Abstract
With trauma, sepsis, cancer, or uremia, animals or patients experience accelerated degradation of muscle protein in the ATP-ubiquitin-proteasome (Ub-P’some) system. The initial step in myofibrillar proteolysis is unknown because this proteolytic system does not break down actomyosin complexes or myofibrils, even though it degrades monomeric actin or myosin. Since cytokines or insulin resistance are common in catabolic states and will activate caspases, we examined whether caspase-3 would break down actomyosin. We found that recombinant caspase-3 cleaves actomyosin, producing a characteristic, approximately 14-kDa actin fragment and other proteins that are degraded by the Ub-P’some. In fact, limited actomyosin cleavage by caspase-3 yields a 125% increase in protein degradation by the Ub-P’some system. Serum deprivation of L6 muscle cells stimulates actin cleavage and proteolysis; insulin blocks these responses by a mechanism requiring PI3K. Cleaved actin fragments are present in muscles of rats with muscle atrophy from diabetes or chronic uremia. Accumulation of actin fragments and the rate of proteolysis in muscle stimulated by diabetes are suppressed by a caspase-3 inhibitor. Thus, in catabolic conditions, an initial step resulting in loss of muscle protein is activation of caspase-3, yielding proteins that are degraded by the Ub-P’some system. Therapeutic strategies could be designed to prevent these events.
Introduction
Loss of muscle mass in catabolic conditions such as uremia, acidosis, cancer, sepsis, and diabetes mellitus contributes substantially to the morbidity of patients. In animal models of these conditions, the mechanism(s) causing muscle atrophy includes activation of the ATP-ubiquitin-proteasome (Ub-P’some) proteolytic system, leading to accelerated degradation of muscle protein (1, 2). In patients as well, there is evidence for involvement of the Ub-P’some system in the muscle atrophy that occurs in response to head injury, sepsis, cancer, AIDS, muscle denervation, or chronic kidney failure (3–8).
Evidence for involvement of the Ub-P’some system in rodent models of catabolic conditions includes increased ubiquitin conjugation to proteins, accumulation of ubiquitin-conjugated proteins in muscle, and inhibition of accelerated muscle proteolysis when proteasome activity is blocked (9–13). However, the sequence of events leading to muscle atrophy is more complicated because the Ub-P’some system does not break down the complexes of proteins contained in actomyosin or myofibrils; these complexes contain the bulk of proteins in muscle (1, 2, 14). Thus, one or more additional proteases must release constituent proteins of actomyosin before the Ub-P’some system can degrade the contractile proteins of muscle.
Increased circulating levels of cytokines and/or insulin resistance are present in many catabolic conditions, and apoptotic pathways are activated in the muscle of rodents with cancer, muscular dystrophy, or heart failure, as well as in patients with muscle denervation or heart failure (15–19). Moreover, apoptotic proteases (i.e., interleukin-1β-converting enzyme) can cleave actin in vitro, suggesting that proteases associated with apoptosis could be involved in muscle protein breakdown (20, 21). A potential mediator in catabolic conditions is decreased PI3K activity, since the insulin resistance associated with catabolic conditions involves impaired PI3K activity, which is a major influence controlling apoptosis (22–24). In these studies, we have examined caspase-3 as a candidate for the initial step in the process of accelerated muscle protein loss stimulated by catabolic conditions. We also examined the influence of PI3K activity as a mediator of protein degradation in muscle cells.
Methods
Cell culture conditions.
L6 skeletal muscle cells (ATCC, Manassas, Virginia, USA) were maintained in DMEM containing 1 g glucose per liter and supplemented with 10% heat-inactivated FCS under a humidified 5% CO2/95% O2 atmosphere. Changing the serum content to 2% horse serum stimulated confluent myocytes to form myotubes.
Preparation of cell extracts and muscle homogenates.
Myotubes were washed in ice-cold PBS, scraped free, and centrifuged. Cell pellets were then resuspended in buffer A, consisting of 20 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT containing 0.1 mM PMSF, antipain (5 μg/ml), leupeptin (5 μg/ml), and aprotinin (5 μg/ml). The cells were kept on ice, and after 30 minutes the swollen cells were gently homogenized with 20 strokes of a Dounce homogenizer. Centrifugation (1,600 g for 10 minutes) removed nuclei and debris; we measured actin cleavage by Western blotting analysis with an affinity-purified, anti-actin antibody that recognizes the carboxy-terminal 11 amino acids of α-actin (Sigma-Aldrich, St. Louis, Missouri, USA).
To obtain evidence of actin degradation in the muscle of rats with accelerated loss of muscle protein because of uremia or diabetes (11, 12), we homogenized their epitrochlearis or gastrocnemius muscles on ice in a hypotonic buffer composed of 5 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM EGTA, 1 mM β-mercaptoethanol, 1% glycerol with 0.1 mM PMSF, leupeptin (5 μg/ml), and aprotinin (5 μg/ml). The homogenates were centrifuged (1,600 g for 10 minutes) before the pellets were resuspended in Laemmli sample buffer. With denaturing conditions, the actin fragments were detected in pellets that contained insoluble myofibrils. Actin fragments were not detected in the soluble fraction of muscle. Actomyosin fragments and actin cleavage were identified by Western blot using the anti-actin antibody.
Evaluation of actomyosin cleavage by caspase-3.
Actomyosin was obtained from Sigma-Aldrich and subjected to electrophoresis in a nondenaturing polyacrylamide gel. The single band obtained indicated no contamination by actin or actin fragments. In 20 μl of buffer A, we added different amounts of actomyosin and recombinant, active caspase-3 (Upstate Biotechnology Inc., Lake Placid, New York, USA) equivalent to 52,000 U/mg (1 U = 1 nmol DEVD-AMC [a peptide substrate of caspase-3] cleaved per minute) (Calbiochem-Novabiochem Corp., San Diego, California, USA). The mixtures were incubated at 37°C and then examined for evidence of actin cleavage as described.
To evaluate the proteolytic cleavage of actin in muscle cells, we incubated L6 muscle cell lysates or a homogenate of psoas muscle (20 μg protein in 20 μl of buffer A) with or without 0.1 μg recombinant caspase-3 for 2–3 hours at 37°C. In some experiments, we evaluated the requirement for ATP by incubating the L6 cell lysates (50 μg protein in 50 μl) with or without 2 mM ATP. Reactions were stopped by adding Laemmli sample buffer, and the proteins were analyzed by Western blotting with the anti-actin antibody.
Measurements of protein degradation.
