Click chemistry describes a powerful set of chemical reactions that are rapid, selective, and produce high yields.[1] The most recognized of these reactions is the copper (I)-catalyzed azide-alkyne cycloaddition, which has been applied to diverse areas, ranging from materials science to chemical biology.[2–8] For biological applications, both azido and alkyne groups are considered to be bioorthogonal chemical reporters because they do not interact with any native biological functional groups. As a result, these bioorthogonal reporters can be incorporated into a target biomolecule using the cell’s biosynthetic machinery to provide chemical handles that can be subsequently tagged with exogenous probes. The bioorthogonal reporters are complementary to genetically encoded tags, such as green fluorescent protein (GFP),[9] and provide a powerful approach to tag biomolecules without the need of direct genetic encoding. Bioorthogonal labeling via click chemistry is highly sensitive with low background despite the complex cellular environment. In practice, however, the sensitivity is constrained by the abundance of the target molecules, the labeling efficiency of the chemical reporters, and the performance of the exogenous probes.[7] In almost all cases, bright and photostable probes are highly desirable, particularly for long-term tracking and sensitive detection of low-abundance biomolecules.
Fluorescent nanoparticles such as quantum dots (Qdots) exhibit improved brightness and photostability over traditional fluorescent dyes.[10–12] In the context of click chemistry, however, the copper catalyst irreversibly quenches Qdot fluorescence and prevents their usage in the various applications based on copper-catalyzed click chemistry.[13] Because of copper’s cytotoxicity, copper-free bioorthogonal approaches, such as the Staudinger ligation and the strain-promoted azide-alkyne cycloaddition, have been developed for live cell and in vivo applications.[7] Qdots can be employed in the copper-free methods,[13,14] where their instability caused by copper is not an issue. However, Qdots’ intrinsic toxicity, caused by the leaching of heavy metal ions, is still a critical concern.
Semiconducting polymer dots (Pdots) represent a new class of ultrabright fluorescent probes,[15–25] which can overcome both issues for click chemistry-based applications. Previous studies showed that Pdots were not cytotoxic in different cellular assays,[16,19,23,25] making them appealing for studies in living system. In this study, we focus on their biological applications involving copper-catalyzed click chemistry. Pdots are brighter fluorescent probes than Qdots, can have up to a thousand-fold faster emission rates than Qdots, and are photostable and do not “blink”.[16] For biological applications, however, a significant problem of Pdots is the control over their surface chemistry and conjugation to biological molecules. This is a significant challenge that has prevented the widespread adoption of Pdots in biological studies.
Here we present a general method that overcomes this challenge by creating functional groups on the Pdot surface. Because the formation of Pdot is driven by hydrophobic interaction, some amphiphilic polymer with hydrophilic functional groups may be co-condensed into a single dot during nanoparticle formation. The hydrophilic groups on the amphiphilic polymer can be used as handles for functionalizing the Pdots for conjugation to biomolecules. We found that a general copolymer, poly(styrene-co-maleic anhydride) (PSMA), successfully functionalized the Pdots for further surface conjugations (Scheme 1). PSMA provides excellent options for Pdot functionalization because it is commercially available in a broad range of molecular weights and maleic anhydride contents. In this study, PSMA was employed to functionalize Pdots made from a highly fluorescent semiconducting polymer Poly[9,9-dioctylfluorenyl-2,7-diyl)-co-1,4-benzo-{2,1′-3}-thiadiazole)] (PFBT), but the method can be applied to any hydrophobic, fluorescent, semiconducting polymers. During Pdot formation, the hydrophobic polystyrene units of PSMA molecules were likely anchored inside the Pdot particles while the maleic anhydride units localized to the Pdot surface and hydrolyzed in the aqueous environment to generate carboxyl groups on the Pdot surface. The carboxyl groups enabled further surface conjugations.
Scheme 1.
