Abstract
Transforming growth factor (TGF)-β is a potent inducer of epithelial to mesenchymal transition (EMT). However, it remains elusive as to which molecular mechanisms determine the cellular capacity to undergo EMT in response to TGF-β. We have found that both epidermal growth factor receptor (EGFR) overexpression and mutant p53 tumor suppressor genes contribute to enrichment of an EMT-competent cellular subpopulation amongst telomerase-immortalized human esophageal epithelial cells during malignant transformation. EGFR overexpression triggers oncogene-induced senescence, accompanied by induction of cyclin dependent kinase inhibitors p15INK4B, p16INK4A and p21. Interestingly, a subpopulation of cells emerges by negating senescence without loss of EGFR overexpression. Such cell populations express increased levels of zinc finger E-box binding (ZEB) transcription factors ZEB1 and ZEB2, and undergo EMT upon TGF-β stimulation. Enrichment of EMT-competent cells was more evident in the presence of p53 mutation, which diminished EGFR-induced senescence. RNA interference directed against ZEB resulted in induction of p15INK4B and p16INK4A, reactivating the EGFR-dependent senescence program. Importantly, TGF-β-mediated EMT did not take place when cellular senescence programs were activated by either ZEB knockdown or activation of wild-type p53 function. Thus, senescence checkpoint functions activated by EGFR and p53 may be evaded through the induction of ZEB, thereby allowing expansion of an EMT-competent unique cellular subpopulation, providing novel mechanistic insights into the role of ZEB in esophageal carcinogenesis.
Keywords: EGFR, EMT, senescence, ZEB1, ZEB2
Introduction
Esophageal squamous cell carcinoma (ESCC) is amongst the deadliest cancers known (1) and is a paradigm for investigation for all types of squamous cell cancers. Its high mortality rate is attributed to diagnosis at an advanced stage characterized by invasion and metastases to local lymph nodes and remote organs, as well as lack of curative therapy. Genetic lesions associated frequently with ESCC include inactivation of tumor suppressors p53 and p16INK4A and overexpression of cyclin D1 and epidermal growth factor receptor (EGFR) (2) in addition to telomerase activation (3). EGFR overexpression and p53 mutations are particularly common in premalignant lesions (4–6). The presence of p53 mutations is positively correlated with EGFR overexpression (7).
Epithelial to mesenchymal transition (EMT) occurs during fundamental biological and disease processes including development and cancer (8). EMT in cancer leads to loss of cell-cell adhesion and cell polarity as well as altered cell-extracellular matrix interactions, resulting in invasion and metastasis (8–9). EMT is associated also with resistance to anti-cancer agents such as EGFR inhibitors (10). Although transforming growth factor (TGF)-β is one of the most potent EMT inducers present in the tumor microenvironment (11), EMT is not the sole consequence of TGF-β mediated stimulation. It remains unknown as to what determines the cellular capacity to undergo EMT in response to TGF-β (12). Amongst the transcription factors essential in EMT are zinc finger E-box binding proteins ZEB1 (a.k.a. deltaEF1) and ZEB2 (a.k.a SIP1)(13). ZEB1 and ZEB2 (ZEB) are critical regulators of TGF-β-mediated signaling through physical interaction with the SMAD proteins to recruit co-activators and co-repressors (14). ZEB are implicated in EMT in several tumor types (9). Zeb1 deficient mouse embryonic fibroblasts undergo premature replicative senescence and ectopic E-cadherin expression (15). However, the precise roles of ZEB in EMT remain to be elucidated.
Cellular senescence is induced by eroded telomeres, oncogene-induced DNA damage and epigenetic derepression of the INK4A/ARF locus (16–17). Senescent cells exhibit flat and enlarged cell morphology as well as proliferative arrest accompanied by increased senescence-associated β-galactosidase (SABG) activity and upregulation of cell cycle inhibitors such as p15INK4B, p16INK4A and p21 (CDKN1A). In primary human esophageal epithelial cells, telomerase (hTERT) activation overcomes replicative senescence, establishing a non-transformed immortalized diploid cell line EPC2-hTERT, which maintains functionally intact p53 and p16INK4A (18). Ectopically expressed p16INK4A alone induces senescence while activation of oncogenes such as Ha-RasG12V and AKT also induce senescence in EPC2-hTERT cells (18–20), indicating senescence as a critical barrier function against oncogene-induced malignant transformation in human esophageal cells. Recently, EMT has been implicated in the early stages of carcinogenesis to bypass oncogene-induced senescence (21). However, it remains unclear how cellular senescence functions may be inactivated during EMT associated with malignant transformation.
We have demonstrated recently that EGFR overexpression and p53 mutations (p53R175H and p53V143A) are necessary and sufficient to transform EPC2-hTERT cells, leading to increased cell motility, anchorage independent growth and tumor formation in nude mice (22). Herein, we have investigated how cells acquire the capacity to undergo EMT in response to TGF-β. We find that EGFR and mutant p53 cooperate to enrich an EMT-competent subpopulation of human esophageal cells expressing ZEB1 and ZEB2, which suppress p15INK4B and p16INK4A to overcome EGFR-mediated senescence.
