Abstract
Although only 1.5% of the human genome appears to code for proteins, much effort in cancer research has been devoted to this minimal fraction of our DNA. However, the last few years have witnessed the realization that a large class of non-coding RNAs (ncRNAs), named microRNAs, contribute to cancer development and progression by acting as oncogenes or tumor suppressor genes. Recent studies have also shown that epigenetic silencing of microRNAs with tumor suppressor features by CpG island hypermethylation is a common hallmark of human tumors. Thus, we wondered whether there were other ncRNAs undergoing aberrant DNA methylation-associated silencing in transformed cells. We focused on the transcribed-ultraconserved regions (T-UCRs), a subset of DNA sequences that are absolutely conserved between orthologous regions of the human, rat and mouse genomes and that are located in both intra- and intergenic regions. We used a pharmacological and genomic approach to reveal the possible existence of an aberrant epigenetic silencing pattern of T-UCRs by treating cancer cells with a DNA-demethylating agent followed by hybridization to an expression microarray containing these sequences. We observed that DNA hypomethylation induces release of T-UCR silencing in cancer cells. Among the T-UCRs that were reactivated upon drug treatment, Uc.160+, Uc283+A and Uc.346+ were found to undergo specific CpG island hypermethylation-associated silencing in cancer cells compared with normal tissues. The analysis of a large set of primary human tumors (n=283) demonstrated that hypermethylation of the described T-UCR CpG islands was a common event among the various tumor types. Our finding that, in addition to microRNAs, another class of ncRNAs (T-UCRs) undergoes DNA methylation-associated inactivation in transformed cells supports a model in which epigenetic and genetic alterations in coding and non-coding sequences cooperate in human tumorigenesis.
Keywords: DNA methylation, CpG island, non-coding RNA, epigenetics
Introduction
The best-studied sequences of the genome are protein-coding genes, but their coding exons account for only 1.5% of the genome (2% if untranslated regions (UTRs) are included) (International Human Genome Sequencing Consortium, 2004). Thus, a large fraction of the genome is constituted by non-protein-coding regions that might have critical biological relevance (Mattick, 2003). The importance of this functional part of the non-protein-coding genome is particularly apparent in a large class of small, non-coding RNAs (ncRNAs), called microRNAs (miRNAs; Ambros, 2004; Bartel, 2004; He and Hannon, 2004; Mendell, 2005). These 19–24-nucleotide transcripts regulate the expression of messenger RNAs at the transcriptional and translational levels by imperfect base pairing with the 3′-UTR, the 5′-UTR and/or the coding region of the target (Miranda et al., 2006; Iwama et al., 2007; Duursma et al., 2008). In human cancer, recent studies have shown that miRNA expression profiles differ between normal tissues and derived tumors, and between tumor types (Calin et al., 2004a; Lu et al., 2005; Volinia et al., 2006). miRNAs can act as oncogenes or tumor suppressors, exerting a key function in tumorigenesis (Esquela-Kerscher and Slack, 2006; Hammond, 2007; Lujambio and Esteller, 2009). Defects in miRNA function have been associated with a failure of miRNA post-transcriptional regulation (Thomson et al., 2006), miRNA transcriptional repression by oncogenic factors (Chang et al., 2008), loss-of-function genetic alterations in miRNA-processing genes (Melo et al., 2009) and, most interestingly, transcriptional silencing associated with hypermethylation of CpG island promoters (Saito et al., 2006; Han et al., 2007; Lujambio et al., 2007, 2008; Toyota et al., 2008; Huang et al., 2009). Thus, as occurs with miRNAs, it is likely that other types of ncRNAs are also involved in human tumorigenesis and undergo epigenetic and genetic defects in this disease. In this paper, we have focused on the possible DNA methylation-associated silencing in human cancer cells of another class of ncRNAs, transcribed-ultraconserved regions (T-UCRs) (Bejerano et al., 2004; Calin et al., 2007; Katzman et al., 2007).
Ultraconserved regions (UCRs) are a subset of conserved sequences that are located in both intra- and intergenic regions (Bejerano et al., 2004; Katzman et al., 2007). They are absolutely conserved (100%) between orthologous regions of the human, rat and mouse genomes (Bejerano et al., 2004) and are strongly constrained functional elements (Katzman et al., 2007). The UCRs exhibit almost no natural variation in the human population (Bejerano et al., 2004). In the human genome, 481 sequences longer than 200 bp that are maximally conserved with orthologous segments in rodents are defined as UCRs (Bejerano et al., 2004). Of these, 56% are non-exonic (without evidence of an encoding protein), 23% are partly exonic (coincide with the messenger RNA of a known human protein-coding gene, including UTRs) and 21% are possibly exonic (inconclusive evidence of overlap with protein-coding genes). Much effort is currently being devoted to unraveling the putative functions of UCRs, such as an antisense regulatory role for protein-coding genes or other ncRNAs, or a regulatory function as enhancers (Nobrega et al., 2003; Pennacchio et al., 2006 ). The majority (93%) of UCRs are transcribed (called T-UCRs) in normal human tissues, both ubiquitously and in a tissue-specific manner (Calin et al., 2007). Most importantly, recent data suggest that T-UCRs are altered at the transcriptional level in human tumorigenesis and the aberrant T-UCR expression profiles can be used to differentiate human cancer types (Calin et al., 2007).
