Abstract
Summary: Despite the availability of effective treatment for several decades, leprosy remains an important medical problem in many regions of the world. Infection with Mycobacterium leprae can produce paucibacillary disease, characterized by well-formed granulomas and a Th1 T-cell response, or multibacillary disease, characterized by poorly organized cellular infiltrates and Th2 cytokines. These diametric immune responses confer states of relative resistance or susceptibility to leprosy, respectively, and have well-defined clinical manifestations. As a result, leprosy provides a unique opportunity to dissect the genetic basis of human in vivo immunity. A series of studies over the past 40 years suggests that host genes influence the risk of leprosy acquisition and the predilection for different clinical forms of the disease. However, a comprehensive, cellular, and molecular view of the genes and variants involved is still being assembled. In this article, we review several decades of human genetic studies of leprosy, including a number of recent investigations. We emphasize genetic analyses that are validated by the replication of the same phenotype in independent studies or supported by functional experiments demonstrating biological mechanisms of action for specific polymorphisms. Identifying and functionally exploring the genetic and immunological factors that underlie human susceptibility to leprosy have yielded important insights into M. leprae pathogenesis and are likely to advance our understanding of the immune response to other pathogenic mycobacteria. This knowledge may inform new treatment or vaccine strategies for leprosy or tuberculosis.
INTRODUCTION
Mycobacterium leprae is the etiological agent of human leprosy, an ancient affliction of humankind that has persisted into contemporary times despite the facts that it is not highly transmissible and that chemotherapy has been available for 60 years (215). M. leprae produces a broad spectrum of illness, and the host factors that regulate susceptibility to its diverse clinical forms are largely unknown. Studies of human genetic variation and its link to leprosy over the past 35 years strongly suggest that genetic factors influence susceptibility to leprosy and its varied clinical forms (9, 53, 88, 247). Because leprosy's divergent clinical forms reflect two distinct immune responses (Th1 versus Th2) to the same pathogen, human infection with M. leprae offers a unique opportunity to link innate and adaptive immune responses to specific host genes. Insight into the genetic determinants of these immune responses has illuminated the immunopathogenesis of leprosy. In addition, this field may broaden our understanding of the host response to Mycobacterium tuberculosis, a related but far more virulent and prevalent pathogen.
Overview of Leprosy
Epidemiology.
M. leprae is a fastidious, acid-fast, intracellular pathogen. In 2008, there were approximately 250,000 new cases reported, predominantly in India, Brazil, and Indonesia (333). Humans were previously thought to be the only important reservoirs of the bacteria, but it is now appreciated that leprosy, or Hansen's disease, may also be acquired from environmental sources (59, 60, 73, 170). A number of reports have linked leprosy to exposure to armadillos (169) or soil exposure (170). Leprosy is likely transmitted by aerosol droplets taken up through nasal or other upper airway mucosa (67, 215), where it has been detected by PCR techniques (148, 221). Large numbers of organisms have been found in the nasal secretions of lepromatous leprosy patients (223, 278, 279). Estimates of the incubation period between exposure and clinically manifest disease vary from months to decades (216), which makes epidemiological assessments of incidence and mechanisms of transmission difficult. Epidemiological studies of leprosy have established several risk factors for the disease, the strongest of which are genetic relatedness (204, 263) and close contact with leprosy patients, especially those with lepromatous disease (128, 204; summarized in reference 203). Other potential risk factors include low education level (156), food insecurity (156), water exposure (156, 167), infrequent changing of bed linen (156), armadillo exposure (59, 60, 73, 169, 311), lack of BCG vaccination (61, 65, 182, 295), and soil exposure (33, 170). Age has been reported to be a risk factor for leprosy or leprosy immune reactions in some studies (204, 243) but not in others (21, 156; see also summary in reference 203). Interestingly, in many, but not all, ethnic groups, there is a 2-to-1 ratio of males to females affected (21, 215; see summary in reference 203).
Natural history.
Leprosy is primarily a disease of the skin and peripheral nervous system. Less commonly, the eyes, bone, lymph nodes, nasal structures, and testes may also be involved (328). The disease's clinical manifestations fall into two poles, tuberculoid (TT) or “paucibacillary” (PB) and lepromatous (LL) or “multibacillary” (MB), with several intermediate forms (indeterminate, borderline tuberculoid [BT], borderline borderline [BB], and borderline lepromatous [BL]) (249) (Fig. 1). According to the WHO classification, multibacillary leprosy includes the LL, BL, and BB forms, and paucibacillary leprosy encompasses the TT and BT forms (140, 332). In some regions, patients with borderline, as opposed to polar, forms of leprosy (BB, BL, and BT) make up the majority of cases (36, 50, 143). Clinically, patients with lepromatous leprosy have a high burden of leprosy bacilli in skin biopsy specimens (“multibacillary”); multiple skin lesions consisting of macules, papules, plaques, or nodules; and thickened peripheral nerves with anesthesia and may eventually develop keratitis, uveitis, loss of eyebrow hair, ulceration of the nose, bone destruction, and thickened, waxy skin (249) due to infiltration by macrophages, lymphocytes, and plasma cells (328). At the opposite end of the spectrum, patients with tuberculoid (TT and BT) leprosy have a low burden of organisms (“paucibacillary”) in skin biopsy specimens and can present with a single, anesthetic skin lesion with or without a thickened peripheral nerve (249). Spontaneous resolution of tuberculoid and indeterminate infections has been observed. In contrast, spontaneous regression of disease does not occur in lepromatous leprosy patients (215).
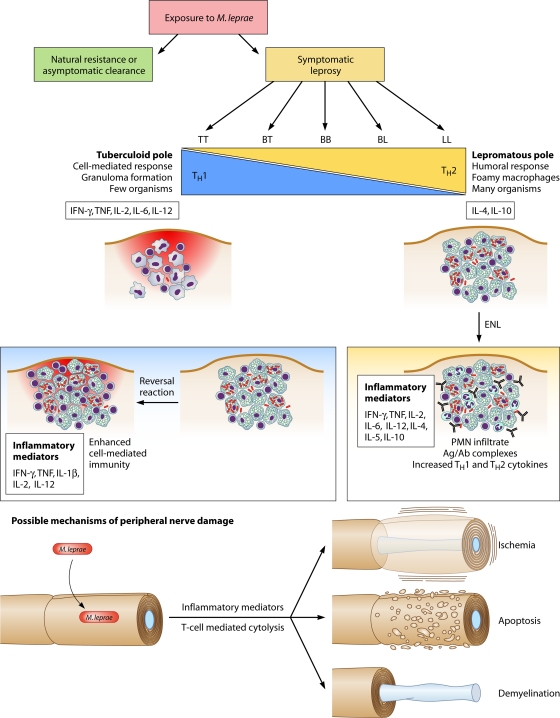
FIG. 1.
The leprosy spectrum and possible mechanisms of tissue damage. Leprosy manifestations are classified along a clinical spectrum of tuberculoid (TT), borderline tuberculoid (BT), borderline borderline (BB), borderline lepromatous (BL), and lepromatous (LL) leprosy. Each pole is associated with a characteristic cell-mediated or humoral immune profile. The cell-mediated (Th1) response of the TT pole features the elimination or containment of the organism in granulomas, while the ineffective humoral response at the LL (Th2) pole allows the proliferation of mycobacteria within and around foamy macrophages. Reversal reactions reflect a sudden shift toward the Th1 pole from the BT, BB, or BL state and can lead to irreversible nerve damage (neuritis). Erythema nodosum leprosum (ENL) reactions occur in patients with BL or LL leprosy and reflect an increase in both cell-mediated and humoral responses to M. leprae. ENL is associated with the systemic release of TNF and IL-4, a brisk polymorphonuclear leukocyte (PMN) influx, and antigen-antibody (Ag/Ab) complex deposition. The mechanism of nerve damage is unclear but may involve immune injury due to the release of inflammatory cytokines or activity of cytotoxic T cells, ischemia due to edema within the perineural sheath, apoptosis, or demyelination (see discussion in the text).
Immunology.
The clinicopathological features of leprosy have distinct immunological correlates (44, 249). Immunologically, lepromatous leprosy is characterized by a Th2 T-cell immune response (interleukin-4 [IL-4] and IL-10), antibody complex formation, the absence of granulomas, and failure to restrain M. leprae growth. Tuberculoid leprosy features a Th1 T-cell cytokine response (gamma interferon [IFN-γ] and IL-2), vigorous T-cell responses to M. leprae antigen, and containment of the infection in well-formed granulomas (44, 273). Lepromatous leprosy lesions are characterized by a lack of CD4+ T cells, numerous CD8+ T cells, and foamy macrophages, whereas tuberculoid leprosy lesions have a predominance of CD4+ T cells and well-formed granulomas (258, 340, 342). In lepromatous leprosy, robust antibody formation occurs but is not protective, and cell-mediated immunity is conspicuously absent (58, 309). In contrast, in tuberculoid leprosy, cell-mediated immunity is relatively preserved, and there is little evidence of M. leprae-specific humoral immunity (44, 258, 340, 342). However, as noted above, the majority of patients are not found at the poles of the leprosy spectrum but in the intermediate categories of BL, BB, and BT disease, which are clinically “unstable.” The immunology of these borderline states is poorly understood (273).
Leprosy reactions.
The Mitsuda reaction is a delayed-type hypersensitivity response (measured 21 to 28 days after inoculation) to intradermally administered leprosy antigens (of which lepromin is one formulation). Patients with tuberculoid leprosy typically have strongly positive Mitsuda reactions, a measure of the presence of functional cell-mediated immunity; in contrast, LL patients commonly have little to no reaction (134, 202, 212, 249, 260, 284). Two types of spontaneous immune reactions, or “reactive states,” can also occur in leprosy (Fig. 1). Reversal reactions (RRs), also known as type 1 reactions, represent the sudden activation of a Th1 inflammatory response to M. leprae antigens. They occur most frequently, although not exclusively, in borderline categories (BL, BT, or BB categories), often after the initiation of treatment, and reflect a switch from a Th2- toward a Th1-predominant response (44, 180, 273, 329). Erythema nodosum leprosum (ENL), also known as a type 2 reaction, is an acute inflammatory condition involving high levels of tumor necrosis factor (TNF) (264), tissue infiltration by CD4 cells and neutrophils (152), and deposition of immune complexes and complement, resulting in immune-complex-associated vasculopathy, panniculitis, and uveitis (44). ENL occurs in LL or BL patients and is more commonly seen in patients with a high bacterial index (249). Numerous investigators have measured intralesional and systemic cytokine production during leprosy reactions (102, 151, 178, 283, 340), but those studies did not consistently show a consistent Th1 versus Th2 cytokine pattern for reversal reaction versus ENL (summarized in references 272 and 273). For example, increased amounts of Th1 cytokines, such as IFN-γ, IL-12, and IL-2, have been demonstrated for both reversal reactions and ENL (summarized in references 44 and 273). A major drawback of these studies is the inability to determine whether the measured cytokine response is the cause or the consequence of inflammation (273). For these reasons, the immune mechanism of these reactions is still poorly understood.
Cellular and immune pathogenesis.
M. leprae is initially recognized by several innate immune receptors, including the Toll-like receptors (TLRs). The TLRs are a family of highly conserved, type 1 transmembrane proteins that orchestrate the innate immune response to microbial motifs, also known as pathogen-associated molecular patterns (PAMPs) (6, 28, 139). The interaction of these ligands with the extracellular domain of TLRs leads to the activation of a signaling pathway and the expression of chemokines and cytokines (6). TLR2 forms a heterodimer with TLR1 to mediate the recognition of several mycobacterial motifs, including the 19-kDa protein and other lipopeptides (26, 123, 233). Functional work by several investigators has shown that TLR2/1 is a critical mediator of the innate immune response to M. leprae (34, 163) and that M. leprae predominantly activates the TLR2/1 heterodimer (163). Based on data from previous studies with M. tuberculosis, several other signaling receptors may also be involved in M. leprae recognition. These receptors include TLRs 4, 6, 8, and 9; NOD2; DC-SIGN (dendritic cell [DC]-specific intercellular adhesion molecule 3-grabbing nonintegrin) (or CD209), Dectin-1, and Mincle (19, 20, 23, 49, 68, 75, 189, 190, 220, 255, 269, 299, 300, 314, 346). TLR2 may also act cooperatively with the lectins Dectin-1 and MBL (mannose binding lectin) (72, 101, 137).
M. leprae is an obligate intracellular pathogen with a distinct tropism for Schwann cells of the peripheral nervous system and for macrophages (37, 267, 293, 297). The pronounced specificity of M. leprae for Schwann cells is related to the tissue-specific expression of laminin-2 on Schwann cells. M. leprae contains a phenolic glycolipid (PGL-1) that has been shown to bind to the G domain of the α2 chain of laminin-2 on the membrane of Schwann cells (214). The uptake of M. leprae into the Schwann cell is thought to occur when the PGL-laminin-2 complex interacts with α-dystroglycan, the laminin-2 receptor located on the Schwann cell membrane (214, 239, 240). Laminin binding protein 21 (LBP21) also mediates the intracellular entry of M. leprae into the Schwann cell (Fig. 2) (238, 281). A variety of other receptors on monocytes and macrophages may also facilitate intracellular entry by M. leprae. On monocytes, PGL-1 mediates M. leprae phagocytosis via the complement receptor CR3 and serum complement 3 (267). On macrophages, complement receptors 1 and 4 help phagocytose M. leprae (266). Another candidate phagocytic receptor on the macrophage is the mannose receptor, which binds mannose and other carbohydrate moieties on mycobacteria (157, 268).
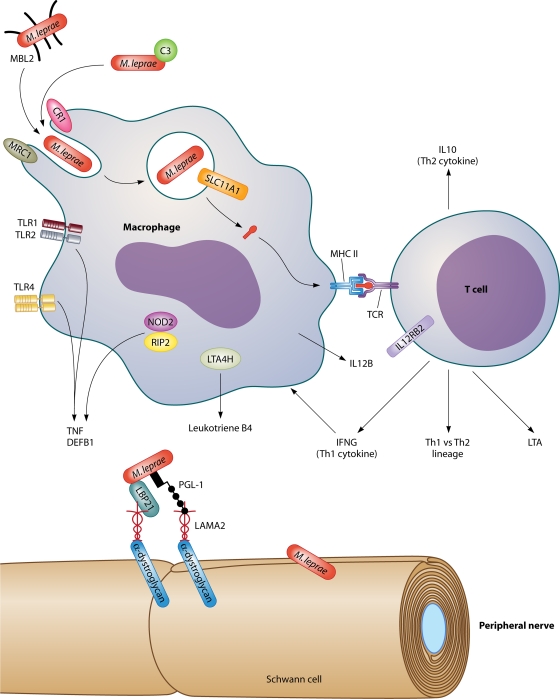
FIG. 2.