Materials and Methods
Cell lines and monolayer culture
EPC1-hTERT and EPC2-hTERT, established from independent primary cultures of normal human esophageal epithelial cells, and their derivatives were grown in Keratinocyte-serum free medium (Invitrogen, Carlsbad, CA) at 37°C in a 5% CO2 atmosphere as described previously (18, 20, 22–23). HCE7, an ESCC cell line was grown as described previously (24). Countess™ Automated Cell Counter (Invitrogen) was used to count cells with 0.2% Trypan Blue dye to exclude dead cells. Cells were treated with 5 ng/ml of recombinant human TGF-β1 (R&D Systems, Minneapolis, MN) reconstituted in 4 mM HCl containing 0.1% bovine serum albumin. AG 1478 (Calbiochem, La Jolla CA) was reconstituted in dimethyl sulfoxide and used at 100 nM. Phase contrast images were acquired using a Nikon Eclipse TS100 microscope. Spindle-shaped cells were scored by counting at least 100 cells per high-power field (n=6) under light microscopy.
Retrovirus- and Lentivirus-mediated gene transfer
Retroviral vectors expressing EGFR in pFB-Neo and/or either p53R175H or p53V143A in pBABE-puro were stably transduced into EPC1-hTERT and EPC2-hTERT cells as described previously (20, 22–23). Stable cell lines were established by drug selection for 7 days with 300 μg/ml of G418 (Invitrogen) for pFB-Neo and 1 μg/ml of Puromycin (Invitrogen) and pBABE-puro.
The lentiviral pGIPZ vectors expressing short hairpin RNA (shRNA) directed against human ZEB1 designated ZEB1-A and ZEB1-B (clone ID # V2LHS_116663 and V2LHS_116659), ZEB2 designated ZEB2-A and ZEB2-B (V2LHS_232431 and V2LHS_268826) or a non-silencing scramble sequence (Open Biosystems) were transfected into HEK-293T cells with Arrest-In Transfection Reagent (Open Biosystems) to produce replication-incompetent viruses. Cells were infected as in retrovirus-mediated gene transfer and flow sorted for the GFP brightest cells (top 20%).
Transient transfection and dual-luciferase assays
Transient transfection was carried out using the FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. Briefly, 1 × 105 cells were seeded per well in 24-wellplates 24 hours before transfection. 400 ng of the luciferase-reporter constructs p15P751-luc (25) containing the p15INK4B promoter (0.75-kb)(gift of Dr. Xiao-Fan Wang) or pGL3-p16 (26) containing the p16INK4A promoter (2.3-kb)(gift of Dr. James W. Rocco) was transfected along with 5 ng of phRL-SV40-renilla luciferase vector (Promega) to calibrate the variation of transfection efficiencies among wells. Cells were incubated for 48 hours before cell lysis. Luciferase activities were determined using the Dual-Luciferase™ Reporter Assay system (Promega) and the ORION Microplate Luminometer (Berthold Detection Systems USA, Oak Ridge, TN). The mean of fire fly luciferase activity was normalized with the co-transfected renilla luciferase activity. Transfection was carried out at least three times, and variation between experiments was not greater than 15%.
5-bromo-2′-deoxyuridine (BrdU) incorporation assays
Cell proliferation was assessed using the Cell Proliferation ELISA kit (Roche, Basel, Switzerland) with BrdU labeling for 2 hrs prior to fixation. All experiments were performed in triplicate.
Senescence-Associated β-galactosidase (SABG) assays
The Senescence β-Galactosidase Staining Kit (Cell Signaling, Danvers, MA) was used to stain senescent cells, which were scored by counting at least 100 cells high-power field (n=6) under light microscopy.
RNA isolation, cDNA synthesis and real-time RT-PCR
RNA extraction and cDNA synthesis were performed as described previously (27). Real-time RT-PCR was done with TaqMan® Gene Expression Assays (Applied Biosystems) for CDH1 (Hs00170423_m1), CDH2 (Hs00983062_m1), ZEB1 (Hs00232783_m1), ZEB2 (Hs00207691_m1), SNAI1 (Hs00195591_m1), SNAI2 (Hs00161804_m1), TWIST1 (Hs00361186_m1) and CDKN1A (Hs00355782_m1) using the ABI PRISM® 7000 Sequence Detection System (Applied Biosystems). SYBR green reagent (Applied Biosystems) was used to quantitate mRNA for β-actin as described (27). The relative level of each mRNA was normalized to β-actin as an internal control.
Immunofluorescence
Cells grown in chamber slides (Nalge Nunc, Naperville, IL) precoated with BD Matrigel™ Matrix (BD Biosciences, San Jose, CA) were fixed in 1:1 methanol/acetone for 10 min at −20°C and blocked with 1% bovine serum albumin for 30 min. Slides were incubated with mouse anti-E-cadherin (1: 200; BD Biosceineces) or mouse anti-vimentin (1:10,000; Novus Biologicals, Littleton, CO) overnight at 4°C, and then with appropriate Cy2- or Cy3-conjugated secondary antibody (1: 400; Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. Nuclei were counterstained by DAPI (1:10,000; Invitrogen). Stained objects were examined with a Nikon Microphot microscope and imaged with a digital camera.
Western blot analysis
Whole cell lysates were prepared as described (20, 22). Nuclear extracts were purified as described previously (28). Briefly, cells were washed twice with PBS, resuspended in buffer A [10 mM HEPES-KOH (pH 7.8), 10 mM KCl, 0.1 mM EDTA (pH 8.0), 0.1 % NP-40, 1 mM DTT, 0.5 mM PMSF, 2 μg/ml Pepstatin] and vortexed vigorously. Following centrifugation at 5,000 rpm for one minute, the nuclear pellets were resuspended in buffer C [50 mM HEPES-KOH (pH 7.8), 0.4 M KCl, 0.1 mM EDTA (pH 8.0), 5 mM MgCl2, 20 % Glycerol, 1 mM DTT, 0.5 mM PMSF, 2 μg/ml Pepstatin] and mixed gently at 4°C for 30 min. Following centrifugation at 15,000 rpm for 15 min, the supernatant was recovered as nuclear extracts.