Protein degradation in L6 cell lysate homogenates was assessed by measuring the net release of tyrosine, since muscle cells neither synthesize nor degrade this amino acid (25). Cells were harvested in a buffer consisting of 5 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM EGTA, 1% glycerol, 1 mM β-mercaptoethanol, and 25 mM chymostatin. Cell lysates were dialyzed against buffer (20 mM Tris-HCl, pH 7.6, 10% glycerol, 5 mM MgCl2, and 2 mM DTT) to remove accumulated tyrosine. Aliquots of each lysate (200 μg protein per sample) were then incubated for 2 hours at 37°C in 0.2 ml of reaction buffer (20 mM Tris, pH 7.6, 2 mM DTT, 10 mM MgCl2, 10% glycerol, 250 μg/ml ubiquitin) with or without adding an ATP-generating system (1 mM ATP, 100 μg/ml creatine phosphokinase, 10 mM creatine phosphate). Reactions were stopped with trichloroacetic acid (5% final concentration), precipitated proteins were removed by centrifugation, and free tyrosine was measured fluorometrically to calculate the rate of protein degradation (26).
Measurement of caspase-3 activity.
Frozen gastrocnemius muscle was pulverized and homogenized on ice in a buffer consisting of 100 mM HEPES (pH 7.5), 10% sucrose, 0.1% NP-40, 10 mM DTT, and protease inhibitor cocktail (Roche Applied Science, Indianapolis, Indiana, USA). Homogenates were subjected to three cycles of freeze-thaw before centrifugation at 18,000 g for 30 minutes. The supernatant (20 μl) was combined with reaction buffer consisting of 100 mM HEPES (pH 7.5), 10% sucrose, and 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonate (CHAPS; Sigma-Aldrich), and the mixture was preincubated for 30 minutes at 30°C with or without 50 μM of the cell-permeable, peptide inhibitor of caspase-3 (Ac-DEVD-CHO) (Calbiochem-Novabiochem). The fluorogenic substrate DEVD-AMC (50 μM) was added, and the reactions were incubated at 30°C for 60 minutes. Fluorescence was measured using excitation and emission wavelengths of 360 and 460 nm, respectively. Results were calculated as the total caspase-3 activity per muscle.
To evaluate the contribution of caspase-3 cleavage of actomyosin to the degradation of muscle proteins by the Ub-P’some system, we incubated 50 μg actomyosin with 0.1 μg recombinant caspase-3 at 37°C for varying periods to produce fragments of actomyosin; the reaction was stopped by adding 5 nM Ac-DEVD-CHO. The presence of the 14-kDa actin fragment in the mixture was confirmed by Western blot using the anti-actin antibody.
To obtain sufficient muscle to evaluate a cell-free preparation of the Ub-P’some system we excised approximately 20 g gastrocnemius muscle tissue from an anesthetized rabbit and immediately froze it in liquid nitrogen before storing at –70°C (14). The frozen muscle was broken into small pieces and placed in 50 ml of ice-cold buffer consisting of 5 mM Tris (pH 7.6), 1% glycerol, 2 mM DTT, 1 mM EDTA, 1 mM EGTA, and the protease inhibitor mixture; it was gently homogenized using a Teflon Dounce. The homogenate was centrifuged at 3,000 g for 5 minutes and then 30,000 g for 30 minutes at 4°C; a final extract was prepared by centrifugation at 100,000 g for 1 hour. The supernatant was concentrated with a Centriprep 3 (Millipore Corp., Bedford, Massachusetts, USA) and reconstituted in 10 ml of storage buffer (20 mM Tris, pH 7.6, 2 mM DTT, 10 mM MgCl2, 10% glycerol, and 20 mM KCl) (14). The muscle extract was stored at –70°C.
The influence of caspase-3 and the Ub-P’some system on total protein degradation was measured after adding an aliquot of the actomyosin/actin fragment preparation to 1 mg of protein in the final extract and incubating the mixture for 120 minutes at 37°C. Identical reactions were performed with and without adding 2 mM ATP to calculate the rate of ATP-dependent protein degradation, which represents proteolysis by the Ub-P’some system (14). Protein degradation was measured as the amount of tyrosine released during the incubation (26).
To evaluate protein degradation in intact rat muscle, the epitrochlearis muscles from diabetic and pair-fed, sham-injected rats were isolated and incubated, after which the rate of protein degradation was measured as described (11, 12). One muscle was incubated in Krebs-Henseleit bicarbonate buffer containing cycloheximide to prevent reincorporation of released tyrosine (26). The contralateral muscle was incubated in the same medium that also contained 10 μM Ac-DEVD-CHO to inhibit caspase-3 activity. The same muscles were then examined for the accumulation of the actin-cleavage products (see above).
Evaluation of PI3K activity and actomyosin cleavage.
To evaluate an insulin-dependent pathway that could mediate actomyosin cleavage, we constructed a dominant-negative p85 subunit of PI3K in an adenovirus vector, AdTrackp85ΔiSH2, that also contained the gene for GFP (27). We infected L6 cells with AdTrackp85ΔiSH2 or the control adenovirus (AdGFP) and evaluated whether suppressing PI3K activity is necessary for caspase-mediated cleavage of actomyosin. The efficiency of infection was examined by the presence of GFP expression using fluorescent microscopy; PI3K activity was measured as described (27). We measured the generation of the 14-kDa actin fragment in response to serum starvation and assessed how varying PI3K activity changed this response.
To identify a link between PI3K activity and activation of caspase-3, we examined whether the amount of the proapoptotic Bax protein in the conformational state that results in activation of caspase is increased in serum-deprived C2C12 muscle cells (28). C2C12 muscle cells were incubated without serum for 6 or 24 hours and compared to cells incubated with 10% FCS serum. Cells were lysed in CHAPS lysis buffer consisting of 10 mM HEPES (pH 7.4), 150 NaCl, 1% CHAPS, and 1 μg/ml protease inhibitors. The lysates were centrifuged (15,000 g, 4°C for 10 minutes), and 500 μg protein was incubated overnight at 4°C with monoclonal antibody 6A7 (Sigma-Aldrich), which recognizes only the activated form of Bax. Then, 100 μl anti–mouse IgG1-conjugated agarose beads (Sigma-Aldrich) were added and incubated overnight at 4°C. After washing the mixture extensively with CHAPS lysis buffer and then boiling it, released proteins were separated by SDS-PAGE (12% gel). The level of activated Bax was detected by a polyclonal antibody that specifically binds to Bax (1:500; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA). HRP-conjugated anti–rabbit IgG (1:4,000; Amersham Biosciences Corp., Piscataway, New Jersey, USA) was used as the secondary antibody. Total Bax in a portion of the lysate was also evaluated using a polyclonal antibody specific to Bax.
Statistical analyses.
Differences in protein degradation measured in cell lysates were analyzed from the mean ± SEM of the percentage increase in ATP-stimulated tyrosine released. This calculation was used to correct for the variability in basal rates of protein degradation in groups of cells. One-way ANOVA followed by pairwise comparisons using the Student-Newman-Kuels test was used to determine whether differences between sample groups (e.g., protein degradation in isolated rat muscles) were significant at the P < 0.05 level.
