Abstract
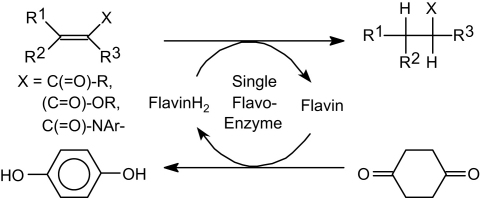
The asymmetric bioreduction of activated C C-bonds catalyzed by a single flavoprotein was achieved via direct hydrogen transfer from a sacrificial 2-enone or 1,4-dione as hydrogen donor without requirement of a nicotinamide cofactor. Due to its simplicity, this system has clear advantages over conventional FAD-recycling systems.
Keywords: Biotransformation, Flavin, Enoate reductase, Disproportionation, Cofactor recycling
Graphical abstract
1. Introduction
Disproportionation reactions generally furnish a 1:1 mixture of products and are often plagued by unfavorable equilibria, hence they are commonly considered as inefficient and are rarely used in organic synthesis.1 In biology, the disproportionation of sulfur yielding hydrogen sulfide and sulfate and of hydrogen peroxide furnishing O2 and H2O represent the most dominant disproportionation processes in Nature.2 The disproportionation of cyclohex-2-enone, forming equimolar amounts of cyclohexanone and phenol has been described for several flavoproteins from the old yellow enzyme (OYE) family, such as OYE isoenzymes 1–3 and estrogen-binding protein.3, (a), (b), (c) In the context of these studies, this phenomenon has been considered either as a minor side reaction demonstrating the catalytic promiscuity4 of OYEs or as ‘aromatase’ activity of enoate reductases catalyzing the formation of the phenolic A-ring in steroids, such as 17β-estradiol, from the corresponding enone-precursor 19-nortestosterone.3, (a), (b), (c) Overall, this reaction constitutes a flavin-dependent hydrogen transfer, during which an equivalent of [2H] is formally transferred from a cyclohex-2-enone (being oxidized) onto another one (being reduced). The oxidized product constitutes a conjugated dienone, which spontaneously tautomerises to form phenol, thereby providing a large driving force of ca. −30 kcal/M for the disproportionation reaction. During this hydrogen-transfer reaction, the flavin-cofactor is recycled internally and no external nicotinamide cofactor is required for the reductive half-reaction.5, (a), (b) In nicotinamide-dependent systems, C C-bonds are reduced at the expense of an external hydride donor,6 such as formate, glucose, glucose-6-phosphate or phosphite, which requires a second dehydrogenase enzyme, such as FDH, GDH, G6PDH7 or phosphite-DH,8 respectively. This technology is generally denoted as ‘coupled-enzyme-approach’, which depends on the concurrent operation of two independent redox enzymes for substrate reduction and co-substrate-oxidation, respectively.9 Aiming to reduce the complexity of these redox systems,10, (a), (b) considerable efforts have recently been devoted to the development of nicotinamide-independent electrochemical and light-driven recycling systems for reduced flavins, which take advantage of the direct transfer of a hydride (or electrons, respectively) from a donor onto the flavin.11 In this context, the nicotinamide-independent disproportionation of enones is of appealing simplicity, since it requires only a single flavoprotein and represents a ‘coupled-substrate-approach’.12
2. Results and discussion
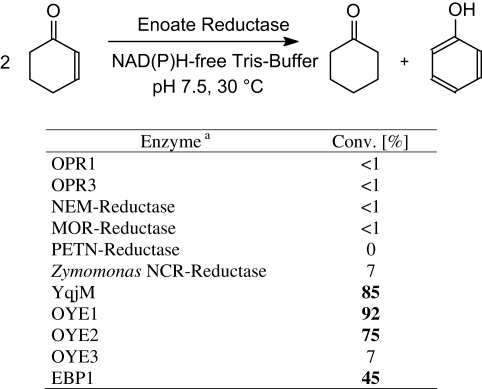
In an initial screening, a set of cloned and overexpressed enoate reductases were tested for their catalytic activity in the disproportionation of cyclohex-2-enone (Scheme 1). To our delight, the desired disproportionation activity was observed in a variety of OYE-homologs. Although all of these enoate reductase-type proteins have been reported to reduce cyclohex-2-enone to cyclohexanone at the expense of NAD(P)H,13 the corresponding disproportionation activity was exceedingly low in 12-oxophytodienoic acid reductase (OPR) isoenzymes 1 & 3 and in N-ethylmaleimide-, morphinone-, and pentaerythritol tetranitrate reductase. Likewise, cyclohexenone reductase from Zymomonas mobilis showed only modest activity, whereas the OYE-homolog YqjM14 and OYE isoenzymes15 1 and 2 from yeast were highly active. Surprisingly, OYE isoenzyme 3 was almost inactive. The divergent behavior of OYE isoenzymes is reflected by their structural relationship: Whereas both highly active isoenzymes 1 and 2 show an amino acid sequence identity of 92%, isoenzyme 3 is a more distant relative (sequence identity 80%). The disproportionation activity of estrogen-binding protein (EBP1)3, (a), (b), (c) could be nicely reproduced using EBP1 cloned into Escherichia coli. By taking the strongest disproportionation activities as a lead, further experiments were performed using YqjM, OYE1, and OYE2.