20 μg of denatured protein was fractionated on a NuPAGE Bis-Tris 4–12% gel (Invitrogen). Following electrotransfer, Immobilon-P membranes (Millipore) were incubated with primary antibodies listed in Table S1, and then with the appropriate HRP-conjugated secondary antibody (GE Healthcare, Piscataway, NJ). β-actin and histone H1 served as loading controls for whole cell lysates and nuclear extracts, respectively.
Statistical Analysis
Data from triplicate and hexaduplicate experiments are presented as mean ± SE and were analyzed by two-tailed Student’s t test. P <0.05 was considered significant.
Results
EGFR overexpression and p53 mutation promote enrichment of EMT-competent subpopulation of cells
EGFR overexpression and concurrent expression of mutant p53 transform EPC2-hTERT cells, conferring invasive characteristics as described previously (22). EMT was suggested by gene expression profiling of EPC2-hTERT-EGFR-p53R175H cells grown in organotypic 3-D culture, a form of human tissue engineering (Michaylira et al. submitted). When cells were treated with TGF-β in monolayer culture, more than 90% of EPC2-hTERT-EGFR-p53R175H cells exhibited of spindle-shaped cell morphology within 3 weeks (Fig. 1, A and B). This was accompanied by loss of E-cadherin as well as induction of mesenchymal markers such as N-cadherin and vimentin (Fig. 1, C and D), indicating EMT. EMT was also induced in EPC2-hTERT-EGFR-puro cells, yet to a limited extent (30–40%)(Fig. 1, A–D). In fact, the frequency of EMT reached plateau despite extended TGF-β treatment (6–8 weeks) in EPC2-hTERT-EGFR-puro cells (Fig. 1B and data not shown). Moreover, the majority of EPC2-hTERT-neo-p53R175H or EPC2-hTERT-neo-puro cells failed to undergo EMT by TGF-β treatment (Fig. 1, A–D). These observations indicated that EPC2-hTERT cells had drifted toward EMT-competent and EMT-incompetent states depending upon genetic alterations induced by retrovirus-mediated gene transduction.
Figure 1. EGFR and mutant p53 enrich EMT-competent subpopulation of cells.
EPC2-hTERT cell derivatives carrying the indicated genotypes were stimulated with or without TGF-β1.
A, Phase contrast images were taken 14 days after TGF-β treatment. Arrows indicate spindle-shaped cells suggesting EMT. Scale bar, 100 μm.
B, Spindle-shaped cells were scored at the indicated time points after TGF-β stimulation. **, P <0.001 vs. neo-puro; *, P <0.05 vs. neo-puro; ns, not significant vs. neo-puro; ‡, P <0.001 vs. EGFR-puro (n=6).
C, Cells were double-stained for E-cadherin (green) and vimentin (red). Note the presence of E-cadherin negative and vimentin positive spindle-shaped cells (arrow), suggesting spontaneous EMT (upper panel). Scale bar, 100 μm.
D, E-cadherin (CDH1) and N-cadherin (CDH2) mRNA levels in A. **, P <0.001 vs. neo-puro with TGF-β (+); #, P <0.001 vs. indicated genotype with TGF-β (−)(n=3).
TGF-β stimulated SMAD2/3 phosphorylation in all of the EPC2-hTERT cell derivatives (Fig. S1A), confirming TGF-β receptor activation. Since TGF-β did not induce apoptosis (Fig. S1B), elimination of EMT-competent cells during TGF-β treatment was an unlikely mechanism for minimal EMT observed in EPC2-hTERT-neo-p53R175H and EPC2-hTERT-neo-puro cells (Fig. 1, A, B and D). TGF-β reduced cell proliferation by 40–60% in all genotypes (data not shown). However, the extent of TGF-β-mediated cytostatic effects was not associated with the frequency of EMT. Although EMT was induced in EGFR overexpressing cells (Fig. 1, A and B), pharmacological inhibition of EGFR by AG1478 did not prevent TGF-β from inducing EMT in EPC2-hTERT-EGFR-p53R175H cells (Fig. S2, A–C), indicating that the EGFR activity per se may be dispensable during EMT. Since parental EPC2-hTERT cells are thought to be a heterogeneous cell population derived from primary culture, we suspected that they may consist of EMT-competent and incompetent subpopulations of cells and that retrovirus-mediated EGFR transduction may select EMT-competent cells preferentially. Consistent with such a notion, spontaneous EMT was observed without TGF-β treatment in the cells with EGFR overexpression, but not without EGFR overexpression (Fig. 1, A and C).
ZEB1 and ZEB2 are associated with TGF-β-mediated EMT in the cells with EGFR overexpression
The EMT-competent nature of the cells with EGFR overexpression prompted us to explore the role of unique transcription factors essential in EMT. Amongst them, ZEB1 and ZEB2, but not SNAI1, SNAI2 and TWIST1 were found upregulated at the mRNA levels prior to TGF-β stimulation in EGFR overexpressing EPC2-hTERT derivatives (Fig. 2A and data not shown). ZEB1 and ZEB2 proteins were also detected without TGF-β treatment in the nuclear extracts, but not whole cell lysates of EGFR overexpressing cells (Fig. 2, B and C), implying ZEB as a master regulator of EMT-competency in human esophageal cells. In addition, ZEB1 and ZEB2 were expressed in HCE7, an ESCC cell line exhibiting full characteristics of EMT (Fig. 2B, and data not shown). In EPC2-hTERT-EGFR-p53R175H cells, TGF-β induced robustly ZEB1 and ZEB2 along with the other factors including SNAI1, SNAI2 and TWIST1 (Fig. 2, A and C, and data not shown). Interestingly, TGF-β failed to induce ZEB1, ZEB2 and SNAI1 in the absence of EGFR overexpression (Fig. 2, A and C), suggesting a role for EGFR overexpression in the altered transcriptional gene expression program in EMT. Nonetheless, neither EGFR stimulation nor inhibition affected ZEB expression (Fig. S2D, and data not shown), in agreement with the premise that the EGFR activity may not be required for TGF-β-mediated EMT (Fig. S2, A–C).
