cambridge, MA), full-length PKCα (#2506, 1:1,000 dilution), GEF-H1 (#4076, 1:1,000 dilution) and GAPDH (14C10, 1:1,000 dilution) were from Cell Signaling (Danvers, MA), Rac1 (clone 23A8, 1:1,000 dilution) and RhoA (clone 55, 1:500 dilution) were from Millipore, fibrillarin (sc-56676, 1:800) and β-actin (sc-69879, 1:1,000 dilution) were from Santa Cruz Biotechnology (Santa Cruz, CA). Blots were washed with TBS containing 0.5% (v/v) Tween®-20 before probing with secondary goat anti-rabbit and anti-mouse horseradish peroxidase antibodies (Santa Cruz Biotechnology) at 1:6,000 dilution. Blots were then exposed to film using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, Pittsburgh, PA).
CELL INVASION ASSAYS
Chemoinvasion assays were performed as previously reported [Reiland et al., 2004]. Briefly, transwell membrane inserts (Corning, Lowell, MA) were coated with Matrigel™ (BD Biosciences, San Jose, CA) at a 1:30 dilution in serum-free media for 16 h before use. Inserts were washed with serum-free media, then 1.5 × 105 cells/insert in DMEM/F-12 plus 0.1% (w/v) BSA were incubated in top chambers, and HBEC conditioned media supplemented with 5 µM N-formyl-Met-Leu-Phe (Sigma) was used in bottom chambers. Assay were performed for 24 h, then inserts were removed, stained with 400 µl staining solution (Cell Biolabs, San Diego, CA) for 10 min, washed, and extracted with 200 µl extraction solution (Cell Biolabs) for 10 min. Absorbance values for 100 µl of extracted solution were read at 560 nm using a SpectraMax Plus384 spectrophotometer (Molecular Devices, Sunnyvale, CA).
CELL PROLIFERATION ASSAYS
BMM (SBCl3, SB1B) cells were suspended at a concentration of 1.0 × 106 cells/ml in growth media, and 100 µl was added to a 96-well plate and treated with 10 µl AlamarBlue (Sigma) for 4 h. Absorbance values were measured at 570 and 600 nm, and percent AlamarBlue reduction was determined using manufacturer’s protocol and the percent reduction equation, # = (117216)(OD 570) − (80586)(OD 600)/(115677)(control OD 600) − (14652)(con-14652)(control OD 570)× 100.
CELL ADHESION ASSAYS
BMM (SBCl3, SB1B) cells were suspended at a concentration of 2.0 × 105 cells/ml in serum-free media, and 100 µl were added to a 96-well plate pre-treated with 0.5 µg/ml fibronectin (Sigma) and allowed to adhere for 4 h at 37°C. Media was then removed, wells washed three times with 250 µl PBS, and stained with 200 µl staining solution for 10 min. Wells were washed four times with 250 µl ddH2O, then 200 µl of extraction solution were added per well for 10 min, and 150 µl of extracted solution were used to measure OD values (560 nm).
Rac1 AND RhoA ACTIVITY ASSAYS
Rac1 and RhoA small GTPase activity assays were performed using the GLISA kits (BK124, BK125) (Cytoskeleton, Denver, CO) according to manufacturer’s instructions. We first determined the cell number to cell lysis buffer volume ratio that would reproducibly result in protein concentrations adjusted to 0.5 µg/µl. Cell lysates were then snap-frozen in liquid nitrogen to preserve GTP bound GTPase (Rac1 or RhoA) activity. GTP-bound Rac1 GTPase was bound by Rac1-GTP-binding protein linked wells of 96-well plates. Bound active Rac1 was detected using a Rac1 specific antibody. The degree of Rac1 activation was determined by comparison to activity values obtained from lysates derived from untreated cells. Similarly, GTP bound RhoA GTPase was bound by RhoA-GTP-binding protein linked wells of 96-well plates. Bound active RhoA was detected with a RhoA specific antibody. Absorbance readings were obtained at 490 nm for both Rac1 and RhoA activities.
DENSITOMETRY
Densitometric analyses were performed using the ImageJ software downloaded from NIH website: http://rsbweb.nih.gov/ij/download.html. Values were calculated by obtaining mean threshold values using the same area for each reading, then comparing the percentages of inputs to samples.
STATISTICAL ANALYSES
Data are represented as mean ± standard deviation. Significance values were obtained by Student’s paired t-test (*P<0.05; **P<0.01) when compared to untreated controls. All figures are representative of at least three independent experiments. P values of <0.05 were considered statistically significant.