The downregulation of protein-coding genes, such as many important tumor suppressor genes in human cancer (Jones and Baylin, 2007; Esteller, 2008) and miRNAs with growth-inhibitory functions (Lujambio et al., 2007, 2008; Huang et al., 2009), has been tightly linked to the presence of CpG island promoter hypermethylation. Therefore, we wondered whether the same mechanism could have a role in the loss of adequate T-UCR expression in tumors. Herein, we present a pharmacological and genomic unmasking of T-UCR CpG islands that undergo cancer-specific hypermethylation-associated transcriptional silencing, such as Uc.160+, Uc283+A and Uc.346+, in the most common types of human cancer. These findings support a model in which epigenetic disruption of emerging new classes of ncRNAs, such as T-UCRs, constitutes a hallmark of human tumorigenesis.
Results
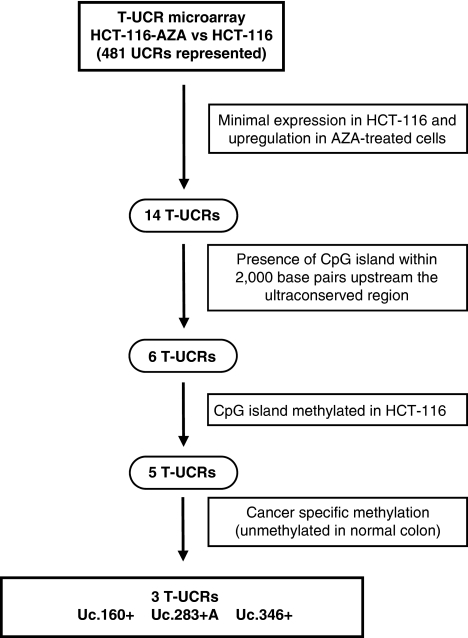
To identify T-UCRs with putative DNA methylation-related inactivation in human tumors, we followed the experimental algorithm shown in Figure 1. First, we treated the human colorectal cancer cell line HCT-116 with the DNA methylation inhibitor 5-aza-2′-deoxycytidine. To measure T-UCR levels, total RNA was extracted before and after treatment with the DNA-demethylating drug in each case, and profiled using our previously validated UCR platform (Calin et al., 2007). This UCR microarray chip includes a total of 481 human UCR sequences longer than 200 bp (Bejerano et al., 2004). T-UCRs can be transcribed from one strand (sense or antisense) or bidirectionally (Calin et al., 2007). In total, two 40-mer probes were designed for each UCR: one corresponding to the sense genomic sequence (named ‘+') and the other to the complementary sequence (named ‘+A'). Thus, 962 possible T-UCRs can be studied using this approach. Our pharmacological screening revealed that 14 of 481 (2.9%) human UCRs printed on the microarray had minimal basal expression in the untreated cells and ⩾2.25 log 2 (ratio between geometric means of intensity) upregulation upon use of 5-aza-2′-deoxycytidine (Supplementary Table 1). Of these 14 significantly upregulated T-UCRs, 6 (42.9%) had one canonical CpG island within a 2000-bp distance upstream in the transcriptional sense of that particular T-UCR: Uc.283+A, Uc.346+, Uc.160+, Uc.282+A, Uc.469+A and Uc.392+A (Supplementary Table 1). The studied length likely covers most of the CpG islands with a possible impact on T-UCR expression because T-UCR length has been described to range between 0.8 and 1.8 kb (Calin et al., 2007).
Figure 1.
Schematic strategy used to unmask DNA methylation-associated repression of T-UCRs in colon cancer cells. AZA, 5-aza-2′-deoxycytidine.
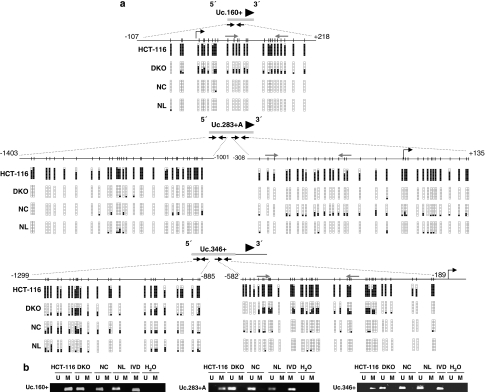
To demonstrate the presence of CpG island methylation in these six upregulated T-UCRs upon DNA demethylation treatment, we undertook bisulfite genomic-sequencing analyses of multiple clones in the HCT-116 colorectal cancer cell line from which the T-UCR expression microarray data were obtained. We found dense DNA methylation in 5 of 6 (83.3%) of the described CpG island-associated T-UCRs: Uc.283+A, Uc.346+, Uc.160+, Uc.282+A and Uc.469+A (Figures 2a and b; Supplementary Figure 1), whereas Uc392+A was found to be unmethylated in HCT-116 cells (Supplementary Figure 2). We wanted to focus on the cancer-specific DNA methylation changes; hence we used bisulfite genomic sequencing to analyze the DNA methylation status of these remaining five T-UCR-associated CpG islands in normal colon mucosa (n=5), to exclude tissue-specific DNA methylation patterns. The expression of many T-UCRs is different according to cell type (Calin et al., 2007); hence it was not surprising to observe that 2 of 5 (40%) of these T-UCR-associated CpG islands, Uc.282+A and Uc.469+A, were also densely methylated in normal colorectal mucosa (Supplementary Figure 1). However, and most importantly, the CpG islands for Uc.160+, Uc.283+A and Uc.346+ were always unmethylated in all normal tissues studied (Figures 2a and b). The DNA methylation results were also confirmed using methylation-specific PCR (Figure 2b).
Figure 2.