Figure 2. ZEB1 and ZEB2 expression is associated with EGFR overexpression.
EPC2-hTERT cell derivatives carrying the indicated genotypes were stimulated with or without TGF-β1 for 14 days.
A, Relative mRNA levels of ZEB1, ZEB2 and SNAI1 were determined. **, P <0.001 vs. neo-puro; *, P <0.05 vs. neo-puro (n=3).
B and C, ZEB1 and ZEB2 proteins were determined by Western blotting using nuclear extracts in B and whole cell lysates in C. Arrow heads indicate specific bands for ZEB1 and ZEB2. Arrows indicate non-specific bands.
ZEB and the microRNA (miR)-205 and miR-200 family negatively regulate each other (29–32). In fact, these microRNA species were sharply suppressed upon TGF-β-induced EMT and that miR-200b, miR-141 and miR-205 were downregulated significantly in EPC2-hTERT-EGFR-p53R175H cells prior to TGF-β treatment (Fig. S3). Thus, these microRNAs likely have a role in ZEB expression in EGFR overexpressing cells. However, we cannot conclude whether suppression of these microRNAs led to induction of ZEB, or vice versa.
ZEB1 and ZEB2 are expressed in the cells negating EGFR-induced senescence
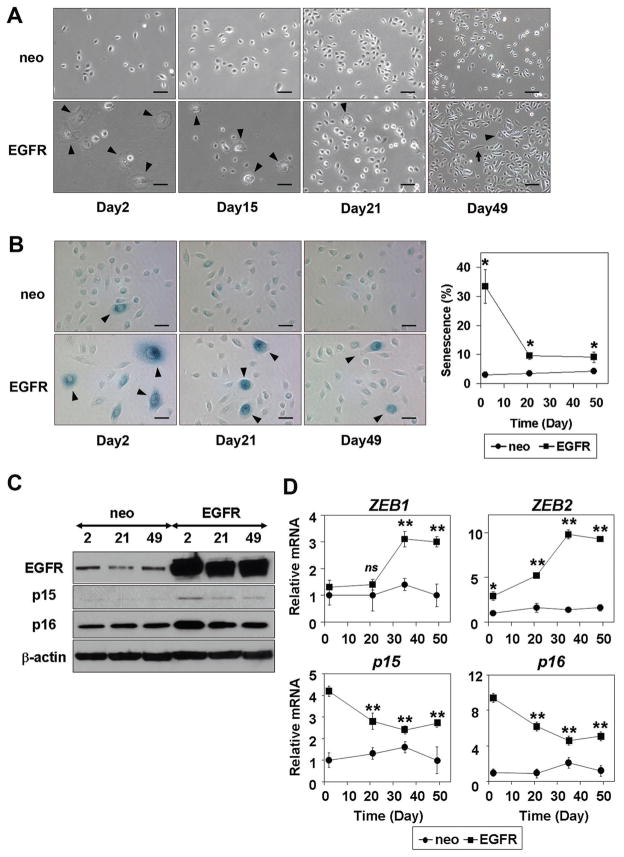
We next aimed at delineating how EGFR overexpression may lead to enrichment of the cells expressing ZEB1 and ZEB2. We have noticed that a small subset of EPC2-hTERT-EGFR-puro cells exhibit proliferative arrest and morphology compatible with senescence corroborated by the SABG activity without TGF-β stimulation (Fig. S4A). In addition, Western blotting detected upregulation of cyclin-dependent kinase inhibitors (CDKI) p15INK4B, p16INK4A and p21 in EPC2-hTERT-EGFR-puro cells (Fig. S4B). We suspected that EGFR overexpression may trigger senescence. In fact, senescence was observed in 30–40% of EPC2-hTERT cells shortly after drug selection upon retrovirus-mediated transduction of EGFR, but not a control empty vector (Fig. 3, A and B). However, actively proliferative cells emerged without losing EGFR and predominated over the senescent cells eventually (Fig. 3, A-C). Interestingly, ZEB 1 and ZEB2 were found to be upregulated as p15INK4B and p16INK4A were downregulated reciprocally in such a cell population (Fig. 3, C and D). Moreover, induction of ZEB1 and ZEB2 was accelerated when EGFR was transduced in EPC2-hTERT cells along with p53R175H, alleviating EGFR-mediated senescence and CDKI upregulation (data not shown, and Fig. S4, A and B). ZEB was also induced following EGFR transduction in EPC1-hTERT, an independently established immortalized human esophageal cell line (Fig. S4C). These observations suggested that EGFR overexpression may allow expansion of a subset of cells negating senescence and expressing ZEB1 and ZEB2, which may have a role in facilitating EMT.
Figure 3. ZEB1 and ZEB2 are expressed in subpopulation of cells negating EGFR-induced senescence.