Results
Activated caspase-3 cleaves actomyosin complexes.
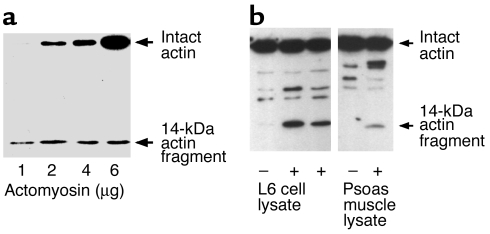
We incubated recombinant caspase-3 with purified, soluble actomyosin complexes to determine whether caspase-3 would cleave proteins in the complexes. Before the incubation, we subjected the actomyosin preparation to nondenaturing PAGE. Actin was detectable only in the purified actomyosin complex (results not shown), excluding the possibility that caspase-3 was simply degrading monomeric actin that had contaminated the actomyosin preparation. After incubating with caspase-3, we used polyclonal antibodies specific to actin to identify the characteristic approximately 42-kDa band corresponding to the size of intact, monomeric actin; we also found an approximately 14-kDa cleaved actin fragment (Figure 1a). Next, we incubated lysates of L6 muscle cells or lysates from rat psoas muscle at 37°C with recombinant caspase-3. Again, we detected the 14-kDa actin fragment (Figure 1b). These results indicate that caspase-3 can cleave actomyosin complexes and actin in an in vitro reconstituted system and in lysates of L6 cells or rat muscle.
Figure 1.
Caspase-3 degrades actomyosin. (a) Recombinant caspase-3 was added to purified actomyosin in vitro, and the mixture was incubated at 37°C for 2 hours. Western blot analysis of the reaction products was performed with polyclonal antibodies against the carboxy terminus of α-actin. The results revealed a characteristic 14-kDa actin fragment. (b) Purified recombinant caspase-3 was added to lysates of L6 skeletal muscle cells or to lysates of rat psoas muscle, and the mixture was incubated at 37°C for 2 hours. Immunoblot analysis of the reaction products demonstrated the appearance of the 14-kDa actin fragment.
Activation of caspase-3 stimulates accumulation of actin fragments in muscle cells.
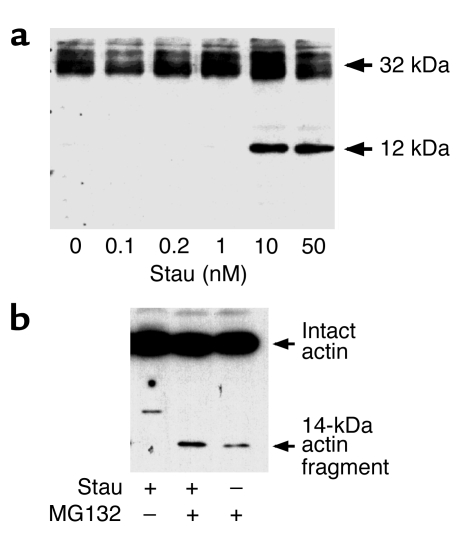
Other investigators found evidence that, in response to heart failure, cancer, denervation, or muscular dystrophy, skeletal muscle can undergo changes consistent with apoptosis, including the presence of apoptotic nuclei in muscle and higher levels of the mRNA for caspase-3 (15–19). To determine whether L6 skeletal muscle cells respond to apoptotic stimuli by activating caspase-3, we treated cells with staurosporine. Staurosporine induced the conversion of procaspase-3 to caspase-3 in a concentration-dependent fashion (Figure 2a). Thus, caspase-3 can be activated in muscle and, therefore, could degrade actomyosin and actin as demonstrated in the reconstituted system (Figure 1).
Figure 2.
Caspase-3-generated actin fragments are degraded by the proteasome. (a) L6 cells were incubated with staurosporine (Stau; 0–50 nM) overnight to activate caspase-3. Cells were lysed and cell proteins were separated by SDS-PAGE. Immunoblot analysis with antibodies against the 12-kDa activated caspase-3 revealed conversion of 32-kDa procaspase-3 to activated caspase-3. (b) We incubated L6 cells with staurosporine and then examined lysates for accumulation of the 14-kDa actin fragment by immunoblotting. The fragment was detected only when the activity of the proteasome was inhibited using MG132 (10 μM).
To examine whether caspase-3 activity and proteolysis in the Ub-P’some system could be acting in sequence, we tested for the generation of actin fragments after incubating L6 cells for 24 hours with staurosporine, with or without the proteasome inhibitor MG132. We did not find the 14-kDa actin fragment in L6 cells when we activated caspase-3 with staurosporine alone, but it was present when we activated caspase-3 and inhibited proteasome activity (Figure 2b). There was a small amount of the actin fragment in control cells treated only with MG132; this could reflect a small number of myoblasts undergoing differentiation, which is dependent on caspase-3 activity. These results indicate that actin degradation is robust when caspase-3 is activated and the resulting actin fragments are degraded by the proteasome.
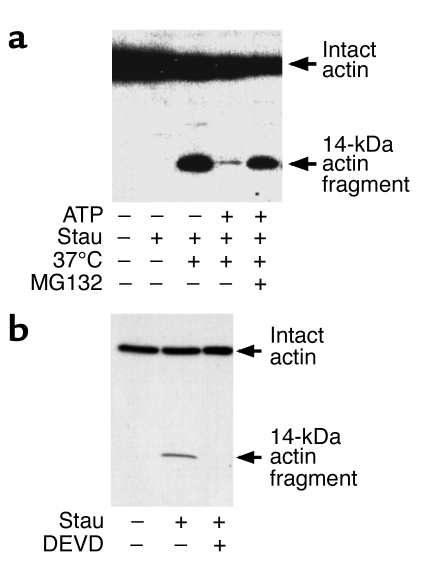
If the Ub-P’some system is rapidly degrading actin, then depleting ATP by incubating cell lysates at 37°C should increase the amount of actin fragment. We examined this possibility and found that 14-kDa actin fragments were generated during incubation of the lysates for 2 hours or more at 37°C (Figure 3a). We attributed this to depletion of ATP, since adding 2 mM ATP to the lysate during the 37°C incubations substantially reduced the concentration of the actin fragment (Figure 3a). We confirmed the link to the Ub-P’some system by examining the effects of adding the proteasome inhibitor, MG132, to lysates that also were supplemented with ATP. In this case, the actin fragment accumulated with activation of caspase-3, even when ATP was added. To identify the influence of caspase-3 in producing the fragment, we added Ac-DEVD-CHO (29, 30) and found that it blocked accumulation of the 14-kDa actin fragment in lysates from staurosporine-treated cells incubated at 37°C (Figure 3b).
Figure 3.