Functionalization and conjugation of fluorescent semiconducting polymer dots for bioorthogonal labeling via click chemistry. A copolymer PSMA was co-condensed with a fluorescent semiconducting polymer PFBT (depicted as green string), thereby forming Pdots with surface carboxyl groups. The carboxyl groups enabled further surface conjugations to functional molecules for copper (I)-catalyzed click reaction. The functionalized Pdots were selectively targeted against newly synthesized proteins or glycoproteins (blue string) in mammalian cells that were metabolically labeled with bioorthogonal chemical reporters.
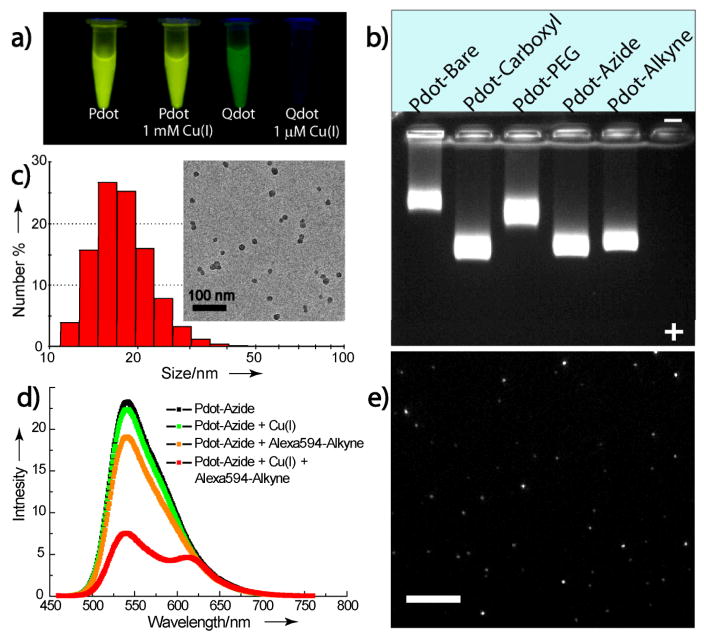
Analysis of the absorption spectrum for ~15 nm-diameter PFBT dots indicated a peak extinction coefficient of 5.0×107 M−1cm−1 (Supplementary Figure 1). Fluorescence quantum yield of the functionalized Pdots was determined to be 0.28 using a dilute solution of Coumarin 6 in ethanol as standard. The large extinction coefficient and high quantum yield indicate much higher per-particle brightness as compared to other fluorescent nanoparticles. Single-particle photobleaching studies of the functionalized Pdots showed that over 109 photons per Pdot were emitted prior to photobleaching, consistent with their excellent photostability.[16] We examined the pH and ion sensitivity of Pdot fluorescence in biological applications, particularly the copper-catalyzed click chemistry. We found that the fluorescence of Pdots was not affected by most biologically relevant ions, including iron, zinc, and copper, three of the most abundant ions in biological organisms. The Pdot fluorescence is also independent of pH in the range of 4 to 9 (Supplementary Figure 2). This fact can be attributed to the hydrophobic organic nature of Pdots, which tend not to have any chemical interaction with ionic species. In contrast, inorganic Qdots are significantly quenched by copper and iron ions.[26] As shown in Figure 1a, PFBT dots remained highly fluorescent in MilliQ water containing a high Cu+ concentration of 1 mM, whereas Qdots were completely quenched at a much lower Cu+ concentration of 1 μM.[13] This property provides a significant advantage for applying Pdots in various studies based on copper (I)-catalyzed click reactions.
Figure 1.
a) Fluorescence photographs of Pdots versus Qdots in the presence of copper (I) under UV illumination. b) Gel electrophoresis of Pdots with different surface functional groups in an 0.7% agarose gel. c) Hydrodynamic diameter of carboxyl funtionalized Pdots measured by dynamic light scattering; inset shows a typical TEM image of functionalized Pdots. d) A fluorescent assay using alkyne-Alexa 594 dye to verify successful functionalization of Pdots with azido groups. e) Single-particle fluorescence images of alkyne-silica nanoparicles coupled to azido-Pdots by click reaction. Scale bar represents 50 μm.