CpG island methylation of T-UCRs in colon cancer cell lines and normal tissues. (a) Bisulfite genomic sequencing of the associated CpG islands for the T-UCRs Uc.160+, Uc.283+A and Uc.346+ in the colorectal cancer cell lines HCT-116 and DKO, normal colon (NC) and normal lymphocytes (NL). CpG dinucleotides are represented as short vertical lines and the location of the T-UCR probe from the expression microarray is represented as a black arrowhead. The locations of the bisulfite genomic sequencing primers are indicated by black arrows. Eight single clones are represented for each sample. Presence of a methylated or unmethylated cytosine is indicated by a black or a white square, respectively. For Uc.283+A and Uc.346+, two regions within the CpG island were analyzed. The transcription start sites identified by RACE are represented by vertical black arrows. (b) Methylation-specific PCR analyses for Uc.160+, Uc.283+A and Uc.346+ methylation in the colorectal cancer cell lines HCT-116 and DKO, normal colon (NC) and normal lymphocytes (NL). Unmethylated (U) or methylated (M) sequences. In vitro methylated DNA (IVD) is shown as a positive control for methylated sequences. The locations of the methylation-specific PCR primers are indicated in panel ‘a' by gray arrows.
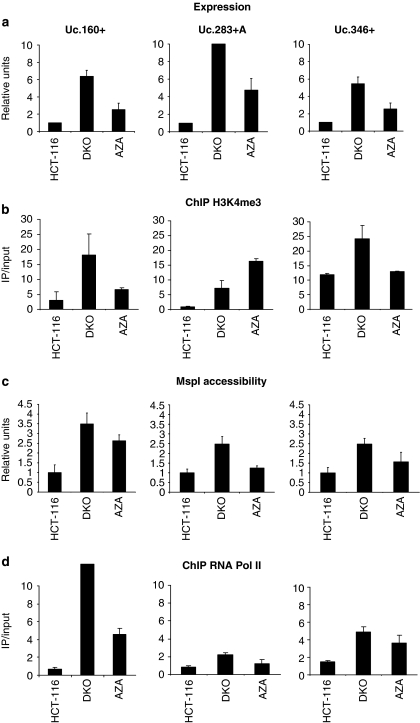
To demonstrate the transcriptional silencing of these T-UCRs in cancer cells in association with the presence of CpG island hypermethylation, we measured T-UCR levels by quantitative reverse transcriptase (RT)–PCR. The expression of Uc.160+, Uc.283+A and Uc.346+ transcripts was not detectable in HCT-116 cells showing CpG island methylation of the corresponding CpG island (Figure 3a). Most importantly, a restoration of T-UCR expression was observed upon treatment with the DNA-demethylating agent (Figure 3a). These results were confirmed using an alternative model, specifically an isogenic HCT-116 cell line in which the two major DNA methyltransferases, DNMT1 and DNMT3b, had been genetically disrupted (HCT116 DKO) (Rhee et al., 2002). The CpG islands for Uc.160+, Uc.283+A and Uc.346+ were unmethylated in DKO cells, although they were hypermethylated in its parental cell line, HCT116 (Figures 2a and b). Most importantly, restoration of Uc.160+, Uc.283+A and Uc.346+ expression was observed in DKO cells (Figure 3a), reinforcing the link between CpG island hypermethylation and T-UCR silencing.
Figure 3.
T-UCR expression and chromatin environment. (a) Expression analyses of T-UCRs by quantitative RT–PCR. Uc.160+, Uc.283+A and Uc.346+ show minimal expression in the hypermethylated HCT-116 cells, while treatment with the DNA-demethylating agent 5-aza-2′-deoxycytidine (AZA) and DKO cells show T-UCR upregulation. (b) Quantitative chromatin immunoprecipitation assay for the histone modification mark trimethylation of lysine 4 of histone H3 (H3K4me3) that is associated with active transcription. The presence of T-UCR CpG island methylation is associated with the lack of H3K4me3 histone, whereas the opposite scenario is observed when DNA demethylation events are present by genetic disruption of the DNMTs (DKO cells) or pharmacological treatment with a DNA-demethylating agent (AZA lane). (c) Chromatin accessibility assay using the MspI restriction enzyme coupled to real-time PCR. The hypermethylated CpG islands of Uc.160+, Uc.283+A and Uc.346+ CpG in HCT-116 are inaccessible to the enzyme, but CpG island demethylation upon 5-aza-2′-deoxycytidine treatment or in DKO cells results in higher accessibility. (d) Quantitative chromatin immunoprecipitation assay for RNA polymerase II (RNA Pol II) shows its absence in T-UCR hypermethylated CpG islands and its enrichment upon DNA hypomethylation events (DKO cells and HCT-116 treated with the DNA-demethylating drug).