Genes and gene products involved in the immune response to M. leprae. Molecular and cellular interactions known or postulated to play a role in the immune response to M. leprae are depicted, as are genes with evidence of an association with susceptibility to leprosy and/or leprosy immune reactions through candidate gene studies, linkage analyses, or genome-wide association studies. Laminin binding protein 21 (LBP21) and phenolic glycolipid 1 (PGL-1) in the M. leprae cell wall bind to the α2 chain of laminin-2 (LAMA2) and α-dystroglycan on the Schwann cell membrane. This permits entry and subsequent damage to the peripheral nerve. Abbreviations: C3, complement factor 3; CR1, complement receptor 1; DEFB1, beta defensin 1; IFNG, gamma interferon; IL10, interleukin-10; IL12B, interleukin-12 subunit p40; IL12RB2, interleukin-12 receptor beta 2; LTA4H, leukotriene A4 hydrolase; LTA, lymphotoxin-α; MHC II, major histocompatibility complex class II; MBL2, mannose binding lectin 2; MRC1, mannose receptor; NOD2, nucleotide oligomerization domain 2; RIP2, receptor-interacting kinase; SLC11A1, solute carrier family 11, member 1 (also known as NRAMP); TCR, T-cell receptor; Th1, T-cell helper type 1; Th2, T-cell helper type 2; TLR, Toll-like receptor; TNF, tumor necrosis factor.
Nerve injury is the hallmark of progressive leprosy infection and involves both myelinated and unmyelinated nerves (115, 146, 147). Biopsy specimens taken from affected nerves of leprosy patients reveal perineural and intraneural inflammation and, in myelinated fibers, eventual demyelination (272). At the tissue level, the influx of immune cells and interstitial fluid (edema) inside inflexible nerve sheaths may cause nerve injury through mechanical compression and ischemia (272). At the cellular level, immunological injury is thought to be a major mechanism of nerve damage. The immune-mediated injury hypothesis has indirect support from in vitro studies in which the stimulation of monocytes or macrophages with M. leprae induces proinflammatory cytokines such as TNF, IL-12, IL-6, IL-1β, IL-18, and IL-15 (102, 151, 163, 198). For example, the 19-kDa protein of M. leprae, which is recognized by the TLR2/1 heterodimer, elicits a robust proinflammatory cytokine response (163) and induces apoptosis in Schwann cells (219). In addition, Schwann cells exposed in vitro to necrotic neurons produce TNF and nitric oxide (172), potent inflammatory mediators. In ex vivo studies, human Schwann cells loaded with M. leprae antigen have been shown to be targeted by cytolytic CD4+ T cells (292). Despite these in vitro and ex vivo observations, the mechanism of nerve injury remains poorly understood, partly due to the lack of good animal models for leprosy and leprosy-induced nerve damage.
ASSESSING THE GENETIC CONTRIBUTION TO LEPROSY RISK
Leprosy has long been observed to be a disease that aggregates in families (47, 118, 145, 232). In the 19th century, the hereditary versus environmental origins of this illness were vigorously debated (48, 118), driven in part by the social stigma attached to leprosy. The discovery of the M. leprae bacillus by Gerhard Henrik Armauer Hansen in 1873 (118, 119) settled the argument for the time being in favor of an environmental etiology. In the modern era it has become clear that while encounter with the M. leprae pathogen is necessary for infection, it is not sufficient, since the majority of exposed individuals do not become infected. Host genetic factors may therefore largely determine which exposed individuals develop disease. Evidence that host genes influence susceptibility to leprosy or its various clinical forms is supported by data from a wide variety of sources. These sources include twin studies, segregation analyses, family-based linkage and association studies, candidate gene association studies, and, most recently, genome-wide association studies (GWASs). The most definitive twin study of leprosy by Chackravartti and Vogel (56) enrolled 62 monozygous and 40 dizygous twin pairs from three different regions in India and found a 3-fold-greater concordance rate for the type of leprosy disease in monozygotic twins than in dizygotic twins (56). Segregation analyses determine whether or not there is a segregation of disease among more closely related individuals (evidence of a “major gene effect”) and what mode of inheritance is at work (dominant, recessive, or additive) (296). A number of segregation studies have been carried out for leprosy (1, 3, 84, 117, 171, 241, 275, 280, 289, 327), several of which have detected the presence of a recessive or codominant mode of inheritance for leprosy overall or for nonlepromatous leprosy (1, 84, 117, 171, 289).
Study Design for Complex Diseases
It has been widely presumed for many infectious diseases, including leprosy, that susceptibility is governed by polygenic inheritance, or the additive effect of multiple genes, each with a modest effect on the infectious phenotype. Two study designs are typically used to examine diseases with complex inheritance patterns: linkage studies of families and association studies (candidate gene or genome wide). Linkage studies look for evidence of the segregation of a genetic marker and a disease trait within families. Genetic association studies assess whether the frequency of a particular genetic variant differs between individuals with a disease compared to unrelated controls.
Linkage studies.
Linkage studies often follow up on the results of segregation analyses (discussed above) in the same study population. Several genome-wide linkage studies of leprosy susceptibility have been performed by using a family-based design (197, 282). A major strength of genome-wide linkage studies is the absence of bias: no hypothesis as to which chromosomal loci or genes might be linked to disease status is required. Linkage studies genotype microsatellites or SNPs (single-nucleotide polymorphisms) spaced evenly throughout the genome, typically every 10 centimorgans (cM). Susceptibility loci identified in these studies are then investigated further by higher-resolution mapping of markers or gene alleles and linkage to disease traits (8, 196, 306). Linkage studies have also been used to evaluate candidate regions (142, 330) and candidate genes (2, 261) in leprosy. Significance in these studies is reported via Z scores, LOD scores (logarithm of odds), or P values (168). The proposed criteria for significance in genome-wide linkage studies are somewhat stringent, given the risk of false positives due to the large number of markers studied. For example, one common genome-wide linkage study design relies on sibling pairs. Suggested threshold levels of significance for “suggestive linkage,” “significant linkage,” and “highly significant linkage” for individual markers in these sibling pair genome-wide linkage studies are P values of 7 × 10−4, 2 × 10−5, and 3 × 10−7, respectively (corresponding to LOD scores of 2.2, 3.6, and 5.4, respectively) (168). The suggested P value for validating linkage in replication studies (which typically focus on a candidate region of ∼20 cM in size) is a P value of 0.01 (168). Linkage studies are also the most powerful study design for identifying rare variants of genes that confer a large risk of disease (18, 296). Conversely, they have reduced statistical power to detect genes with modest or weak effects on disease risk, even when hundreds of families are included (13, 237, 251, 296). Linkage studies that have identified major susceptibility loci or genes for leprosy or leprosy immune reactions are described below.
Genetic association studies.
In contrast to linkage studies, association studies evaluate whether common polymorphisms in candidate genes are associated with susceptibility to disease, usually in unrelated individuals. These studies are hypothesis driven and often focus on genetic variants that are predicted to alter protein structure or function. The most common study design is a case-control format with comparisons of one or more polymorphism (single nucleotide, insertions, deletions, or microsatellite [MS] markers) frequencies between cases and controls. A major strength of this study design is the power to find relatively modest effects, generally with smaller sample sizes than family-based studies (251, 296). One disadvantage is the problem of population stratification or admixture, where differences in ethnic compositions of the cases and controls can lead to spurious disease associations. Methods to control for population stratification include matching cases and controls for ethnicity and adjusting for ethnicity as a possible confounder in a multivariate logistic regression model. An alternative study design that is robust to population stratification is the transmission disequilibrium test (TDT). This approach looks for evidence of nonrandom transmission of the candidate allele from a heterozygous parent to an affected child and can be used to corroborate findings of either linkage studies or association studies.
It is important to remember that association, even down to the SNP level, is not necessarily causation. Genetic associations at specific loci may derive from neighboring alleles in linkage disequilibrium (LD) with the candidate gene that is being studied. In these situations, the candidate gene SNP serves as a proxy for the association. However, if the haplotype structure of the region surrounding the candidate gene is not explored, the alleles most responsible for the disease association will remain unascertained. The haplotype structures at specific genetic loci often differ between populations (population-specific linkage disequilibrium). This variability can make it difficult to replicate disease associations when the underlying LD structure has not been evaluated for both populations. Both linkage and genetic association study designs are also vulnerable to the generation of false-positive results from multiple comparisons. This problem is especially relevant in the current era of genome-wide linkage scans and high-throughput genotyping strategies, as used in GWASs. As a result, replication and validation of findings in independent populations coupled with investigation of the underlying haplotype structure of each population are an essential part of a careful study design. The candidate gene approach can also be linked to functional studies of the polymorphisms to determine whether there is a biological mechanism relevant to disease pathogenesis.
Finally, although neither association nor linkage studies are designed to detect rare alleles with weak effects (296), adequate power has been a particular problem for candidate gene association studies and probably accounts in part for these studies' rather poor track record for replicating SNP associations. The need to include adequate numbers of cases and controls is particularly important when the frequency of the allele(s) being studied is low (≤5%). Numerous candidate gene association studies have relied on sample sizes of 50 to 100 cases. There is not adequate power to detect a disease association of a variant allele with a population frequency of 5% in these small studies unless the odds ratio (OR) rises to the level of 3.0 to 4.0 (for example, for an α of 0.05 with 100 cases, 100 controls, and a minor allele frequency [MAF] of 0.05, power equals 0.17 for an OR of 1.5, 0.44 for an OR of 2, 0.87 for an OR of 3, and 0.98 for an OR of 4). However, many candidate genes have disease associations with ORs in the range of 1.5 to 2.5 or lower. To have power to detect associations of low-frequency SNPs with a more modest influence on disease susceptibility, study investigators would need to recruit 300 to 600 cases and an equal number of controls (for example, for an α of 0.05 with 500 cases, 500 controls, and an MAF of 0.05, power equals 0.58 for an OR of 1.5, 0.97 for an OR of 2, and 1.00 for an OR of 3 to 4). Such large numbers have been the exception rather than the rule in candidate gene association studies (Table 1).
TABLE 1.