Actin fragment degradation requires ATP, proteasome activity, and caspase-3. (a) L6 cells were incubated with staurosporine to activate caspase-3 as in Figure 2. Cell lysates were incubated on ice or at 37°C for 3 hours as indicated. We then tested for production of the 14-kDa actin fragment. When we added 2 mM ATP, the accumulation of the fragment was largely eliminated, and this outcome was blocked by adding MG132 (10 μM). (b) Lysates from control or staurosporine-treated cells were incubated for 3 hours at 37°C with Ac-DEVD-CHO (DEVD) (5 μM), and Western blot analysis revealed that caspase-3 was responsible for the appearance of the 14-kDa actin fragment.
Others have proposed that calcium-activated proteases (calpains) are involved in the initial breakdown of muscle protein in catabolic conditions (31–33). By contrast, when calcium-activated proteases are inhibited in isolated muscles of rats subjected to starvation, muscle denervation, cancer, acidosis, uremia, or diabetes, there is no suppression of the accelerated muscle proteolysis associated with these conditions (11, 12, 34–36). Still, we examined whether calpains are involved in the proteolytic responses we observed. We activated caspase-3 in L6 cells, added the calpain inhibitor ALLN (Calbiochem-Novabiochem Corp., San Diego, California, USA) to the lysates and incubated them for 3 hours at 37°C. There was no decrease in the accumulation of the 14-kDa actin fragment. These results indicate that calpains are not required for the muscle cell responses that follow activation of caspase-3.
Actin cleavage and ATP-dependent proteolysis.
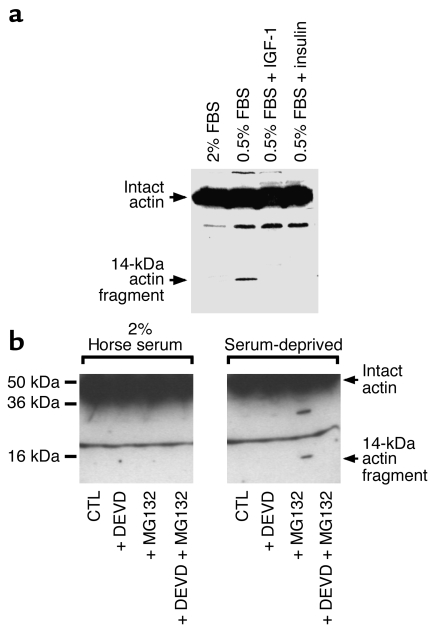
To determine whether catabolic conditions activate similar proteolytic responses, we incubated L6 cells in serum-deprived medium and tested for actin cleavage. We also investigated whether insulin or IGF-1 would block actin fragment accumulation, since these hormones/growth factors can inhibit caspase-3 activation as well as proteolysis in skeletal muscle (12, 37–39). We found that serum deprivation stimulated production of the 14-kDa actin fragment in cell lysates incubated for 3 hours at 37°C (Figure 4a). Accumulation of the actin fragment was blocked when we added either 10 nM insulin or IGF-1 to the medium (Figure 4a). These results are consistent with the antiapoptotic role of these hormones (38, 40). Thus, physiologic stimuli that activate apoptotic proteases in muscle cells lead to generation of actin-breakdown products.
Figure 4.
Catabolic stimuli generate 14-kDa actin fragments. (a) L6 cells were subjected to serum deprivation (0.5% serum) for 24 hours, and cell lysates were prepared. Incubation of the lysates at 37°C for 2 hours led to the generation of the 14-kDa actin fragment. Supplementing serum-deprived cells with IGF-1 (1 nM) or insulin (1 nM) prevented the generation of the actin fragment. (b) Cells were incubated in 2% serum (CTL) or serum-deprived medium (CTL) overnight with or without the proteasome inhibitor MG132 (10 μM) or Ac-DEVD-CHO (5 μM). Cells were lysed and the lysates examined immediately for the presence of the 14-kDa actin fragment by immunoblot analysis. Ac-DEVD-CHO prevented the accumulation of the actin fragment even when MG132 was added to inhibit proteasome activity.
We then tested whether such stimuli yield sequential action of caspase-3 and the Ub-P’some system. Since ATP-dependent protein degradation corresponds closely to proteolysis in the Ub-P’some system (1, 2, 14), we examined whether serum deprivation would stimulate ATP-dependent proteolysis in skeletal muscle cells. We incubated L6 cells overnight in medium containing 2% horse serum, in serum-free medium, or in serum-free medium supplemented with 100 nM insulin. The cells were lysed, and lysates were incubated at 37°C for 2 hours with or without an ATP-generating system. We calculated ATP-dependent proteolysis as tyrosine released from lysates incubated with ATP, minus tyrosine released from lysates incubated without ATP. ATP-dependent proteolysis in control cells incubated overnight in serum-supplemented medium increased 72% ± 1% as compared with cells incubated without ATP. In serum-deprived cells, the ATP-dependent increase in proteolysis was 149% ± 17% (P < 0.05 vs. control cells). This was about twofold higher than ATP-stimulated protein degradation in control cells. When we added insulin, serum deprivation increased ATP-dependent protein degradation by only 53% ± 4%. Thus, insulin significantly suppressed ATP-stimulated proteolysis (P < 0.05 vs. serum-deprived cells). Insulin also suppressed ATP-dependent protein degradation stimulated by serum deprivation to a level below that measured in serum-supplemented cells. We conclude that serum deprivation not only increases production of the actin fragment but also stimulates ATP-dependent proteolysis in muscle cells, suggesting that one mechanism for the antiproteolytic effect of insulin in muscle (37) is to block caspase-3 activity.
Catabolic signals induce actin cleavage in skeletal muscle cells and rat muscle.
To examine these relationships in vivo, we incubated L6 cells in 2% horse serum or serum-free medium for 24 hours and immediately tested for the presence of the 14-kDa actin fragment. We did not find the actin fragment in muscle cells subjected to serum deprivation unless proteasome activity was inhibited (Figure 4b). The fragment also did not accumulate if caspase-3 was blocked by Ac-DEVD-CHO (Figure 4b). Notably, this occurred even though the cells were subjected to serum deprivation and incubated with the proteasome inhibitor MG132.
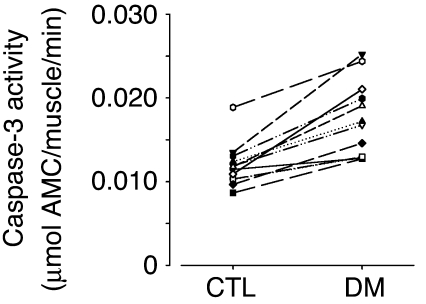
In rats, we find that acute diabetes accelerates muscle proteolysis in the Ub-P’some system and insulin reverses these responses (12, 37). We measured caspase-3 activity in gastrocnemius muscles of rats with acute diabetes and compared it to the value measured in muscles of the respective pair-fed, control rat (Figure 5). There was a 47% ± 7% increase in caspase-3 activity (P < 0.01).