RESULTS
SYNDECAN PROFILING IN BRAIN METASTATIC MELANOMA CELLS
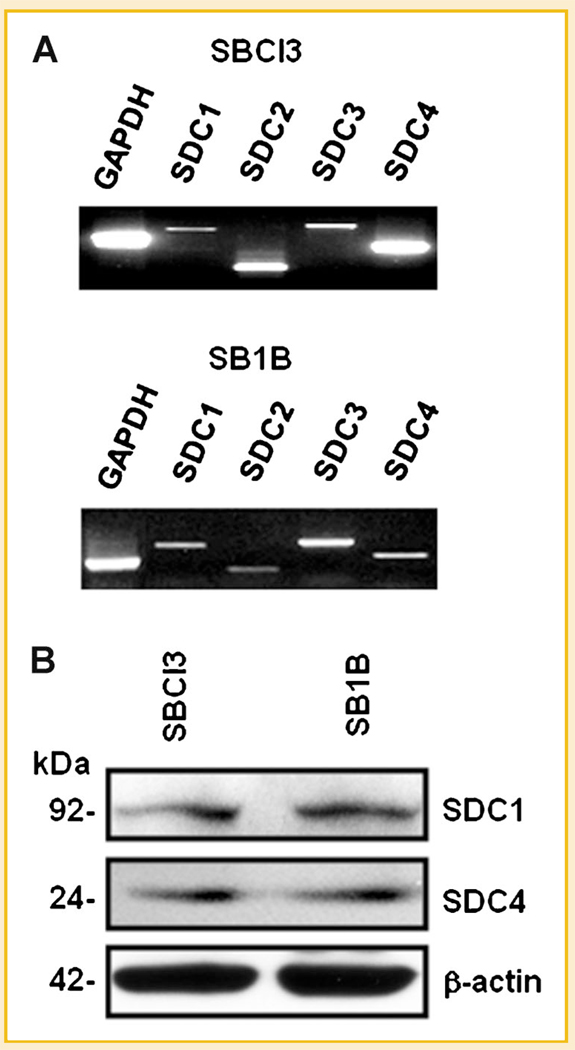
We initially sought to determine the pattern of syndecan gene expression in our human BMM cells (tumorigenic but non-metastatic SBCl3, and highly brain metastatic SB1B). Accordingly, we performed RT-PCR and identified distinct levels for all four mammalian SDC members in these cell lines (Fig. 1A). Since SDC1 and SDC4 have established roles in cell invasion, adhesion, and cytoskeletal dynamics [Alexander et al., 2000; Beauvais and Rapraeger, 2004; Tkachenko et al., 2005; Dovas et al., 2006; Sanderson and Yang, 2008], we investigated their protein expression, detecting it in SBCl3 and SB1B cell lines (Fig. 1B).
Fig. 1.
Syndecan profiling in melanoma cells. A: Total RNA was extracted from brain metastatic SB1B or syngeneic non-metastatic SBCl3 melanoma cells, and levels of the four mammalian syndecans (SDCs) were determined by RT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a positive control. B: Protein expression of SDC1/4 in BMM cells. Whole cell lysates treated with heparitinase and chondroitinase from SBCl3 and SB1B cells were Western blotted for SDC1/4 to determine syndecan core protein levels. β-Actin was used as a loading control. See Materials and Methods Section for additional details.
HEPARANASE REGULATES SDC1/4 EXPRESSION
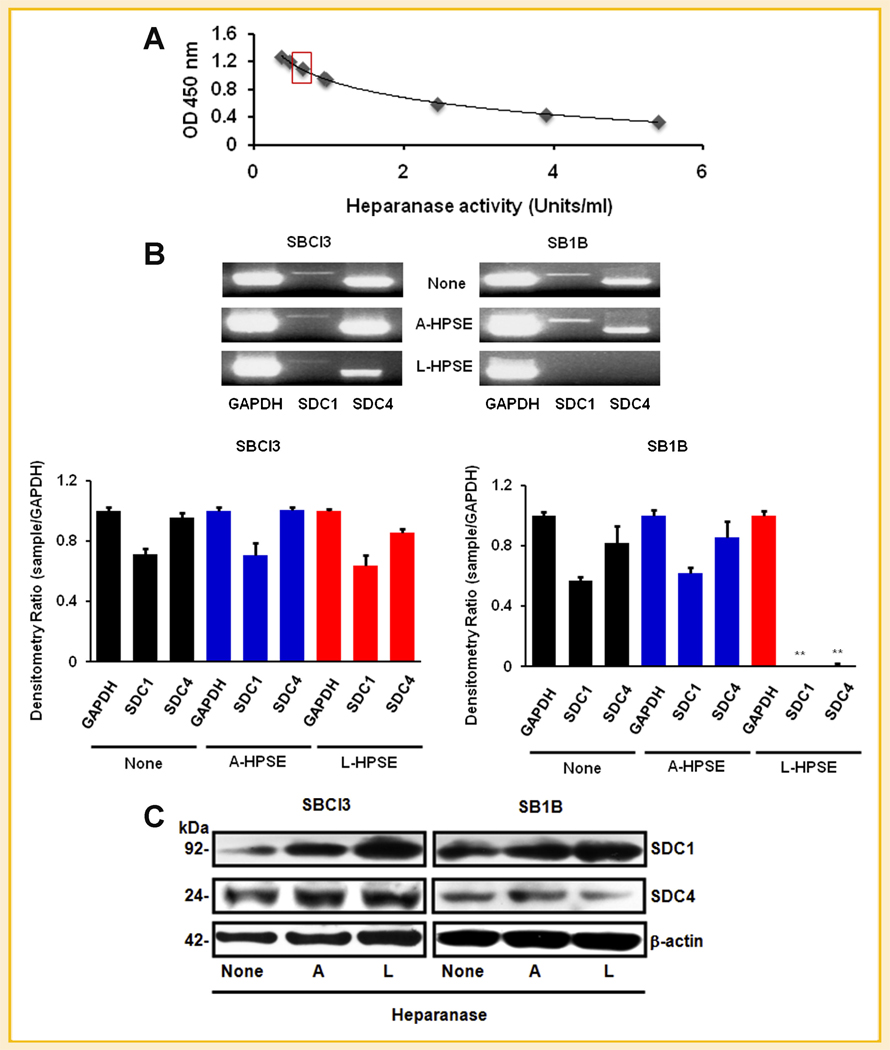
We have previously demonstrated that cell surface HSPG, notably SDC1, are targets of heparanase [Reiland et al., 2004]. We aimed to determine what roles exogenous heparanase had on SDC1/4 gene expression. Subsequent to assessing the activity of our recombinant HPSE preparations (Fig. 2A), we treated SBCl3 and SB1B cells with or without active or latent heparanase (A-HPSE or L-HPSE respectively) for 18 h. We performed semi-quantitative RT-PCR analysis, and detected similar SDC1/4 gene expression patterns from SBCl3 and SB1B cells (Fig. 2B). While treatment with A-HPSE did not significantly alter SDC1/4 gene expression from either cell line (Fig. 2B), L-HPSE exposure resulted in a significant reduction of SDC1/4 gene expression from the SB1B cells (Fig. 2B). From these data, it appears that SDC1 and SDC4 gene expressions is differentially regulated in response to exogenous latent heparanase in brain metastatic SB1B compared to non-brain metastatic SBCl3 melanoma cells.