Summary of non-HLA leprosy candidate gene association studiesc
| Gene | Reference(s) (yr) | Population | Sample size | Variant(s)a | Phenotype | OR (95% CI), rel. risk, or χ2 value | P valueb |
|---|---|---|---|---|---|---|---|
| CFB (B factor, Bf) | 114 (1980) | Thailand (Chinese) | 24 controls, 38 leprosy | Bf S allele | NE: lep | ||
| Thailand (Thai) | 184 controls, 198 leprosy | Bf S allele | NE: lep | ||||
| 69 (1993) | Brazil | 172 controls, 109 leprosy (73 LL [46 with ENL], 36 non-TL) | Bf F allele (“Bf-F1”) | S: ENL | ND | <0.03 | |
| C2 | 114 (1980) | Thailand Chinese | 24 controls, 38 leprosy | C2 C allele | NE: lep | ||
| Thailand (Thai) | 184 controls, 198 leprosy | C2 C allele | NE: lep | ||||
| C3 | 83 (1972) | Angola | 439 controls, 468 leprosy (97 LL, 180 TL, 191 IL) | S (major allele) vs F (minor allele) | NE: lep or lep type | ||
| 15 (1973) | Angola | 439 controls, 470 leprosy (97 LL, 182 TL, 191 IL) | S (major allele) vs F (minor allele) | NE: lep or lep type | |||
| 4 (1974) | Ethiopia and Mali | 66 controls, 152 leprosy | S (major allele) vs F (minor allele) | S: lep | [OR, 2.33 (0.19-0.98) (dom)] | [0.041] | |
| [OR, 2.43 (0.17-0.98) (het)] | [0.041] | ||||||
| 294 (1975) | Ethiopia | 55 related controls, 91 leprosy | S (major allele) vs F (minor allele) | NE: lep | |||
| C4A | 114 (1980) | Thai and Chinese individuals combined | 123 controls, 201 leprosy (27 TT, 103 BL, 71 LL) | C4A F1 allele (“functionally inactive”) | S: con vs. TT vs BB vs LL (4 × 2 chi square) | χ2 = 13.7 (ND) | <0.01 |
| S: LL vs con | χ2 = 12.6 (ND) | <0.001 | |||||
| C4B | 69 (1993) | Brazil | 172 controls, 109 leprosy (36 non-LL, 73 LL [46 ENL, 27 no ENL]) | C4B*Q0 absent allele | S: lep vs con | ND | 4.4 × 10−5 |
| S: ENL vs no ENL | Rel. risk = 5.3 | [0.017] | |||||
| S: ENL | ND | 3.6 × 10−7 | |||||
| C4B*1 | S: ENL | ND | 3.9 × 10−3 | ||||
| CR1 | 93 (2004) | Malawi | 166-252 controls, 186-399 leprosy (>90% PB) | G3093T | NE: lep | ||
| A4795G “McCoy” | R: lep | OR, 0.3 (0.1-0.8) (rec) | 0.02 | ||||
| G4828A “Swain-Langley” | NE: lep | ||||||
| A4870G | NE: lep | ||||||
| C5507G | NE: lep | ||||||
| DEFB1 (β-defensin 1) | 230 (2009) | Mexico | 151 controls, 75 leprosy | G668C | S: lep | OR, 2.42 (1.37-4.28) (dom) | 0.009 |
| (46 LL) | S: LL vs con | OR, 3.06 (1.47-6.04) (dom) | 0.024 | ||||
| A692G | NE: lep or lep type | ||||||
| A1836G | NE: lep or lep type | ||||||
| Haplotype (668/692/1836) CGA | S: LL | 2.25 (1.23-4.03) | 0.009 | ||||
| FCN2 | 70 (2009) | Brazil, mixed ethnicity | 210 controls, 158 leprosy (92 LL, 14 TT, 22 BL, 27 IL) | 20 SNPs | NE: lep | ||
| Haplotype AGA (−986/−602/−4) | R: lep | OR, 0.13 (0.03-0.43) | <0.013 | ||||
| Haplotype AGAG (−986/−602/−4/+6424) | R: lep | OR, 0.10 (0.11-0.43) | <0.011 | ||||
| IFNG | 248 (2003) | Brazil | 98 controls, 96 leprosy (10 TT, 59 BL, 27 LL) | Intron 1 CA repeat (short [fewer than 122 bp]→long [122-126 bp]) | S: lep | [OR, 2.62 (1.29-5.32)] | 0.01 |
| 93 (2004) | Malawi | 236 controls, 402 cases | Intron 1 T874A | NE: lep | |||
| IL10 | 262 (2002) | Brazil | 62 controls, 202 leprosy (143 MB, 59 PB) | C−819T | S: PB vs MB | OR, 2.28 (1.1-4.5) (allele) | <0.01 |
| S: PB vs con | OR, ↑ [ND] (allele) | <0.05 | |||||
| S: lep | OR, 2.64 (0.93-8.04) (rec) | 0.04 | |||||
| C−592A | NE: lep or lep type | ||||||
| 206 (2004) | Brazil | 283 controls, 297 leprosy (131 PB, 166 MB) | C−819T | NE: lep or lep type | |||
| A−1082G | NE: lep or lep type | ||||||
| C−2763A | NE: lep or lep type | ||||||
| G−2849A | NE: lep or lep type | ||||||
| T−3575A | NE: lep or lep type | ||||||
| Haplotype −3575A/−2849G/−2763C | R: lep | OR, 0.35 (0.13-0.91) | 0.005 | ||||
| R: lep vs PB vs MB | OR, 0.32 (0.12-0.83) (ordinal trait) | 0.006 | |||||
| Haplotype −3575T/−2849A/−2763C | S: lep | OR, 2.37 (1.04-5.39) | 0.027 | ||||
| 93 (2004) | Malawi | 191-215 controls, 349-362 leprosy (>90% PB) | C−819T | NE: lep | |||
| C−592A | NE: lep | ||||||
| A−1082G | NE: lep | ||||||
| 185 (2005) | India | 266 controls, 282 leprosy (144 MB, 142 PB) | C−819T | S: lep | OR, ↑ (ND) (allele) | “Significant” [ND] | |
| S: lep | OR, 2.50 (1.49-4.00) (rec) | <0.001 | |||||
| S: MB vs con | OR, 2.63 (1.51-4.76) (rec) | 0.001 | |||||
| S: PB vs con | OR, 2.32 (1.29-4.16) (rec) | 0.005 | |||||
| C−592A | S: lep | OR, ↑ [ND] (allele) | “Significant” [ND] | ||||
| S: lep | OR, 2.43 (1.47-4.00) (rec) | <0.001 | |||||
| S: MB vs con | OR, 2.56 (1.44-4.54) (rec) | 0.001 | |||||
| S: PB vs con | OR, 2.32 (1.29-4.16) (rec) | 0.005 | |||||
| G−1082A | NE: lep or lep type | ||||||
| C−2763A | NE: lep or lep type | ||||||
| G−2849A | NE: lep or lep type | ||||||
| T−3575A | NE: lep or lep type | ||||||
| Haplotype −3575T/−2849G/−2763C/−1082A/−819C/−592C | R: lep | OR, 0.58 (0.37-0.89) | 0.01 | ||||
| Haplotype −3575T/−2849G/−2763C/−1082A/−819T/−592A | S: MB > PB > con; NE: lep | ND (ordinal trait) | 0.0002 | ||||
| Diplotype of proximal promoter SNPs G−1082A, C−819T, and C−592A (ACC/ACC) | R: lep | [OR, 0.35 (0.19-0.63)] | 0.001 | ||||
| R: MB vs con | [OR, 0.26 (0.11-0.60)] | 0.002 | |||||
| R: PB vs con | [OR, 0.44 (0.22-0.88)] | 0.02 | |||||
| Diplotype of proximal promoter SNPs, G−1082A, C−819T, and C−592A (ATA/ATA) | S: lep | [OR, 1.82 (1.05-3.23)] | 0.03 | ||||
| S: MB vs con | [OR, 1.96 (1.03-3.85)] | 0.04 | |||||
| 225 (2008) | Brazil | 368-380 controls, 321-369 leprosy | C−819T | S: lep | OR, 1.60 (ND) (TT vs CC) | 0.05 | |
| OR, 1.44 (ND) (dom) | 0.026 | ||||||
| A−1082G | ND (not in HWE) | ||||||
| C−2763A | NE: lep | ||||||
| G−2849A | NE: lep | ||||||
| T−3575A | NE: lep | ||||||
| Haplotype −3575T/−2849G/−2763C/−819T | S: lep | OR, 1.35 (ND) | 0.03 | ||||
| 225 (2008) | Meta-analysise | 1,347 controls, 1,355 leprosy | C−819T | S: lep | OR, 1.30 (1.13-1.49) (allele) | 0.0003 | |
| S: lep | OR, 1.28 (1.03-1.60) (dom) | 0.0237 | |||||
| S: lep | OR, 1.66 (1.29-2.15) (rec) | 0.0001 | |||||
| 96 (2009) | Brazil | 240 controls, 156 leprosy | Diplotype G−1082A/C−819T/ C−592C | NE: lep | |||
| IL12B (IL-12p40) | 207 (2007) | India | 89 controls, 80 leprosy | 3′ UTR TaqI site SNP (genotype 2.2 vs 1.1 or 1.2) | R: lep | [OR, 0.13 (0.03-0.58) (rec)] | 0.002 |
| NE: lep type | |||||||
| 14 (2008) | Mexico | 51 controls, 44 LL | 3′ UTR 1188 A/C (TaqI site) | S: lep | [OR, 7.2 (2.38-21.61) (rec)] | <0.05 | |
| IL12RB2 | 218 (2005) | Japan | 68 controls, 176 leprosy (130 LL, 46 TL) | A−1035G | S: LL vs TL | OR, 3.97 (ND) (allele) | <0.001 |
| A−1023G | S: LL vs TL | OR, 2.95 (ND) (allele) | <0.01 | ||||
| −650delG | S: LL vs TL | OR, 3.74 (ND) (allele) | <0.001 | ||||
| A−464G | S: LL vs TL | OR, 3.64 (ND) (allele) | <0.01 | ||||
| Haplotype −1035A/−1023A/−650G/ −464A | R: LL vs TL | ND | 0.0002 | ||||
| R: LL vs con | ND | 0.039 | |||||
| 4 other haplotypes tested | NE: lep or lep type | ||||||
| KIR | 97 (2008) | Brazil | 289 controls, 165 leprosy (65 LL, 49 BB, 42 TT) | KIR2DL1-5 | NE: lep or lep type | ||
| KIR3DL1-3 | NE: lep or lep type | ||||||
| KIR2DS3 | NE: lep type | ||||||
| S: TT vs LL | 2.72 (1.12-6.58) | 0.04 | |||||
| KIR2DS1-2, KIR2DS4-5 | NE: lep or lep type | ||||||
| KIR2DP1 | NE: lep or lep type | ||||||
| KIR3DP1 | NE: lep or lep type | ||||||
| LAMA2 (laminin α-2) | 334 (2002) | Indonesia | 58 controls, 53 leprosy (26 TL, 27 LL) | T7809C | S: TL vs LL | χ2 = 8.07 (genotype) | <0.025 |
| OR, 6.73 (1.94-23.36) (TC vs. TT) | <0.005 | ||||||
| LTA (lymphotoxin-α) | 93 (2004) | Malawi | 333 controls, 184 leprosy (85% PB) | 5′ UTR microsatellite (AC/GT)n at −3.5-kb 105-bp allele | S: lep | OR, 1.6 (1.1-2.4) | 0.03 |
| 5′ UTR microsatellite (AC/GT)n at −3.5-kb 101-bp allele | S: MB vs con | χ2 = 11.7 (ND) | 0.003 | ||||
| 8 (2007) | India | 371 controls, 364 leprosy | LTA−294 univariate analysis | S: lep | 1.78 (1.29-2.45) | 0.0004 | |
| LTA−294 multivariate analysis | NE: lep | ||||||
| LTA−293 | NE: lep | ||||||
| LTA+10 | NE: lep | ||||||
| LTA+80 (A allele) univariate analysis | NE: lep | ||||||
| LTA+80 (A allele) multivariate analysis | S: lep | 1.60 (1.10-2.33) (dom) | 0.01 | ||||
| LTA+80 (A allele) subgroup ages 16-25 yr | S: lep | 2.95 (1.32-6.58) (dom) | 0.006 | ||||
| LTA+252 | NE: lep | ||||||
| LTA+368 | NE: lep | ||||||
| MCCD1 rs2259435 (A allele) univariate analysis | S: lep | 1.59 (1.24-2.04) | 0.0002 | ||||
| MCCD1 rs2259435 (A allele) multivariate analysis | S: lep | 1.82 (1.38-2.41) | 0.0004 | ||||
| Vietnamd | 194 simplex leprosy families | LTA−294 | NE: lep | ||||
| LTA−293 univariate analysis | S: lep | 1.97 (1.30-2.99) | 0.0009 | ||||
| LTA−293 multivariate analysis | S: lep | 1.97 (1.30-2.99) | 0.0009 | ||||
| LTA+10 | NE: lep | ||||||
| LTA+80 univariate analysis | S: lep | 1.74 (1.16-2.60) | 0.007 | ||||
| LTA+252 | NE: lep | ||||||
| LTA+368 univariate analysis | S: lep | 1.63 (1.09-2.43) | 0.02 | ||||
| Brazil | 192 controls, 209 leprosy | LTA−294 | NE: lep | ||||
| LTA−293 | NE: lep | ||||||
| LTA+10 | NE: lep | ||||||
| LTA+80 (A allele) | NE: lep | ||||||
| LTA+80 (A allele) subgroup ages 16-25 yr | Trend: lep | 2.76 (0.74-10.32) | NS [ND] | ||||
| LTA+252 | NE: lep | ||||||
| LTA+368 | NE: lep | ||||||
| LTA4H | 304 (2010) | Nepal | 899 leprosy (443 MB without ENL, 335 PB) | rs1978331 T/C | R: MB vs PB | OR, 0.62 (0.46-0.82) (het) | 0.001 |
| rs2660898 T/G | R: MB vs PB | OR, 0.70 (0.52-0.95) (het) | 0.021 | ||||
| MBL2 | 93 (2004) | Malawi | 261 controls, 231 leprosy | G239A | NE: lep | ||
| 71 (2007) | Brazil | 214 controls, 264 leprosy (150 LL, 36 TL, 37 BL, 17 IL) | Haplotype LYPA (−550L/221Y/+4P/exon 1 A) | S: lep | OR, 2.25 (1.31-3.88) | 0.020 | |
| S: LL vs con | OR, 2.22 (1.21-4.05) | 0.060 | |||||
| S: BL vs con | OR, 2.98 (1.29-6.87) | 0.060 | |||||
| “Low”-MBL-producing genotypes | R: LL vs TL | OR, 0.31 (0.13-0.71) | 0.012 | ||||
| MICA/MICB | 331 (1999) | China | 112 controls, 69 leprosy | MICA-A5 | R: lep | Rel. risk, 0.62 (allele) | 0.06 |
| R: MB vs con | Rel. risk, 0.56 (allele) | <0.05 | |||||
| Haplotype MICA-A5 with HLA-B46 | R: lep | Rel. risk, 0.37 | <0.03 | ||||
| R: MB vs con | Rel. risk, 0.22 | <0.01 | |||||
| MRC1 | 12 (2010) | Vietnamd | 490 simplex families (53% MB) + 90 multiplex families (55% MB) for a total of 704 leprosy patients (325 PB, 374 MB) | rs1926736 G/A A allele (396S) | R: lep | OR: 0.76 (0.60-0.96) (best dom) | 0.035 |
| R: MB | OR: 0.71 (0.51-0.99) (best dom) | 0.034 | |||||
| Brazil | 384 leprosy (∼162 MB, ∼167 PB), 399 controls, | rs1926736 A/G G allele (396G) | S: lep | OR, 1.34 (1.06-1.70) (additive) | 0.016 | ||
| S: MB | OR, 1.42 (1.05-1.93) (additive) | 0.023 | |||||
| rs2437257 C/G (407L) | NE: lep | ||||||
| R: MB | OR, 0.63 (0.41-0.97) (dom) | 0.04 | |||||
| Haplotype G396-F407 | S: lep | OR, 1.41 (1.13-1.76) | 0.003 | ||||
| S: MB | OR, 1.61 (1.21-2.14) | 0.001 | |||||
| Haplotype S396-F407 | R: lep | OR, 0.75 (0.59-0.95) | 0.019 | ||||
| R: MB | OR, 0.68 (0.50-0.93) | 0.015 | |||||
| NOD2 | 93 (2004) | Malawi | ND | C802T | Not present in population | ||
| C2104T | |||||||
| G2722C | |||||||
| 3020insC 1007fs | |||||||
| 27 (2010) | Nepal | 101 controls, 933 leprosy | rs12448797 T/C (C allele) | S: lep | OR, 2.18 (1.06-5.23) (allele) | 0.031 | |
| OR, 2.47 (1.12-6.44) (dom) | 0.021 | ||||||
| rs2287195 A/G (G allele) | S: lep | OR, 1.51 (1.08-2.14) (allele) | 0.013 | ||||
