Abstract
The crystal structure of the title complex, [Yb2(C8H4O4)2(C2O4)(H2O)2]n, features an extended three-dimensional framework made up of Yb3+ ions coordinated by terephthalate ligands, oxalate ligands and water molecules. The Yb3+ ion has a distorted square-antiprismatic coordination formed by one aqua ligand, two O atoms from an oxalate ligand and five O atoms belonging to four terephthalate anions. Two symmetry-independent terephthalate anions, as well as the oxalate anion, occupy special positions on inversion centers. The water molecule participates in O—H⋯O hydrogen bonding with both terephthalate anions.
Related literature
For isotypic structures, derivatives of Lu and Dy, see: Li & Wang (2009 ▶) and Li et al. (2009 ▶), respectively.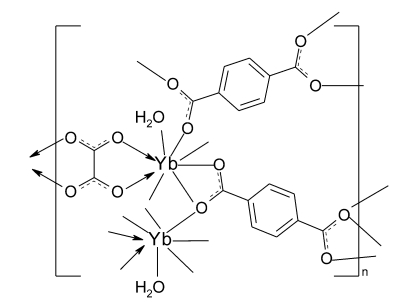
Experimental
Crystal data
[Yb2(C8H4O4)2(C2O4)(H2O)2]
M r = 798.36
Triclinic,
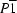
a = 7.034 (2) Å
b = 7.583 (2) Å
c = 10.213 (3) Å
α = 75.372 (4)°
β = 70.851 (4)°
γ = 88.126 (4)°
V = 497.2 (2) Å3
Z = 1
Mo Kα radiation
μ = 9.43 mm−1
T = 295 K
0.24 × 0.15 × 0.05 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2003 ▶) T min = 0.211, T max = 0.629
2630 measured reflections
1882 independent reflections
1785 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.120
S = 1.06
1882 reflections
154 parameters
H-atom parameters constrained
Δρmax = 4.45 e Å−3
Δρmin = −4.21 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810033052/ya2126sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810033052/ya2126Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Yb—O1 | 2.790 (6) |
| Yb—O1i | 2.315 (6) |
| Yb—O2 | 2.314 (6) |
| Yb—O3 | 2.264 (6) |
| Yb—O4ii | 2.209 (6) |
| Yb—O5 | 2.310 (6) |
| Yb—O6iii | 2.321 (6) |
| Yb—O7 | 2.293 (5) |
Symmetry codes: (i) 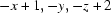 ; (ii)
; (ii) 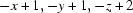 ; (iii)
; (iii) 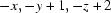 .
.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7—H7A⋯O3i | 0.85 | 1.92 | 2.751 (6) | 167 |
| O7—H7B⋯O2iv | 0.85 | 1.91 | 2.754 (6) | 178 |
Symmetry codes: (i) 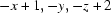 ; (iv)
; (iv) 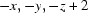 .
.
Acknowledgments
This work was supported by the Jiangxi Provincial Educational Foundation (GJJ09227).
supplementary crystallographic information
Comment
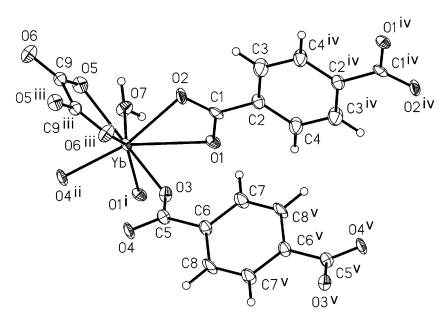
The title compound is isostructural with its Lu and Dy analogues (Li & Wang, 2009; Li et al., 2009). As illustrated in Fig. 1, the Yb atoms is coordinated by two oxalate O atoms and five O atoms of four terephthalate anions; an aqua ligand completes a distorted square antiprismatic geometry. The Yb-O distances are in the range of 2.208 (6)–2.790 (6) Å, with an average Yb-O bond of 2.352 Å; these parameters are similar to M-O bonds found in the above mentioned previously reported isostructural complexes.