DNA methylation silencing of tumor suppressor genes (Nguyen et al., 2002; Ballestar et al., 2003; McGarvey et al., 2008; Jacinto et al., 2009) and microRNAs with growth-inhibitory functions (Saito et al., 2006; Lujambio et al., 2007) is closely linked to chromatin modifications. To determine active vs inactive expression, we analyzed by chromatin immunoprecipitation the presence of a histone modification mark associated with active transcription, trimethylation of lysine 4 of histone H3 (Esteller, 2007), in these T-UCRs in HCT-116 cells. Uc.160+, Uc.283+A and Uc.346+ CpG island hypermethylation was accompanied by the absence or minimal presence of trimethylation of lysine 4 of histone H3 (Figure 3b). This histone mark was present in the studied T-UCR CpG islands in DKO cells or upon treatment of HCT-116 cells with the DNA-demethylating agent (Figure 3b). Furthermore, because DNA methylation and histone tail modifications crosstalk with nucleosome positioning in transcriptional regulation (Henikoff, 2007, 2008), we wondered whether T-UCR CpG island methylation silencing is also associated with a less accessible local chromatin conformation, as it occurs for hypermethylated CpG islands of tumor suppressor genes (Lin et al., 2007). We examined chromatin accessibility in the Uc.160+, Uc.283+A and Uc.346+ CpG islands using nuclei isolated from HCT-116 cells with increasing concentrations of the MspI restriction enzyme coupled to real-time PCR. We observed that when Uc.160+, Uc.283+A and Uc.346+ CpG islands are hypermethylated, these sites are inaccessible to the enzyme (Figure 3c), but restoration of T-UCR expression associated with CpG island demethylation upon 5-aza-2′-deoxycytidine treatment or in DKO cells results in higher accessibility (Figure 3c). In addition, the unmethylated T-UCR CpG islands were occupied by RNA polymerase II that was not present in the hypermethylated CpG islands (Figure 3d). In DKO cells or upon the use of the DNA-demethylating drug, RNA polymerase II occupancy for the studied T-UCR CpG islands was restored (Figure 3d). Most importantly, we characterized the transcription start sites for the studied T-UCRs using rapid amplification of cDNA ends (RACE). For Uc.160+ and Uc.283+A, the transcripts were originated within the studied corresponding CpG islands (vertical black arrows in Figure 2a), while for Uc.346+ transcription started just 189 bp downstream of the CpG island. Thus, our data suggest that expression of the Uc.160+, Uc.283+A and Uc.346+ transcripts occurs in the context of an unmethylated CpG island, and that cancer-specific hypermethylation of the described T-UCR CpG islands is associated with transcriptional silencing in a chromatin environment characteristic of gene repression.
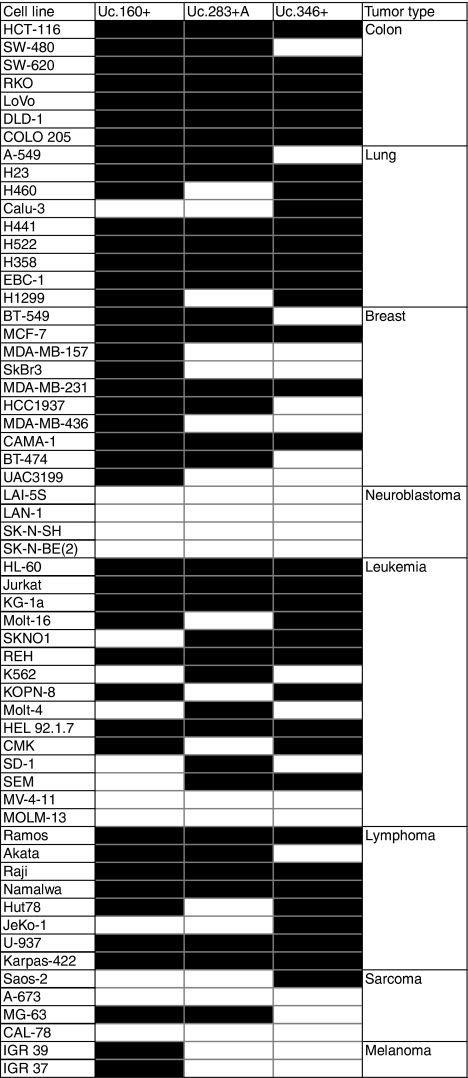
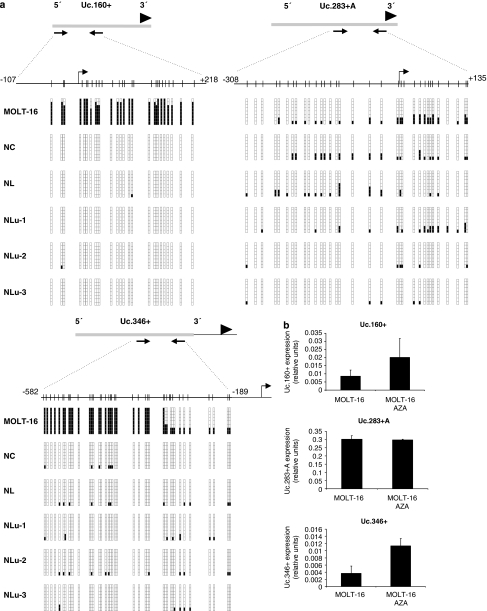
The presence of Uc.160+, Uc.283+A and Uc.346+ cancer-specific CpG island hypermethylation and transcriptional silencing was not a unique feature of the particular colorectal cancer cell line HCT-116, but upon analyzing a large set of human cancer cell lines (n=59) from eight different tumor types, we observed that it was commonly found in colon, breast, lung, lymphoma and leukemia cells (Table 1). The T-UCR CpG island methylation status was not associated with the K-ras and p53 mutational status or the presence of microsatellite instability (Fisher's exact test, P>0.05). Quantitative RT–PCR studies in the leukemia cell line MOLT-16 confirmed that Uc.160+, Uc.283+A and Uc.346+ CpG island hypermethylation was associated with loss of the corresponding T-UCR, while cells with unmethylated CpG islands expressed these transcripts (Figures 4a and b). We also confirmed in MOLT-16 cells that treatment with the demethylating agent restored T-UCR expression (Figure 4b). Moreover, we used the neuroblastoma cell line LAN-1, which was unmethylated for all the three studied T-UCR CpG islands, to further confirm the association between an unmethylated Uc.160+, Uc.283+A and Uc.346+ CpG island and expression of the corresponding transcript (Supplementary Figure 3). We also performed chromatin immunoprecipitation to obtain a detailed map of RNA polymerase II occupancy in the three characterized T-UCRs in LAN-1 cells. We observed an enrichment in RNA polymerase II in the corresponding transcription start sites previously identified by RACE (Supplementary Figure 3), reinforcing the link between T-UCR-unmethylated 5′-ends and active transcription. Furthermore, we also used the T-UCR-unmethylated neuroblastoma cell lines to study the cellular localization of these transcripts. If mature microRNAs are mostly located in the cytosol (Ambros, 2004; Bartel, 2004; He and Hannon, 2004), other non-coding RNAs have been shown to localize in nucleus (Chen and Carmichael, 2009). Using quantitative RT–PCR in isolated nuclear and cytoplasmic fractions of the neuroblastoma cell lines LAN-1, SK-N-SH and SK-N-EB1, we have observed that Uc.160+, Uc.283+A and Uc.346+ CpG are almost exclusively localized in the cytosol (Supplementary Figure 4). This finding might be an important clue for further research focused on the physiological roles of T-UCRs.