Scheme 1.
Disproportionation of cyclohex-2-ene catalyzed by enoate reductases.aOPR1, OPR3=oxophytodienoate reductase isoenzymes 1 and 3, respectively, from tomato;16 NEM-reductase=N-ethylmaleimide reductase;18 MOR-reductase=morphinone reductase;17 PETN-reductase=pentaerythritol tetranitrate reductase;18Z. mobilis NCR reductase=nicotinamide-dependent cyclohexenone reductase;19 YqjM=OYE-homolog from Bacillus subtilis;14 OYE1–3=Old Yellow Enzyme isoenzymes from yeasts;15 EBP1=estrogen-binding protein,3 employed as cell-free extract of E. coli expressing EBP1.
In order to convert the scrambling-like hydrogen-transfer reaction between two identical cyclohexenone molecules into a useful directed redox process, a pair of suitable enone substrate/co-substrate—one only being reduced, the other only being oxidized—have to be coupled. During our previous studies on NAD(P)H-dependent enone reduction, we observed that α-substituted cyclic enones were quickly reduced, whereas an alkyl-substituent in the β-position severely impeded the reaction rate.10, (a), (b), 15 Hence, we envisaged that an α-substituted enone might act as H-acceptor, while a β-substituted analog would serve as H-donor.
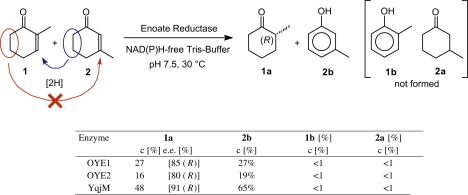
In order to test the viability of this concept, an equimolar amount of α- (1) and β-methylcyclohex-2-enone (2) were subjected to OYE1, OYE2, and YqjM in a nicotinamide-free buffer system (Scheme 2). The results of these experiments provided a clear proof-of-concept: Depending on the enzyme, the desired reduced α-methyl derivative 1a was formed in up to 48% conversion, while the oxidized β-methyl analog 2b was detected in approximately equimolar amounts.20 In contrast, only trace amounts of the corresponding cross-hydrogen-transfer products 1b and 2a, which would arise from undesired oxidation of 1 and reduction of 2 were found, indicating that the directed hydrogen transfer indeed worked as envisaged.
Scheme 2.
Asymmetric hydrogen transfer between α- (1) and β-methylcyclohex-2-enone (2) catalyzed by enoate reductases.
Investigation of the optical purity and absolute configuration of 1a revealed that the product was formed in the same highly selective fashion as in the classic reduction-mode using NAD(P)H-recycling, ensuring that the chiral induction process of the enzymes remained unchanged.(b), 15
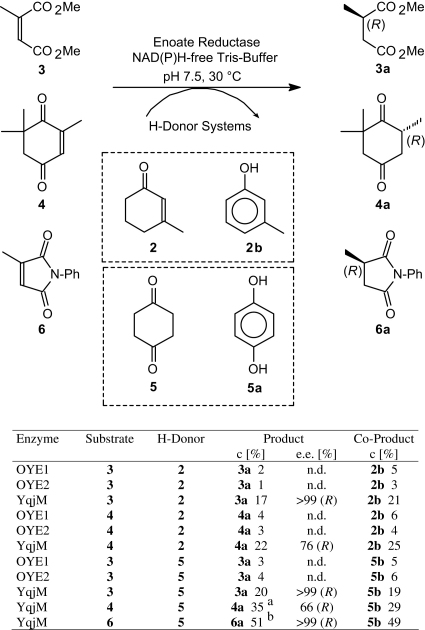
In order to test the applicability of this nicotinamide-free C C-bond reduction system, we subjected two activated alkenes (3, 4), which are known to be readily reduced by enoate reductases in presence of NAD(P)H,10, (a), (b), 15 to the hydrogen-transfer protocol in presence of equimolar amounts of β-methylcyclohex-2-enone (2) as hydrogen donor (Scheme 3). Whereas OYE1 and OYE2 showed only modest conversion, YqjM furnished the corresponding reduction products 3a and 4a in up to 22% conversion together with a stoichiometric amount of 3-methylphenol 2b. Within experimental errors, the enantiomeric excess of (R)-configurated products 3a and 4a was identical to that of the nicotinamide-driven process, indicating that the enzymatic chiral induction remained intact.10, (a), (b), 15
Scheme 3.
Asymmetric bioreduction of activated alkenes via a coupled-substrate approach. n.d.=not determined. aA trace of 2,3-epoxy-2,6,6-trimethylcyclohexane-1,4-dione was formed (≤3%), presumably due to spontaneous epoxidation of the C C-bond by H2O2 derived via enzyme-catalyzed reduction of O2.21bA trace of aniline was detected as side product (≤3%).
Since the use of equimolar amounts of 3-methylcyclohex-2-enone (2) as co-substrate would be economically unfavorable, a cheaper hydrogen donor was sought. After attempts using 1-indanone and hydroquinone failed, cyclohexane-1,4-dione (5)—yielding 1,4-dihydroxybenzene (hydroquinone, 5a) as oxidation product—was found to provide a suitable alternative. Using YqjM, all substrates showed enhanced conversion as compared to β-methylcyclohex-2-enone (2) as co-substrate.