In the title complex, both symmetry independent terephthalate (tp) anions occupy special positions on the inversion centers; nevertheless they exhibit different modes of coordination of Yb atoms. The tp1 anion (O1 to O2, C1 to C4) functions as chelating-bridging tridentate ligand: two carboxylate oxygen atoms (O1 and O2) chelate one Yb atom; the O1 atom is additionally bonded to another Yb atom; the Yb···Yb separation is 4.232 (1) Å. Two edge-sharing [YbO8] polyhedra are bridged by the bidentate tp2 (O3 to O4, C5 to C8) ligands thus generating chains along the [010] direction. The chains are further linked by the tp1 and tp2 ligands into three-dimensional framework. The oxalate anion is also located on the inversion center and acts as a tetradentate ligand connecting the edge-sharing [YbO8] polyhedra along the [100] direction thus even further stabilizing the three-dimensional framework. The aqua ligand provides H-bond donors which participate in H-bonds with terephthalate oxygen atoms O2 and O3.
Experimental
A mixture of YbCl3.6H2O (1.00 mmol, 0.39 g), oxalic acid (0.50 mmol, 0.05 g), terephthalic acid (0.50 mmol, 0.09 g), NaOH (2.00 mmol, 0.08 g) and H2O (10.0 ml) was heated in a 23 ml stainless steel reactor with a Teflon liner at 453 K for 72 h. A small amount of colorless plate-like crystals were filtered and washed with water and acetone.
Refinement
H atoms attached to C atoms were included at calculated positions and treated as riding atoms [C–H = 0.93 Å and Uiso(H) = 1.2Ueq(C)]. Water H atoms were located in difference Fourier maps placed at the idealized positions with O-H distance of 0.85 Å and included in the final refinement as fixed contribution with Uiso(H) = 1.5Ueq(O). The highest density peak and deepest hole are located at 0.87 Å and 1.09 Å from the Yb atom respectively.
Figures
Fig. 1.
The fragment of the structure of the title compound, with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level and H atoms are shown as small circles of arbitrary radii. Symmetry codes: (i) 1 -x, -y, 2 - z; (ii) 1 - x, 1 - y, 2 - z; (iii) -x, 1 - y, 2 - z; (iv) 1 - x, -y, 1 - z; (v) 2 - x, 1 - y, 1 - z.
Crystal data
| [Yb2(C8H4O4)2(C2O4)(H2O)2] | Z = 1 |
| Mr = 798.36 | F(000) = 372 |
| Triclinic, P1 | Dx = 2.666 Mg m−3 |
| Hall symbol: -p 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.034 (2) Å | Cell parameters from 196 reflections |