Figure 5.
Stimulation of caspase-3 activity in muscle of rats with acute diabetes. Caspase-3 activity was measured in gastrocnemius muscles of 12 pairs of rats with acute diabetes (DM) and their respective pair-fed, sham-injected controls. Activity was calculated as the amount of DEVD-AMC hydrolyzed per gastrocnemius muscle, and caspase-3 activity in the muscle of the diabetic and respective pair-fed control rat is connected by a line. On average there was a 47% ± 7% increase with acute diabetes (P < 0.01).
To test for involvement of caspase-3 in the muscle atrophy caused by diabetes, we measured the rate of protein degradation in muscles incubated with and without Ac-DEVD-CHO (Table 1). Acute diabetes stimulated (P < 0.01) muscle protein degradation, but the caspase-3 inhibitor suppressed (P < 0.05) this response. Protein degradation in muscles of control rats was not significantly changed in response to the caspase-3 inhibitor (Table 1). Using the same muscles, we tested for the presence of the 14-kDa actin fragment; it was increased in response to diabetes. Muscles incubated with Ac-DEVD-CHO had less accumulation of the actin fragment (Figure 6a). There was no actin fragment accumulation in muscles of control rats.
Table 1.
An inhibitor of caspase-3 activity suppresses the accelerated muscle proteolysis stimulated by acute diabetes
Figure 6.
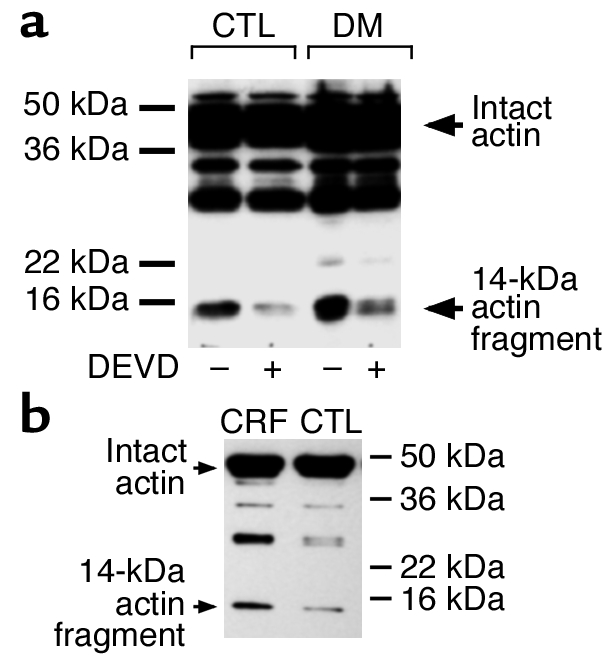
Catabolic conditions generate actin fragments in rat muscle. (a) Epitrochlearis (mixed-fiber) muscles from rats with acute diabetes (DM) and pair-fed, sham-operated control rats were incubated in Kreb’s Ringer bicarbonate buffer with or without 10 μM Ac-DEVD-CHO for 2 hours before homogenization in hypotonic buffer. After centrifugation as described in Methods, the pellets were resuspended in SDS-PAGE sample buffer and subjected to immunoblotting to detect the 14-kDa actin fragment. Positions of the molecular weight markers are indicated on the right side of the panel. (b) Gastrocnemius (mixed-fiber) muscles from rats with chronic uremia (CRF) and pair-fed, sham-operated control rats were subjected to immunoblotting to detect the 14-kDa actin fragment as described in the upper panel.
In vivo models of catabolic conditions.
To determine whether actin cleavage also occurs in response to other conditions characterized by accelerated muscle proteolysis, we examined chronically uremic rats (11). There was substantially more of the 14-kDa actin fragment in muscles of rats with uremia compared with control rats (Figure 6b).
Actin cleavage stimulates proteolysis.
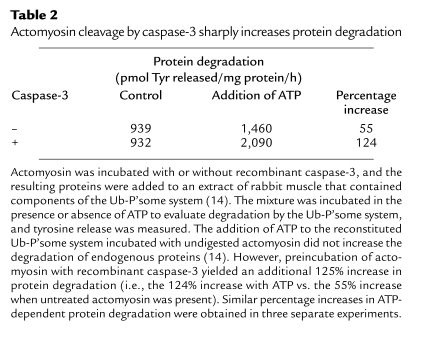
To evaluate the contribution of caspase-3–mediated degradation of actomyosin to total protein degradation in muscle, we prepared a cell-free extract from rabbit muscle that contains the components of the Ub-P’some system as described by Solomon and Goldberg (14). We evaluated the impact of incubating intact actomyosin or caspase-3–digested actomyosin complexes to total protein degradation by the cell-free preparation. Basal rates of tyrosine release (26) varied in the extracts of rabbit muscle, so we compared the percentage increase in ATP-stimulated tyrosine release because it represents degradation by the Ub-P’some system (14). As shown in Table 2, if approximately 1% of actomyosin was cleaved by recombinant caspase-3, the rate of ATP-dependent protein degradation measured as tyrosine release more than doubled (124% increase) compared with the 55% increase in ATP-dependent protein degradation when undigested actomyosin was added. Similar values were obtained in three separate experiments.
Table 2.
Actomyosin cleavage by caspase-3 sharply increases protein degradation
Activation of muscle protein catabolism.
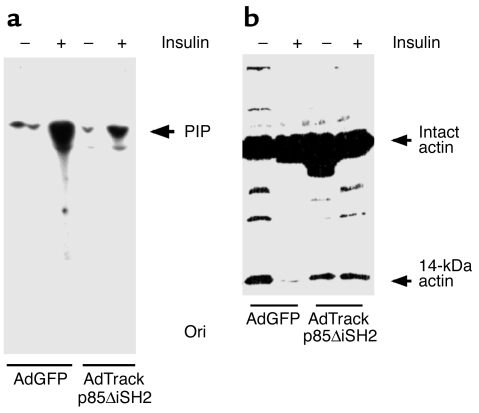
Conditions such as chronic uremia or sepsis that cause muscle atrophy are frequently characterized by insulin resistance (22, 23), and there is decreased activity of PI3K in models of insulin resistance (24). Since PI3K is an antiapoptotic mediator, it could regulate the activity of caspase-3 (41, 42). Therefore, we examined PI3K as a common mediator of protein breakdown in muscle via the sequence: activation of caspase-3, actomyosin cleavage, and ultimately, protein degradation. L6 cells were transfected with adenovirus constructs containing the cDNAs for GFP or GFP plus the dominant-negative p85 protein in which the inner SH2 domain had been deleted (AdTrackp85ΔiSH2). This mutation prevents interaction between the PI3K p85 regulatory and p110 catalytic subunits, thereby inhibiting PI3K activity. As shown in Figure 7, serum starvation of cells infected with the control adenovirus (AdGFP) led to the appearance of the 14-kDa actin fragment; insulin (10 nM) blocked this response. When L6 cells were infected with AdTrackp85ΔiSH2 and subjected to serum starvation, insulin did not eliminate the appearance of the actin fragment.