EPC2-hTERT cells were stably transduced with EGFR or a control vector (neo). A, Phase contrast images were taken at the indicated time points after drug selection. Arrowheads point enlarged cells consistent with senescence. Arrows indicate spindle-shaped cells implying spontaneous EMT occurring at a low frequency. Scale bar, 100 μm. B, SABG staining in A. Arrowheads identify SABG positive cells. Line graph (right panel) represents SABG positive rates (%) at each time point. *, P <0.05 vs. neo (n=6). C, Protein levels for EGFR, p15INK4B and p16INK4A at the indicated time points in A.
D, mRNA levels for indicated genes at the indicated time points in A. **, P <0.001 vs. neo; *, P <0.05 vs. neo; ns, not significant vs. neo (n=3).
ZEB1 and ZEB2 promote TGF-β-mediated EMT by suppressing senescence
To address the role of ZEB1 and ZEB2 in TGF-β-mediated EMT, we targeted ZEB in EPC2-hTERT-EGFR-p53R175H cells by RNA interference. Stable knockdown of either ZEB1 or ZEB2 resulted in upregulation of p15INK4B and p16INK4A, accompanied by transcriptional activation of the respective promoters (Fig. 4, A and B), and senescence in a subset of the cells (Fig. 4C), indicating the possibility that ZEB may mitigate EGFR-induced senescence. Furthermore, TGF-β triggered massive senescence in ZEB knockdown cells, preventing induction of spindle-shaped cell morphology and a cadherin class switch (Fig. 4, C and D). By contrast, TGF-β induced EMT in scrambled shRNA-transduced control cells (Fig. 4, C and D). Interestingly, ZEB1 knockdown resulted in partial inhibition of ZEB2, despite the lack of homology between the ZEB1 shRNAs and ZEB2 mRNA (Fig. 4A). Such an effect has been observed by others (29, 33), and may be accounted for in part by de-repression of the miR-200 family (32). Thus, our data indicate the possibility that ZEB1 may influence the ZEB2 expression level. Therefore, ZEB1 and/or ZEB2 is/are required for EPC2-hTERT-EGFR-p53R175H cells to undergo EMT in response to TGF-β, and that ZEB may prevent EGFR from activating cellular senescence checkpoint functions through suppression of p15INK4B and p16INK4A.
Figure 4. ZEB1 and ZEB2 promote TGF-β-mediated EMT by suppressing senescence mediated by p15INK4B and p16INK4A.
EPC2-hTERT-EGFR-p53R175H cells expressing independent shRNA sequences directed against ZEB1 (ZEB1-A and ZEB1-B), ZEB2 (ZEB2-A and ZEB2-B) or a scrambled non-silencing control shRNA were characterized.
A, Western blotting determines the effect of RNA interference. Note that TGF-β1 treatment was done for 14 days to induce the detectable level of ZEB proteins in the control cells which underwent EMT as shown in C.
B, Western blotting determines the impact of ZEB1 or ZEB2 knockdown upon p15INK4B and p16INK4A expression in the absence of TGF-β treatment (upper panel). Luciferase assays determined the impact of ZEB silencing upon the p15INK4B and p16INK4A promoters (lower panel). **, P <0.001 vs. scramble; *, P <0.05 vs. scramble (n=4).
C, Representative phase-contrast images of the cells treated with or without TGF-β1 for 14 days. Arrowheads point enlarged cells consistent with senescence. Arrows indicate spindle-shaped cells compatible with EMT. Scale bar, 100 μm. Histogram represents SABG positive rates. *, P <0.001 vs. scramble with TGF-β (+); †, P <0.05 vs. scramble with TGF-β (−)(n=6).
D, Relative mRNA levels of E-cadherin (CDH1) and N-cadherin (CDH2) in C. **, P <0.001 vs. scramble with TGF-β (+); ‡, P <0.001 vs. scramble with TGF-β (−)(n=3).
Senescence prevents TGF-β from inducing ZEB in the EMT-competent cells
Cellular senescence upon ZEB knockdown was associated with reactivation of CDKI (Fig. 4, B and C). Impaired EMT in such ZEB knockdown cells (Fig. 4, C and D), thus can be attributed to suppressed ZEB-dependent transcriptional regulation of EMT markers such as E-cadherin, N-cadherin and vimentin. However, it remains unclear whether senescence per se affects the EMT processes including TGF-β-stimulated ZEB augmentation observed in EPC2-hTERT-EGFR-p53R175H cells (Fig. 2, A and C).
To determine whether senescence can block EMT, we established EPC2-hTERT-EGFR-p53V143A cells, where temperature sensitive mutant p53V143A gains a tertiary conformation similar to wild-type p53 and DNA binding as well as transcriptional activities at 32.5°C (34). When EPC2-hTERT-EGFR-p53V143A cells were exposed to 32°C, massive senescence was induced as determined by SABG assays (Fig. 5, A and B). Cell proliferation was suppressed greatly (Fig. 5C) along with upregulation of p21 (Fig. 5D). This supported the notion that mutant p53 may alleviate EGFR-induced senescence by suppressing p21 as observed in EPC2-hTERT-EGFR-p53R175H cells (Fig. S4, A and B), thus contributing to expansion of the EMT-competent cells during EGFR-transduction. By contrast, senescence was minimally induced in EPC2-hTERT-EGFR-p53R175H cells (Fig. 5), corroborating that p53R175H does not have wild-type p53 activity.
Figure 5. Activation of the wild-type p53 functions of temperature sensitive mutant p53V143A results in senescence.
The wild-type p53 activity of p53V143A was induced by exposing EPC2-hTERT-EGFR-p53V143A or EPC2-hTERT-EGFR-p53R175H (control) cells to 32°C for 72 hrs. A, Phase contrast and bright field images were taken for SABG stained cells. Arrowheads point SABG positive cells. Scale bar, 100 μm.