At this early stage, this novel NAD(P)H-independent cofactor recycling system has not yet been fully optimized, particularly in view of the (co)-substrate concentrations22 and the overall conversions. Incomplete conversions might be attributed to a certain degree of enzyme inhibition, most presumably caused by the co-product phenol(s), which are known to form charge-transfer complexes with flavins.23 Since the latter process is reversible, the removal of phenols by in-situ (co)-product removal (ISPR)24 using biphasic aqueous-organic solvent systems25 is the first choice. This strategy has been successfully applied to industrial scale using (flavin-dependent) enzymatic Beyer–Villiger oxidation26 and (cytochrome P450-dependent) alkene epoxidation mediated by whole viable cells.27
3. Conclusion
An novel substrate-coupled C C-bond bioreduction system was developed, which depends only on a single flavoprotein and neither requires a second (dehydrogenase) recycling enzyme, nor a nicotinamide cofactor. Due to its simplicity, it has clear advantages over other nicotinamide-independent alternative systems, such as light-driven and electrochemical FAD-recycling systems.11
4. Experimental
4.1. General
GC–MS analyses were performed with an Agilent 7890A GC system equipped with an Agilent 5975C mass-selective detector (electron impact, 70 eV) using a (5%-phenyl)-methylpolysiloxane phase column (Agilent HP-5 ms, 30 m, 250 μm, 0.25 μm). Helium was used as carrier gas (column flow: 2 mL/min). GC–FID analyses were carried out with a Varian 3800 by using H2 as a carrier gas (14.5 psi). HPLC analyses were performed by using a Shimadzu system equipped with a Chiralcel OD-H column (25 cm, 0.46 cm). NMR spectra were measured on a Bruker AMX spectrometer at 360 MHz.
2-Methyl-2-cyclohexen-1-one (1), 2-methylcyclohexan-1-one (1a), 3-methylcyclohexan-1-one (2a) and rac-levodione were provided by BASF (Ludwigshafen). Cyclohexanone, 2-cyclohexen-1-one, phenol, 3-methyl-2-cyclohexen-1-one (2), 1,4-cyclohexanedione (5), hydroquinone (5a), and N-phenyl-2-methylmaleimide (6) were purchased from Aldrich. Citraconic acid was purchased from Alfa Aesar and 4-ketoisophorone was from ABCR Co.
4.2. Synthesis of substrates and reference materials
4.2.1. rac-Methylsuccinic acid
Citraconic acid (105 mg, 0.81 mmol) was dissolved in THF/EtOH 50:50 (10 mL) and was hydrogenated at atmospheric pressure and room temperature in the presence of 10% Pd/C (5 mg) as catalyst. After 24 h, the mixture was filtered through Celite and evaporated yielding 99% of rac-1b (106 mg, 0.80 mmol). Mp=110–115 °C. 1H NMR (D2O): δ 3.62–3.64 (d, 3H, J=7.2 Hz), 4.96–5.15 (m, 2H), 5.29–5.34 (m, 1H). 13C NMR (D2O): δ 18.7, 38.2, 39.6, 179.0, 182.8.15
4.2.2. rac-Dimethyl-2-methylsuccinate (3a)
A solution of rac-methylsuccinic acid (32 mg, 0.24 mmol) in BF3/MeOH (0.5 mL, 14%) was stirred at 100 °C for 1 h. H2O (0.5 mL) was added and the reaction mixture was extracted with n-hexane (3×1 mL). The combined organic layers were dried over Na2SO4, filtered and evaporated, yielding 46% of rac-(3a) (17 mg, 0.11 mmol). 1H NMR (CDCl3): δ 1.22–1.24 (d, 3H, J=7.2 Hz), 2.39–2.45 (dd, 1H, J=6.06 Hz, J=16.52 Hz), 2.72–2.79 (dd, 1H, J=8.15 Hz, J=16.51 Hz), 2.91–2.94 (m, 1H), 3.69 (s, 3H), 3.71 (s, 3H). 13C NMR (CDCl3): δ 17.0, 35.7, 37.4, 51.7, 51.9, 172.3, 175.7.15
4.2.3. Citraconic acid dimethylester (3)
Substrate 3 was synthesized according to the procedure described above starting from citraconic acid. 1H NMR (CDCl3): δ 2.06–2.07 (d, 3H, J=1.6 Hz), 3.73 (s, 3H), 3.83 (s, 3H), 5.86–5.87 (d, 1H, J=1.6 Hz). 13C NMR (CDCl3): δ 20.5, 51.8, 52.4, 120.6, 145.7, 165.4, 169.4.15
4.2.4. rac-N-Phenyl-2-methylsuccinimide (6a)
N-Phenyl-2-methylmaleimide (6, 50 mg, 0.27 mmol) was dissolved in EtOAc (5 mL) and was hydrogenated at atmospheric pressure at room temperature using 10% Pd/C (2.8 mg) as catalyst. After 24 h, the mixture was filtered through Celite and evaporated yielding 94% of rac-6a (48 mg, 0.25 mmol). 1H NMR (CDCl3): δ 1.47 (d, 3H, J=7 Hz), 2.52 (dd, 1H, J=17.4 Hz, J=4 Hz), 3.01–3.10 (m, 1H), 3.11 (dd, 1H, J=17.3 Hz, J=9.2 Hz), 7.29–7.51 (m, 5H).15
4.3. General procedure for the screening for enzymatic disproportionation of cyclohex-2-enone
An aliquot of the isolated enzyme OPR1, OPR3, YqjM, OYE1, OYE2, OYE3, Z. mobilis ER, NEM-Red, MOR-Red, and PETN-Red (protein purity >90%, protein content 90–110 μg/mL) was added to a Tris–HCl buffer solution (0.8 mL, 50 mM, pH 7.5) containing cyclohex-2-enone (10 mM). The mixture was shaken at 30 °C and 120 rpm for 24 h and the products were extracted with EtOAc (2×0.5 mL). The combined organic phases were dried (Na2SO4) and the resulting samples were analyzed on achiral GC. Products were identified by comparison with authentic reference materials via co-injection on GC–MS and achiral GC. Column: 6% Cyanopropyl-phenyl phase capillary column (Varian CP-1301, 30 m, 0.25 mm, 0.25 μm), detector temperature 250 °C, split ratio 30:1; temperature program: 80 °C; hold 2 min.; rise to 120 °C with 5 °C/min. TRet: cyclohex-2-enone 2.97 min, cyclohexanone 2.43 min, phenol 4.98 min.