Figure 7.
Phosphatidylinositol 3-kinase activity and actin cleavage. (a) L6 cells were infected with an adenovirus that expresses GFP (AdGFP) or a dominant-negative PI3K (AdTrackp85ΔiSH2) and GFP. The cells were incubated in 0.5% FBS with or without 10 nM insulin, and PI3K activity was measured. The dominant-negative PI3K suppressed the ability of insulin to stimulate PI3K activity. PIP, phosphatidylinositol phosphate; Ori, origin of loading. (b) L6 cells were treated similarly and then subjected to Western blot to detect the actin fragment. The inhibition of PI3K activity abolished the ability of insulin to suppress the cleavage of actomyosin.
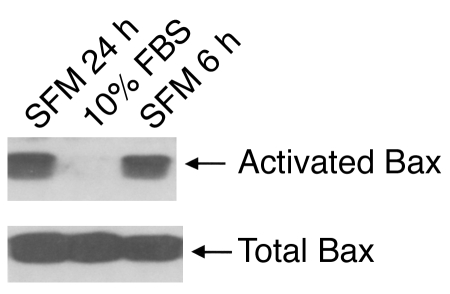
What mediator could activate caspase-3 to generate the 14-kDa actin fragment? Yamaguchi and Wang reported that the PI3K pathway regulates the change in Bax conformation that leads to release of cytochrome c from mitochondria and activation of caspase-3 (28). We evaluated whether conditions that reduce PI3K activity such as serum starvation (Figure 7) will also cause a conformational change in Bax. As shown in Figure 8, serum withdrawal for 6 or 24 hours significantly increases the conformational change in Bax present in C2C12 muscle cells. This occurred when the total amount of Bax was unchanged. These results provide a link between PI3K activity and regulation of caspase-3 activity.
Figure 8.
Serum starvation induces activation of Bax. C2C12 muscle cells were cultured without serum for 6 or 24 hours before being lysed with buffer (Methods). SFM, serum-free medium. The change in Bax conformation was detected after immunoprecipitation using anti-Bax 6A7 antibody and detected by immunoblotting with anti-Bax rabbit antiserum. Total Bax was evaluated using anti-Bax rabbit antiserum.
Discussion
The bulk of muscle proteins (50–70%) exists in actomyosin complexes and myofibrils, and in different catabolic conditions, contractile proteins in these complexes are degraded by the Ub-P’some system (1, 2). The response is complicated, however, as shown by Solomon and Goldberg, who constructed a reconstituted system containing the components of the Ub-P’some system and found that it degrades monomeric actin or myosin but does not degrade actomyosin complexes (14). They concluded that disassociation of actomyosin complexes is the rate-limiting step in muscle protein degradation.
Our results indicate that caspases play a critical, initial role in muscle protein degradation. We found that caspase-3 cleaves actomyosin in vitro and in muscle cells; products of caspase-3 action are rapidly removed by the ATP-dependent, Ub-P’some system. Moreover, the characteristic 14-kDa actin fragments are present in atrophying muscles of animals with diabetes or chronic uremia. In response to diabetes, caspase-3 activity in muscle increases. In addition, Ac-DEVD-CHO, the inhibitor of caspase-3, suppresses both the accelerated muscle proteolysis and the accumulation of actin fragments that is induced by acute diabetes. The finding that Ac-DEVD-CHO suppresses the accelerated rate of protein degradation in muscle of rats with acute diabetes is enlightening for two reasons. First, exposure of muscle to Ac-DEVD-CHO for only a relatively brief period did suppress total protein degradation. Inhibiting caspase-3 activity during the incubation could only block the breakdown of additional actomyosin complexes and would not suppress the degradation of proteins already released from previously cleaved actomyosin complexes. Thus, over a short period, the caspase-3 inhibitor could only exert limited suppression of total protein breakdown compared with an inhibitor of the proteasome (11, 12), and the results in Table 2 are consistent with caspase-3 acting as an initial step triggering the breakdown of muscle protein complexes. We also found that the caspase-3 inhibitor did not significantly suppress protein degradation in muscles of paired, control rats. This result suggests that the stimulation of caspase-3 activity is a response to a catabolic condition rather than regulating basal protein turnover in muscle.
How much does actomyosin breakdown contribute to total protein degradation in muscle? To examine this question, we prepared a cell-free homogenate of rabbit skeletal muscle containing the elements of the Ub-P’some system (14). When we activated the Ub-P’some system in the muscle homogenate by adding ATP, we confirmed that there was an increase in the degradation of endogenous proteins (Table 2). In contrast, when we added the breakdown products of actomyosin resulting from its digestion by recombinant caspase-3, there was an additional 125% increase in ATP-stimulated protein degradation over that measured when undigested actomyosin was present. The large increase in total protein degradation indicates that actomyosin cleavage converts actomyosin to individual myofibrillar proteins that are degraded by the Ub-P’some system. This response occurred in conditions in which we limited the cleavage of actin in actomyosin to about 1%, based on its detection by the anti-actin antibody. Notably, acute diabetes or chronic uremia is associated with accumulation of the actin fragment in muscle to approximately the same degree (Figure 6). Since a somewhat small accumulation of the actin fragment is associated with a substantial increase in total protein degradation, we conclude that the cleavage of actomyosin in the muscle of diabetic or uremic rats is a major contributor to the increase in total protein breakdown.
What other systems could be involved in accelerating muscle proteolysis? Calpains are a possibility, in light of the reports of others that calpains clip myofibrillar proteins into fragments in a septic rat model (32). In addition, Huang and Forsberg reported that expression of the endogenous inhibitor of calpains, calpastatin, or a dominant inhibitor of calpains in L8 muscle cells suppressed the cleavage of fodrin (31). They also found a lower rate of total protein degradation, suggesting that the calpain inhibitors had suppressed actomyosin degradation. Tidball and Spencer reported that muscle atrophy due to hindlimb unloading was attenuated in transgenic mice that overexpress calpastatin in a muscle-specific manner (43). In contrast, we and others find that inhibiting calcium-dependent proteases does not suppress either total or myofibrillar protein degradation in muscles isolated from rats with acute diabetes or chronic uremia (1, 2). In the present study as well, we found no evidence that calpains are critical mediators of the accelerated muscle protein degradation found in the conditions we studied, although we cannot exclude the possibility that activation of calpains in some other pathologic conditions could lead to muscle atrophy.