Table 1. Profile of T-UCR CpG island methylation status in human cancer cell lines.
Figure 4.
CpG island methylation and expression of T-UCRs in cancer cell lines and normal tissues. (a) Bisulfite genomic sequencing of the associated CpG islands for the T-UCRs Uc.160+, Uc.283+A and Uc.346+ in the leukemia cell line MOLT16 and normal tissues (colon, NC; lymphocytes, NL; lung, Nlu-1 to Nlu-3). CpG dinucleotides are represented as short vertical lines and the location of the T-UCR probe from the expression microarray is represented as a black arrowhead. The locations of the bisulfite genomic sequencing primers are indicated by black arrows. Eight single clones are represented for each sample. Presence of a methylated or unmethylated cytosine is indicated by a black or white square, respectively. The transcription start sites identified by RACE are represented by vertical black arrows. (b) Expression analyses of T-UCRs by quantitative RT–PCR. Uc.160+ and Uc.346+ show minimal expression in the hypermethylated MOLT-16 cells, while treatment with the DNA-demethylating agent 5-aza-2′-deoxycytidine (AZA) induces T-UCR upregulation. The Uc.283+A CpG island is unmethylated in MOLT-16 and the T-UCR is expressed at similar levels in the untreated and demethylating agent-treated cells.
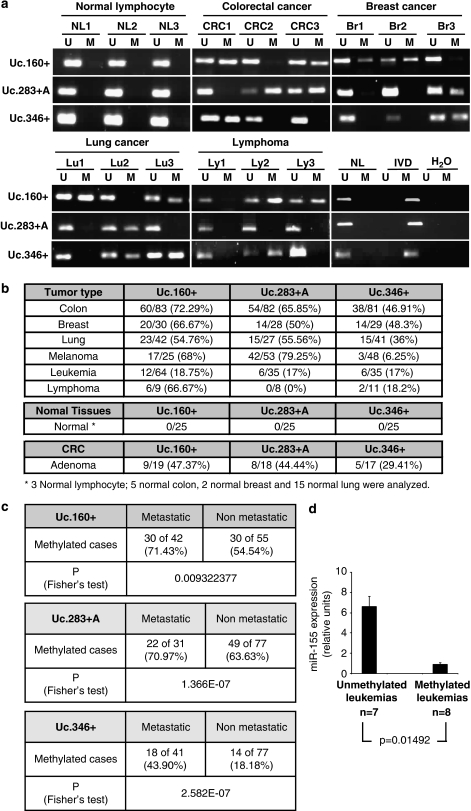
Most importantly, the CpG island hypermethylation of Uc.160+, Uc283+A and Uc.346+ was not an in vitro phenomenon, because when we examined 283 human primary malignancies corresponding to six different tissue types, we observed that Uc.160+, Uc.283+A and Uc.346+ CpG island hypermethylation was commonly found in primary colon, breast and lung tumors, in addition to melanomas, leukemias and lymphomas (Figures 5a and b). We confirmed in a set of corresponding normal tissues (fifteen lung, two breast and three lymphocyte samples), in addition to the previously analyzed five colon mucosas, the absence of hypermethylation of the described T-UCRs (Figures 4a and 5b). For colorectal tumors, where K-ras and p53 mutational and microsatellite instability data were available (Lujambio et al., 2007, 2008), the T-UCR CpG island methylation status was not associated with any of the described genetic alterations (Fisher's exact test, P>0.05). To determine whether hypermethylation of the described T-UCRs might represent an early lesion in tumorigenesis, we examined the CpG island methylation status of Uc.160+, Uc.283+A and Uc.346+ in benign colorectal adenomas (n=19), a lesion that is a precursor to invasive colorectal tumors. We observed that T-UCR hypermethylation is already present in these premalignant lesions, although in a lower frequency to that of established colon carcinomas (Figure 5b). However, the most compelling data regarding the role of Uc.160+, Uc.283+A and Uc.346+ epigenetic silencing in tumor progression were obtained when the T-UCR CpG island hypermethylation status was determined with respect to the existence or non-existence of lymph node metastasis in the tumors for which data were available (Figure 5c). The presence of Uc.160+, Uc.283+A and Uc.346+ CpG island hypermethylation in the primary tumor was significantly associated with those tumors that were positive for metastatic cancer cells in the corresponding lymph nodes (Fisher's exact test; P=0.0093, P=1.366E−07 and P=2.5818E−07, respectively) (Figure 5c), which highlights the importance of the in vivo role of T-UCR epigenetic silencing in tumorigenesis.