| OR, 1.85 (1.19-2.88) (dom) | 0.004 | ||||||
| rs8044354 A/G (G allele) | S: lep | OR, 1.53 (1.11-2.10) (allele) | 0.006 | ||||
| OR, 2.07 (1.33-3.22) (dom) | 0.0006 | ||||||
| rs13339578 G/A (A allele) | S: lep | OR, 1.71 (1.10-2.68) (dom) | 0.012 | ||||
| rs4785225 C/G (G allele) | S: lep | OR, 1.67 (1.07-2.60) (dom) | 0.017 | ||||
| rs751271 A/C (C allele) | S: lep | OR, 1.65 (1.07-2.58) (dom) | 0.016 | ||||
| rs1477176 T/C (C allele) | R: lep | OR, 0.44 (0.28-0.71) (allele) | <0.001 | ||||
| OR, 0.43 (0.26-0.73) (dom) | 0.0005 | ||||||
| 124 ENL | rs2287195 A/G (G Allele) | S: ENL vs no ENL | OR, 1.82 (1.12-3.00) (dom) | 0.011 | |||
| 428 no ENL | rs8044354 A/G (G Allele) | S: ENL vs no ENL | OR, 1.34 (0.99-1.82) (allele) | 0.046 | |||
| OR, 2.17 (1.26-3.88) (dom) | 0.003 | ||||||
| rs7194886 C/T (T allele) | S: ENL vs no ENL | OR, 1.66 (1.07-2.57) (dom) | 0.016 | ||||
| rs6500328 A/G (G allele) | S: ENL vs no ENL | OR, 1.69 (1.09-2.62) (dom) | 0.012 | ||||
| rs17312836 A/C (C allele) | S: ENL vs no ENL | OR, 1.43 (1.00-2.04) (allele) | 0.039 | ||||
| OR, 1.70 (1.09-2.64) (dom) | 0.013 | ||||||
| rs1861759 A/C (C allele) | S: ENL vs no ENL | OR, 1.42 (1.00-2.01) (allele) | 0.037 | ||||
| OR, 1.78 (1.15-2.73) (dom) | 0.006 | ||||||
| rs1861758 C/T (T allele) | S: ENL vs no ENL | OR, 1.41 (0.99-1.99) (allele) | 0.047 | ||||
| OR, 1.69 (1.09-2.63) (dom) | 0.014 | ||||||
| 240 RR | rs2287195 A/G (G allele) | R: RR vs no RR | OR, 0.74 (0.58-0.95) (allele) | 0.013 | |||
| 693 no RR | OR, 0.69 (0.50-0.96) (dom) | 0.023 | |||||
| rs8044354 A/G (G allele) | R: RR vs no RR | OR, 0.74 (0.59-0.92) (allele) | 0.005 | ||||
| OR, 0.69 (0.49-0.96) (dom) | 0.021 | ||||||
| rs8043770 C/G (G allele) | R: RR vs no RR | OR, 0.73 (0.57-0.94) (allele) | 0.012 | ||||
| OR, 0.67 (0.49-0.92) (dom) | 0.010 | ||||||
| rs7194886 C/T (T allele) | R: RR vs no RR | OR, 0.74 (0.55-0.98) (allele) | 0.032 | ||||
| OR, 0.69 (0.49-0.96) (dom) | 0.023 | ||||||
| rs1861759 A/C (C allele) | R: RR vs no RR | OR, 0.75 (0.57-0.99) (allele) | 0.041 | ||||
| OR, 0.69 (0.49-0.95) (dom) | 0.019 | ||||||
| rs4785225 C/G (G allele) | R: RR vs no RR | OR, 0.71 (0.52-0.97) (dom) | 0.027 | ||||
| rs751271 A/C (C allele) | R: RR vs no RR | OR, 0.73 (0.53-0.99) (dom) | 0.038 | ||||
| PARK2/PACRG | 196 (2004) | Brazil | 388 controls, 587 leprosy (38% PB, 62% MB) | rs2803104 (A allele) | NE: lep | ||
| 10 kb_target_5_2 (T allele) | S: lep | ND | 0.019 | ||||
| PARK2_e01(−697) (G allele) | S: lep | ND | 0.001 | ||||
| PARK2_e01(−2599) (T allele) | S: lep | ND | 0.003 | ||||
| PARK2_e01(−3024) (C allele) | NE: lep | ||||||
| PARK2_e01(−3800) (G allele) | S: lep | ND | 0.009 | ||||
| 28 kb_target_2_1 (T allele) | NE: lep | ||||||
| 28 kb_target_4_1 (A allele) | S: lep | ND | 0.003 | ||||
| rs1514343 (T allele) | S: lep | ND | 0.045 | ||||
| rs1333955 (C allele) | S: lep | ND | 0.034 | ||||
| rs1040079 (C allele) | S: lep | ND | 0.001 | ||||
| 40 kb_target_8_F60 (A allele) | S: lep | ND | 0.032 | ||||
| 40 kb_target_8_F706 (G allele) | NE: lep | ||||||
| 184 (2006) | India | 350 controls, 286 leprosy (144 MB, 142 PB) | PARK2_e01(−2599) | NE: lep or lep type | |||
| PARK2_e01(−697) | NE: lep or lep type | ||||||
| 28 kb_target_2_1 | NE: lep or lep type | ||||||
| 10 kb_target_5_2 | NE: lep or lep type | ||||||
| rs1040079 | NE: lep or lep type | ||||||
| SLC11A1 (NRAMP) | 256 (1999) | India | 165 controls, 227 leprosy (105 TL, 122 LL) | 5′ MS [(GT)n or (CA)n] | NE: lep or lep type | ||
| 3′ UTR TGTG del/ins | NE: lep or lep type | ||||||
| (469 + 14 G/C) intron 4 SNP | NE: lep or lep type | ||||||
| Exon 2 polymorphism | NE: lep or lep type | ||||||
| 191 (2001) | Mali | 201 controls, 273 leprosy (92 PB, 181 MB) | 3′ UTR TGTG deletion | S: MB vs PB (het vs del/del) | OR, 5.79 (1.46-24.61) | 0.003 | |
| 89 (2004) | Brazil | 61 controls (30 positive for MR, 24 negative for MR), 90 leprosy (45 MB, 45 PB, 18 positive for MR, 64 negative for MR) | Alleles of 5′ promoter microsatellite [(GT)n or (CA)n] polymorphisms | NE: lep, lep type, MR | |||
| 321 (2007) | Thailand | 140 controls, 37 leprosy (13 PB, 24 MB) | (469 + 14 G/C) intron 4 SNP | NE: lep or lep type | |||
| D543N SNP | NE: lep or lep type | ||||||
| 3′ UTR TGTG del/ins | NE: lep or lep type | ||||||
| 93 (2004) | Malawi | 283-429 controls, 244-258 leprosy (>90% PB) | Promoter microsatellite(GT)n genotypes 199/201, 199/199, 201/201 | NE: lep | |||
| Other promoter microsatellite(GT)n genotypes | NE: lep | ||||||
| Exon 2, 9-bp del | NE: lep | ||||||
| 3′ UTR TGTG del/ins | NE: lep | ||||||
| 3′ UTR CAAA del/ins | NE: lep | ||||||
| TLR1 | 149 (2007) | Turkey | 90 controls, 57 leprosy | T1805G G allele (602S) | R: lep | OR, 0.48 (0.29-0.80) | 0.004 |
| T1805G GG genotype (602SS) | R: lep | ND | 0.02 | ||||
| 198 (2008) | Nepal | 933 leprosy (311 TL, 490 LL) | T1805G G allele (602S) | NE: lep | |||
| NE: TL vs LL | |||||||
| R: RR | OR, 0.51 (0.29-0.87) (allele) | 0.01 | |||||
| 271 (2009) | Bangladesh | 543 controls, 842 leprosy (702 PB, 140 MB) | T1805G G allele (602S) | NE: lep | |||
| A743G GG genotype (248SS) | S: lep | OR, 1.34 (1.06-1.70) (rec) | [0.016] | ||||
| A743G G allele (248S) | R: ENL | OR, 0.40 (0.16-0.99) (allele) | [0.04] | ||||
| A743G GG genotype (248SS) | Trend: RR | OR, 1.57 (0.97-2.55) (rec) | NS | ||||
| TLR2 | 93 (2004) | Malawi | 379 controls, 210 cases (26 MB, 184 PB) | Intron 2 microsatellites 216 bp, 222 bp, 224 bp, 226 bp | NE: lep | ||
| Intron 2 microsatellite 224 bp | MB vs PB | χ2= 6.3 (ND) | 0.042 | ||||
| 35 (2008) | Ethiopia | 197 controls, 441 leprosy (298 LL, 138 TL, 5 uncharacterized, 66 RR, 150 no RR) | Microsatellite (280 bp) | S: RR | OR, 5.83 (1.98-17.15) (rec) | 0.001 | |
| Microsatellite (288 bp) | R: LL vs. TL | OR, 0.49 (0.27-0.90) (dom) | 0.02 | ||||
| Microsatellite (290 bp) | R: lep | OR, 0.62 (0.41-0.93) (additive) | 0.02 | ||||
| Other microsatellites | NE: lep, lep type, RR | ||||||
| C597T (N199N) | R: RR | OR, 0.34 (0.17-0.68) (dom) | 0.002 | ||||
| T1350C (S450S) | NE: lep, lep type, RR | ||||||
| “280 C-T haplotype” (280 bp, 297C, 1350T) | S: RR | OR, 6.39 (2.14-19.07) (rec) | 0.001 | ||||
| TLR4 | 36 (2009) | Ethiopia | 197 controls, 441 leprosy (298 lepromatous [199 BL, 81 LL, 18 MB], 138 tuberculoid [128 BT, 3 TT, 7 PB], 5 uncharacterized, 133 neuritis, 66 RR, 17 ENL) | G896A | R: lep | OR, 0.34 (0.20-0.57) (additive) | <0.001 |
| T1196C | R: lep | OR, 0.15 (0.06-0.39) (dom) | <0.001 | ||||
| G1530T | R: lep | OR, 0.38 (0.14-1.01) (dom) | 0.05 | ||||
| A1976G | NE: lep or lep type | ||||||
| Haplotype 896G/1196T/1530G/1976A | R: lep | OR, 0.12 (0.05-0.34) | <0.001 | ||||
| Haplotype 896G/1196C/1530T/1976A | R: lep | OR, 0.23 (0.08-0.69) | 0.008 | ||||
| 93 (2004) | Malawi | 288 controls, 235 leprosy | G896A | NE: lep | |||
| TNF | 257 (1997) | India | 160 controls, 228 leprosy (121 LL, 107 TT) | G−308A | S: LL vs con | Rel. risk, 2.5 (1.1-6.5) (allele) | 0.03 |
| 261, 262 (2000 and 2002, respectively) | Brazil | 92 controls, 300 leprosy (MB [70 LL, 85 BL, 55 BB], PB [63 BT, 2 TT], 10 indeterminant, 15 pure neuritis) | G−308A | R: lep | [OR, 0.63 (0.39-1.0) (allele)] | <0.05 | |
| R: MB vs con | [OR, 0.53 (0.30-0.91) (allele)] | <0.01 | |||||
| S: PB vs MB | OR, 1.65 (0.9-2.9) (allele) | <0.05 | |||||
| G−238A | NE: lep or lep type | ||||||
| 93 (2004) | Malawi | 258-283 controls, 216-243 leprosy (>90% PB) | G−238A | NE: lep | |||
| G−308A | NE: lep | ||||||
| G−376A | NE: lep | ||||||
| C−863A | NE: lep | ||||||
| 321 (2007) | Thailand | 140 controls, 37 leprosy (13 PB, 24 MB) | G−308A | S: MB vs con (het) | OR, 2.69 (ND) | 0.04 | |
| G−238A | NE: lep or lep type | ||||||
| VDR | 256 (1999) | India | 166 controls, 231 leprosy (107 TL, 124 LL) | TaqI T→C (“T”→“t”) | S: TL vs con | OR, 3.22 (1.47-7.13) (rec) | 0.001 |
| R: LL vs con | [OR, 0.60 (0.37-0.96) (dom)] | [0.03] | |||||
| [OR, 0.54 (0.33-0.86) (het)] | [0.01] | ||||||
| 93 (2004) | Malawi | 328-398 controls, 168-247 leprosy (>90% PB) | TaqI T→C (“T”→“t”) | Controls violate HWE | |||
| ApaI G→T (“a”→“A”) | NE: lep | ||||||
| BsmI C→T (“b”→“B”) | NE: lep | ||||||
| 113 (2006) | Brazil | 68 controls, 102 leprosy (55 PB, 47 MB) | TaqI T→C (“T”→“t”) genotypes alone | NE: lep or lep type | |||
| 85 (2009) | Mexico | 144 controls, 71 LL | TaqI T→C (“T”→“t”) | R: lep (LL) | OR, 0.55 (0.31-0.98) (dom) | 0.04 | |
| [OR, 0.50 (0.27-0.92) (het)] | [0.03] |
By convention, the variant SNP (a) is designated second (e.g., “A→a,” “A/a,” or “A[nucleotide position]a”).
When present, corrected P values are reported.
Abbreviations: con, controls, lep, leprosy (multiple forms or unspecified forms); MB, multibacillary; PB, paucibacillary; BL, borderline (depending on the study, this may indicate BB, BL, BT, or any combination of these); IL, indeterminate leprosy; LL, lepromatous leprosy (depending on study, this may indicate LL only or LL plus BL or plus BB); TL, tuberculoid leprosy (depending on the study, this may indicate TT or TT plus BT); ENL, erythema nodosum leprosum; RR, reversal reaction; MR, Mitsuda reaction; S, susceptible; R, resistant; dom, dominant model of genetic analysis (compares aa plus Aa versus AA); rec, recessive genetic model (compares aa versus AA plus Aa); het, heterozygotes or heterozygous model of analysis (Aa versus AA plus aa); HWE, Hardy-Weinberg equilibrium; del, deletion; ins, insertion; ND, no data (data not shown or not available); NE, no effect (no association found); NE: lep or lep type, phenotypes investigated and relevant variant not associated with either phenotype (i.e., not with leprosy overall or a specific type of leprosy, such as MB, PB, TT, or LL); NS, not significant; rel risk, relative risk. Data in brackets indicate where P values, ORs, or 95% confidence intervals were calculated by these reviewers or where ORs have been inverted to simplify comparison of the same risk allele across different studies.
The Vietnamese component of this study was a family-based association study.
Advances in genomic technology and immunology have accelerated the number of candidate gene association studies of infectious diseases. Table 1 summarizes both positive (association found) and negative (no association detected) association studies of leprosy for non-HLA candidate genes. A series of linkage and candidate gene studies has demonstrated associations of the major histocompatibility complex (MHC) class I and II loci (especially with HLA-DR2 alleles) with leprosy susceptibility (30, 39, 277, 305, 316, 317, 331, 347). Those and other studies of the MHC region were extensively reviewed elsewhere previously (108, 315) and will not be included in this review. This review prioritizes a discussion of the genes that have been most thoroughly examined with consistent and validated genetic findings and well-established functional effects. We will first describe family-based linkage studies that have identified regions or genes involved in susceptibility to leprosy. We will then review candidate gene association studies, which typically use a case-control study design with unrelated individuals. Finally, we will summarize findings from two recent genome-wide association studies of leprosy.