| b = 7.583 (2) Å | θ = 2.1–27.3° |
| c = 10.213 (3) Å | µ = 9.43 mm−1 |
| α = 75.372 (4)° | T = 295 K |
| β = 70.851 (4)° | Plate, colorless |
| γ = 88.126 (4)° | 0.24 × 0.15 × 0.05 mm |
| V = 497.2 (2) Å3 |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 1882 independent reflections |
| Radiation source: fine-focus sealed tube | 1785 reflections with I > 2σ(I) |
| graphite | Rint = 0.025 |
| φ and ω scans | θmax = 26.0°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2003) | h = −8→8 |
| Tmin = 0.211, Tmax = 0.629 | k = −8→9 |
| 2630 measured reflections | l = −7→12 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.120 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0995P)2] where P = (Fo2 + 2Fc2)/3 |
| 1882 reflections | (Δ/σ)max < 0.001 |
| 154 parameters | Δρmax = 4.45 e Å−3 |
| 0 restraints | Δρmin = −4.21 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Yb | 0.31071 (4) | 0.22186 (3) | 1.01498 (3) | 0.01504 (19) | |
| C1 | 0.3854 (12) | 0.0294 (11) | 0.7912 (9) | 0.0203 (16) | |
| C2 | 0.4409 (12) | 0.0093 (11) | 0.6438 (9) | 0.0202 (16) | |
| C3 | 0.3229 (16) | 0.0805 (15) | 0.5592 (11) | 0.033 (2) | |
| H3 | 0.2048 | 0.1355 | 0.5976 | 0.039* | |
| C4 | 0.6198 (14) | −0.0700 (13) | 0.5823 (10) | 0.030 (2) | |
| H4 | 0.7015 | −0.1163 | 0.6370 | 0.036* | |
| C5 | 0.7144 (12) | 0.4806 (10) | 0.7821 (9) | 0.0189 (16) | |
| C6 | 0.8607 (13) | 0.4930 (11) | 0.6352 (10) | 0.0211 (18) | |
| C7 | 0.8292 (13) | 0.3852 (12) | 0.5521 (10) | 0.0282 (19) | |
| H7 | 0.7149 | 0.3066 | 0.5875 | 0.034* | |
| C8 | 1.0353 (13) | 0.6070 (12) | 0.5820 (10) | 0.0275 (19) | |
| H8 | 1.0602 | 0.6780 | 0.6374 | 0.033* | |
| C9 | −0.0833 (11) | 0.4309 (10) | 1.0574 (9) | 0.0194 (16) | |
| O1 | 0.4818 (8) | −0.0459 (8) | 0.8752 (6) | 0.0228 (12) | |
| O2 | 0.2462 (9) | 0.1353 (8) | 0.8314 (6) | 0.0228 (12) | |
| O3 | 0.6140 (8) | 0.3311 (8) | 0.8501 (6) | 0.0224 (12) | |
| O4 | 0.6994 (9) | 0.6187 (8) | 0.8296 (7) | 0.0248 (13) | |
| O5 | −0.0295 (9) | 0.2697 (8) | 1.0944 (7) | 0.0246 (12) | |
| O6 | −0.2511 (8) | 0.4911 (8) | 1.1074 (7) | 0.0254 (13) | |
| O7 | 0.1351 (8) | −0.0452 (7) | 1.1590 (7) | 0.0242 (13) | |
| H7B | 0.0166 | −0.0740 | 1.1644 | 0.036* | |
| H7A | 0.1979 | −0.1429 | 1.1672 | 0.036* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Yb | 0.0191 (3) | 0.0156 (3) | 0.0129 (3) | 0.00119 (15) | −0.00688 (17) | −0.00586 (17) |