Regarding the cellular mediator that could activate caspase-3 in the conditions associated with accelerated muscle protein degradation, insulin resistance is common to many of these disorders (e.g., sepsis or chronic uremia), while other conditions are characterized by a low level of insulin (e.g., acute diabetes or starvation) (12, 22, 23, 37, 44). Not only is PI3K signaling integral for insulin responses, but it also is critical for antiapoptotic responses (41, 42). Consequently, dysfunction of the PI3K signaling process could be a common initiator of muscle protein degradation. In support of this possibility, we found evidence that serum withdrawal (which reduces PI3K activity; Figure 7) stimulates actomyosin cleavage in muscle cells and insulin blocks this response. But when PI3K activity is blocked, the ability of insulin to suppress actomyosin cleavage is abolished (Figure 7). These in vitro results indicate that an essential role of PI3K signaling is to suppress actomyosin cleavage. Regarding the relevance of PI3K activity in vivo, certain catabolic conditions including those we studied are associated with decreased PI3K activity in muscle. For example, we found that the PI3K activity associated with insulin receptor substrate-1 (IRS-1) is low in the muscle of rats with chronic uremia (J.L. Bailey et al., unpublished observations). Rats with chronic uremia exhibit a marked increase in the rate of muscle protein degradation (11) as well as evidence of actin cleavage (Figure 6). We also find that acute diabetes stimulates muscle proteolysis (12) and is associated with decreased IRS-1–associated PI3K activity and evidence of caspase-3 activation (S.W. Lee et al., unpublished observations). Since PI3K is an essential regulator of apoptotic pathways (41, 42), it would provide a link between a deficiency of insulin or impaired insulin-signaling processes and activation of caspase-3 resulting in loss of muscle protein. We extended these results by examining whether the proapoptotic protein, Bax, is activated. In serum-starved muscle cells that exhibit suppressed PI3K activity (Figure 7), the level of Bax in the conformational state that activates caspase-3 is increased (Figure 8). Serum starvation increases the appearance of the 14-kDa actin fragment (Figure 4). These results are consistent with the reported sequence of events that link decreased PI3K activity to caspase-3 (28).
We have emphasized that caspase-3 is involved in the catabolic response because we found that recombinant caspase-3 will degrade actomyosin complexes and because Ac-DEVD-CHO not only prevents the generation of the actin fragment but also suppresses protein breakdown in muscle of rats with acute diabetes. Since the specificity of an inhibitor may not be absolute, we do not exclude the possibility that other proteases could be involved in initiating muscle proteolysis. Others have reported that caspases can cleave actin in vitro (20), but Song et al. (45) did not find actin fragments in leukemia or lymphoma cells that were undergoing apoptosis. They concluded that actin is not degraded by apoptosis-associated proteases. A potential reason for the difference from our results is that actin fragments in their experiments were rapidly degraded by the Ub-P’some system. Alternatively, the cells they were examining may have different pathways of protein degradation than those in muscle cells.
In summary, muscle atrophy is a frequent complication of many catabolic conditions. Our findings indicate that muscle protein loss occurs in two steps: (i) catabolic stimuli cause the breakdown of actomyosin to release monomeric actin and myosin; (ii) the resulting fragments and monomeric proteins are then degraded by the Ub-P’some system. Blocking caspase activity in intact muscle of rats with accelerated muscle proteolysis reduces both the accumulation of actin and the acceleration of protein degradation. Armstrong and colleagues published preliminary results suggesting a beneficial, cardioprotective effect of infused caspase inhibitors following a myocardial infarction (46). Their results, obtained in an intact rat, suggest that the responses we identified could provide new targets for preventing muscle atrophy in catabolic conditions.
Acknowledgments
This project was supported by the NIH through grants HL70762 (J. Du), DK37175 (W.E. Mitch), DK63658 (S.R. Price), and DK50740 (S.R. Price), and a Scientist Development grant to J. Du from the American Heart Association.
Footnotes
Conflict of interest: Jie Du has applied for a patent to use the 14 kDa actin fragment as a sign of protein catabolism in muscle.
Nonstandard abbreviations used: ATP-ubiquitin-proteasome (Ub-P’some); 3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonate (CHAPS); insulin receptor substrate-1 (IRS-1).
References
- 1.Mitch WE, Goldberg AL. Mechanisms of muscle wasting: the role of the ubiquitin-proteasome system. N. Engl. J. Med. 1996;335:1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 2.Lecker SH, Solomon V, Mitch WE, Goldberg AL. Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. J. Nutr. 1999;129:227S–237S. doi: 10.1093/jn/129.1.227S. [DOI] [PubMed] [Google Scholar]
- 3.Mansoor O, et al. Increased mRNA levels for components of the lysosomal, Ca++-activated and ATP-ubiquitin-dependent proteolytic pathways in skeletal muscle from head trauma patients. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2714–2718. doi: 10.1073/pnas.93.7.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiao G, et al. Sepsis is associated with increased mRNAs of the ubiquitin-proteasome proteolytic pathway in human skeletal muscle. J. Clin. Invest. 1997;99:163–168. doi: 10.1172/JCI119143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickering WP, et al. Nutrition in CAPD: serum bicarbonate and the ubiquitin-proteasome system in muscle. Kidney Int. 2002;61:1286–1292. doi: 10.1046/j.1523-1755.2002.00276.x. [DOI] [PubMed] [Google Scholar]
- 6.Llovera M, et al. Ubiquitin gene expression is increased in human muscle undergoing neurogenic involvement. Neurochem. Int. 1999;34:137–140. doi: 10.1016/s0197-0186(98)00080-1. [DOI] [PubMed] [Google Scholar]
- 7.Llovera M, et al. Ubiquitin and proteasome gene expression is increased in skeletal muscle of slim AIDS patients. Int. J. Mol. Med. 1998;2:69–73. [PubMed] [Google Scholar]
- 8.Williams AB, Sun X, Fischer JE, Hasselgren P-O. The expression of genes in the ubiquitin-proteasome proteolytic pathway is increased in skeletal muscle from patients with cancer. Surgery. 1999;126:744–749. [PubMed] [Google Scholar]
- 9.Lecker SH, et al. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J. Clin. Invest. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solomon V, Lecker SH, Goldberg AL. The N-end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J. Biol. Chem. 1998;273:25216–25222. doi: 10.1074/jbc.273.39.25216. [DOI] [PubMed] [Google Scholar]