Fig. 2.
Differential expression of SDC by exogenous heparanase. A: Heparanase activity plot. Concentration of active HPSE used in experiments is indicated by the red box. Heparanase was prepared as described in the Materials and Methods Section. B: Heparanase regulates SDC gene expression. RT-PCR was performed on RNA from BMM cells to determine changes in SDC gene expression level. Cells were treated with or without 500 ng/ml of active or latent heparanase (A-HPSE or L-HPSE, respectively) for 18 h at 37°C, then RNA was isolated, followed by semi-quantitative RT-PCR. Densitometric values represent expression of SDC1/4 normalized to GAPDH. C: Effects of HPSE on SDC expression. Cells were treated with or without active or latent (A- or L-HPSE) (500 ng/ml) for 16 h at 37°C, then whole cell lysates were treated with hepartinase and chondroitinase for 4 h at 37°C, then Western blotted with antibodies against SDC1/4, heparanase, and β-actin (loading control). See Materials and Methods Section for additional details.
We also investigated whether treatment with exogenous active or latent HPSE could modulate SDC1/4 core protein expression. Cell lysates were treated with heparitinase and chondroitinase to evaluate syndecan core protein levels. We noticed that reducing the serum concentration (10–5%) (v/v) in the growth media for optimal enzyme performance [Reiland et al., 2006], and utilizing for A/L-HPSE treatments resulted in increased SDC1 protein expression from the SBCl3 cells (Figs. 1B and 2C). Exposing SB1B cells to A-HPSE did not change SDC1 protein expression, but treatment with L-HPSE resulted in decreased SDC1 protein expression. Notably, we detected SDC4 expression in both cell lines but was not significantly altered by HPSE exposure (Fig. 2C).
GEF-H1 ASSOCIATES WITH SDC4 INTRACELLULAR CARBOXY TERMINUS
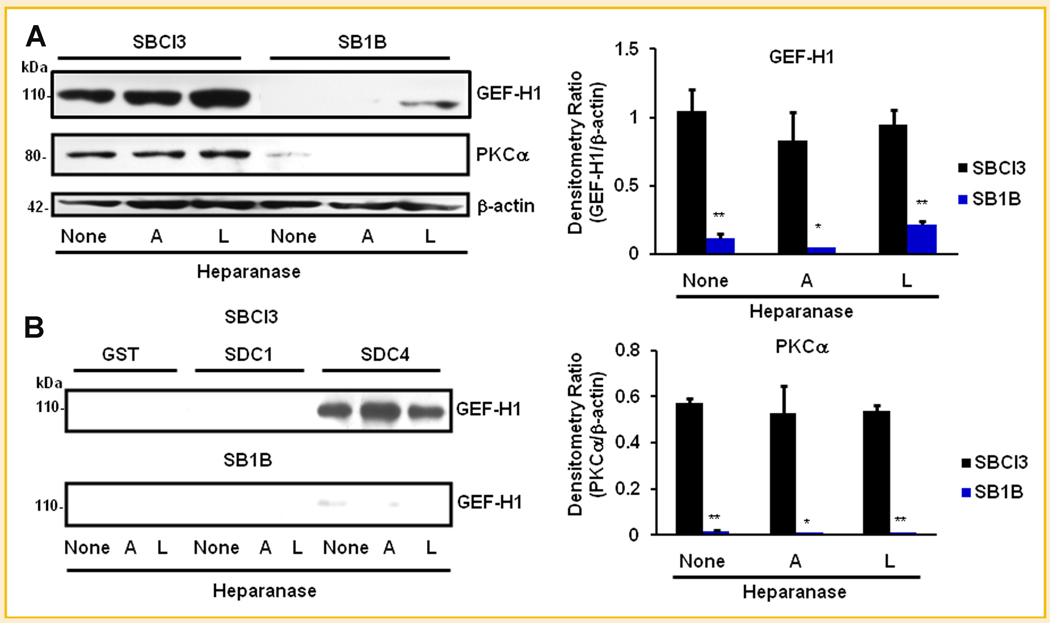
GEF-H1 is a RhoA GEF that can bind microtubules and regulate Rac1 and RhoA GTPase activity, and binds Rac1 as an inhibitor [Beauvais and Rapraeger, 2004; Sanz-Moreno et al., 2008; Avalos et al., 2009; Sanz-Moreno and Marshall, 2009; Symons and Segall, 2009]. We hypothesized that GEF-H1 may be a component of SDC4 intracellular signaling. Accordingly, we sought to identify roles of heperanase in the regulation of signaling proteins via SDCs and affecting BMM cell metastatic properties. We constructed GST fusion proteins of SDC1 and SDC4 CT, and performed affinity chromatography pulldowns using BMM whole cell lysates, followed by Western blotting analyses to detect the presence of GEF-H1 and PKCα. We observed greater GEF-H1 and PKCα expression, both of which regulated RhoA activity levels [Avalos et al., 2009] in SBCl3 compared to SB1B cells (Fig. 3A). Under control conditions, the two cell lines expressed endogenous GEF-H1, although SB1B had very low levels (Fig. 3A). Treatment with A-HPSE resulted in decreased GEF-H1 expression in SB1B cells. Furthermore, PKCα, which is known to bind SDC4 and regulate RhoA activity along with GEF-H1 [Dovas et al., 2006], was preferentially expressed in non-brain metastatic SBCl3 cells, but was not detectable in highly brain metastatic SB1B cells treated with either A-HPSE or L-HPSE (Fig. 3A). Importantly, we observed that endogenously expressed GEF-H1 could associate with SDC4 CT and that the association was modulated by exogenous HPSE (A-HPSE vs. L-HPSE) (Fig. 3B). Specifically, we observed increased GEF-H1/SDC4 CT association subsequent to treatment with A-HPSE. This association is preferential to SDC4 CT, as we did not detect it between GEF-H1 and SDC1 CT (Fig. 3B).