4.4. Hydrogen transfer between 2- (1) and 3-methylcyclohex-2-enone (2)
An aliquot of the isolated enzyme YqjM, OYE1, OYE2 (protein purity >90%, protein content 90–110 μg/mL) was added to a Tris–HCl buffer solution (0.8 mL, 50 mM, pH 7.5) containing the substrate 1 (10 mM) and the co-substrate 2 (10 mM). The mixture was shaken at 30 °C and 120 rpm for 24 h and products were extracted with EtOAc (2×0.5 mL). The combined organic phases were dried (Na2SO4) and the resulting samples were analyzed on achiral GC. Products were identified by comparison with authentic reference materials via co-injection on GC–MS and achiral GC. Column: 14% cyanopropyl-phenyl phase capillary column (J&W Scientific DB-1701, 30 m, 0.25 mm, 0.25 μm), detector temperature 250 °C, split ratio 30:1. Temperature program: 110 °C, hold 5 min, rise to 200 °C with 10 °C/min, hold 2 min. TRet: 2-methylcyclohexenone (1) 4.38 min; 2-methylcyclohexanone (1a):3.70 min; 3-methylcyclohexenone (2) 6.27 min; 3-methylphenol (2b) 7.90 min; 2-methylphenol (1b) 7.02 min; 3-methylcyclohexanone (2a) 3.63 min.
4.5. Source of enzymes
The open reading frame of Lycopersicon esculentum OPR1 was cloned into pET-21a and expressed as a C-terminal hexahistidine tagged protein in E. coli BL21 cells. The expressed recombinant protein was purified on a Ni-NTA affinity column (Invitrogen) according to the manufacturer's protocol. L. esculentum OPR3 and YqjM from Bacillus subtilis were expressed and purified as reported.14, 28 OYE 1–3 from Saccharomyces sp. and NCR reductase from Z. mobilis were provided by BASF (Ludwigshafen).15 PETNr from Enterobacter cloacae, NemA from E. coli and MorR from Pseudomonas putida M10 were provided by N.C. Bruce (Department of Biology, University of York, York, UK).29
4.6. Cloning of estrogen-binding protein EBP1
Two synthetic genes encoding the Estrogen-Binding Protein EBP1 from Candida albicans,3b the original sequence and one with codon optimization for expression in E. coli, were sub-cloned into the Multiple Cloning Site of a pDHE-1650 vector (BASF vector). The pDHE-1650 vector as well as the synthetic genes were digested with NdeI and HindIII (New England Biolabs) in the corresponding buffer 2 (NEB) for 16 h at 37 °C. After purification of the digested fragments from an agarose gel (GFX PCR, DNA, gel-band purification kit, GE Healthcare), the fragments were ligated with T4 DNA ligase, provided by Roche. The resulting plasmids were transformed in E. coli TG1.
4.7. Sequences of the synthetic genes
4.7.1. Original gene
atgactattgaatcaactaattcatttgttgtcccatcagatactaaattaattgatgttactccattaggttcaacaaaattatttcaaccaattaaagtcggtaacaatgttttacctcaacgtattgcttatgtcccaaccaccagatttagagcttctaaagatcatattccaagtgatttacaattaaattattataatgctcgttctcaatatccaggtacattgattattactgaagcaacatttgcatctgaaagaggtggtattgatttacatgttccaggtatttataatgacgctcaagctaaaagttggaagaaaatcaatgaagcaattcatggcaatggaagtttcagttcagttcaattatggtatttaggtagagttgctaatgctaaagatttgaaagattctggattacctcttattgcgccatcagcagtttattgggatgagaatagtgaaaaattggccaaagaagctggaaatgaattgagagcattaactgaagaagaaattgatcatattgttgaagttgaatatcctaatgctgctaaacatgcacttgaagcaggatttgattatgttgaaatccatggtgctcatggttacttgttggatcagtttttaaatcttgcctctaataaaagaaccgataaatatggttgtggtagtattgaaaatcgtgcacgattattattaagagtggttgataaattaattgaagttgttggtgctaatagattggcattacgtttatcaccatgggctagtttccaaggtatggaaattgaaggtgaagaaatccattcatatattttacaacaattacaacaacgtgctgataatggtcaacaattggcttatatttctcttgttgaacctcgtgttactggtatttatgatgtttctttaaaagatcaacaaggtcgtagtaatgaatttgcttataagatttggaaaggaaattttattcgtgctggtaattatacttatgatgctccagaatttaaaactttgattaatgatttaaagaatgatcgtagtattattggattttctagatttttcacttcaaatcctgatttagtggaaaaattgaaattgggtaaaccattgaattattataatcgtgaagaattttataagtactacaactatggttataattcttatgatgaatcagaaaagcaagtcattggtaaaccattggcatagaagctt.