GENOME-WIDE LINKAGE STUDIES
Chromosome 10p13
The first genome-wide linkage analysis of leprosy was reported by Siddiqui et al. (282) and involved 224 families in India consisting of 245 sibling pairs, all but 4 of whom had exclusively paucibacillary disease. Three hundred eighty-eight microsatellite markers covering the entire genome were used in an initial screen to identify regions associated with leprosy susceptibility (maximum LOD score of ≥1, or P < 0.10) in an initial set of 103 sibling pairs (93 families). This screen produced 28 regions of interest (weak suggestive linkage), which were further assessed by using 37 markers in a separate set of 142 sibling pairs (131 families). In the second screen, one region on chromosome 10p showed significant linkage. This region was then fine-mapped, and significant linkage with paucibacillary leprosy was found for marker D10S1661 at 10p13 (LOD score, 4.09; P < 2 × 10−5) (282). Interestingly, this locus was confirmed as a risk factor for paucibacillary leprosy, but not overall leprosy susceptibility, in a separate linkage study by Mira et al. (197). In a follow-up study performed with families from Vietnam and cases and controls from Brazil (see below for details), two SNPs in the mannose receptor 1 gene (MRC1), located in the 10p13 region, were found to be associated with multibacillary leprosy and leprosy overall but not with paucibacillary disease (12). The lack of an association with paucibacillary disease in that study suggests that the causative gene at the 10p13 locus has not yet been identified.
Chromosome 6q25-26: PARK2 and PACRG
To identify genes that control susceptibility to leprosy, Mira and colleagues genotyped 388 microsatellite markers across the entire genome of 86 families in southern Vietnam with either multibacillary (MB) (56.1%) or paucibacillary (PB) (43.9%) disease (197). Chromosomal sites showing preliminary evidence of linkage were then fine-mapped with additional markers, and a region on chromosome 6q25-q27 was linked to leprosy (LOD score, 4.31; P = 5 × 10−6). In a separate group of 208 families, a transmission disequilibrium test (TDT) confirmed that two markers in the 6q25-27 region were strongly linked to leprosy susceptibility. In addition, linkage analysis performed on subsets of the families categorized as having either the PB or MB type of leprosy showed that 6q25-27 was not linked to one particular form of leprosy but seemed to be a determinant of leprosy risk overall (197). Evidence of the linkage of 10p13 with paucibacillary leprosy was also noted in this study (maximum LOD score, 1.74; P < 0.003), validating the findings of Siddiqui et al. in India (282). In addition, evidence for linkage at chromosome 6p21, the HLA locus, was also found (multipoint maximum likelihood binomial [MLB] LOD score, 2.62; P = 2.5 × 10−4), consistent with data from a previous report (277). Subsequently, that same group examined 81 SNPs in the 6.4-megabase region of 6q25-27 (196) that had been linked to leprosy in their previous study (197). In this scan, 17 SNPs that were associated with leprosy susceptibility were in or near the core promoter region of PARK2 and PACRG and were in strong linkage disequilibrium with each other. PARK2 (also known as PARKIN), a gene associated with Parkinson's disease, encodes an E3 ubiquitin ligase, and PACRG (also known as the Parkin-coregulated gene) is a neighboring gene of unknown function. Two SNPs, PARK2_e01(−2599) and rs1040079, accounted for the entire association at this locus. These results were validated in a separate set of 975 unrelated individuals in Brazil (587 with leprosy and 388 controls) (Table 1). Nine SNPs were confirmed to be significantly associated with leprosy risk in this population, of which the most highly significant were again the PARK2_e01(−2599) and rs1040079 alleles and a third SNP, PARK2_e01(2697) (196). In a separate case-control study in India, Malhotra et al. did not find a significant association (after conservative Bonferroni correction) between leprosy and SNPs in the PARK2 or PACRG coregulatory region, including PARK2_e01(−2599) and rs1040079, despite adequate power (184) (Table 1). The identification of PARK2 and PACRG as major leprosy risk genes in two populations, but not a third, highlights the heterogeneity of risk alleles for infectious diseases across different ethnic groups.
Chromosome 6p21: Lymphotoxin-α
In a study published in 2007 (8), the original PARK2 investigators (196, 197) revisited a second linkage peak that was found in the 6p21 chromosomal region on the initial genome-wide scan performed with Vietnamese families. This second peak fine-mapped to lymphotoxin-α (LTA), a T-cell cytokine gene located in the HLA class III region. Eight SNPs in this gene region (LTA−293, rs3131628, rs2523500, LTA+80, LTA+368, rs2516479, rs2844484, and rs2256965) showed linkage to leprosy. Notably, 7 of 8 SNPs were in strong linkage disequilibrium (LD) with each other, and the causative SNP could not be conclusively identified (8). Since the LTA+80 polymorphism was known to have a functional effect, it was considered to be the most likely candidate SNP. The LTA+80 polymorphism is located in a regulatory E2 box motif, C(A/C)GCAG, of the gene (8). The A allele allows the binding of the activated B-cell factor 1 transcriptional repressor and is associated with decreased LTA expression. The C allele alters the binding site of the transcriptional repressor (158). The abrogation of the LTA signaling pathway in mice is associated with increased susceptibility to intracellular pathogens (38, 116). Attempts to validate LTA SNP associations were made in two additional case-control studies in India (364 patients with leprosy and 371 controls) and Brazil (209 leprosy patients and 192 healthy controls) (Table 1). In addition, a second Vietnam sample set was studied (104 families). In these populations, no consistent association was seen between disease status and the SNPs LTA+80, LTA−294, or LTA−293 in unadjusted analyses. However, in all three ethnic groups, the risk conferred by the LTA+80 allele showed age dependence, with the highest ORs seen for the youngest age group. For example, the association of the LTA+80[A] allele with leprosy risk in Vietnam was almost entirely due to the effect on patients under 16 years of age (OR, 5.76; P = 4 × 10−5) (8). Since the LTA+80 variant is within 200 to 1,000 kb from neighboring HLA class I and II loci, theoretically, the susceptibility effect of this variant could be due to linkage disequilibrium with alleles in the HLA class I or II loci (108, 305, 339). However, there was no evidence of LD between alleles in HLA classes I and II and LTA+80 in Vietnamese and Indian subjects (8), strong evidence that the LTA gene is an independent risk factor. Subsequently, Fitness et al. used a candidate gene approach to investigate several alleles of a 5′ untranslated region (UTR) microsatellite polymorphism of LTA in Malawi (184 leprosy cases and 333 controls) and found one allele with an association with leprosy (93) (Table 1).
Overall, data from several different study populations provide good evidence that the LTA gene is implicated in leprosy susceptibility in some populations (Vietnam and India), although it is less clear which specific SNP accounts for the association. The LTA+80 SNP, for example, appears to exert its effect mostly on younger subjects. Its association with leprosy in older individuals or age-unspecified individuals is inconsistent (findings for a Brazilian population were negative). Interestingly, the age-specific incidence rates for leprosy in countries where the disease is endemic, such as India, show a peak in incidence in children aged 10 to 14 years, followed by a decline and then a second rise at around the age of 30 years that levels off (215). The reasons for this variable incidence are unknown but suggest that distinct risk factors may operate for individuals in different age groups.
Chromosomes 20p12 and 20p13
Two studies have reported linkage between chromosomal regions 20p12 and 20p13 and leprosy susceptibility (195, 306). The first study (306) was an extension of the genome-wide scan by Siddiqui et al. (282) that had identified a major susceptibility locus on chromosome 10p13 and a second region of weaker linkage on chromosome 20. In the initial screen, 388 microsatellite markers were genotyped in 93 families (103 sibling pairs) from Tamil Nadu, India, to identify regions associated with leprosy susceptibility (306). In the follow-up study, 11 markers in the chromosome 20 region with suggestive evidence of linkage (maximum LOD score of ≥1, or P < 0.10) in the initial scan were examined for a second set of 82 families in Tamil Nadu and 58 families in the neighboring state of Andhra Pradesh (140 families total). Except for 10 families, all siblings had paucibacillary leprosy. One marker (D20S115) showed strong evidence for linkage, with a multipoint maximum logarithm of odds score (MLS) of 2.17, although the effect was seen only within the Tamil Nadu families. To confirm these results, transmission disequilibrium testing of D20S115 and eight flanking markers was carried out for the families from Tamil Nadu. A microsatellite marker flanking D20S115 (D20S835) was associated with leprosy (P = 0.021) (306).
The second study to find an association with leprosy susceptibility in the chromosome 20 region was reported by Miller et al. for a Brazilian population (195). In the first stage, 21 families were genotyped for 405 markers, and nine regions with preliminary linkage to leprosy susceptibility were identified. In a second stage examining 50 new families, a linkage peak was found at marker D20S889, located on chromosome 20p13. This marker also showed linkage (LOD score, 1.51; P = 0.004) in the combined analysis (stage 1 and stage 2). The 20p13 site is about 3.5 megabases distal to the 20p12 linkage peak at D20S115 discussed above for paucibacillary leprosy susceptibility in India (306). Interestingly, among the families with tuberculoid leprosy, there was a nonsignificant linkage peak near marker D20S115 that was associated with the D20S835 allele (195). This study also replicated the finding of linkage with the chromosome 6p21 region (HLA and LTA loci) but failed to find linkage with chromosome 6q25 or 10p13. In addition, the authors noted evidence of a linkage at chromosome 17q22 (195).
CANDIDATE REGION LINKAGE STUDIES
Chromosome 17q11-21
A study by Jamieson and coworkers (142) selectively explored linkage between loci on chromosome 17 and leprosy susceptibility in a Brazilian population. Human chromosome 17q is syntenic to mouse chromosome 11, a region previously associated with increased susceptibility to cutaneous leishmaniasis in the mouse (31, 201, 253). Jamieson and colleagues therefore genotyped 16 microsatellite markers across chromosome 17q11.1-21.31 in 72 multicase leprosy families (208 affected individuals) and observed a broad region of linkage with two peaks at markers D17S250 (maximum Z score for likelihood ratio [Zlr], 2.34; P = 0.01) and D17S1795 (Zlr, 2.67; P = 0.02) (142). No data are presented for SNPs in regional candidate genes, which include multiple innate immune genes such as NOS2A, MCP-1, MIP1-α, MIP1-β, RANTES, CCR7, STAT3, STAT5A, and STAT5B.
Chromosome 21q22
Wallace and coworkers performed a search for genes associated with tuberculoid or lepromatous leprosy on chromosome 21 using an identity-by-descent (IBD) regression analysis (330). This method of analysis compares the likelihood of linkage for a given allele in type-concordant sibling pairs (both siblings have the same form of leprosy) to that in type-discordant sibling pairs (those in which siblings have different forms of leprosy). In the first stage of the analysis, 83 families in Malawi were genotyped for markers for the selected region. Makers for regions showing preliminary evidence of linkage (P < 0.05) were genotyped in 185 extended pedigrees. In the first stage of the analysis, regions on chromosomes 10q23, 15q21, and 21q22 were singled out for further study. In the second stage of the analysis, only the region on chromosome 21q22 retained a suggestive association with leprosy susceptibility (P ∼ 0.001) that did not meet genome-wide linkage study criteria for significance (330). Those authors speculated that the most likely candidate gene for this region is ITGB2, a gene that encodes the β2 subunit of leukocyte integrins (330). Interestingly, the 21q22 region was more likely to be shared by type-concordant pairs and less likely to be shared by type-discordant sibling pairs, suggesting that this locus affects susceptibility to a specific form of leprosy rather than leprosy overall.
SUMMARY OF LINKAGE STUDIES
The strongest evidence for the linkage of non-HLA genes with leprosy and/or leprosy type susceptibility exists for chromosome 6q25, subsequently linked to the PARK2/PCRG gene regulatory region (196, 197); chromosome 6p21, subsequently mapped to the lymphotoxin-α gene (8, 195, 197); and chromosome 10p13, for which the candidate gene has not yet been identified (197, 282). Each of these regions has been validated for separate populations and/or alternate ethnic groups studied by the same or other investigators, and in two of the three cases, a causative gene has been identified. While the causative gene has not been identified for chromosomal region 10p13, this region's specific association with paucibacillary leprosy was validated in a subsequent study (197).
CANDIDATE GENE ASSOCIATION STUDIES
Innate Immune Receptors
TLR1.
Toll-like receptor 1 (TLR1) forms a heterodimer with TLR2 to mediate the recognition of M. leprae (163). A TLR1 polymorphism, T1805G (I602S), encodes a nonsynonymous SNP in the transmembrane domain of TLR1 that regulates signaling in response to Pam3CysK4, a synthetic ligand of TLR1 (121, 302, 338). Individuals homozygous for the 1805G variant are functionally TLR1 deficient (121). In previous work, we and others have observed that the T1805G SNP strongly regulates NF-κB signaling via the TLR1/2 receptor such that leukocytes from 1805G homozygous donors have a 2-fold-or-greater reduction in responses to Pam3CysK4 compared to 1805T homozygous donors (121, 149, 198, 338). In HEK293 cells stimulated with Pam3CysK4, NF-κB signaling was deficient in 1805G compared to 1805T transfectants (121). Additionally, we found that the T1805G SNP regulates in vitro responses to whole irradiated M. leprae bacteria and to cell wall extracts of M. leprae (198). Both Johnson et al. (149) and Wurfel et al. (338) demonstrated that leukocytes from individuals with the 1805GG genotype lack surface expression of TLR1, in contrast to leukocytes from 1805TT individuals. Therefore, the 1805G variant of human TLR1 confers a state of functional TLR1 deficiency in which hyporesponsiveness to TLR1 ligands appears linked to a TLR1 trafficking defect. Interestingly, this polymorphism has extreme variation in frequency among different populations worldwide and suggests that TLR1 could have different impacts on susceptibility to leprosy and other diseases depending on the population studied (22, 121). The 1805G SNP has been shown to be under positive selection, suggesting that this allele, which confers a hyporesponsive immune response to M. leprae and possibly other bacterial pathogens, offers a selective advantage in certain populations (22, 79).
Johnson and coworkers examined the role of the TLR1 T1805G SNP in modulating leprosy susceptibility in 57 patients and 90 controls in Turkey and found that the 1805G SNP was associated with protection from leprosy (OR, 0.48; P = 0.004) (149). We examined the relationship of TLR1 T1805G with tuberculoid versus lepromatous leprosy and two leprosy immune reactions, erythema nodosum leprosum (ENL) and reversal reaction (RR), in 933 leprosy cases in Nepal, including 238 cases of RR (198). TLR1 1805G was not associated with leprosy risk overall, nor was it associated with tuberculoid or lepromatous leprosy, although there was a trend toward an association with lepromatous leprosy (OR, 4.76; P = 0.11). However, the 1805G SNP was significantly associated with protection from a reversal reaction, a Th1-mediated event (OR, 0.51; P = 0.01). The frequency of 1805G was low in this population and decreased the overall power to detect associations.