- 11.Bailey JL, et al. The acidosis of chronic renal failure activates muscle proteolysis in rats by augmenting transcription of genes encoding proteins of the ATP-dependent, ubiquitin-proteasome pathway. J. Clin. Invest. 1996;97:1447–1453. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price SR, et al. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome pathway by a mechanism including gene transcription. J. Clin. Invest. 1996;98:1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tawa NE, Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J. Clin. Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 15.Belizario JE, Lorite MJ, Tisdale MJ. Cleavage of caspases-1, -6, -8 and -9 substrates by proteases in skeletal muscle from mice undergoing cancer cachexia. Br. J. Cancer. 2001;84:1135–1140. doi: 10.1054/bjoc.2001.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vescovo G, et al. Apoptosis in the skeletal muscle of patients with heart failure: investigation of clinical and biochemical changes. Heart. 2000;84:431–437. doi: 10.1136/heart.84.4.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard I, et al. Loss of calpain 3 proteolytic activity leads to muscular dystrophy and to apoptosis-associated I-kappa Balpha/nuclear factor kappa B pathway perturbation in mice. J. Cell Biol. 2000;151:1583–1590. doi: 10.1083/jcb.151.7.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalla Libera L, Zennaro R, Sandri M, Ambrosio GB, Vescovo G. Apoptosis and atrophy in rat slow skeletal muscles in chronic heart failure. Am. J. Physiol. 1999;277:C982–C986. doi: 10.1152/ajpcell.1999.277.5.C982. [DOI] [PubMed] [Google Scholar]
- 19.Sandri M, Minetti C, Pedemonte M, Carraro U. Apoptotic myonuclei in human Duchenne muscular dystrophy. Lab. Invest. 1998;78:1005–1016. [PubMed] [Google Scholar]
- 20.Mashima T, et al. Actin cleavage by CPP-32/apopain during the development of apoptosis. Oncogene. 1997;14:1007–1012. doi: 10.1038/sj.onc.1200919. [DOI] [PubMed] [Google Scholar]
- 21.Kayalar C, Ord T, Testa MP, Zhong LT, Bredesen DE. Cleavage of actin by interleukin-1 beta-converting enzyme to reverse DNAase I inhibition. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2234–2238. doi: 10.1073/pnas.93.5.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasselgren P-O, Warner BW, James H, Takehara H, Fischer JE. Effect of insulin on amino acid uptake and protein turnover in skeletal muscle from septic rats: evidence for insulin resistance of protein degradation. Arch. Surg. 1987;122:228–233. doi: 10.1001/archsurg.1987.01400140110015. [DOI] [PubMed] [Google Scholar]
- 23.Cecchin F, Ittoop O, Sinha MK, Caro JF. Insulin resistance in uremia: insulin receptor kinase activity in liver and muscle from chronic uremic rats. Am. J. Physiol. 1988;254:E394–E401. doi: 10.1152/ajpendo.1988.254.4.E394. [DOI] [PubMed] [Google Scholar]
- 24.Yu C, et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- 25.Baracos V, Langman M, Mak A. An in vitro preparation of the extensor digitorum communis muscle from the chick (Gallus domesticus) for studies of protein turnover. Comp. Biochem. Physiol. A. 1989;92:555–563. doi: 10.1016/0300-9629(89)90365-4. [DOI] [PubMed] [Google Scholar]
- 26.Clark AS, Mitch WE. Comparison of protein synthesis and degradation in incubated and perfused muscle. Biochem. J. 1983;212:649–653. doi: 10.1042/bj2120649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franch HA, Wang X, Sooparb S, Brown NS, Du J. Phosphatidylinositol 3-kinase activity is required for epidermal growth factor to suppress proteolysis. J. Am. Soc. Nephrol. 2002;13:903–909. doi: 10.1681/ASN.V134903. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi H, Wang H-G. The protein kinase PKB/Akt regulates cell survival and apoptosis by inhibiting Bax conformational change. Oncogene. 2001;20:7779–7786. doi: 10.1038/sj.onc.1204984. [DOI] [PubMed] [Google Scholar]
- 29.Nicholson DW, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 30.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Forsberg NE. Role of calpain in skeletal-muscle protein degradation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12100–12105. doi: 10.1073/pnas.95.21.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams AB, et al. Sepsis stimulates release of myofilaments in skeletal muscle by a calcium-dependent mechanism. FASEB J. 1999;13:1435–1443. doi: 10.1096/fasebj.13.11.1435. [DOI] [PubMed] [Google Scholar]
- 33.Deval C, et al. Identification of cathepsin L as a differentially expressed message associated with skeletal muscle wasting. Biochem. J. 2001;360:143–150. doi: 10.1042/0264-6021:3600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baracos VE, DeVivo C, Hoyle DHR, Goldberg AL. Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am. J. Physiol. 1995;268:E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- 35.Medina R, Wing SS, Haas A, Goldberg AL. Activation of the ubiquitin-ATP-dependent proteolytic system in skeletal muscle during fasting and denervation atrophy. Biomed. Biochim. Acta. 1991;50:347–356. [PubMed] [Google Scholar]
- 36.Mitch WE, et al. Metabolic acidosis stimulates muscle protein degradation by activating the ATP-dependent pathway involving ubiquitin and proteasomes. J. Clin. Invest. 1994;93:2127–2133. doi: 10.1172/JCI117208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitch WE, et al. Evaluation of signals activating ubiquitin-proteasome proteolysis in a model of muscle wasting. Am. J. Physiol. 1999;276:C1132–C1138. doi: 10.1152/ajpcell.1999.276.5.C1132. [DOI] [PubMed] [Google Scholar]
- 38.Kulik G, Klippel A, Weber MJ. Antiapoptotic signaling by the insulin-like growth factor-1 receptor, phosphatidylinositol 3-kinase and Akt. Mol. Cell. Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomas FM, et al. Increased weight gain, nitrogen retention and muscle protein synthesis following treatment of diabetic rats with insulin-like growth factor (IGF)-I and des(1–3)IGF-I. Biochem. J. 1991;276:547–554. doi: 10.1042/bj2760547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertrand F, et al. Insulin antiapoptotic signaling involves insulin activation of the nuclear factor kappaB-dependent survival genes encoding tumor necrosis factor receptor-associated factor 2 and manganese-superoxide dismutase. J. Biol. Chem. 1999;274:30596–30602. doi: 10.1074/jbc.274.43.30596. [DOI] [PubMed] [Google Scholar]
- 41.Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 42.Kennedy SG, et al. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 43.Tidball JG, Spencer MJ. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J. Physiol. 2002;545:819–828. doi: 10.1113/jphysiol.2002.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wing SS, Goldberg AL. Glucocorticoids activate the ATP-ubiquitin-dependent proteolytic system in skeletal muscle during fasting. Am. J. Physiol. 1993;264:E668–E676. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- 45.Song Q, et al. Resistance of actin to cleavage during apoptosis. Proc. Natl. Acad. Sci. U. S. A. 1997;94:157–162. doi: 10.1073/pnas.94.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong RC, et al. Caspase inhibitors reduce infarct size when dosed post perfusion in a rodent cardiac ischemia/reperfusion model. Circulation. 2001;104:11–12. . [Google Scholar]