| C1 | 0.024 (4) | 0.017 (4) | 0.023 (5) | −0.008 (3) | −0.009 (3) | −0.007 (3) |
| C2 | 0.026 (4) | 0.020 (4) | 0.016 (4) | 0.003 (3) | −0.008 (3) | −0.004 (3) |
| C3 | 0.036 (5) | 0.043 (6) | 0.023 (5) | 0.015 (4) | −0.014 (4) | −0.012 (4) |
| C4 | 0.032 (4) | 0.039 (5) | 0.025 (5) | 0.015 (4) | −0.015 (4) | −0.015 (4) |
| C5 | 0.026 (4) | 0.017 (4) | 0.013 (4) | 0.001 (3) | −0.007 (3) | −0.002 (3) |
| C6 | 0.026 (4) | 0.017 (4) | 0.020 (5) | −0.001 (3) | −0.006 (4) | −0.007 (3) |
| C7 | 0.030 (4) | 0.028 (4) | 0.024 (5) | −0.015 (3) | 0.000 (4) | −0.010 (4) |
| C8 | 0.034 (4) | 0.024 (4) | 0.028 (5) | −0.014 (3) | −0.007 (4) | −0.014 (4) |
| C9 | 0.022 (4) | 0.017 (3) | 0.021 (4) | 0.004 (3) | −0.009 (3) | −0.006 (3) |
| O1 | 0.025 (3) | 0.031 (3) | 0.018 (3) | 0.002 (2) | −0.014 (2) | −0.008 (2) |
| O2 | 0.029 (3) | 0.028 (3) | 0.015 (3) | 0.007 (2) | −0.008 (3) | −0.013 (2) |
| O3 | 0.024 (3) | 0.022 (3) | 0.020 (3) | 0.002 (2) | −0.004 (2) | −0.007 (2) |
| O4 | 0.032 (3) | 0.026 (3) | 0.019 (3) | −0.003 (2) | −0.005 (3) | −0.016 (3) |
| O5 | 0.030 (3) | 0.020 (3) | 0.023 (3) | 0.004 (2) | −0.011 (3) | −0.003 (2) |
| O6 | 0.023 (3) | 0.026 (3) | 0.019 (3) | 0.007 (2) | −0.004 (2) | 0.003 (2) |
| O7 | 0.020 (3) | 0.017 (3) | 0.029 (3) | −0.001 (2) | −0.004 (3) | −0.001 (2) |
Geometric parameters (Å, °)
| Yb—O1 | 2.790 (6) | C3—H3 | 0.9300 |
| Yb—O1i | 2.315 (6) | C4—H4 | 0.9300 |
| Yb—O2 | 2.314 (6) | C5—O4 | 1.249 (10) |
| Yb—O3 | 2.264 (6) | C5—O3 | 1.264 (10) |
| Yb—O4ii | 2.209 (6) | C5—C6 | 1.497 (12) |
| Yb—O5 | 2.310 (6) | C6—C7 | 1.387 (13) |
| Yb—O6iii | 2.321 (6) | C6—C8 | 1.395 (11) |
| Yb—O7 | 2.293 (5) | C7—C8v | 1.377 (12) |
| Yb—Ybi | 4.2315 (10) | C7—H7 | 0.9300 |
| C1—O2 | 1.274 (10) | C8—H8 | 0.9300 |
| C1—O1 | 1.277 (10) | C9—O6 | 1.245 (10) |
| C1—C2 | 1.473 (12) | C9—O5 | 1.269 (9) |
| C2—C3 | 1.390 (13) | C9—C9iii | 1.550 (15) |
| C2—C4 | 1.401 (12) | O7—H7B | 0.85 |
| C3—C4iv | 1.390 (13) | O7—H7A | 0.85 |
| O4ii—Yb—O3 | 98.9 (2) | O1—Yb—Ybi | 30.57 (12) |
| O4ii—Yb—O7 | 102.6 (2) | C9iii—Yb—Ybi | 163.60 (15) |
| O3—Yb—O7 | 141.9 (2) | O2—C1—O1 | 119.6 (8) |
| O4ii—Yb—O5 | 79.5 (2) | O2—C1—C2 | 118.4 (8) |
| O3—Yb—O5 | 145.4 (2) | O1—C1—C2 | 121.8 (7) |
| O7—Yb—O5 | 70.19 (19) | C3—C2—C4 | 118.5 (8) |
| O4ii—Yb—O2 | 159.4 (3) | C3—C2—C1 | 120.3 (8) |
| O3—Yb—O2 | 85.5 (2) | C4—C2—C1 | 121.1 (8) |