A third group examined another TLR1 SNP, A743G (N248S), for leprosy associations using a large group of 842 patients and 543 controls in Bangladesh. The 248SS genotype and S allele were associated with an increased risk of leprosy (OR, 1.34; P = 0.02, recessive model) and a decreased risk of ENL (OR, 0.40; P = 0.04), respectively, although the subgroup analysis for ENL had a sample size of only 11 (271). In addition, the 248S allele was associated with a trend toward an increased risk of RR (subgroup of 75 cases), but this was not statistically significant. Interestingly, in our prior functional studies of TLR1 (121), we found that SNP 248S (743G) was in strong LD with 602I (1805T), a hyperfunctional variant of TLR1 associated with increased signaling in response to triacylated lipopeptide and M. leprae (198). The T1805G SNP was also investigated in this Bangladeshi population and was found to have no association with leprosy susceptibility (271). In summary, these studies suggest that TLR1 influences susceptibility to leprosy immune reactions and leprosy susceptibility, with slightly more evidence in favor of T1805G, rather than A743G, as the causative SNP. In addition, the T1805G SNP was recently investigated in a GWAS in India, and the G allele was associated with protection against leprosy (336).
NOD2.
The Nod-like receptors (NLRs) are a family of cytosolic receptors that detect microbial cell wall products. Nucleotide oligomerization domain 2 (NOD2) recognizes muramyl dipeptide (MDP) (98) as well as mycobacteria (90, 91). The cell wall from M. leprae contains a unique MDP structure, which differs from that seen for M. tuberculosis and most Gram-negative organisms, and may elicit distinct NOD2-mediated immune responses (64, 183). To our knowledge, no functional studies of the role of NOD2 in the immune response to M. leprae have been reported. However, in mouse models of M. tuberculosis infection, the deletion of NOD2 is associated with impaired in vivo and in vitro immune responses (75, 220, 344). Human mutations in NOD2 have been associated with altered susceptibility to tuberculosis (TB) in African-Americans (19), inflammatory bowel disease (131), and Blau's disease (192), the latter two of which are diseases with dysregulated granuloma formation. Three recent genetic association studies (including two genome-wide association studies [GWASs]) have also examined the role of NOD2 polymorphisms in leprosy (27, 337, 348).
A recent GWAS identified two polymorphisms in NOD2, rs9302752 and rs7194886, associated with susceptibility to leprosy (348). That group also reported associations with polymorphisms in RIP2 kinase, a molecule in the NOD2 signaling pathway (see below for a full discussion of this GWAS). In contrast, Wong et al. did not note associations with NOD2 pathway genes (NOD2 and Rip2K) in an association study in India and Mali (337). When those authors fine-mapped the NOD2 gene region, they identified five polymorphisms in the NOD2 gene region, including rs7194886, with leprosy associations that became insignificant after correction for multiple comparisons (337). In a separate study by Fitness and colleagues, several uncommon NOD2 polymorphisms associated with Crohn's disease were examined in leprosy cases from Malawi, but no association was observed (93). In a recent case-control study of 933 leprosy patients (124 with ENL and 240 with RR) and 101 controls from Nepal, we identified common noncoding polymorphisms in the region around NOD2 by searching a region on chromosome 16q12 from 50 kb upstream to 50 kb downstream of the NOD2 and CYLD genes for haplotype-tagging SNPs. Eight SNPs were associated with increased susceptibility to leprosy (odds ratios ranging from 1.7 to 2.5) (27) (Table 1). Ten of these SNPs were also associated with leprosy reactions (see Table 1 for details), especially ENL. Of note, some polymorphisms included in this study were in genes adjacent to NOD2 (SLIC-1 [or SNX20] and CYLD) and may therefore implicate these genes in the host response to M. leprae.
Taken together, those studies provide evidence that the NOD2 gene region and intracellular immunity may be important in the host response to M. leprae. However, a number of questions remain. First, strong associations in the NOD2 gene in one population have been marginal or absent in other populations (337). This result could be due to population-specific linkage disequilibrium, which occurs when the true causative SNP is in LD with the identified SNP in one population but not in another (11). Second, NOD2 polymorphisms may be important for different aspects of the disease in different ethnicities. Ethnic variation in the NOD2 association, for example, also occurs for Crohn's disease (343). This result can arise for diseases governed by complex or polygenic inheritance patterns, where susceptibility is a cumulative effect of epistatic interactions among several or many genes. Last, the mechanism by which these noncoding region SNPs might alter the innate immune response to leprosy is unclear. Overall, therefore, data from NOD2 genetic studies suggest that this gene may play a role in leprosy. However, the lack of consistency among genetic association studies in different populations and the absence of a functional mechanism for these SNPs require further exploration.
TLR2.
Bochud et al. studied a number of Toll-like receptor 2 gene (TLR2) variants in an Ethiopian case-control study (441 cases and 197 controls) (35). A 290-bp microsatellite (MS) polymorphism composed of two adjacent variable-number tandem repeats (VNTRs) in the TLR2 promoter region was less frequent in cases than in controls (OR, 0.62; P = 0.02, additive model). When comparing lepromatous and tuberculoid patients, another MS variant (288 bp) was less frequent in those with lepromatous leprosy (OR, 0.49; P = 0.02, dominant model). In a small subgroup of patients (n = 216) monitored for 8 years to assess leprosy reactions, the 288-bp MS variant was also strongly associated with an increased risk of RR (OR, 5.83; P = 0.001, recessive model). In addition, a synonymous-coding-region SNP, C597T, was associated with protection from RR (OR, 0.34; P = 0.002, dominant model). The C597T association with RR remained significant even after a conservative Bonferroni adjustment (e.g., 0.002 × 21 = 0.042) (35).
Fitness and colleagues also investigated an intron 2 MS polymorphism previously associated with altered receptor function (322, 345) in a Malawi population (∼210 leprosy patients and ∼379 controls), but no association was found with disease (93). They identified a borderline significant (P = 0.042) difference in the genotype frequency of the 224-bp microsatellite between PB and MB patients, although the small number of MB patients (n = 26) makes this result possibly due to sampling error.
One study reported the association of a putative TLR2 polymorphism, C2029T (R677W), with leprosy susceptibility in a South Korean population (154). Subsequent investigators convincingly demonstrated that R677W is an artifact that arises when genotyping primers amplify both TLR2 and a nearby TLR2 pseudogene and that this SNP does not exist in the authentic TLR2 gene (186, 194).
TLR4.
Toll-like receptor 4 (TLR4) is present on many cell types, including macrophages, monocytes, and dendritic cells, and recognizes lipopolysaccharide (LPS) from Gram-negative bacteria (228). While M. leprae signals predominantly through TLR1/2 heterodimers (163), there is evidence that live M. tuberculosis may signal through TLR4 (190). Several polymorphisms have been described for the coding region of TLR4 (G896A, C1196T, G1530T, and A1976G), and some (G896A and C1196T) have been shown to impair signaling through TLR4, as evidenced by decreased cytokine production in patients given inhaled LPS (17). The G896A (D299G) and C1196T (T399I) polymorphisms have been extensively studied and are associated with many bacterial illnesses, including Gram-negative infections and Legionnaires' disease (5, 109, 122, 270). Functional studies of these polymorphisms have suggested a possible signaling defect, although this was not validated by other groups and remains unclear (17, 52, 80, 82, 193, 222, 265, 313, 326).
In an Ethiopian population (441 cases and 197 controls), we identified two TLR4 SNPs, G896A and C1196T, that were associated with protection from leprosy (OR of 0.34 [P < 0.001, additive model] and OR of 0.16 [P < 0.001, dominant model], respectively) (36). Additionally, we found that signaling in monocytes through TLR4 by endotoxin was inhibited by heat-killed M. leprae (36). In another study in Malawi of 235 patients and 88 controls, Fitness et al. found no association of the G896A polymorphism with leprosy (93). Overall, the association of TLR4 with leprosy is inconclusive, since the associations in Ethiopia were not validated in Malawi, and requires further investigation in other populations. For example, the different conclusions of these two studies regarding the association of TLR4 with leprosy overall could be due to different patterns of linkage disequilibrium between the two populations or to a true lack of an effect of TLR4 (and/or causative genes in LD with TLR4) on leprosy susceptibility. Alternatively, it is possible that TLR4 is associated predominantly with lepromatous leprosy, since 298 of 441 (67.5%) of the Ethiopian cases were at the lepromatous pole (classified as BL, LL, or multibacillary), compared to only 26 of 270 (9.6%) cases in Malawi. In fact, the authors of the Malawi study stated that the results for most SNPs studied did not change when the 28 multibacillary patients were excluded. From this, it can be inferred that the Malawi study likely lacked the power to detect genetic associations specific to multibacillary disease.
MRC1.
Mannose receptor 1 (MRC1) is a phagocytic receptor that recognizes mannose, fucose, N-acetylglucosamine (GlcNAC) (157), and mannose-capped lipoarabinomannan, a mycobacterial cell wall lipoprotein (144, 268). MRC1 localizes to chromosome 10p13 (77), a previously identified leprosy susceptibility locus (282). A study by Alter and colleagues examined polymorphisms in exon 7 of the MRC1 gene in a family-based association study of 580 Vietnamese families and a case-control study of 783 Brazilians (12). Exon 7 was the focus of investigation, since unpublished reports implicated this region of the MRC1 gene as a risk factor for PB leprosy (Tosh et al., unpublished data cited in reference 127 and referenced in reference 12). Of three previously described nonsynonymous SNPs in MRC1, only one, G396S, was found after sequencing healthy individuals in Vietnam. In Vietnamese families, weak evidence for an association of the 396S variant was observed with protection against leprosy (OR, 0.76; P = 0.035) and against MB disease (OR, 0.71; P = 0.034). To replicate these findings, exon 7 SNPs were then investigated in a genetic association study of 384 leprosy patients and 399 controls in Brazil. In Brazil, the 396S allele was again associated with protection from leprosy overall (OR, 0.75; P = 0.016) and from MB disease (OR, 0.70; P = 0.023). When MRC1 haplotypes containing the 396G or 396S SNP were ectopically expressed in HEK293 cells, no differences in the phagocytosis of zymosan or ovalbumin were observed between the two variants (12). Interestingly, the transfected HEK cells were unable to bind M. leprae or BCG, suggesting that the mannose receptor may cooperate with an unknown second receptor to internalize M. leprae (12).
Although only one genetic study has been performed on MRC1 and leprosy, the concordant findings for the two different ethnic groups are encouraging and suggest a possible role for MRC1 variants in leprosy susceptibility. However, functional studies of the MRC1 SNPs are inconclusive. This gene was considered to be the strongest candidate gene at the chromosome 10p13 locus, a region that has been linked to susceptibility to PB leprosy. The lack of an association between MRC1 and PB leprosy in the current study suggests that an alternate candidate gene specific for PB leprosy may exist in the 10p13 region.
VDR.
Vitamin D modulates diverse effects on the immune system, which can be inhibitory or stimulatory depending on the cell type and the nature of the immune response. In dendritic cells, vitamin D inhibits maturation by blocking the expression of MHC class II, CD40, CD80, and CD86 (208) and reducing the expression of IL-12 (224). In macrophages, vitamin D also downregulates IL-12 expression (66). However, in monocytes and macrophages infected with M. tuberculosis, vitamin D augments the TLR1/2-stimulated expression of cathelicidin and thereby enhances intracellular killing (179). In contrast, the net effect of vitamin D on the adaptive immune response is an enhancement of Th2 T-cell responses at the expense of Th1 responses (208). Vitamin D blocks Th1 responses by inhibiting the expression of the Th1 cytokines IL-2, IFN-γ, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (29, 246, 308) and suppressing lymphocyte proliferation (174, 250). Vitamin D also enhances the generation of nonspecific T suppressor cells, inhibits the induction of CD8+ T cells, and reduces class II antigen expression in mixed-lymphocyte reactions (174, 208).
Several polymorphisms located near the 3′ UTR of the vitamin D receptor gene (VDR) (BsmI, ApaI, and TaqI) have been reported to regulate the stability or transcriptional activity of VDR mRNA (209). The TaqI C (or t, as it is sometimes represented) polymorphism has been associated with higher mRNA transcript levels of VDR (209), although this finding is controversial (200, 323).
The TaqI VDR polymorphism was studied in a population in Kolkata, India, in a case-control genetic association study involving 231 patients with leprosy (107 with tuberculoid disease and 124 with lepromatous disease) and 166 controls matched for ethnicity (256). No differences were seen in the distribution of TaqI genotypes between all leprosy patients and controls. However, the CC (tt) genotype was found significantly more frequently in tuberculoid leprosy than in controls (OR, 3.22; P = 0.001, adjusted), and conversely, the TT genotype was enriched for patients with lepromatous leprosy compared to controls (OR, 1.67; P = 0.04, adjusted) (256). Interestingly, although not reported by those authors, individuals with TC genotypes were protected from lepromatous leprosy, suggesting a heterozygous advantage (Table 1).
A second study in Malawi examining ∼247 leprosy cases and ∼398 controls also found a significant association of the TaqI CC (tt) genotype with an increased risk of leprosy (OR, 4.3; P = 0.004, unadjusted) (93). All but 26 of the leprosy cases were PB or tuberculoid leprosy. However, as those authors pointed out, the controls were not in Hardy-Weinberg equilibrium (HWE) for this polymorphism, suggesting that this finding may be due to population stratification or genotyping error rather than a bona fide association.
TaqI receptor polymorphisms were also investigated in a study of 71 Mexican patients with lepromatous leprosy compared to 144 healthy blood bank donor controls. The TC (Tt) or CC (tt) genotypes combined were associated with protection from leprosy (in this case, lepromatous leprosy), with marginal significance (OR, 0.55; P = 0.04) (85), an effect similar in direction and magnitude to that reported in the Kolkata study (256). Of note, the patients included in that study appeared to have lepromatous leprosy exclusively. In another study in Brazil of 102 Brazilian patients (55 with PB and 47 with MB) and 68 nonconsanguineous household contacts, no association was found between TaqI genotypes and overall leprosy susceptibility or leprosy subtype (113). It is quite likely that this study lacked the power to find an association of TaqI with either PB or MB disease, given 1.5- to 2.5-fold-higher numbers of patients with lepromatous or tuberculoid leprosy in other studies reporting an association for this SNP (85, 256). This negative study illustrates the problem of comparing genetic association studies carried out with populations with different ratios of multibacillary to paucibacillary disease, which could be circumvented by carrying out replication studies of populations with a similar bias along the leprosy clinical spectrum or by ensuring the recruitment of adequate numbers of patients in the relevant categories.