4.7.2. Synthetic gene
atgaccattgaaagcaccaacagctttgtggtgccgagcgataccaaactgattgatgtgaccccgctgggcagcaccaaactgtttcagccgattaaagtgggcaacaacgtgctgccgcagcgcattgcgtatgtgccgaccacccgctttcgcgcgagcaaagatcatattccgagcgatctgcagctgaactattataacgcgcgcagccagtatccgggcaccctgattattaccgaagcgacctttgcgagcgaacgcggcggcattgatctgcatgtgccgggcatttataacgatgcgcaggcgaaaagctggaaaaaaattaacgaagcgattcatggcaacggcagctttagcagcgtgcagctgtggtatctgggccgcgtggcgaacgcgaaagatctgaaagatagcggcctgccgctgattgcgccgagcgcggtgtattgggatgaaaacagcgaaaaactggcgaaagaagcgggcaacgaactgcgcgcgctgaccgaagaagaaattgatcatattgtggaagtggaatatccgaacgcggcgaaacatgcgctggaagcgggctttgattatgtggaaattcatggcgcgcatggctatctgctggatcagtttctgaacctggcgagcaacaaacgcaccgataaatatggctgcggcagcattgaaaaccgcgcgcgcctgctgctgcgcgtggtggataaactgattgaagtggtgggcgcgaaccgcctggcgctgcgcctgagcccgtgggcgagctttcagggcatggaaattgaaggcgaagaaattcatagctatattctgcagcagctgcagcagcgcgcggataacggccagcagctggcgtatattagcctggtggaaccgcgcgtgaccggcatttatgatgtgagcctgaaagatcagcagggccgcagcaacgaatttgcgtataaaatttggaaaggcaactttattcgcgcgggcaactatacctatgatgcgccggaatttaaaaccctgattaacgatctgaaaaacgatcgcagcattattggctttagccgcttttttaccagcaacccggatctggtggaaaaactgaaactgggcaaaccgctgaactattataaccgcgaagaattttataaatattataactatggctataacagctatgatgaaagcgaaaaacaggtgattggcaaaccgctggcgtagctaagctt.
4.8. Expression of estrogen-binding protein EBP1
The enzymes were expressed in E. coli TG1 (Stratagene). LB medium (2 ml) with antibiotic supplement (100 μg/μL ampicillin) were inoculated with a single colony of E. coli TG1 pDHE-EBP1 and incubated with shaking (220 rpm) for 5 h at 37 °C. This culture was diluted 1:100 with fresh LB medium (100 μg/μL, 2 g/L rhamnose, the pDHE vector has a rhamnose-inducible promoter) and incubated with shaking (220 rpm) for 16 h at 37 °C. Then the cells were harvested by centrifugation (5 min, 10,000 rpm) and washed with 50 mM Tris–HCl pH 7.5. The cell pellet was used for further experiments.
4.9. General procedure for the bioreduction of activated alkenes 3, 4, and 6 using 3-methyl-2-cyclohexen-1-one (2) or 1,4-cyclohexanedione (5) as H-donor
An aliquot of isolated enzyme OYE1, OYE2, and YqjM (protein purity >90%, protein content 90–110 μg/mL) was added to a Tris–HCl buffer solution (0.8 mL, 50 mM, pH 7.5) containing substrate (3, 4, or 6, 10 mM) and 3-methyl-2-cyclohexen-1-one (2) or 1,4,-cyclohexanedione (5) (10 mM). The mixture was shaken at 30 °C and 120 rpm for 24 h and the products were extracted with EtOAc (2×0.5 mL). The combined organic phases were dried (Na2SO4) and the resulting samples were analyzed on achiral GC–MS and achiral GC. Column: 14% cyanopropyl-phenyl phase capillary column, J&W Scientific DB-1701, 30 m, 0.25 mm, 0.25 μm, detector temperature 250 °C, split ratio 30:1. Temperature program: 110 °C, hold 5 min, rise to 200 °C with 10 °C/min, hold 4 min. TRet: 3-Methylcyclohexenone (2) 6.27 min; 3-methylcyclohexanone (2a) 3.63 min; citraconic acid dimethylester (3) 6.91 min; dimethyl-2-methylsuccinate (3a) 5.89 min; 4-ketoisophorone (4) 8.22 min; levodion (4a) 9.26 min; 1,4,-cyclohexanedione (5) 8.38 min; hydroquinone (5a) 13.51 min; N-phenyl-2-methylmaleimide (6) 15.84 min; N-phenyl-2-methylsuccinimide (6a) 14.12 min;
4.10. Determination of enantiomeric excess and absolute configuration
The enantiomeric excess of 1a and 3a was determined using a modified β-cyclodextrin capillary column (Chiraldex B-TA, 40 m, 0.25 mm). Detector temperature 200 °C, injector temperature 180 °C, split ratio 25:1. Temperature program for 1a: 80 °C hold 2 min, 5 °C/min to 105 °C, 10 °C/min, hold 4 min. Retention times: (R)-1a 6.34 and (S)-1a 6.47 min. Temperature program for 3a: 90 °C hold 4 min, 3 °C/min to 115 °C, 30 °C/min to 180 °C. Retention times: (S)-3b 7.33 min; (R)-3b 7.45 min; The enantiomeric excess of 4a was determined using a β-cyclodextrin capillary column (CP-Chirasil-DEX CB, 25 m, 0.32 mm, 0.25 μm film). Temperature program for 4a: 90 °C hold 2 min, 4 °C/min to 115 °C, 20 °C/min to 180 °C, hold 2 min. Retention times: (R)-4a 6.42; (S)-4a 6.74 min. The enantiomeric excess of 8b was determined on HPLC using n-heptane/i-PrOH 95:5 (isocratic) at 18 °C and 1 mL/min. Retention times: (R)-8b 25.10 min; (S)-8b 29.15 min.15
Acknowledgements
This work was partially financed by the Fonds zur Förderung der wissenschaftlichen Fortschung (FWF, Vienna, project no. 18689); N.C. Bruce and H. Housden (University of York) are cordially thanked for providing samples of N-ethylmaleimide-, morphinone- and PETN-reductase.