Fig. 3.
GEF-H1 as a SDC-associated proteins. A: Expression levels of GEF-H1 and PKCα in SBCl3 and SB1B BMM cells. Whole cell lysates from BMM cells treated with A-HPSE or L-HPSE were Western blotted for GEF-H1 or PKCα to determine protein expression levels in different BMM cell lines. β-Actin was used as a loading control. Densitometric analyses represent GEF-H1 or PKCα levels normalized to β-actin. *P-value <0.05, **P-value <0.01, comparing SBCl3 to SB1B expression levels. B: GST pulldown of GEF-H1 by SDC1 and SDC4. BMM cells were treated with or without 500 ng/ml of A-HPSE or L-HPSE for 18 h at 37°C, then whole cell lysates were generated, passed over GST-SDC1/4 CT fusion protein affinity columns. This was followed by immunoblotting for GEF-H1. See Materials and Methods Section for additional details.
EFFECTS OF SDC-ASSOCIATED GEF-H1 ON BMM CELL INVASION
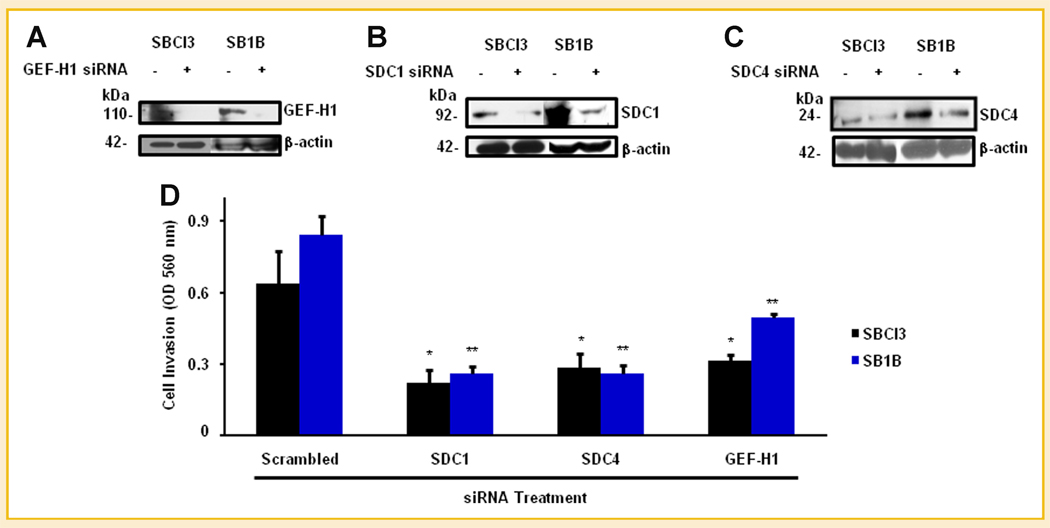
To determine biological outcomes of SDC-associated GEF-H1 on BMM cell invasive phenotype, we interrogated effects of knocking down either GEF-H1 or SDC1/4 on BMM cell phenotype. Transfections of SBCl3 and SB1B cells with siRNA against SDC1/4, GEF-H1, or scrambled controls resulted in knockdown of each individual protein (Fig. 4A–C). Knockdown of GEF-H1 or SDC 1/4 significantly decreased cell invasive abilities (Fig. 4D). These results demonstrate that GEF-H1 and SDC1/4 have defined roles in BMM cell invasion. Conversely, we did not observe any significant differences in BMM cell proliferation and adhesive properties in response to GEF-H1 or SDC1/4 knockdowns (Supplemental Table I).
Fig. 4.
Effects of GEF-H1 and SDC1/4 on BMM cell invasion. A: Knockdown of GEF-H1 in BMM cells. SBCl3 cells were treated with SmartPool siRNA (Dharmacon) for GEF-H1 or scrambled control, and protein level was determined by Western blotting. Knockdown of SDC1 (B) and SDC4 (C) in BMM cells. BMM cells (SBCl3 and SB1B) were treated with siRNA for SDC1 or scrambled control and Western blotted to determine SDC1 knockdown. β-Actin was used as a loading control for all samples. C: Effects of inhibition of GEF-H1 and SDCs on BMM cell invasion. Cells were treated with siRNA for GEF-H1, SDC1, or SDC4, or scrambled control for 36 h, then plated on Matrigel™ coated inserts and allowed to invade for 24 h. Invasion was determined by crystal violet staining followed by extraction and OD readings were made at 490 nm. Data are representative of three independent experiments and presented as means ± standard deviations. *P-value <0.05, **P-value <0.01. Statistical analyses for each sample were made by comparing GEF-H1, SDC1, or SDC4 siRNA treated cells versus scrambled control for each cell line. See Materials and Methods Section for additional details.