The somewhat contradictory findings with respect to the effect of the CC (tt) genotype or C (t) allele on leprosy susceptibility in different studies could be due to differences in ethnic background, variations in sample size affecting study power to investigate associations with different types of leprosy, or other aspects of study design. Another consideration is that the 3′ end of the VDR gene also contains several polymorphisms in various degrees of linkage disequilibrium in different populations (135, 256). As a result, the effect of a TaqI polymorphism might be attributable not to TaqI per se but to other alleles (including ApaI or BsmI) that are in population-specific linkage disequilibrium with the TaqI SNP. It is interesting that of the two studies that reported an association of this SNP, both displayed a dominant protective effect of the t allele against lepromatous leprosy as well as a heterozygous advantage (85, 256).
Summary of association studies of innate immune receptors.
The most consistent findings among the different studies of receptors are for the effect of TLR1 (SNP T1805G) on leprosy susceptibility and/or an altered risk of a reversal reaction and for NOD2 associations with leprosy. The most clear functional effect is that of TLR1 1805G, which has been shown by several groups to impart a signaling defect for ligands operating through the TLR1/2 heterodimer (121, 149, 198, 338).
Cytokines
TNF.
TNF is a multifunctional proinflammatory cytokine produced by monocytes and macrophages and is important for the control of mycobacterial and other infectious diseases. Treatment with TNF inhibitors may be associated with the development of leprosy, and the withdrawal of TNF inhibitor treatment in leprosy patients may also enhance the formation of a type I reversal reaction (274). There is also evidence that TNF inhibitors may be effective for the treatment of recurrent erythema nodosum leprosum (81). While some studies have documented the association of the TNF promoter region polymorphisms with susceptibility to different infections, the validity of these findings is unclear due to a lack of reproducible data in reports from other populations (24, 51, 187).
The first association study of the TNF promoter region SNP G-308A was carried out by Roy et al. in a population in Kolkata, India (257). In that study, the TNF −308A allele was noted to be significantly increased for patients with LL (n = 121) compared to TT (n = 107 patients) (RR = 2.5; P = 0.03) (257). This finding was not replicated in a linkage study of six French Polynesian families (176). In a Brazilian population of 300 leprosy cases and 92 controls, the −308A allele was associated with paucibacillary disease (OR, 1.65; P < 0.05) (261, 262), a finding opposite of that seen for the Indian population (257). In addition, the −308A allele was associated with protection from leprosy acquisition (OR, 0.63; P < 0.05) and MB leprosy (OR, 0.53; P < 0.01). These findings were more pronounced for the female subgroup, where the TNF −308A allele was associated with protection from leprosy overall (OR, 0.4; P = 0.01) and from MB leprosy (OR, 0.21; P < 0.01). In this Brazilian study, a second SNP, G−238A, was not associated with leprosy overall or leprosy type. A separate TDT analysis in Brazil found that the −308G allele was more frequent for patients with leprosy or lepromatous leprosy (277). In a case-control association study in Thailand, the −308A allele was associated with multibacillary leprosy (OR, 2.69; P = 0.04) (321). A linkage study of 223 families and 230 sibling pairs in southern India also demonstrated associations of a TNF MS polymorphism with leprosy, but the associations failed the bootstrapping test used in this study (P = 0.089 after bootstrapping) (307). In this Indian study, the G−308A polymorphism was also not associated with susceptibility to leprosy. Last, in a case-control study of 933 patients with leprosy and 101 control patients in Nepal, we found that the −308A allele was associated with protection against leprosy (OR, 0.52; P = 0.016) (W. R. Berrington, unpublished data).
The biological mechanism of action of G−308A and other promoter variants remains uncertain. Overall, functional investigations of the −308A and the −238A promoter SNPs through a variety of approaches (in vitro, in vivo, and ex vivo) have not shown a consistent association of either variant with increased or decreased TNF production (40, 87, 104, 165, 181, 291, 301, 303). The TNF −308A allele, for example, has been shown to have increased transcriptional activity in some studies in which this variant has been cloned upstream of a reporter construct (162, 335) but not in others (43, 159). Knight et al. investigated the transcriptional activity of the TNF −308A and −308G variants in vivo in Epstein-Barr virus (EBV)-transformed human B-cell lines and did not find differences in TNF transcription (by levels of phosphorylated RNA polymerase II) between the two alleles (159). The variable results of these functional studies of the −308A variant may be due partly to different experimental approaches, such as the reliance on in vitro assays of plasmid reporter gene expression versus in vivo assays with human cell lines, the use of specific cell lines, or variable stimulation conditions (335). Even if it were clear which TNF alleles were associated with altered gene transcription or cytokine production, the role of TNF (and other cytokines) in the pathogenesis of leprosy and leprosy immune reactions is still not well understood (272, 273).
Collectively, the genetic association data for TNF are also inconsistent. While some studies support an association of the SNP G−308A with altered susceptibility to leprosy overall or leprosy type, the SNP appears to have opposite effects on leprosy polarity (tuberculoid versus lepromatous) in different populations. The TNF gene is located on chromosome 6q23-12 in a region in close linkage with HLA-DR (51, 187, 213), a gene locus that has also been associated with leprosy (39, 40, 108, 315, 317). Similarly, the TNF −308 SNP is in LD with the LTA gene, and a haplotype containing the −308A allele and LTA SNPs +10A, +252G, and +723A has been shown to be associated with increased LTA transcriptional activity (159). Population-specific differences in the LD between TNF and either or both of these two candidate gene regions, if unaccounted for, could potentially confound the results of association studies of this candidate gene.
IL10.
Interleukin-10 (IL-10) is an anti-inflammatory cytokine that inhibits the production of IL-1, IL-6, and TNF in LPS- and IFN-γ-activated macrophages (92, 129, 138, 166, 205). IL-10 also impairs the host response to mycobacteria, as shown for IL-10-overexpressing mice that failed to clear mycobacterial infection (211). In humans, elevated mRNA expression levels of the Th2 cytokines IL-4, IL-5, and IL-10 has been demonstrated for LL lesions in contrast to TT lesions, where the Th1 cytokines gamma interferon and IL-2 predominate (341). Data from clinical studies suggest that higher levels of IL-10 and lower TNF-to-IL-10 ratios correlate with multibacillary leprosy, T-cell anergy in patients with lepromatous leprosy, and progression from leprosy exposure to symptomatic illness (177, 199).
Santos et al. investigated SNPs in the IL10 promoter region in 143 patients with MB leprosy, 59 patients with PB leprosy, and 62 healthy controls (262). A greater frequency of the −819TT genotype was found among leprosy patients (MB and PB combined) than controls (OR, 2.64; P = 0.04, recessive model) (262). The promoter polymorphism −819T was also more frequent for patients with PB leprosy (262). Those investigators also studied the association between haplotypes of five promoter polymorphisms in IL10 and multibacillary or paucibacillary leprosy in a separate group of approximately 297 patients from Rio de Janeiro, Brazil (206). No association was found between any of the individual polymorphisms, including C−819T, and leprosy susceptibility, although there were some haplotype associations. In addition, one haplotype was more strongly associated with PB leprosy (Table 1) (206). Franceschi and colleagues also investigated promoter SNPs A−1082G, C−819T, and C−592A in 156 leprosy patients (65 LL, 49 TT, and 45 BB patients) and 240 controls in Brazil (Parana state) and found a lower frequency of the IL10 promoter haplotype −1082G/−819C/−592C among patients with lepromatous leprosy. However, this finding lost significance after correction for multiple comparisons (96).
Malhotra and colleagues also studied IL10 SNPs and haplotypes for 282 leprosy patients and 266 controls in India and found associations for both the C−819T and the C−592A polymorphisms (185). Individuals with the −819TT genotype were at a significantly increased risk of leprosy (OR, 2.50; P < 0.001), and individuals with the −592CC genotype were significantly protected from leprosy (OR, 0.60; P = 0.006) (185). That group also examined the effect of the individual IL10 SNPs T−3575A, G−2849A, C−2763A, A−1082G, C−819T, and C−592A and haplotypes of these SNPs on leprosy type (185). In contrast to the report by Santos et al., they found that the −819TT genotype was more frequent in patients with multibacillary (MB) leprosy than in controls (OR, 2.63; P = 0.001). In addition, the −592CC genotype was associated with protection from MB leprosy (OR, 0.48; P = 0.002) (185) (Table 1).
Fitness et al. examined three IL10 proximal promoter polymorphisms, A−1082G, C−819T, and C−592A, in a Malawian population (∼362 leprosy cases and ∼215 controls) (93). No association was found between any of these IL10 variants and leprosy risk, although there was a trend toward an association of the −592CC genotype with leprosy resistance compared to AA homozygotes (OR, 0.58; P = 0.06) (93).
In a large case-control study (∼369 leprosy patients and ∼380 controls), Pereira et al. investigated the C−819T SNP and four other IL10 promoter SNPs (225). Carriers of the −819T allele and individuals with the −819TT genotype were found to be at an elevated risk of leprosy compared to controls (OR, 1.44 for the comparison of TT/CT versus CC; P = 0.026, adjusted analysis) (225). Those authors subsequently performed a meta-analysis of that study and four prior studies performed in Brazil, India, and Malawi (93, 185, 206, 262). A fifth, negative study from Brazil was not included (96). The meta-analysis showed that the −819T allele and the TT genotype were both significantly associated with an elevated risk of leprosy (P = 0.0001 to 0.024), although the magnitude of the risk was modest (OR, 1.28 to 1.66) (225).
A number of functional studies have been carried out on disease-associated polymorphisms or haplotypes of IL10, with inconsistent results. A high-frequency IL10 haplotype containing SNP −819T has been described for healthy Dutch Caucasians and African-Americans and was associated with elevated levels of IL-10 production (110). However, other studies found lower IL-10 levels associated with the −819T SNP or in haplotypes containing this SNP (225, 312). In one study, cytokine responses of peripheral blood mononuclear cells (PBMCs) from healthy donors with differing genotypes were examined. Donors with the −819 TT or CT genotypes produced significantly less IL-10 in response to stimulation with high doses of M. leprae than did individuals with the CC genotype (225).
Elevated IL-10 production in individuals with the −819T SNP is a biologically plausible mechanism to account for enhanced leprosy susceptibility but leads to the prediction that this SNP would be enriched in MB rather than PB leprosy. Consistent with this expectation, elevated IL-10 production has been associated with MB or lepromatous forms of leprosy in clinical studies (199, 341). In the genetic association studies reviewed here, one group reported an association of −819T with MB leprosy (185), and another reported an association with PB leprosy (262). One potential explanation for these conflicting data could be that “distal” promoter SNPs closer to the 5′ end of the promoter than −819T may be the actual regulatory SNPs for IL-10 production (110).
In summary, the majority of studies of the IL10 promoter SNP C−819T support a role for its association with leprosy susceptibility, and functional studies of this SNP in human primary cells suggest that it mediates IL-10 production in response to M. leprae. However, there is no consistent evidence that the C−819T SNP is associated with any particular form of leprosy. There is suggestive evidence that the IL10 SNP C−592A alters leprosy risk, with significance found in one study (185) and a consistent trend found in another study (93). For the IL10 polymorphisms T−3575A, G−2849A, C−2763A, and A−1082G, different haplotype combinations have been studied for different ethnic groups, and the data are difficult to evaluate.
IFNG.
Gamma interferon (IFN-γ) is a prototypic Th1 cytokine produced by activated CD4+ T cells or CD8+ T cells that is essential for the effective control of intracellular pathogens. IFN-γ from T cells stimulates a mycobactericidal response in macrophages involving the production of NO and other reactive species (94). IFN-γ produced by dendritic cells, macrophages, and NK cells also upregulates the expression of the signaling subunit of the IL-12 receptor on T cells, allowing T cells to respond to IL-12 from innate immune cells, undergo Th1 differentiation, and produce additional IFN-γ (130, 310). Mice deficient in IFN-γ develop disseminated M. tuberculosis after aerosol or intravenous challenge (63, 95). Similarly, in an experimental leprosy infection model using mouse footpads, macrophages from heavily infected tissue are refractory to gamma interferon activation and fail to kill unrelated intracellular pathogens or to produce normal levels of superoxide (160).
Gamma interferon gene (IFNG) polymorphisms have been extensively investigated in tuberculosis association studies, and several common SNPs are associated with TB susceptibility (62, 99, 325). The IFNG gene contains a microsatellite polymorphism in intron 1 with a variable number of CA repeats. Pravica and coworkers have shown that certain alleles of this MS repeat are associated with elevated levels of IFN-γ production in healthy individuals (231). Seven alleles at the intron 1 MS polymorphic site were investigated in an association study of 192 leprosy patients and 196 controls (248). Although no allele was individually associated with an altered risk of leprosy or leprosy subtypes, there was a significant difference in the distribution of short versus long CA repeats among tuberculoid leprosy patients (P = 0.013) compared to controls. In addition, when individuals were divided into groups of those with longer alleles and those with shorter alleles, a significantly higher percentage of leprosy patients had the longer alleles (17.1% versus 6.5%; P = 0.01) (248). Another IFNG intron 1 polymorphism, T874A, had no association with leprosy in Malawi (93).
Transport Molecules
SLC11A1.
The solute carrier family 11 member 1 gene (SLC11A1), also known as NRAMP1 (natural resistance-associated macrophage protein 1), was first described as a gene known as Ity/Lsh/Bcg that controlled the susceptibility of inbred mice to intracellular pathogens, including M. leprae, Mycobacterium bovis (BCG), Leishmania donovani, and Salmonella enterica serovar Typhimurium (41, 42, 46, 226, 227, 286, 287). SC11A1 has since been shown to have diverse effects on macrophage function, but the mechanism by which this gene regulates the killing of intracellular pathogens remains unsettled. Ity/Lsh/Bcg has been shown to control macrophage activation and to reside in the endosome and lysosome, where it functions as an iron transporter (285, 288). SLC11A1 removes iron essential for the survival of intracellular bacteria from the phagolysosome, where these organisms reside, into the cytoplasm (32). Mice with a mutation in Nramp1 are also deficient in other aspects of innate immune function, including antigen presentation, the oxidative burst, NOS expression and NO production, TNF production, and IL-1β production. These diverse effects may or may not be linked to iron and free-radical metabolism (32). In addition, the adaptive immune phenotype of NRAMP-mutated mice is biased toward Th2, rather than Th1, T-cell responses (32). An SLC11A1 promoter polymorphism has also been associated with increased susceptibility to tuberculosis in two different studies (25, 31).