Starting from the carboxyl-functionalized Pdots, we were able to react them with either amine-azido or amine-alkyne groups using the standard carboxyl-amine coupling catalyzed by 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC).[27] Gel electrophoresis was performed to characterize the formation of different functional groups on the Pdot surface using a 0.7% agarose gel (Figure 1b). Compared with unfunctionalized, bare Pdots, the carboxyl-functionalized Pdots exhibited an apparent increase in mobility in the gel. Dynamic light scattering and transmission electron microscopy (TEM) measurements showed that both the bare and the functionalized Pdots had comparable particle sizes, with an average of ~15 nm in diameter (Figure 1c). Therefore, the high mobility of PSMA-functionalized Pdots indicated the formation of negatively charged carboxyl groups on the Pdot surface. Surface conjugation was performed with different amine-containing molecules (amine-terminated polyethylene glycol (PEG), azide, and alkyne). Dynamic light scattering of the conjugated Pdots showed no obvious change in particle size because the conjugation was with small molecules. However, they exhibited shifted migration bands in the gel as anticipated, due to the reduced charges of the Pdot conjugates compared to the carboxyl-functionalized Pdots. These results clearly indicate successful carboxyl functionalization of the Pdots as well as all the subsequent surface modifications.
Figure 1d shows a fluorescence assay examining the reactivity of azido-Pdots towards a terminal alkyne group via copper (I)-catalyzed click reaction. When mixed with a copper solution, the azido-Pdots exhibited an emission intensity similar to that of the pure Pdots, confirming that their fluorescence is insensitive to copper ions. A slight decrease in intensity was observed in the mixture of Pdots and alkyne-Alexa 594 (no Cu (I)), but this was primarily due to the inner filter effect rather than direct quenching caused by fluorescence resonance energy transfer (FRET). In contrast, when directly linked to alkyne-Alexa 594 in the presence of Cu (I), the azido-Pdots showed remarkable fluorescence quenching accompanied by an emission peak from the Alexa dye. This spectroscopic change was a direct result of efficient FRET from the PFBT dots to the Alexa dye in close proximity and indicated the effective azide-alkyne click reaction. In addition, we also clicked the azido-Pdots onto alkyne-functionalized silica nanoparticles to convert the optically inert silica particles into highly fluorescent probes. The Pdot-silica conjugates were clearly visible at the single-particle level even on a mercury lamp-illuminated, low-magnification (4×) fluorescent microscope (Figure 1e).
To demonstrate cellular labeling with Pdots and click chemistry, we visualized newly synthesized proteins that were modified by bioorthogonal non-canonical amino-acid tagging (BONCAT). In the BONCAT technique, newly synthesized proteins in cells are metabolically labeled with an azido- (or alkyne-) bearing artificial amino acid. The artificial amino acid endows the proteins with unique chemical functionality that subsequently can be tagged with exogenous probes for detection or isolation in a highly selective manner.[28] Azidohomoalanine (AHA) and homopropargylglycine (HPG) are two artificial amino acids commonly used in this method.[29,30] They are effective surrogates for methionine, an essential amino acid; in the absence of methionine, the cellular synthesis machinery straightforwardly incorporates them into proteins. This approach is operationally similar to the traditional metabolic labeling with radioactive amino acid 35S-methionine. After incorporation, AHA and HPG are susceptible to tagging with exogenous probes, which in our case are the highly fluorescent Pdots for in situ imaging.
Our first experiment for protein imaging was to target the AHA-labeled proteins with Pdot-alkyne probes. MCF-7 human breast cancer cells were grown to confluence before passage into serum-free medium lacking methionine. After incubation to deplete any residual methionine, cell cultures were supplemented with AHA for four hours. Then the cells were washed and fixed before carrying out the click reaction with alkyne-Pdots in the presence of CuSO4, a reducing agent (sodium ascorbate), and a triazole ligand. The Pdot-tagged cells were viewed immediately on a confocal fluorescence microscope. Identical settings were used to acquire images from the Pdot-labeled cells and the negative controls.