Figure 5.
T-UCR CpG island hypermethylation patterns in human tumorigenesis. (a) Methylation-specific PCR analyses for Uc.160+, Uc.283+A and Uc.346+ methylation in primary human tumors from different types. In vitro methylated DNA (IVD) and normal lymphocytes (NLs) are shown as positive and negative controls for methylated and unmethylated sequences, respectively. (b) Frequency of Uc.160+, Uc.283+A and Uc.346+ hypermethylation in primary human tumors from different types, normal tissues and premalignant lesions (colorectal adenomas). (c) Distribution of Uc.160+, Uc.283+A and Uc.346+ CpG island hypermethylation in primary tumors according to the presence or absence of lymph node metastasis. The presence of hypermethylation of the T-UCRs is significantly associated with the existence of metastasis in human malignancies. (d) Observation of an unmethylated Uc.160+ CpG island in all leukemias that overexpress the miR-155 transcript determined by quantitative RT–PCR in comparison with the presence of Uc.160+ hypermethylation in all leukemias with low levels of miR-155 (P=0.01492, Student's t-test). CRC, colorectal carcinoma.
One of the main challenges in research into UCR (and ncRNAs generally) is the identification of a particular cellular function. If tumor suppressor roles have been described for coding genes (Jones and Baylin, 2007; Esteller, 2008) and microRNAs (Davalos and Esteller, 2010) undergoing cancer-specific CpG island hypermethylation-associated silencing, T-UCRs might also act in a similar manner. We have briefly addressed this issue by cloning the full length of the T-UCR Uc.283+A (2227 bp) and restoring its expression by transfection in the hypermethylated and silenced HCT-116 colorectal cancer cell line. Upon re-introduction of Uc.283+A, we have observed a marked increase in the cell-doubling time and a major enhancement in the number of dead cells, assessed by the trypan blue and cell cycle assays (Supplementary Figure 5). These results suggest that the described hypermethylated T-UCRs might have potential growth-inhibitory features that will warrant further investigation.
From a molecular genetics standpoint, untranscribed UCRs might have regulatory functions as enhancers (Nobrega et al., 2003; Pennacchio et al., 2006), while myriad functions can be postulated for T-UCRs, such as an antisense inhibitory role for protein-coding genes or other ncRNAs, including miRNAs. We have preliminarily studied the possible effect of T-UCRs in the expression of protein-coding genes located upstream and downstream of the transcription start sites of Uc.160+, Uc.283+A and Uc.346+ (Supplementary Table 3). The available microarray expression data that compare HCT-116 vs DKO cells (Schuebel et al., 2007) show that the expression of three T-UCR-surrounding genes included in the described genomic platform (AP3B1 for Uc.160+ RFX4 for Uc.346+ and ERCC6 for Uc.283+A) was unchanged in the DNA-hypomethylated cells. Thus, because DKO cells re-express the T-UCRs characterized in our study and there is no effect in the expression levels of the neighboring genes AP3B1, RFX-4 and ERCC6, we can hypothesize that T-UCRs exert their function at another level. The precise molecular targets of these T-UCRs will be the focus of future research in this area.
On the other hand, instead of T-UCRs acting on protein-coding genes and miRNAs, it is possible that miRNAs exert control over T-UCRs. Evidence supporting this contention comes from the demonstration that many T-UCRs have significant antisense complementarity with particular miRNAs, the negative correlation between expression of specific T-UCRs and predicted interactor miRNAs, and the demonstration in a small subset of T-UCRs of an interaction in vitro (luciferase assays) and in vivo (miR transfection) with miR/T-UCR (Calin et al., 2007). The latter observation is particularly relevant to Uc.160+, in which transfection of the oncogenic miR-155 into leukemia cells significantly reduced the expression level of Uc.160+ (Calin et al., 2007). Thus, if it is widely accepted that the probability of simultaneous hits in the same molecular pathway in a given tumor is very low, simultaneous overexpression of miR-155 and CpG island hypermethylation-associated inactivation of Uc.160+ in the same leukemia sample should be a rare event. To test whether this inverse association exists, we analyzed the association between miR-155 expression and Uc.160+ CpG island methylation status in our 15 human leukemia cell lines. We observed an unmethylated Uc.160+ CpG island in the leukemias with high levels of the miR-155 transcript (n=7) compared with the presence of Uc.160+ hypermethylation in the leukemias with low levels of miR-155 (n=8) (P=0.01492, Student's t-test) (Figure 5d). Thus, the mutual exclusivity of the epigenetic alteration of Uc.160+ and the overexpression of the oncogenic miR-155 suggests that they may have a critical and cooperative role in human leukemogenesis.