B, Histogram represents SABG positive rates (%) in A. ** P <0.001 vs. 37 °C (n=6).
C, Cell proliferation assessed by BrdU uptake in A. **, P <0.001 vs. 37°C (n=3).
D, Relative p21 mRNA levels in A. **, P <0.001 vs. 37°C (n=3).
When stimulated by TGF-β, EPC2-hTERT-EGFR-p53V143A cells were prone to undergo EMT at 37°C (Fig. 6, A and B). When senescence was induced fully, however, EPC2-hTERT-EGFR-p53V143A cells no longer underwent EMT upon TGF-β treatment, as indicated by lack of cadherin class switch at 32°C (Fig. 6B). Despite p53 activation, apoptosis was not induced with or without TGF-β treatment (Fig. S5A and data not shown), excluding apoptosis as a potential mechanism preventing EMT. Interestingly, TGF-β stimulation neither augmented ZEB1 and ZEB2 levels nor induced TWIST1, SNAI1 and SNAI2 in senescent EPC2-hTERT-EGFR-p53V143A cells (Fig. 6C and Fig. S5B), indicating that senescence abates the induction of downstream transcription factors crucial for EMT. Nonetheless, senescence per se did not block TGF-β receptor activation in EPC2-hTERT-EGFR-p53V143A cells (Fig. S5C). Thus, activation of cellular senescence program appeared to prevent TGF-β from inducing transcription factors essential in EMT.
Figure 6. Senescence prevents TGF-β from inducing EMT.
Senescence was induced in EPC2-hTERT-EGFR-p53V143A as shown in Figure 5. Cells were further stimulated with or without TGF-β1 for 14 days at either 32°C or 37°C. A, Phase contrast images. Arrows indicate spindle-shaped cells. Arrowheads indicate enlarged cells consistent with senescence. Scale bar, 100 μm. Histogram represents spindle-shaped cell rate (%). ** P <0.001 vs. 37°C (n=6).
B, E-cadherin and N-cadherin levels determined by Western blotting in A.
C, Relative mRNA levels for ZEB1, ZEB2 and SNAI1 in EPC2-hTERT-EGFR-p53V143A cells. **, P <0.001 vs. TGF-β (−) at 37°C; ns, not significant vs. TGF-β (−) at 32°C (n=3).
In aggregate, our data indicate that EGFR overexpression and p53 mutation in non-transformed human esophageal cells may lead to enrichment of EMT competent subpopulation of cells with ZEB upregulation. ZEB1 and ZEB2 may negatively regulate p15 INK4B and p16INK4A to facilitate cells overcoming EGFR-induced senescence. Mutant p53 may also alleviate EGFR-induced senescence by suppressing p21. In the EMT-competent cells with suppressed senescence checkpoint functions, TGF-β induces ZEB and other factors to promote EMT.
Discussion
TGF-β is a potent inducer of EMT. However, EMT is not necessarily a common outcome of TGF-β treatment, especially in human cell lines (35). However, there are carcinoma cell lines with mesenchymal traits suggestive of EMT. Such cell lines have been attributed to specific molecular states, such as acquisition of K-Ras independency (36) and ZEB1 and ZEB2 upregulation through suppression of the miR-200 family of microRNAs (29). We now demonstrate that EGFR and mutant p53, essential for malignant transformation of human esophageal cells (22) may promote selective expansion of an EMT-competent subpopulation of cells expressing ZEB1 and ZEB2 (Fig. 1 and 2). Our data also suggest that EMT competent cells may be capable of negating oncogene-activated senescence checkpoint functions through ZEB1 and/or ZEB2 (Fig. 3 and 4) whereas cellular senescence may prevent TGF-β from inducing ZEB and other transcription factors to activate the EMT program (Fig. 5 and 6). In our proposed model, p53 as well as the INK4 locus-encoded CDKI p15 INK4B and p16INK4A serve as barrier functions against EGFR oncogene-mediated cellular stress (Fig. S6). Interestingly, ZEB1 and ZEB2 expression was associated with EGFR overexpression (Fig. 2) and implicated in suppression of p15 INK4B and p16INK4A (Fig. 4).
EMT confers cancer cells resistance to EGFR inhibitors (37). ZEB1 knockdown resulted in mesenchymal to epithelial transition and increased sensitivity to Erlotinib, an EGFR inhibitor in head and neck squamous cell carcinoma cell lines (38). Thus, EMT influences EGFR activities in transformed cells. However, the EGFR kinase activity did not appear to be required for ZEB expression or TGF-β-induced EMT in established EPC2-hTERT cell derivatives with EGFR overexpression (Fig. S2). Nonetheless, ZEB1 and ZEB2 expression was increased in the EGFR overexpressing cells without TGF-β stimulation (Fig. 2). We speculate that a small subset of parental EPC2-hTERT cells expressing ZEB1 and ZEB2 were selected as a result of EGFR-induced senescence (Fig. 3), eliminating cells without ZEB expression. Alternatively, ZEB may be induced through a cellular reprogramming event in a unique subset of cells, acquiring an EGFR-independent status. In agreement with such a notion, ZEB1 has been implicated in stemness-maintenance through miR-200 family-mediated regulation of Sox2, Klf4 and Bmi1 (39). Given downregulation of p15INK4B and p16INK4A following EGFR-induced senescence (Fig. 3), it is tempting to speculate that EGFR triggered an epigenetic reprogramming event involving microRNAs such as miR-200b and miR-141 (Fig. S3), resulting in induction of ZEB as well as Bmi1, a Polycomb factor essential in transcriptional repression of p16INK4A, leading to repression of these CDKI. Thus, cellular reprogramming events may take place during malignant transformation of EPC2-hTERT cells selecting EMT-competent cells with ZEB expression.