Figure 2 shows confocal fluorescence and bright-field images of the Pdot-labeled cells and the control samples. We observed very bright fluorescence for the AHA-labeled cells tagged with Pdot-alkyne via click reaction (Figure 2a). When the cells were incubated under identical conditions but in absence of the reducing agent (sodium ascorbate) that forms copper (I) from CuSO4, cell labeling by Pdots was not observed (Figure 2b), indicating that Pdot-alkyne was selective for the copper (I)-catalyzed reaction. In a different control, copper (I)-catalyzed Pdot-alkyne tagging was performed under identical conditions as those in Figure 2a but in cells not exposed to AHA. In this control, cell labeling also was not observed (Supplementary Figure 3), indicating Pdot-alkyne tagging was highly specific for the cellular targets of interest. In addition, we also used Pdot-azide to detect newly synthesized proteins in MCF-7 cells incubated with HPG. In this case, the Pdot-azide also specifically and effectively labeled the targets (Supplementary Figure 4). In comparison with the Pdot-alkyne labeling (AHA-treated cells), we did not observe obvious difference in the fluorescence brightness of the Pdot-azide labeling (HPG-treated cells). This is consistent with the literature results that HPG and AHA show very similar activities in the synthesis of nascent proteins in mammalian cells.[28,29]
Figure 2.
Fluorescence imaging of newly synthesized proteins in the AHA-treated MCF-7 cells tagged with Pdot-alkyne probes. a) Positive Pdot labeling in the presence of copper (I). b) Negative control for Pdot labeling carried out under identical conditions as in (a) but in the absence of the reducing agent (sodium ascorbate) that generates copper (I) from copper (II). The left four panels show fluorescence images; green fluorescence is from Pdots and blue fluorescence is from the nuclear stain Hoechst 34580. The right four panels show Nomarski (DIC) and combined DIC and fluorescence images. Scale bar represents 20 μm.
Next, we used Pdot-alkyne probes to selectively target glycoproteins, a subset of proteins extensively involved in various biological functions.[31] The bioorthogonal chemical reaction strategy has been previously developed for probing glycans on cultured cells and in various living organisms.[32–35] The method involves metabolic labeling of glycans with a monosaccharide precursor that is functionalized with an azido group, after which the azido sugars are covalently tagged with imaging probes. We incubated MCF-7 cells with N-azidoacetylgalactosamine (GalNAz) for three days in order to enrich O-linked glycoproteins with the azido groups. The GalNAz-treated cells were tagged with Pdot-alkyne via click reaction and subsequently viewed on a confocal microscope. Bright cell-surface labeling was observed for the cells positively tagged with Pdot-alkyne (Figure 3a). In the negative control, where cells were incubated with Pdot-alkyne in the absence of the reducing agent, cell labeling was not observed (Figure 3b). As an additional control, we carried out Pdot tagging under identical conditions but in cells lacking azides; in this case, cell labeling was not observed, again indicating Pdot labeling was highly specific for the cellular targets of interest.
Figure 3.
Fluorescence imaging of glycoproteins in GalNAz-treated MCF-7 cells tagged with Pdot-alkyne probes. a) Positive Pdot labeling in the presence of copper (I). b) Negative control for Pdot labeling carried out under identical conditions as in (a) but in the absence of copper (I). The left four panels show fluorescence images; green fluorescence is from Pdots and blue fluorescence is from the nuclear stain Hoechst 34580. The right four panels show Nomarski (DIC) and combined DIC and fluorescence images. Scale bar represents 20 μm.
For the Pdot labeling described in this study, we applied very low Pdot concentrations (~50 nM), which was orders of magnitude less than the general concentration used for small dye molecules (typically in the μM range). We emphasize that Pdot tagging via click chemistry was highly specific in our experiments with virtually no background labeling in all the control samples. Finally, we used the Pdot probes to detect glycoproteins and newly synthesized proteins in a different cell line, 3T3 fibroblast. In all these cases, the Pdots also specifically and effectively labeled the targets, demonstrating the strategy was equally efficient and successful in different cell lines.