Discussion
The increasing number of cancer exonomes and genomes sequenced exhibits a low rate of true genetic mutations (Velculescu, 2008). Thus, there is a large fraction of the genome constituted by non-coding DNA sequences that is likely to contain genetic and epigenetic defects in malignancies. The best example is the most widely studied type of ncRNA, the microRNAs. miRNA dysregulation has been found in many human diseases, cancer being the paradigmatic example (Lu et al., 2005; Calin and Croce, 2006; Volinia et al., 2006). miRNA genetic alterations, such as deletions, mutations and amplifications, have been increasingly found in human tumors (Calin et al., 2004b; Zhang et al., 2006; Rossi et al., 2008; Spizzo et al., 2009), and the miRNA processing machinery can also be altered in transformed cells (Zhang et al., 2006; Rossi et al., 2008; Merritt et al., 2008; Hill et al., 2009; Melo et al., 2009). Most importantly, miRNAs can also undergo cancer-specific 5′-CpG island hypermethylation-associated silencing (Saito et al., 2006; Han et al., 2007; Lujambio et al., 2007, 2008; Toyota et al., 2008; Huang et al., 2009). We have now provided data that suggest that DNA methylation-associated silencing of miRNAs in human cancer may be just the tip of the iceberg and that it might be many more different classes of ncRNAs undergoing epigenetic inactivation in transformed cells, T-UCRs being the second prototypical example.
UCRs were discovered by bioinformatic comparisons of the genomes of mouse, rat and human (Bejerano et al., 2004). In total, 481 UCR sequences show 100% identity among the three species. Some of them contain protein-coding sequences, but over half are not predicted to code for any protein (Bejerano et al., 2004). Using a custom-designed microarray (Calin et al., 2007), a new class of ncRNAs encoded from a subset of UCRs was identified: transcribed UCRs (T-UCRs). These T-UCRs (also called ultraconserved genes, UCGs) are expressed in normal tissues with housekeeping and tissue-specific patterns (Calin et al., 2007), in a similar manner as protein-coding genes and miRNAs do. Interestingly, we observed that while almost half of the T-UCR-associated CpG islands are unmethylated in all tissues, the other half show tissue-specific T-UCR CpG island methylation, as occurs with promoter CpG islands of coding genes (Jones and Baylin, 2007; Esteller, 2008) and miRNAs (Lujambio and Esteller, 2007, 2009). The genome-wide expression profile of T-UCRs reveals that these ncRNAs have distinct signatures in normal tissues and human cancers, and between tumor types (Calin et al., 2007). Many genetic defects could cause T-UCR dysregulation in transformed cells, and, in fact, several of these T-UCRs are located at fragile sites, amplified regions and loss-of-heterozygosity loci (Calin et al., 2004b; Zhang et al., 2006; Rossi et al., 2008). However, it is reasonable to propose that they could also undergo CpG island hypermethylation-associated silencing. Our finding that epigenetic inactivation by CpG island hypermethylation of a subset of T-UCRs, such as Uc160+, Uc.283+A and Uc.346+, occurs in a wide spectrum of human cancer cell lines and primary tumors of varying cellular and tissue origin provides clear support for the concept that major disruption of ncRNA programming is a common feature of cancer cells.
An enormous task still lies ahead. New classes of ncRNAs are emerging, such as piwi-interacting RNAs, small-nucleolar RNAs, long ncRNAs, telomere-specific small RNAs and transcription-initiation RNAs, that might be subject to transcriptional regulation of neighboring CpG islands and DNA methylation. The particular functions of these ncRNAs are beginning to be elucidated, but little is currently known about their alteration in diseases such as cancer. However, it seems likely that aberrations in the DNA methylation pattern of their underlying genomic sequences will turn out to have a role, and therefore merit further investigation. For T-UCRs, once we have demonstrated their epigenetic disturbance in human neoplasms, we will be in a position to begin to understand their biological function better. For untranscribed UCRs, previous studies have suggested important roles in promoting the expression of several genes and in regulating alternative splicing (Bejerano et al., 2004, 2006; Lareau et al., 2007). However, for T-UCRs, which have a chromatin context (histone modifications, chromatin accessibility and RNA polymerase II occupancy) similar to that observed in protein-coding genes and miRNAs depending on their transcriptional status, several functions are still being studied.
In summary, our findings demonstrate that, in addition to miRNAs, epigenetic silencing of other classes of ncRNAs, such as T-UCRs, is a common feature of human cancer. Our results not only provide further valuable knowledge about the mechanisms involved in normal and aberrant regulation of ncRNA transcription, but also indicate that the tumor-specific CpG island hypermethylated T-UCRs identified might be useful biomarkers of the disease. Above all, the reactivation of those T-UCRs that have undergone aberrant CpG island hypermethylation in the transformed cells might provide a new molecular basis for the therapeutic use of pharmacological compounds with DNA-demethylating activity in the treatment of cancer patients.
Materials and methods
Cell lines, culture conditions and primary study samples
The 59 human cancer cell lines examined in this study were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). HCT-116 and DKO cells were a generous gift from Dr Bert Vogelstein (Johns Hopkins Kimmel Comprehensive Cancer Center, Baltimore, MD, USA). Cell lines were treated with 1 μ 5-aza-2′-deoxycytidine (Sigma-Aldrich, St Louis, MO, USA) for 48 h to achieve demethylation (Herman et al., 1998). Cell-doubling time, trypan blue assay and cell cycle analysis were performed as described in Supplementary Materials and methods. DNA samples from primary tumors (n=283), benign colorectal adenomas (n=19) and normal tissues (n=25) were obtained at the time of the clinically indicated surgical procedures. All patients gave written consent to participate in the study and the Ethics Committee of the different hospitals cleared the study protocol.