Induction of senescence by wild-type human EGFR is a novel finding. However, EGFR activation is known to trigger cell cycle arrest (40–41), which is antagonized by human papilloma virus E6 and E7 proteins (42), implicating the pRB and p53 pathways. EGFR overexpression led to upregulation of p15INK4B, p16INK4A and p21 in EPC2-hTERT cells (Fig. S4B). ZEB-mediated suppression of CDKI in our cells (Fig. 4B) is reinforced by premature replicative senescence associated with upregulation of p15INK4B and p21 in Zeb1 knockout mouse embryonic fibroblasts (15), although ZEB knockdown did not result in derepression of p21 in our cell systems (data not shown). TWIST was not upregulated in EGFR-transduced EPC2-hTERT cells without TGF-β treatment (data not shown). However, Twist suppresses cellular senescence through negative regulation of p14ARF and MDM2/p53 and Chk1/2 DNA damage response pathways in human prostate epithelial cells (43). Twist proteins also prevent ErbB2 and H-RasV12 oncogenes from inducing senescence through suppression of p21 and p16INK4A (21). Thus, our findings extend these paradigms of cohesive regulation of senescence and EMT programs.
The role of p53 in EMT is largely unknown. Mutant p53 may stabilize Slug protein by preventing MDM2-mediated proteasomal degradation of Slug (44). However, this is an unlikely mechanism in our cell lines as EMT was only minimally induced without SNAI2 induction in EPC2-hTERT-neo-p53R175H cells (Fig. 1 and data not shown). Our experiments using temperature sensitive p53V143A (Fig. 5 and 6) demonstrated that wild-type p53 activity triggers senescence, preventing TGF-β from inducing EMT without compromising the TGF-β receptor activity. Adorno et al. demonstrated that TGF-β induces formation of a ternary complex comprising of mutant p53, Smad2 and ΔNp63α in a Ras-MAPK-dependent fashion and facilitates cell invasion by suppressing ΔNp63α-mediated inhibition of the TGF-β-induced promigratory responses (45). Since ZEB1 has been implicated in transcriptional repression of p53 family members, ΔNp63, TAp73 and ΔNp73, it is tempting to speculate that ΔNp63 may be a target for ZEB1 upon TGF-β-induced EMT.
ZEB1 and ZEB2 were both expressed in EGFR overexpressing cells without TGF-β stimulation (Fig. 2), raising the possibility that ZEB may have a function independent of TGF-β receptor activation. However, this may be unlikely since SMAD2 phosphorylation was detected without TGF-β stimulation in EPC2-hTERT cell derivatives (Fig. S1A). TGF-β-induced EMT involved robust induction of both ZEB1 and ZEB2 expression (Fig. 2, A and C). Moreover, TGF-β greatly enhanced senescence in ZEB knockdown cells (Fig. 4). These observations are consistent with the biological functions of ZEB as key downstream molecules in the TGF-β pathway (13). ZEB1 and ZEB2 proteins may exert opposing effects in TGF-β-mediated SMAD-dependent transcriptional regulation (46). Therefore, further study is required to determine the role of each ZEB protein in regulation of its transcriptional target genes including E-cadherin, vimentin and p15INK4b.
In conclusion, our novel data underscore the role of EGFR overexpression and p53 mutations in enrichment of a subset of esophageal cells that is capable of undergoing EMT in response to TGF-β through ZEB transcription factors, shedding new insights upon invasive cell growth and inactivation of senescence checkpoint functions during malignant transformation.
Supplementary Material
Acknowledgments
This study was supported in part by NIH Grants R01DK077005 (to MN, SO, HN), P01-CA-098101 (Mechanisms of Esophageal Carcinogenesis to HN, SO, MN, GW, KG, DBS, JK, CZM, TO, AJPK, MH, JAD, AKR), K01-DK-066205 (to HN), Uehara Memorial Foundation Research fellowship (to SO), NIH/K99-CA138498 and NIH/NRSA F32-CA103085 (to DBS), NIH/NRSA F32-DK075230 (to CZM), NIH/NRSA F32-DK082149 (to KDG), NIH/NCI T32-CA115299 (to JK and GW), P30-DK050306 (to RK), American Gastroenterological Association Foundation Student Research Fellowship Award (to Momo Nakagawa), and the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and its core facilities. We thank Dr. Charles H. Pletcher (Flow Cytometry & Cell Sorting Facility), Dr. Gary P. Swain and Ms. Daniela Budo (Morphology Core), Dr. Gary D. Wu and Dr. Sue Keilbaugh (Molecular Biology/Gene Expression Core) and Dr. Richard Carroll (Cell Culture Core). We thank Ben Rhoades and Christie M. Gutierrezfor technical assistance, Catrina King, Drs. Takaomi Okawa, Johannes von Burstin, Maximilian Reichert and Perry S. Mongroo (Rustgi laboratory) and Claudia D. Andl (Vanderbilt University) for helpful discussions. We thank Drs Xiao-Fan Wang (Duke University) and James W. Rocco (Massachusetts General Hospital) for reagents.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Nakagawa H, Katzka D, Rustgi AK. Biology of esophageal cancer. In: Rustgi AK, editor. Gastrointestinal Cancers. London: Elsevier; 2003. pp. 241–51. [Google Scholar]
- 3.Hiyama T, Yokozaki H, Kitadai Y, et al. Overexpression of human telomerase RNA is an early event in oesophageal carcinogenesis. Virchows Arch. 1999;434:483–7. doi: 10.1007/s004280050372. [DOI] [PubMed] [Google Scholar]