References and notes
- 1.Banks R.L. J. Mol. Catal. 1980;8:269–276. Notable exceptions are the Cannizzaro reaction (2R–CHO→R–CH2OH+R–CO2H), Tishchenko reaction (2R–CHO→R–CO2R), Kornblum–DeLaMare rearrangement (R′2CH–O–O–CR′3→R′–CO–R′+R'3C–OH), Meerwein–Ponndorf–Verley/Oppenauer reduction/oxidation (R′R′CO+2-PrOH↔R′R′CH–OH+acetone), Boudouard reaction (2CO→CO2+C) and the catalytic disproportionation of toluene (2MePh→benzene+xylene), see: [Google Scholar]; Abdal Kareem M.A., Chand S., Mishra I.M. J. Sci. Ind. Res. 2001;60:319–327. [Google Scholar]
- 2.Finster K. J. Sulfur Chem. 2008;29:281–292. [Google Scholar]; Grinbergs A. Latvian Chem. J. 2003;3:292. [Google Scholar]; Chem. Abstr. 2004;140:419639. It should be noted that in the biochemical literature, ‘disproportionation’ is often denoted as ‘dismutation’. [Google Scholar]
- 3.(a) Vaz A.D.N., Chakraborty S., Massey V. Biochemistry. 1995;34:4246–4256. doi: 10.1021/bi00013a014. [DOI] [PubMed] [Google Scholar]; (b) Buckman J., Miller S.M. Biochemistry. 1998;37:14326–14336. doi: 10.1021/bi981106y. [DOI] [PubMed] [Google Scholar]; (c) Karplus P.A., Fox K.M., Massey V. FASEB J. 1995;9:1518–1526. doi: 10.1096/fasebj.9.15.8529830. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien P.J., Herschlag D. Chem. Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]; Kazlauskas R.J. Curr. Opin. Chem. Biol. 2005;9:195–201. doi: 10.1016/j.cbpa.2005.02.008. [DOI] [PubMed] [Google Scholar]; Bornscheuer U.T., Kazlauskas R.J. Angew. Chem., Int. Ed. 2004;43:6032–6040. doi: 10.1002/anie.200460416. [DOI] [PubMed] [Google Scholar]; Hult K., Berglund P. Trends Biotechnol. 2007;25:231–238. doi: 10.1016/j.tibtech.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 5.(a) Walsh C. Acc. Chem. Res. 1980;13:148–155. [Google Scholar]; Kohli R.M., Massey V. J. Biol. Chem. 1998;273:32763–32770. doi: 10.1074/jbc.273.49.32763. [DOI] [PubMed] [Google Scholar]; (b) Williams R.E., Bruce N.C. Microbiology. 2002;148:1607–1614. doi: 10.1099/00221287-148-6-1607. [DOI] [PubMed] [Google Scholar]
- 6.Stuermer R., Hauer B., Hall M., Faber K. Curr. Opin. Chem. Biol. 2007;11:201–213. doi: 10.1016/j.cbpa.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto H., Matsuyama A. In: Biocatalysis in the Pharmaceutical and Biotechnology Industry. Patel R.N., editor. CRC; Boca Raton, FL: 2007. pp. 623–644. [Google Scholar]; Wandrey C. Chem. Rec. 2004;4:254–265. doi: 10.1002/tcr.20016. [DOI] [PubMed] [Google Scholar]; Kragl U., Vasic-Racki D., Wandrey C. Indian J. Chem., Sect. B. 1993;32B:103–117. [Google Scholar]
- 8.Vrtis J.M., White A.K., Metcalf W.W., van der Donk W.A. Angew. Chem., Int. Ed. 2002;41:3391–3393. doi: 10.1002/1521-3773(20020902)41:17<3257::AID-ANIE3257>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]; Johannes T.W., Woodyer R.D., Zhao H. Biotechnol. Bioeng. 2006;96:18–26. doi: 10.1002/bit.21168. [DOI] [PubMed] [Google Scholar]; Torres Pazmino D.E., Snajdrova R., Baas B.-J., Ghobrial M., Mihovilovic M.D., Fraaije M.W. Angew. Chem., Int. Ed. 2008;47:2275–2278. doi: 10.1002/anie.200704630. [DOI] [PubMed] [Google Scholar]
- 9.Faber K. 5th ed. Springer; Heidelberg: 2004. Biotransformations in Organic Chemistry. pp 178–182. [Google Scholar]
- 10.During the bioreduction of conjugated enones and enals using enoate reductases, carbonyl reduction and substrate-racemisation have frequently been observed as side reactions.; (a) Hall M., Stueckler C., Kroutil W., Macheroux P., Faber K. Angew. Chem., Int. Ed. 2007;46:3934–3937. doi: 10.1002/anie.200605168. [DOI] [PubMed] [Google Scholar]; (b) Hall M., Stueckler C., Ehammer H., Pointner E., Oberdorfer G., Gruber K., Hauer B., Stuermer R., Macheroux P., Kroutil W., Faber K. Adv. Synth. Catal. 2008;350:411–418. [Google Scholar]