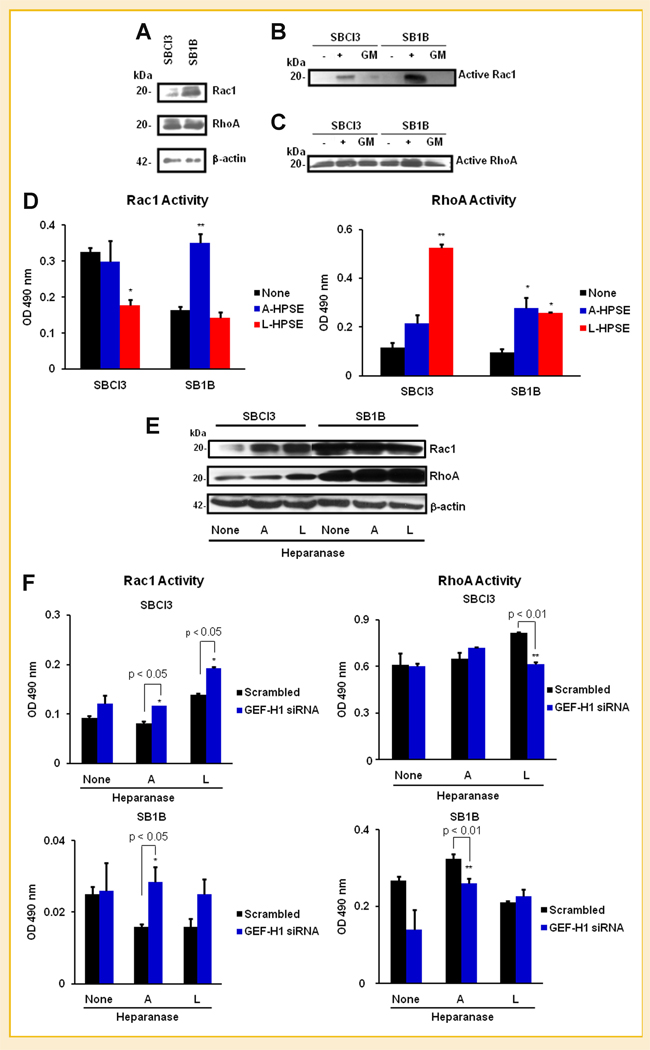
EFFECTS OF HEPARANASE ON Rac1 AND RhoA ACTIVITY
Since we identified significant effects on cell invasion subsequent to GEF-H1 knockdown, and because cytoskeletal dynamics are fundamental to this process, we investigated Rac1 and RhoA GTPase expression and activity from SBCl3 and SB1B cell lysates. SBCl3 cells possessed less Rac1 expression than the SB1B cells, while RhoA expression was comparable between the two cell lines in our model system (Fig. 5A). Further, the Rac1 and RhoA protein expressed from regular cell lysates had inducible GTPase activity when non-hydrolizable GTP was added as a positive control. Serum-containing growth media did not efficiently induce GTPase activity (Fig. 5B and C). Next, because we observed inhibition of invasion upon SDC and GEF-H1 knockdown (Fig. 4C), we investigated the effects of GEF-H1 and heparanase on Rac1/RhoA GTPase activities in BMM cells. Treatment of SBCl3 cells with L-HPSE (500 ng/ml) significantly decreased Rac1 activity while increasing RhoA activity (Fig. 5D). Conversely, we detected an increase in Rac1 activity in SB1B cells upon treatment following exposure to A-HPSE, while L-HPSE had no effect. Both forms of heparanase resulted in increased RhoA activity in SB1B cells (Fig. 5D). We also studied the effects of active/latent heparanase (A/L-HPSE) on total Rac1 and RhoA expression levels. Highly metastatic SB1B cells displayed higher levels of both Rac1 and RhoA expression than non-metastatic SBCl3 cells (Fig. 5E). SBCl3 cells displayed low levels of basal Rac1, which was slightly increased by A/L-HPSE, while neither form had an effect on SB1B Rac1 expression. In contrast, A/L-HPSE treatment either did not alter RhoA expression (SBCl3 cells) or slightly increased it (SB1B cells) (Fig. 5E). These results indicate that both forms of heparanase regulate intracellular GTPase activity and expression in brain metastatic versus non-metastatic BMM cells.
Fig. 5.
Roles of GEF-H1, SDCs and Heparanase in BMM cell Rac1/RhoA activities. A: Expression of Rac1 and RhoA small GTPases in BMM cells. Differential activation of Rac1 (B) and RhoA (C) in BMM cells in response to 10% (v/v) serum-containing media (GM). Positive and negative controls of non-hydrolyzed GTP were from the Rac and Rho Activation Assays (Millipore) are indicated by + and −, respectively. The upper band represents the phosphorylated form of the protein. D: Effects of HPSE on Rac1 and RhoA. SBCl3 and SB1B cells were treated with or without 500 ng/ml A-HPSE or L-HPSE for 16 h at 37°C, then serum-starved for 24 h. Rac and Rho activities were determined using the Cytoskeleton G-LISA activation kits. E: Effects of HPSE on Rac1 and RhoA expression. SBCl3 and SB1B cells were treated with or without A-HPSE or L-HPSE, and whole cell lysates were analyzed for Rac1 and RhoA expression levels. F: Effects of GEF-H1 knockdown and heparanase on Rac1/RhoA activity. SBCl3 and SB1B cells were transfected with siRNA for GEF-H1 or scrambled control, then cells were treated with or without active or latent (A- or L-HPSE) (500 ng/ml) for 16 h at 37°C. Cells were then lysed and screened for Rac1/RhoA activity using the Cytoskeleton G-LISA activation kits. All data are represented as means ± standard deviations for three experiments. *P-value <0.05, **P-value <0.01. See Materials and Methods Section for additional details.