| O7—Yb—O2 | 85.2 (2) | C4iv—C3—C2 | 120.5 (9) |
| O5—Yb—O2 | 85.5 (2) | C4iv—C3—H3 | 119.8 |
| O4ii—Yb—O1i | 82.5 (2) | C2—C3—H3 | 119.8 |
| O3—Yb—O1i | 80.7 (2) | C3iv—C4—C2 | 121.0 (8) |
| O7—Yb—O1i | 71.5 (2) | C3iv—C4—H4 | 119.5 |
| O5—Yb—O1i | 132.4 (2) | C2—C4—H4 | 119.5 |
| O2—Yb—O1i | 118.1 (2) | O4—C5—O3 | 124.2 (7) |
| O4ii—Yb—O6iii | 79.2 (2) | O4—C5—C6 | 117.8 (7) |
| O3—Yb—O6iii | 75.4 (2) | O3—C5—C6 | 118.0 (7) |
| O7—Yb—O6iii | 139.4 (2) | C7—C6—C8 | 118.8 (8) |
| O5—Yb—O6iii | 70.34 (19) | C7—C6—C5 | 120.4 (8) |
| O2—Yb—O6iii | 82.5 (2) | C8—C6—C5 | 120.7 (8) |
| O1i—Yb—O6iii | 147.1 (2) | C8v—C7—C6 | 121.0 (7) |
| O4ii—Yb—O1 | 150.1 (2) | C8v—C7—H7 | 119.5 |
| O3—Yb—O1 | 70.77 (19) | C6—C7—H7 | 119.5 |
| O7—Yb—O1 | 74.9 (2) | C7v—C8—C6 | 120.2 (9) |
| O5—Yb—O1 | 125.1 (2) | C7v—C8—H8 | 119.9 |
| O2—Yb—O1 | 50.14 (19) | C6—C8—H8 | 119.9 |
| O1i—Yb—O1 | 68.4 (2) | O6—C9—O5 | 127.2 (8) |
| O6iii—Yb—O1 | 122.4 (2) | O6—C9—C9iii | 117.0 (8) |
| O4ii—Yb—C9iii | 72.4 (2) | O5—C9—C9iii | 115.7 (8) |
| O3—Yb—C9iii | 96.2 (2) | O5—C9—Ybiii | 166.7 (6) |
| O7—Yb—C9iii | 120.2 (2) | C9iii—C9—Ybiii | 76.1 (5) |
| O5—Yb—C9iii | 50.05 (19) | C1—O1—Ybi | 165.8 (6) |
| O2—Yb—C9iii | 87.2 (2) | C1—O1—Yb | 82.4 (5) |
| O1i—Yb—C9iii | 154.0 (2) | Ybi—O1—Yb | 111.6 (2) |
| O6iii—Yb—C9iii | 21.0 (2) | C1—O2—Yb | 104.7 (5) |
| O1—Yb—C9iii | 135.19 (19) | C5—O3—Yb | 140.5 (5) |
| O4ii—Yb—Ybi | 120.04 (17) | C5—O4—Ybii | 157.0 (6) |
| O3—Yb—Ybi | 72.20 (14) | C9—O5—Yb | 117.5 (5) |
| O7—Yb—Ybi | 69.81 (14) | C9—O6—Ybiii | 117.0 (5) |
| O5—Yb—Ybi | 138.46 (14) | Yb—O7—H7B | 123.8 |
| O2—Yb—Ybi | 80.49 (15) | Yb—O7—H7A | 118.8 |
| O1i—Yb—Ybi | 37.81 (15) | H7B—O7—H7A | 107.3 |
| O6iii—Yb—Ybi | 144.33 (15) | ||
| O2—C1—C2—C3 | −10.1 (13) | C2—C1—O2—Yb | −155.3 (6) |
| O1—C1—C2—C3 | 174.7 (9) | O4ii—Yb—O2—C1 | 161.8 (6) |
| O2—C1—C2—C4 | 165.3 (8) | O3—Yb—O2—C1 | 58.4 (5) |
| O1—C1—C2—C4 | −9.8 (12) | O7—Yb—O2—C1 | −84.5 (5) |
| C4—C2—C3—C4iv | 1.1 (16) | O5—Yb—O2—C1 | −155.0 (5) |
| C1—C2—C3—C4iv | 176.7 (8) | O1i—Yb—O2—C1 | −18.5 (6) |
| C3—C2—C4—C3iv | −1.1 (16) | O6iii—Yb—O2—C1 | 134.3 (5) |
| C1—C2—C4—C3iv | −176.7 (8) | O1—Yb—O2—C1 | −10.2 (5) |
| O4—C5—C6—C7 | −152.4 (9) | C9iii—Yb—O2—C1 | 154.9 (5) |
| O3—C5—C6—C7 | 27.6 (12) | Ybi—Yb—O2—C1 | −14.2 (5) |
| O4—C5—C6—C8 | 30.4 (13) | O4—C5—O3—Yb | 30.8 (14) |