- 11.Massey V., Stankovich M., Hemmerich P. Biochemistry. 1978;17:1–8. doi: 10.1021/bi00594a001. For light-driven cofactor-regeneration see: [DOI] [PubMed] [Google Scholar]; Hollmann F., Taglieber A., Schulz F., Reetz M.T. Angew. Chem., Int. Ed. 2007;46:2903–2906. doi: 10.1002/anie.200605169. [DOI] [PubMed] [Google Scholar]; Taglieber A., Schulz F., Hollmann F., Rusek M., Reetz M.T. ChemBioChem. 2008;9:565–572. doi: 10.1002/cbic.200700435. [DOI] [PubMed] [Google Scholar]; Hollmann F., Hofstetter K., Habicher T., Hauer B., Schmid A. J. Am. Chem. Soc. 2005;127:6540–6541. doi: 10.1021/ja050997b. for electrochemical methods see: [DOI] [PubMed] [Google Scholar]; Ruinatscha R., Höllrigl V., Otto K., Schmid A. Adv. Synth. Catal. 2006;348:2015–2026. [Google Scholar]; Hollmann F., Schmid A. Biocatal. Biotransform. 2004;22:63–88. [Google Scholar]; Hollmann F., Hofstetter K., Schmid A. Trends Biotechnol. 2006;24:163–171. doi: 10.1016/j.tibtech.2006.02.003. [DOI] [PubMed] [Google Scholar]; de Gonzalo G., Ottolina G., Carrea G., Fraaije M.W. Chem. Commun. 2005:3724–3726. doi: 10.1039/b504921k. [DOI] [PubMed] [Google Scholar]
- 12.Despite the impressive advances in nicotinamide-cofactor recycling using specific dehydrogenases, such as FDH, GDH, and phosphite-DH, the use of a single dehydrogenase for concomitant substrate reduction and NADH-recycling via oxidation of 2-propanol is the preferred technique in the asymmetric bioreduction of ketones catalyzed by ADHs on industrial scale, see Ref. 7.
- 13.For Old Yellow Enzyme isoenzymes 1–3 see Ref. 3a; for the OYE homolog YqjM from Bacillus subtilis see:; Fitzpatrick T.B., Amrhein N., Macheroux P. J. Biol. Chem. 2003;278:19891–19897. doi: 10.1074/jbc.M211778200. [DOI] [PubMed] [Google Scholar]; Straßer J., Fürholz A., Macheroux P., Amrhein N., Schaller A. J. Biol. Chem. 1999;274:35067–35073. doi: 10.1074/jbc.274.49.35067. for 12-oxophytodienoic acid reductase isoenzyme OPR1 see: [DOI] [PubMed] [Google Scholar]; Messiha H.L., Bruce N.C., Sattelle B.M., Sutcliffe M.J., Munro A.W., Scrutton N.S. J. Biol. Chem. 2005;280:27103–27110. doi: 10.1074/jbc.M502293200. for nicotinamide-dependent cyclohexenone reductase (NCR) see Ref. 19; for morphinone reductase (MorR) see: [DOI] [PubMed] [Google Scholar]; Khan H., Harris R.J., Barna T., Craig D.H., Bruce N.C., Munro A.W., Moody P.C.E., Scrutton N.S. J. Biol. Chem. 2002;277:21906–21912. doi: 10.1074/jbc.M200637200. for pentyerythritol tetranitrate reductase (PETNR) see: for estrogen-binding protein EBP1 see Ref. 3b. No data are available for NemA and OPR3. [DOI] [PubMed] [Google Scholar]
- 14.Kitzing K., Fitzpatrick T.B., Wilken C., Sawa J., Bourenkov G.P., Macheroux P., Clausen T. J. Biol. Chem. 2005;280:27904–27913. doi: 10.1074/jbc.M502587200. [DOI] [PubMed] [Google Scholar]
- 15.Hall M., Stueckler C., Hauer B., Stuermer R., Friedrich T., Breuer M., Kroutil W., Faber K. Eur. J. Org. Chem. 2008:1511–1516. [Google Scholar]
- 16.Breithaupt C., Strassner J., Breitinger U., Huber R., Macheroux P., Schaller A., Clausen T. Structure. 2001;9:419–429. doi: 10.1016/s0969-2126(01)00602-5. [DOI] [PubMed] [Google Scholar]
- 17.Barna T., Messiha H.L., Petosa C., Bruce N.C., Scrutton N.S., Moody P.C.E. J. Biol. Chem. 2002;277:30976–30983. doi: 10.1074/jbc.M202846200. [DOI] [PubMed] [Google Scholar]; Messiha H.L., Munroe A.W., Bruce N.C., Barsukov I., Scrutton N.S. J. Biol. Chem. 2005;280:10695–10709. doi: 10.1074/jbc.M410595200. [DOI] [PubMed] [Google Scholar]