Due to the inhibition of invasion observed after knockdown of SDC1, SDC4, or GEF-H1 (Fig. 4C), we investigated the effects of GEF-H1 knockdown on Rac1/RhoA GTPase activation. Since previous studies have linked GEF-H1 to Rac1/RhoA GTPase activities [Dovas et al., 2006], and considering that we detected the physical association of a GEF-H1 signaling complex with SDC4 CT by GSTPD (Fig. 3B), we assessed whether GEF-H1 knockdown, in conjunction with recombinant human heparanase exposure, altered Rac1/RhoA activities. We observed that GEF-H1 knockdown with subsequent active or latent heparanase treatment promoted Rac1 activity in SBCl3 cells, while latent heparanase decreased RhoA activity. In SB1B cells, GEF-H1 knockdown followed by A-HPSE treatment slightly increased Rac1 activity while decreasing RhoA activity (Fig. 5F). These results indicate that GEF-H1 is important in altering heparanase-induced changes in GTPase activity.
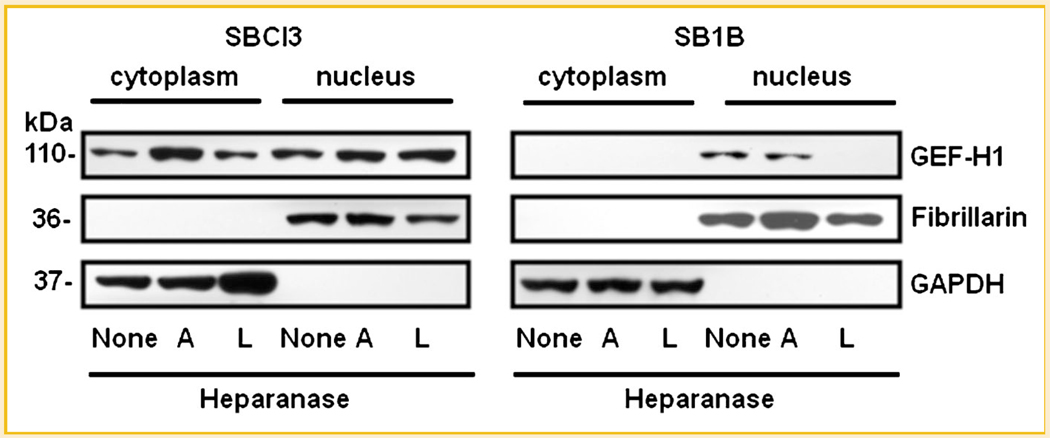
ROLE OF HEPARANASE ON GEF-H1 SUBCELLULAR LOCALIZATION
We performed subcellular localization on cells treated with active or latent heparanase by obtaining cytosolic and nuclear fractions (Fig. 6). We observed contrasting GEF-H1 levels when comparing non-metastatic BMM SBCl3 with the brain metastatic BMM SB1B cells. GEF-H1 was distributed between cytoplasm and nucleus in SBCl3 cells (Fig. 6), while GEF-H1 expression from SB1B cells was restricted to the nucleus. Treatment of the SBCl3 cells with A-HPSE resulted in increased cytosolic GEF-H1 (Fig. 6). However, we could not detect any GEF-H1 expression from the SB1B cells subsequent to treatment with L-HPSE (Fig. 6). These results indicate that metastatic and non-metastatic cells exhibit distinct subcellular localization of GEF-H1, along with different responses to A/L-HPSE treatment.
Fig. 6.
Roles of heparanase on GEF-H1 subcellular localization. Cells were treated with or without of A-HPSE or L-HPSE (500 ng/ml), then Western blotted for GEF-H1. Fibrillarin and GAPDH were used as specific nuclear and cytosolic markers respectively. See Materials and Methods Section for additional details.
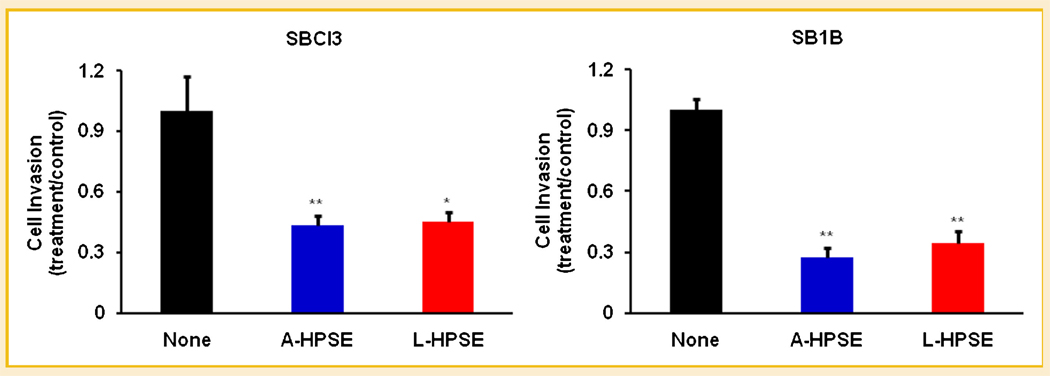
EFFECTS OF HEPARANASE ON BMM CELL METASTATIC PROPERTIES
To distinguish the abilities of heparanase to modulate the metastatic properties of BMM cells, we treated them with active or latent heparanase (500 ng/ml), then performed cell adhesion, proliferation, and invasion analyses. Both A-HPSE and L-HPSE resulted in decreased cell invasiveness, suggesting that the mere presence of exogenous heparanase deters cell invasion independently of its enzymatic activity (Fig. 7). We observed no significant changes in BMM cell adhesion or proliferation regardless of heparanase treatment (Supplemental Table II). These results suggest that exogenous heparanase is an important modulator of BMM cell invasiveness, and can alter it independent of its enzymatic activity.