| O3—C5—C6—C8 | −149.6 (9) | C6—C5—O3—Yb | −149.2 (7) |
| C8—C6—C7—C8v | −1.5 (16) | O4ii—Yb—O3—C5 | −46.7 (9) |
| C5—C6—C7—C8v | −178.8 (8) | O7—Yb—O3—C5 | −170.7 (8) |
| C7—C6—C8—C7v | 1.5 (16) | O5—Yb—O3—C5 | 37.6 (11) |
| C5—C6—C8—C7v | 178.8 (8) | O2—Yb—O3—C5 | 113.0 (9) |
| O2—C1—O1—Ybi | 171.7 (17) | O1i—Yb—O3—C5 | −127.6 (9) |
| C2—C1—O1—Ybi | −13 (3) | O6iii—Yb—O3—C5 | 29.6 (9) |
| O2—C1—O1—Yb | −16.0 (7) | O1—Yb—O3—C5 | 162.2 (9) |
| C2—C1—O1—Yb | 159.1 (7) | C9iii—Yb—O3—C5 | 26.3 (9) |
| O4ii—Yb—O1—C1 | −164.4 (5) | Ybi—Yb—O3—C5 | −165.5 (9) |
| O3—Yb—O1—C1 | −90.6 (5) | O3—C5—O4—Ybii | 50.2 (19) |
| O7—Yb—O1—C1 | 106.3 (5) | C6—C5—O4—Ybii | −129.7 (12) |
| O5—Yb—O1—C1 | 54.5 (5) | O6—C9—O5—Yb | −166.9 (7) |
| O2—Yb—O1—C1 | 9.9 (4) | C9iii—C9—O5—Yb | 9.8 (12) |
| O1i—Yb—O1—C1 | −178.0 (6) | Ybiii—C9—O5—Yb | 160 (2) |
| O6iii—Yb—O1—C1 | −33.1 (5) | O4ii—Yb—O5—C9 | 69.9 (6) |
| C9iii—Yb—O1—C1 | −11.5 (6) | O3—Yb—O5—C9 | −20.5 (8) |
| Ybi—Yb—O1—C1 | −178.0 (6) | O7—Yb—O5—C9 | 177.6 (7) |
| O4ii—Yb—O1—Ybi | 13.6 (5) | O2—Yb—O5—C9 | −95.9 (6) |
| O3—Yb—O1—Ybi | 87.4 (3) | O1i—Yb—O5—C9 | 139.5 (6) |
| O7—Yb—O1—Ybi | −75.7 (3) | O6iii—Yb—O5—C9 | −12.3 (6) |
| O5—Yb—O1—Ybi | −127.5 (2) | O1—Yb—O5—C9 | −128.6 (6) |
| O2—Yb—O1—Ybi | −172.1 (4) | C9iii—Yb—O5—C9 | −5.8 (7) |
| O1i—Yb—O1—Ybi | 0.000 (2) | Ybi—Yb—O5—C9 | −166.1 (5) |
| O6iii—Yb—O1—Ybi | 144.9 (2) | O5—C9—O6—Ybiii | −169.3 (7) |
| C9iii—Yb—O1—Ybi | 166.5 (2) | C9iii—C9—O6—Ybiii | 14.1 (12) |
| O1—C1—O2—Yb | 19.9 (9) |
Symmetry codes: (i) −x+1, −y, −z+2; (ii) −x+1, −y+1, −z+2; (iii) −x, −y+1, −z+2; (iv) −x+1, −y, −z+1; (v) −x+2, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7A···O3i | 0.85 | 1.92 | 2.751 (6) | 167 |
| O7—H7B···O2vi | 0.85 | 1.91 | 2.754 (6) | 178 |
Symmetry codes: (i) −x+1, −y, −z+2; (vi) −x, −y, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: YA2126).
References
- Bruker (2004). APEX2 andSAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Li, Z.-F., Cheng, Q., Chen, J.-Z. & Chen, Y. (2009). Z. Kristallogr. New Cryst. Struct.224, 483–484.
- Li, Z.-F. & Wang, C.-X. (2009). Acta Cryst. E65, m1157. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2003). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810033052/ya2126sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810033052/ya2126Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report