RNA isolation and T-UCR expression microarray analysis
Total RNA was isolated from the colorectal cancer cell line HCT-116, before and after 5-aza-2′-deoxycytidine treatment, by Trizol extraction (Invitrogen, Carlsbad, CA, USA). T-UCR microarray profiling was conducted as described (Calin et al., 2007). Microarray chips were developed with a total of 481 human UCR sequences (Calin et al., 2007). For each UCR, two 40-mer probes were designed, one corresponding to the sense genomic sequence (named ‘+') and the other to the complementary sequence (named ‘+A'). The design criteria were as described (Liu et al., 2004). Each oligo was printed in duplicate in two different slide locations, and therefore quadruplicate numerical values were available for analysis. Several thousand (3484) blank spots were used for background subtraction. RNA extraction and microarray experiments, consisting of the UCR microarray assembly, target preparation and array hybridization, were performed as described in detail elsewhere (Calin et al., 2004a; Liu et al., 2004). Further details are provided in Supplementary Materials and methods.
DNA methylation analyses
The CpG Island Searcher Program (Takai and Jones, 2003) was used to determine which T-UCRs were located within a ±2000-bp proximity of a CpG island, as it has been predicted that more than 90% of the human miRNA promoters are located 1000 bp upstream of the mature miRNA (Saini et al., 2007; Zhou et al., 2007). DNA methylation status was established by PCR analysis of bisulfite-modified genomic DNA, which induces chemical conversion of unmethylated, but not methylated, cytosine to uracil. Two procedures were used. First, methylation status was analyzed by bisulfite genomic sequencing of the corresponding CpG islands. Eight independent clones were analyzed. The second analysis used methylation-specific PCR with primers specific for either the methylated or the modified unmethylated DNA. The primers used are described in the Supplementary Table 2.
Quantification and cellular localization of T-UCRs and miR-155 with real-time PCR
Quantitative RT–PCR was performed to quantify the level of T-UCRs as described previously (Calin et al., 2007). Each RT reaction contained 1 μg of purified and DNase-treated (turbo DNA-free, Ambion, Austin, TX, USA) total RNA, which was reverse transcribed using Thermoscript RT and gene-specific (sense or antisense) primers. cDNA was amplified using real-time PCR and SYBR (Applied Biosystems, Foster City, CA, USA) green detection using PCR primers designed to amplify the same 40-bp regions as the oligo probe on the microarray. The primers used are described in Supplementary Table 2. TaqMan assays were used to quantify the level of miR-155 as described previously (Lujambio et al., 2008). The relative amounts of each UCR and miRNA to the control (U6 and GAPDH, and RNU19, respectively) were determined using the 2(DDCt) method (Livak and Schmittgen, 2001). Further details are provided in Supplementary Materials and methods.
Quantitative chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as previously described (Jacinto et al., 2009) using the anti-trimethyl-K4 histone H3 antibody (ab8580/ab1220; Abcam, Cambridge, MA, USA) and anti-RNA polymerase II (N20, SC899; Santa Cruz Biotechnology, Santa Cruz, CA, USA). The primers used are described in Supplementary Table 2. Further details are provided in Supplementary Materials and methods.
Chromatin accessibility
Nuclei from each cell line were digested in a series of increasing MspI restriction enzyme (Roche Molecular Biochemicals, Mannheim, Germany) concentrations (0, 100 and 400 U/ml) at 37 °C for 1 h. We used the 2(DDCt) method (Livak and Schmittgen, 2001) to determine relative levels of amplification, using a region in the RIZ gene without MspI targets for normalization. Values were expressed as the difference relative to the controls. The primers used are described in Supplementary Table 2. Further details are provided in Supplementary Materials and methods.
RACE, cloning and transfection
Transcription start sites for the three studied T-UCRs (Uc.160+, Uc.283+A and Uc.346+) were identified by RACE in the 5′ direction. The sequences were obtained independently from DKO and LAN-1 cell lines according to the manufacturer's protocol (SMARTerTM RACE cDNA Amplification Kit; Clontech, Palo Alto, CA, USA). The 3′ end of Uc.283+A was localized by primer walking within a region of ∼350 bp. The primers used in both approaches are described in Supplementary Table 2. For transfection, the Uc.283+A expression vector (pCDNA3.1 Uc283+A) was constructed by cloning the DNA corresponding to Uc.283+A into a pCDNA3.1 (+) vector. An empty pCDNA3.1 (+) vector was used as a control. HCT-116 cells were transfected by electroporating 107 cells in 0.8 ml phosphate-buffered saline with 40 μg of the vector at 250 V and 975 mF. After electroporation, cells were washed with phosphate-buffered saline and seeded in fresh medium.
Acknowledgments
This work was supported by Grants SAF2007-00027-65134, Consolider CSD2006-49, CANCERDIP HEALTH-F2-2007-200620 and the Dr Josef Steiner Cancer Research Foundation. AL is supported by a FPU Fellowship. AP is a Sara Borell Postdoctoral Fellow. ME is an ICREA Research Professor.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004a;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004b;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. OncomiRs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nucleosomes at active promoters: unforgettable loss. Cancer Cell. 2007;12:407–409. doi: 10.1016/j.ccr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Iwama H, Masaki T, Kuriyama S. Abundance of microRNA target motifs in the 3′-UTRs of 20527 human genes. FEBS Lett. 2007;581:1805–1810. doi: 10.1016/j.febslet.2007.03.066. [DOI] [PubMed] [Google Scholar]
- Jacinto FV, Ballestar E, Esteller M. Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene. 2009;28:4212–4224. doi: 10.1038/onc.2009.267. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, et al. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. BioEssays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Rossi S, Sevignani C, Nnadi SC, Siracusa LD, Calin GA. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm Genome. 2008;19:526–540. doi: 10.1007/s00335-008-9119-8. [DOI] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the protooncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: microRNAs in cancer. Cell. 2009;137:586–586.e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. Plos Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.