Fig. 7.
Roles of heparanase in BMM cell invasiveness. SBCl3 and SB1B cells were treated with or without A-HPSE or L-HPSE (500 ng/ml) for 16 h at 37°C, then serum-starved for 24 h before being used for invasion assays. All data are represented as means ± standard deviations and are indicative of three independent experiments. *P-value <0.05, **P-value <0.01. See Materials and Methods Section for additional details.
DISCUSSION
The work presented provides first-time evidence demonstrating: (1) an association between GEF-H1 and the CT domain of SDC4; (2) a differential SDC gene and protein expression differ between syngeneic brain-metastatic and non-metastatic melanoma cells; (3) active and latent heparanase modulate Rac1 and RhoA GTPase activities; (4) active and latent heparanase differentially altered SDC gene expression and GEF-H1 subcellular localization; and (5) exogenous heparanase inhibited BMM cell invasiveness.
Just as syndecan 1/4 are central to cell invasion and focal adhesion [Levy-Adam et al., 2008], GTPase activity is important in cell motility [Sanderson and Yang, 2008; O’Connell et al., 2009]. Recent work indicates that Rac1 and RhoA activities can be interchangeable, and are important in tumor cell migration and phenotypic plasticity [Sanz-Moreno et al., 2008; Sanz-Moreno and Marshall, 2009]. GEF-H1 seems to be uniquely positioned to participate in this interplay since it has been proven to bind and regulate GTPases [Ren et al., 1998; Birkenfeld et al., 2007]. Equally relevant, heparanase is well known as a potent pro-tumorigenic, pro-angiogenic, and pro-metastatic enzyme [Ilan et al., 2006]. In addition, heparanase is involved in cancer cell signaling independent of its enzymatic activity [Levy-Adam et al., 2008]. We demonstrate that both exogenous active (58 kDa) and particularly latent (65 kDa) heparanase regulate Rac1/RhoA GTPases. Active heparanase promoted RhoA activity, while not inhibiting Rac1 (Fig. 5D). Latent heparanase modulated Rac1/RhoA by inhibiting Rac1 activity, particularly in the high GEF-H1 expressing SBCl3 cells, while activating RhoA activity in these cells (Fig. 5D). Significantly, we show that exogenous heparanase treatment reduces cell invasiveness (Fig. 7), which refutes the expectation that heparanase would promote cell invasiveness by driving RhoA activity. We demonstrate lower GEF-H1 levels in invasive cells (Fig. 3A), along with regulation of Rac1/RhoA activity by GEF-H1 knockdown (Fig. 5F), suggesting that GEF-H1 is playing a role in invasion by altering GTPase activity which is further augmented by heparanase. These findings are in agreement with other reports using either glioma or other BMM cell models [Zetser et al., 2003; Reiland et al., 2006], indicating that moderate heparanase levels stimulate tumor growth and invasion, while higher heparanase concentrations, such as those used in this study, produce opposite effects. This could be due to disrupted growth factor mediated activities [Reiland et al., 2006]. Interestingly, these inhibitory effects by high heparanase levels have only been reported in brain tumor models, suggesting a more complex regulation of this molecule in the brain [Zhang et al., 2010].
It has been proven that signal transduction pathways stemming from SDC4 CT to PKCα to RhoA exist. SDC4 clustering brings together bound PKCα, which activates RhoA signaling [Dovas et al., 2006]. We demonstrated that endogenously expressed GEF-H1 and PKCα associate with SDC4 CT (Fig. 3B). Importantly, we demonstrate that in a syngeneic system that the highly invasive and brain metastatic SB1B cell line expresses less GEF-H1 and PKCα than non-metastatic counterparts (Fig. 3A). This suggest that GEF-H1 is a critical component of the SDC4 CT-PKCα signaling complex, and disruption of this complex can promote cell metastatic propensities, since highly metastatic cells possess low SDC4 CT–GEF–H1 interactions (Fig. 3B).
We observed that SDC1 and SDC4 are expressed in brain–metastatic and non-metastatic cells, and demonstrated that cells can respond to exposure with exogenous active or latent heparanase differently, according to respective invasive phenotype, which can result in altered SDC gene expression and altered GEF-H1 protein expression/distribution (Figs. 2B,C, 3, and 6). This could be related to differences in signal transduction abilities inherently associated with GEF-H1 expression. Interestingly, we report that exogenous active and latent heparanase resulted in a reduction of nuclear GEF-H1 in highly invasive cells, and are able to promote RhoA activity and inhibit cell invasion. This suggests possible important new functions for heparanase because it can alter cancer cell invasive action and GTPase activity by binding to a cell surface receptor, regardless of its enzymatic function [Fux et al., 2009].
In conclusion, we demonstrate association between SDC4 and GEF-H1, the importance of GEF-H1 in regulating BMM cell Rac1 and RhoA activities in conjunction with heparanase, and that heparanase is an important determinant in the invasive capabilities of BMM cells by affecting GTPase activity and GEF-H1 down-regulation. These findings are of potential relevance because they demonstrate new roles for heparanase independent of its enzymatic activity, and provide impetus for targeting latent heparanase in cancer therapy, particularly melanoma.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Neta Ilan and Dr. Israel Vlodavsky (The Bruce Rappaport Medical Faculty, Technion, Israel) for kindly providing human recombinant latent (65 kDa) heparanase. This work was supported by NIH grant 2 R01 CA086832 to D.M.
Grant sponsor: NIH; Grant number: 2 R01 CA 086832.