- 18.Williams R.E., Rathbone D.A., Scrutton N.S., Bruce N.C. Appl. Environ. Microbiol. 2004;70:3566–3574. doi: 10.1128/AEM.70.6.3566-3574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller A., Hauer B., Rosche B. Biotechnol. Bioeng. 2007;98:22–29. doi: 10.1002/bit.21415. [DOI] [PubMed] [Google Scholar]
- 20.Hirano J., Miyamoto K., Ohta H. Appl. Microbiol. Biotechnol. 2008;80:71–78. doi: 10.1007/s00253-008-1535-x. The slight hydrogen-imbalance is presumed to derive from oxidation of reduced flavin by O2 (producing H2O2); see: [DOI] [PubMed] [Google Scholar]; Riebel B.R., Gibbs P.R., Wellborn W.B., Bommarius A.S. Adv. Synth. Catal. 2002;344:1156–1168. [Google Scholar]
- 21.Mueller N.J., Stueckler C., Hall M., Macheroux P., Faber K. Org. Biomol. Chem. 2009;7:1115–1119. doi: 10.1039/b819057g. [DOI] [PubMed] [Google Scholar]
- 22.The (co)-substrate concentrations used in this study were typically 10 mM. From related studies it can be deduced that increasing of the (co)-substrate concentrations to ca. 100 mM is feasible.
- 23.Abramovitz A.S., Massey V. J. Biol. Chem. 1976;251:5327–5336. [PubMed] [Google Scholar]
- 24.Stark D., von Stockar U. Adv. Biochem. Eng. Biotechnol. 2003;80:149–175. doi: 10.1007/3-540-36782-9_5. [DOI] [PubMed] [Google Scholar]; Lye G.J., Woodley J.M. Trends Biotechnol. 1999;17:395–402. doi: 10.1016/s0167-7799(99)01351-7. [DOI] [PubMed] [Google Scholar]; Etschmann M.M.W., Sell D., Schrader J. Biotechnol. Bioeng. 2005;92:624–634. doi: 10.1002/bit.20655. [DOI] [PubMed] [Google Scholar]
- 25.Among a series of water-immiscible organic solvents, methyl tert-butyl ether has been shown to be an ideal co-solvent for OYE-type enoate reductases; Stueckler, C.; Mueller, N. J.; Winkler, C. K.; Glueck, S. M.; Faber, K., in preparation.
- 26.Alphand V., Carrea G., Wohlgemuth R., Furstoss R., Woodley J.M. Trends Biotechnol. 2003;21:318–323. doi: 10.1016/S0167-7799(03)00144-6. [DOI] [PubMed] [Google Scholar]; Hilker I., Baldwin C., Alphand V., Furstoss R., Woodley J., Wohlgemuth R. Biotechnol. Bioeng. 2006;93:1138–1144. doi: 10.1002/bit.20829. [DOI] [PubMed] [Google Scholar]; Hilker I., Wohlgemuth R., Alphand V., Furstoss R. Biotechnol. Bioeng. 2005;92:702–710. doi: 10.1002/bit.20636. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi O., Umezawa J., Furuhashi K., Takagi M. Tetrahedron Lett. 1989;30:1583–1584. [Google Scholar]; Furuhashi K. In: Chirality in Industry. Collins A.N., Sheldrake G.N., Crosby J., editors. Wiley; New York, NY: 1992. pp. 167–186. [Google Scholar]; White R.F., Birnbaum J., Meyer R.T., ten Broeke J., Chemerda J.M., Demain A.L. Appl. Microbiol. 1971;22:55–60. doi: 10.1128/am.22.1.55-60.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breithaupt C., Kurzbauer R., Lilie H., Schaller A., Strassner J., Huber R., Macheroux P., Clausen T. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14337–14342. doi: 10.1073/pnas.0606603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.French C.E., Nicklin S., Bruce N.C. J. Bacteriol. 1996;178:6623–6627. doi: 10.1128/jb.178.22.6623-6627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miura K., Tomioka Y., Suzuki H., Yonezawa M., Hishinuma T., Mizugaki M. Biol. Pharm. Bull. 1997;20:110–112. doi: 10.1248/bpb.20.110. [DOI] [PubMed] [Google Scholar]; French C.E., Bruce N.C. Biochem. J. 1994;301:97–103. doi: 10.1042/bj3010097. [DOI] [PMC free article] [PubMed] [Google Scholar]