In conclusion, we presented a facile conjugation method that covalently links functional molecules to Pdots for click chemistry-based bioorthogonal labeling of cellular targets. These functionalized Pdots were selectively targeted against newly synthesized proteins and glycoproteins in mammalian cells that were metabolically labeled with bioorthogonal chemical reporters. The highly efficient, specific, and bright protein labeling using Pdots and click chemistry demonstrate the potential of this method for visualizing various cellular processes. We anticipate the method described here will enable Pdots to be used in a wide range of cellular studies and fluorescence applications.
Experimental Section
Functionalized Pdots in aqueous solution were prepared by using a modified nano-precipitation method. In a typical preparation, PFBT and PSMA were dissolved in tetrahydrofuran (THF) to produce a solution mixture with a PFBT concentration of 50 μg/mL and a PSMA concentration of 10 μg/mL. The mixture was sonicated to form a homogeneous solution. A 5 mL quantity of the solution mixture was quickly added to 10 mL of MilliQ water in a bath sonicator. The THF was removed by nitrogen stripping. The solution was concentrated by continuous nitrogen stripping to 5 mL on a 90 °C hotplate followed by filtration through a 0.2 micron filter.
Surface conjugation was performed by utilizing the EDC-catalyzed reaction between carboxyl Pdots and the respective amine-containing molecules. 11-Azido-3,6,9-trioxaundecan-1-amine was used to form azido-Pdots. Propargylamine was used to produce alkyne-Pdots. Amine-terminated poly(ethylene glycol) was used to form PEG-Pdots. In a typical conjugation reaction, 60μL of polyethylene glycol (5% w/v PEG, MW 3350) and 60 μL of concentrated HEPES buffer (1 M) were added to 3 mL of carboxyl Pdot solution (50 μg/mL in MilliQ water), resulting in a Pdot solution in 20 mM HEPES buffer with a pH of 7.3. Then, 30 μL of amine-containing molecules (1 mg/mL) was added to the solution and mixed well on a vortex. Last, 60 μL of freshly-prepared EDC solution (5 mg/mL in MilliQ water) was added to the solution, and the above mixture was magnetically stirred for 4 hours at room temperature. Finally, the resulting Pdot conjugates were separated from free molecules by Bio-Rad Econo-Pac® 10DG columns (Hercules, CA, USA).
For metabolic labeling of newly synthesized proteins, MCF-7 cells were grown to confluence before passage into serum-free medium lacking methionine. After one hour incubation to deplete any residual methionine, cultures were supplemented with 0.1 mM AHA or HPG for four hours. The cells were washed by 1× PBS, fixed with 4% paraformaldehyde/PBS, and blocked using a blocking buffer. The AHA- or HPG-labeled cells were incubated for one hour with a mixture of 1 mM CuSO4, 5 mM sodium ascorbate, 0.5 mM tris((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)amine (TBTA, triazole ligand), and 50 nM alkyne-Pdots (for AHA-labeled cells) or azido-Pdots (for HPG-labeled cells). For metabolic labeling of glycoproteins, MCF-7 cells were cultured using the general EMEM medium containing 50 μM GalNAz for three days in order to enrich the azido groups in O-linked glycoproteins. The GalNAz-labeled cells were washed by 1X PBS, fixed with 4% paraformaldehyde/PBS, and blocked. Then the GalNaz-labeled cells were incubated for one hour with a mixture of 1 mM CuSO4, 5 mM sodium ascorbate, 0.5 mM TBTA, and 50 nM alkyne-Pdots. The Pdot-tagged cells were then counterstained with Hoechst 34580, washed three times by 1× PBS, and imaged immediately on a fluorescence confocal microscope (Zeiss LSM 510).
The materials, cell culture, experimental details of Pdot characterizations, and supplementary Figures are provided in Supporting Information.