Abbreviations used
- BMM
brain metastatic melanoma
- CT
carboxy-terminal
- SDC
syndecan
- GEF
guanine nucleotide exchange factor
- HPSE
heparanase
- A-HPSE
active heparanase
- L-HPSE
latent heparanase
- HSPG
heparan sulfate proteoglycan
- HS
heparan sulfate glycosaminoglycan chain
- RT-PCR
reverse transcriptase-polymerase chain reaction
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
Footnotes
The authors declare no competing conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Alexander CM, Reichsman F, Hinkes MT, Lincecum J, Becker KA, Cumberledge S, Bernfield M. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25:329–332. doi: 10.1038/77108. [DOI] [PubMed] [Google Scholar]
- Avalos AM, Valdivia AD, Munoz N, Herrera-Molina R, Tapia JC, Lavandero S, Chiong M, Burridge K, Schneider P, Quest AF, Leyton L. Neuronal Thy-1 induces astrocyte adhesion by engaging syndecan-4 in a cooperative interaction with alphavbeta3 integrin that activates PKCalpha and RhoA. J Cell Sci. 2009;122:3462–3471. doi: 10.1242/jcs.034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais DM, Rapraeger AC. Syndecan-1-mediated cell spreading requires signaling by alphavbeta3 integrins in human breast carcinoma cells. Exp Cell Res. 2003;286:219–232. doi: 10.1016/s0014-4827(03)00126-5. [DOI] [PubMed] [Google Scholar]
- Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;2:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenfeld J, Nalbant P, Bohl BP, Pertz O, Hahn KM, Bokoch GM. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenfeld J, Nalbant P, Yoon SH, Bokoch GM. Cellular functions of GEF-H1, a microtubule-regulated Rho-GEF: Is altered GEF-H1 activity a crucial determinant of disease pathogenesis? Trends Cell Biol. 2008;18:210–219. doi: 10.1016/j.tcb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Bradley KA, Mehta MP. Management of brain metastases. Semin Oncol. 2004;31:693–701. doi: 10.1053/j.seminoncol.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: Correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–10085. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas A, Yoneda A, Couchman JR. PKCbeta-dependent activation of RhoA by syndecan-4 during focal adhesion formation. J Cell Sci. 2006;119:2837–2846. doi: 10.1242/jcs.03020. [DOI] [PubMed] [Google Scholar]
- Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sci. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Ilina O, Friedl P. Mechanisms of collective cell migration at a glance. J Cell Sci. 2009;122:3203–3208. doi: 10.1242/jcs.036525. [DOI] [PubMed] [Google Scholar]
- Iozzo RV. Heparan sulfate proteoglycans: Intricate molecules with intriguing functions. J Clin Invest. 2001;108:165–167. doi: 10.1172/JCI13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Adam F, Feld S, Suss-Toby E, Vlodavsky I, Ilan N. Heparanase facilitates cell adhesion and spreading by clustering of cell surface heparan sulfate proteoglycans. PLoS One. 2008;3:e2319. doi: 10.1371/journal.pone.0002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, Turner P, Stamps A, McMillan D, Saville G, Ng S, Mason S, Snell D, Schofield D, Gong H, Townsend R, Gallagher J, Page M, Parekh R, Stubberfield C. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–435. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry BP, Blust BE, Singh A, Foster TP, Marchetti D. Heparanase mechanisms of melanoma metastasis to the brain: Development and use of a brain slice model. J Cell Biochem. 2006;97:217–225. doi: 10.1002/jcb.20714. [DOI] [PubMed] [Google Scholar]
- Mythreye K, Blobe GC. Proteoglycan signaling co-receptors: Roles in cell adhesion, migration and invasion. Cell Signal. 2009;21:1548–1558. doi: 10.1016/j.cellsig.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MP, Fiori JL, Kershner EK, Frank BP, Indig FE, Taub DD, Hoek KS, Weeraratna AT. Heparan sulfate proteoglycan modulation of Wnt5A signal transduction in metastatic melanoma cells. J Biol Chem. 2009;284:28704–28712. doi: 10.1074/jbc.M109.028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: Functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- Reiland J, Kempf D, Roy M, Denkins Y, Marchetti D. FGF2 binding, signaling, and angiogenesis are modulated by heparanase in metastatic melanoma cells. Neoplasia. 2006;8:596–606. doi: 10.1593/neo.06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- Ridgway LD, Kim EY, Dryer SE. MAGI-1 interacts with Slo1 channel proteins and suppresses Slo1 expression on the cell surface. Am J Physiol Cell Physiol. 2009;297:C55–C65. doi: 10.1152/ajpcell.00073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Sanderson RD, Yang Y. Syndecan-1: A dynamic regulator of the myeloma microenvironment. Clin Exp Metastasis. 2008;25:149–159. doi: 10.1007/s10585-007-9125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V, Marshall CJ. Rho-GTPase signaling drives melanoma cell plasticity. Cell Cycle. 2009;8:1484–1487. doi: 10.4161/cc.8.10.8490. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Symons M, Segall JE. Rac and Rho driving tumor invasion: Who’s at the wheel? Genome Biol. 2009;10:213. doi: 10.1186/gb-2009-10-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko E, Rhodes JM, Simons M. Syndecans: New kids on the signaling block. Circ Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- Verschraegen CF, Giovanella BC, Mendoza JT, Kozielski AJ, Stehlin JS., Jr Specific organ metastases of human melanoma cells injected into the arterial circulation of nude mice. Anticancer Res. 1991;11:529–535. [PubMed] [Google Scholar]
- Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- Zhang L, Sullivan P, Suyama J, Marchetti D. Epidermal growth factor-induced heparanase nucleolar localization augments DNA topoisomerase I activity in brain metastatic breast cancer. Mol Cancer Res. 2010;8:278–290. doi: 10.1158/1541-7786.MCR-09-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.