- 4.Itakura Y, Sasano H, Shiga C, et al. Epidermal growth factor receptor overexpression in esophageal carcinoma. An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer. 1994;74:795–804. doi: 10.1002/1097-0142(19940801)74:3<795::aid-cncr2820740303>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 5.Parenti AR, Rugge M, Frizzera E, et al. p53 overexpression in the multistep process of esophageal carcinogenesis. Am J Surg Pathol. 1995;19:1418–22. doi: 10.1097/00000478-199512000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Volant A, Nousbaum JB, Giroux MA, et al. p53 protein accumulation in oesophageal squamous cell carcinomas and precancerous lesions. J Clin Pathol. 1995;48:531–4. doi: 10.1136/jcp.48.6.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteve A, Lehman T, Jiang W, et al. Correlation of p53 mutations with epidermal growth factor receptor overexpression and absence of mdm2 amplification in human esophageal carcinomas. Mol Carcinog. 1993;8:306–11. doi: 10.1002/mc.2940080414. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 10.Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: Mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 12.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–87. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. Embo J. 2003;22:2453–62. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–88. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 18.Harada H, Nakagawa H, Oyama K, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–38. [PubMed] [Google Scholar]
- 19.Oyama K, Okawa T, Nakagawa H, et al. AKT induces senescence in primary esophageal epithelial cells but is permissive for differentiation as revealed in organotypic culture. Oncogene. 2007;26:2353–64. doi: 10.1038/sj.onc.1210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takaoka M, Harada H, Deramaudt TB, et al. Ha-Ras(G12V) induces senescence in primary and immortalized human esophageal keratinocytes with p53 dysfunction. Oncogene. 2004;23:6760–8. doi: 10.1038/sj.onc.1207923. [DOI] [PubMed] [Google Scholar]
- 21.Ansieau S, Bastid J, Doreau A, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Okawa T, Michaylira CZ, Kalabis J, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andl CD, Mizushima T, Nakagawa H, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–30. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 24.Okano J, Gaslightwala I, Birnbaum MJ, Rustgi AK, Nakagawa H. Akt/protein kinase B isoforms are differentially regulated by epidermal growth factor stimulation. J Biol Chem. 2000;275:30934–42. doi: 10.1074/jbc.M004112200. [DOI] [PubMed] [Google Scholar]
- 25.Li JM, Nichols MA, Chandrasekharan S, Xiong Y, Wang XF. Transforming growth factor beta activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem. 1995;270:26750–3. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 26.Mroz EA, Baird AH, Michaud WA, Rocco JW. COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res. 2008;68:6049–53. doi: 10.1158/0008-5472.CAN-08-1279. [DOI] [PubMed] [Google Scholar]
- 27.Takaoka M, Harada H, Andl CD, et al. Epidermal growth factor receptor regulates aberrant expression of insulin-like growth factor-binding protein 3. Cancer Res. 2004;64:7711–23. doi: 10.1158/0008-5472.CAN-04-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akakura N, Kobayashi M, Horiuchi I, et al. Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 2001;61:6548–54. [PubMed] [Google Scholar]
- 29.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 32.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Guo XY, Hu GY, Liu WB, Shay JW, Deisseroth AB. A temperature-sensitive mutant of human p53. Embo J. 1994;13:2535–44. doi: 10.1002/j.1460-2075.1994.tb06543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown KA, Aakre ME, Gorska AE, et al. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 2004;6:R215–31. doi: 10.1186/bcr778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr S, Thomson S, Buck E, et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin Exp Metastasis. 2008;25:685–93. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haddad Y, Choi W, McConkey DJ. Delta-crystallin enhancer binding factor 1 controls the epithelial to mesenchymal transition phenotype and resistance to the epidermal growth factor receptor inhibitor erlotinib in human head and neck squamous cell carcinoma lines. Clin Cancer Res. 2009;15:532–42. doi: 10.1158/1078-0432.CCR-08-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 40.Gill GN, Lazar CS. Increased phosphotyrosine content and inhibition of proliferation in EGF-treated A431 cells. Nature. 1981;293:305–7. doi: 10.1038/293305a0. [DOI] [PubMed] [Google Scholar]
- 41.Fan Z, Lu Y, Wu X, DeBlasio A, Koff A, Mendelsohn J. Prolonged induction of p21Cip1/WAF1/CDK2/PCNA complex by epidermal growth factor receptor activation mediates ligand-induced A431 cell growth inhibition. J Cell Biol. 1995;131:235–42. doi: 10.1083/jcb.131.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akerman GS, Tolleson WH, Brown KL, et al. Human papillomavirus type 16 E6 and E7 cooperate to increase epidermal growth factor receptor (EGFR) mRNA levels, overcoming mechanisms by which excessive EGFR signaling shortens the life span of normal human keratinocytes. Cancer Res. 2001;61:3837–43. [PubMed] [Google Scholar]
- 43.Kwok WK, Ling MT, Yuen HF, Wong YC, Wang X. Role of p14ARF in TWIST-mediated senescence in prostate epithelial cells. Carcinogenesis. 2007;28:2467–75. doi: 10.1093/carcin/bgm185. [DOI] [PubMed] [Google Scholar]
- 44.Wang SP, Wang WL, Chang YL, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 45.Adorno M, Cordenonsi M, Montagner M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 46.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. Embo J. 2003;22:2443–52. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.