Supplementary Material
Footnotes
This work was supported by the National Institutes of Health (NS062725, CA147831, and AG029574). We thank the Keck Imaging Center and the Center of Nanotechnology at the University of Washington for use of their facility.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Moses JE, Moorhouse AD. Chem Soc Rev. 2007;36:1249. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 3.Kolb HC, Sharpless KB. Drug Discov Today. 2003;8:1128. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 5.Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 6.Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 7.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem Biol. 2006;1:644. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 8.Sletten EM, Bertozzi CR. Angew Chem Int Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsien RY. Annu Rev Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 10.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 11.Chan WCW, Nie SM. Science. 1998;281:2016. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 12.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S, Devaraj NK, Lee J, Hilderbrand SA, Weissleder R, Bawendi MG. J Am Chem Soc. 2010;132:7838. doi: 10.1021/ja101677r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardin A, Cazet A, Guyon L, Delannoy P, Vinet F, Bonnaffe D, Texier I. Bioconjugate Chem. 2010;21:583. doi: 10.1021/bc900564w. [DOI] [PubMed] [Google Scholar]
- 15.Wu C, Szymanski C, Cain Z, McNeill J. J Am Chem Soc. 2007;129:12904. doi: 10.1021/ja074590d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. ACS Nano. 2008;2:2415. doi: 10.1021/nn800590n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. Angew Chem Int Ed. 2009;48:2741. doi: 10.1002/anie.200805894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Wu C, Sahu S, Fernando L, Szymanski C, McNeill J. J Am Chem Soc. 2009;131:18410. doi: 10.1021/ja907228q. [DOI] [PubMed] [Google Scholar]
- 19.Moon JH, McDaniel W, MacLean P, Hancock LE. Angew Chem Int Ed. 2007;46:8223. doi: 10.1002/anie.200701991. [DOI] [PubMed] [Google Scholar]
- 20.Moon JH, MacLean P, McDaniel W, Hancock LF. Chem Commun. 2007:4910. doi: 10.1039/b710807a. [DOI] [PubMed] [Google Scholar]
- 21.Baier MC, Huber J, Mecking S. J Am Chem Soc. 2009;131:14267. doi: 10.1021/ja905077c. [DOI] [PubMed] [Google Scholar]
- 22.Abbel R, van der Weegen R, Meijer EW, Schenning APHJ. Chem Commun. 2009:1697. doi: 10.1039/b822943k. [DOI] [PubMed] [Google Scholar]
- 23.Pu KY, Li K, Shi JB, Liu B. Chem Mater. 2009;21:3816. [Google Scholar]
- 24.Howes P, Green M, Levitt J, Suhling K, Hughes M. J Am Chem Soc. 2010;132:3989. doi: 10.1021/ja1002179. [DOI] [PubMed] [Google Scholar]
- 25.Rahim NAA, McDaniel W, Bardon K, Srinivasan S, Vickerman V, So PTC, Moon JH. Adv Mater. 2009;21:3492. [Google Scholar]
- 26.Xie HY, Liang HG, Zhang ZL, Liu Y, He ZK, Pang DW. Spectrochimica Acta Part A. 2004;60:2527. [Google Scholar]
- 27.Hermanson GT. Bioconjugate Techniques. 2. Academic Press; San Diego: 2008. [Google Scholar]
- 28.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Proc Natl Acad Sci USA. 2006;103:9482. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beatty KE, Liu JC, Xie F, Dieterich DC, Schuman EM, Wang Q, Tirrell DA. Angew Chem Int Ed. 2006;45:7364. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 30.Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Nat Protoc. 2007;2:532. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 31.Prescher JA, Bertozzi CR. Cell. 2006;126:851. doi: 10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Prescher JA, Dube DH, Bertozzi CR. Nature. 2004;430:873. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 33.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Proc Natl Acad Sci USA. 2006;103:4819. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laughlin ST, Bertozzi CR. Proc Natl Acad Sci USA. 2009;106:12. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breidenbach MA, Gallagher JEG, King DS, Smart BP, Wu P, Bertozzi CR. Proc Natl Acad Sci USA. 2010;107:3988. doi: 10.1073/pnas.0911247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.