Meisner and colleagues studied several SLC11A1 variants in 273 (181 MB and 92 PB) leprosy patients and 201 controls in Mali (191). A 4-allele CA microsatellite in the 5′ region of the gene, an SNP in intron 4 (469 + 14 G/C), and a TGTG deletion/insertion in the 3′ UTR (1729 + 55del4) were examined. Homozygotes for the TGTG 3′ UTR deletion were much more commonly found in the PB group than in the MB group in comparison to heterozygotes (OR, 5.79; P = 0.003) (191). Ferreira et al. investigated the frequency of the CA (=GT) repeat promoter polymorphism in Brazil in 90 cases (45 MB and 45 PB cases) compared to 61 nonconsanguineous household contacts but found no association with PB or MB disease (89).
Two other candidate gene studies also failed to detect any associations between SLC11A1 polymorphisms and leprosy. Vejbaesya and colleagues examined three polymorphisms in SLC11A1; a polymorphism in intron 4 (469 + 14G/C); a coding SNP, G1627A (D543N); and the TGTG deletion in the 3′ UTR (1729 + 55del4) in a very small study (24 MB, 13 PB, and 140 controls) and found no association with leprosy in a Thai population (321). A second study in India (107 tuberculoid versus 124 lepromatous patients) examined the 4-allelic CA repeat promoter polymorphism and several other variants and found no associations with leprosy (256).
A number of investigations of the linkage between leprosy and SLC11A1 have been reported. Abel et al. examined the evidence for linkage between polymorphisms in SLC11A1 and neighboring genes and leprosy in 16 Vietnamese and 4 Chinese families (2). They found evidence of linkage of both the SLC11A1 intragenic haplotypes and the extended haplotypes (formed from SNPs in SLC11A1 and flanking genes) with leprosy in the Vietnamese families and all families combined but not in the Chinese families. That study also looked for associations of SLC11A1 haplotypes with leprosy among unrelated affected and unaffected parents but was not able to identify a haplotype associated with altered leprosy risk, likely due to the small sample size (2). The number of individuals with tuberculoid or lepromatous leprosy was not provided.
Subsequently, that same group used a segregation analysis to investigate genetic determinants of the Mitsuda reaction in 168 Vietnamese and Chinese families (89 lepromatous versus 159 nonlepromatous cases). Those authors found that the quantitative Mitsuda reaction is under the control of a second major gene distinct from SLC11A1 that operates as a recessive trait (241). In a follow-up genome-wide scan to identify chromosomal regions in linkage with quantitative Mitsuda reactivity in 19 families (25 tuberculoid versus 29 lepromatous cases) in Vietnam, two regions of linkage were identified (242). One was located on chromosome 2q35 and corresponded to the SLC11A1 locus, while the second was found on chromosome 17q21-25 and corresponded to a diverse group of immune genes. Of interest, chromosome 17q11-21 was also identified in an unrelated linkage study in Brazil (195) (see above). Other groups have reported no evidence of linkage between SLC11A1 and the Mitsuda reaction and/or leprosy (120, 175, 254, 276).
Despite an in vitro biological mechanism linking SLC11A1 to critical host defenses against mycobacteria, the SLC11A1 genetic data overall are inconsistent with respect to the effects of specific SLC11A1 polymorphisms, such as the TGTG 3′ UTR deletion. Nonetheless, a subset of these data derived from linkage studies (2, 241, 242) suggests that SLC11A1 may modulate either leprosy susceptibility or the immune recognition of M. leprae (Mitsuda reaction) in some populations. Given the observation that Slc11a1 mutations in mice create a bias toward Th2 immune responses (32), it is possible that human SC11A1 polymorphisms alter susceptibility to leprosy type rather than leprosy overall. The lack of an association with SLC11A1 in some genetic studies might then arise from a lack of power due to the failure to include enough individuals with polar disease (TT or LL).
Tissue-Specific Markers
LAMA2.
Wibawa et al. investigated three polymorphisms in the coding region of the G3 domain of the laminin α-2 gene (LAMA2), T7809C, C7879G, and G7894A, in a small study with 53 leprosy cases and 58 controls (334). Only the T7809C variant encoded an amino acid substitution (V2587A). Genotypes for T7809C did not differ between leprosy patients and healthy contacts (defined as those with daily exposure to an individual with leprosy and presumably not family members). However, among patients with tuberculoid leprosy, 19/26 (73%) had the TC genotype, compared to 8/27 (30%) of those with lepromatous leprosy. Conversely, the TT genotype was enriched in patients with lepromatous leprosy (63%) compared to patients with tuberculoid leprosy (23.1%). Those authors noted that patients with tuberculous leprosy tended to experience peripheral nerve damage (neuritis) earlier than those with lepromatous leprosy (140) and speculated that the valine-to-alanine substitution at position 7809 in laminin α-2 allows the enhanced binding of M. leprae, facilitating rapid intracellular entry in hosts with the TC genotype and earlier peripheral nerve damage. The major difficulty with this hypothesis is the fact that the nerve lesions of lepromatous leprosy also contain an abundance of bacilli inside Schwann cells (272), suggesting that the peripheral nerve damage seen for tuberculoid leprosy requires more than the mere invasion of Schwann cells. It is also unclear how homozygosity versus heterozygosity at the C allele would affect the ability of laminin α-2 to bind M. leprae, since no functional data were presented. Nonetheless, this is an intriguing association detected in a small group of cases and controls that awaits replication in future leprosy studies.
Innate Immune Effector Molecules and Serum Proteins
LTA4H.
Leukotriene A4 hydrolase (LTA4H) is an enzyme that converts leukotriene A4 (LTA4) into the proinflammatory leukotriene B4 (LTB4). LTB4 serves as a potent leukocyte chemoattractant and promotes the production of TNF (100, 111), a cytokine critical to M. tuberculosis control. LTA4 production is closely tied to the activation of the eicosanoid pathway, whereas LTA4H expression is fairly ubiquitous (reviewed in reference 259). Interestingly, the chemical inhibition of LTA4H diverts the pathway toward an alternative LTA4 product, the anti-inflammatory lipoxin A4 (LXA4) (244), suggesting that the regulation of the pro- and anti-inflammatory effects of this pathway may be LTA4H dependent (57, 74). In mice, infection with virulent M. tuberculosis strain H37Rv is associated with higher levels of LXA4 than an attenuated strain and promotes necrosis in an LXA4-dependent manner (57, 74). Mice deficient in 5-lipooxygenase are unable to make LXA4 and are resistant to M. tuberculosis infection (57).
In an unbiased genetic screen for susceptibility to Mycobacterium marinum in zebrafish, we identified lta4h as a hypersusceptible mutant (304). Additional characterization showed that increased lipoxins in these mutants blocked the LTB4-induced TNF production critical for mycobacterial resistance (304). Using SNPs derived from a haplotype previously associated with altered LTB4 production and risk of myocardial infarction (124), we next explored associations with a case-control study of 899 Nepalese leprosy patients. Two intronic SNPs in the LTA4H gene (LTA4H), rs1978331 and rs2660898, were associated with protection from multibacillary leprosy (ENL excluded) in a heterozygous model only (OR, 0.62 [P = 0.001] and OR, 0.70 [P = 0.021], respectively). The same SNPs were also associated with protection from tuberculosis in a heterozygous advantage model in a second association study in Vietnam (304).
The fact that LTA4H is a susceptibility locus for three distinct mycobacterial diseases (infection with M. marinum, M. tuberculosis, and M. leprae) in two divergent vertebrates (zebrafish and humans) is compelling evidence that this gene influences susceptibility to mycobacterial disease via a broad and common mechanism, such as the regulation of eicosanoids. The additional finding of an apparent heterozygous advantage in TB and leprosy is intriguing, since it is consistent with the hypothesis that LTA4H regulation is critical to balancing the potentially destructive effects of an unrestrained LTB4-mediated inflammatory response on host tissues versus the hypersusceptibility of an unrestrained LXA4-mediated response in which TNF production is blocked.
MBL2.
Mannose binding lectin (MBL) is a pattern recognition receptor, but unlike the TLRs, it is a soluble serum protein that binds sugar groups on bacteria. A complex containing MBL, MASP-I (mannose binding lectin-associated serine protease I), and MASP-II bound to a pathogen cleaves C2 and C4, leading to complement activation and opsonization (136). MBL may also cooperate with TLR2 in pathogen recognition (137). Baseline levels of MBL can vary between individuals by 2 to 3 logs, and lower levels have been associated with heightened susceptibility to extracellular pathogens, such as Streptococcus pneumoniae and Neisseria meningitidis (78, 298). However, variation in response to infection is minimal, perhaps suggesting that baseline rather than induced MBL levels influence host defense. Paradoxically, while lower levels of MBL are associated with susceptibility to extracellular pathogens, they may also confer resistance to intracellular pathogens like mycobacteria that rely on complement opsonization for cellular entry (290). In some studies, higher levels of MBL have been associated with an increased risk of lepromatous leprosy (105, 112), while frank MBL deficiency (defined as serum levels <100 ng/ml or ∼10-fold lower than average) is associated with protection from lepromatous leprosy (76, 105, 112). Those studies measured MBL levels in patients with active disease, an approach vulnerable to confounding if MBL levels are modulated by leprosy-associated inflammation (105, 324).
MBL deficiency was previously correlated with homozygosity or compound heterozygosity for six well-described SNPs of the MBL2 gene: two promoter SNPs (at positions −550 [H/L] and −221 [Y/X]), a 5′ untranslated region SNP (position +4 [P/Q]), and three nonsynonymous SNPs in exon 1 (allele D, R52C; allele B, G54D; and allele C, G57E) (106). These loci are in strong linkage disequilibrium, resulting in only seven common haplotypes from combinations of these six alleles (71, 76, 106, 136).
To date, one of two studies has reported an association between MBL2 gene polymorphisms and clinical leprosy (71). de Messias-Reason and colleagues investigated haplotypes of the three upstream polymorphisms (H/L, Y/X, and P/Q) and the exon 1 polymorphisms (D, B, C, or A [wild type]) in 264 leprosy cases and 214 controls in Brazil (71). Haplotypes were identified by the direct sequencing of a single PCR product covering all the variant alleles. In a subset of individuals, MBL levels were measured, allowing the correlation of genotypes with MBL expression. Three haplotypes were associated with higher levels of MBL (HYPA, LYPA, and LYQA). Consistent with previous reports that higher MBL expression is a risk factor for mycobacterial disease, the “high”-expressing haplotype LYPA was also more frequent in patients than in controls (OR, 2.25; P = 0.02, adjusted analysis). There was also a nonsignificant association of this haplotype with lepromatous and borderline leprosy (71). In a second study, Fitness et al. analyzed a single exon 1 polymorphism but found no association in Malawi (93).
The association of “high” MBL levels with increased susceptibility to leprosy observed by de Messias-Reason et al. was previously reported (76, 106). What is novel in this study is the strong and direct association of specific MBL haplotypes with altered leprosy susceptibility, where previous studies reported either associations of haplotypes with MBL levels or MBL levels with leprosy only.
FCN2.
The ficolin-2 (also known as “L ficolin”) gene (FCN2) encodes a soluble receptor with structural resemblance to MBL that binds to pathogen molecular motifs (PAMPs), such as lipoteichoic acid, acetyl groups, peptidoglycan, and lipopolysaccharide, and enhances opsonization and phagocytosis of microbes (133, 161). Three promoter polymorphisms of FCN2 have been associated with alterations in levels of circulating ficolin-2, A−986G, G−602A, and A−4G (132). In addition, a fourth polymorphism in exon 8, G6424T (A258S) has been shown to have greater binding capacity for the N-acetylglucosamine (GlcNAC) motif in peptidoglycan than wild-type ficolin-2 and to be associated with lower levels of protein (126, 132, 210).
de Messias-Reason and colleagues investigated FCN2 haplotypes in 158 individuals with leprosy in southern Brazil compared to 210 matched controls (70). A haplotype containing the −986A, −602G, and −4A polymorphisms was associated with a significant decrease in the risk for leprosy (OR, 0.13; P < 0.013, adjusted analysis) (70). In addition, when the −6424G allele was included, the resultant haplotype, AGAG, was also associated with a significantly reduced risk of leprosy (OR, 0.10; P < 0.011, adjusted analysis). Preceding functional studies by another group showed that the AGAG haplotype is associated with normal circulating levels of ficolin-2, while the GGAT and GGAG haplotypes are associated with lower levels of ficolin-2 (210). de Messias-Reason et al. therefore proposed that the mechanism for reduced susceptibility to leprosy in carriers of the AGA or AGAG haplotype in this Brazilian population is due to normal, as opposed to low, levels of the ficolin-2 protein. An additional mechanism conferred by the inclusion of the +6424G allele in the AGAG haplotype may be normal, as opposed to reduced, binding activity of ficolin-2 to a specific PAMP on M. leprae.
Other Candidate Gene Studies
Miscellaneous candidate genes.
A number of other candidate genes not discussed above have reported associations with leprosy (Table 1). These genes include DEFB1, encoding β-defensin 1 (230); MICA and MICB (307, 331), KIR (killer immunoglobulin-like receptor) (97); IL-12 subunit p40 (IL12B) (14, 207); IL-12 receptor β2 (IL12RB2) (218); complement receptor 1 (CR1) (93); and a number of complement factors (C2, C3, C4A, C4B, and CFB [also known as properdin]) (4, 15, 69, 83, 114, 252, 294). A number of complement factor polymorphisms were investigated in the 1970s and 1980s in small groups of patients using older techniques (e.g., migration of DNA fragments on electrophoretic gels) to identify genetic variants (4, 15, 69, 83, 114, 252, 294). C2, C4, and CFB are located on chromosome 6 in close linkage to HLA-B loci (229). Those early studies did not adequately address the possible effects of linkage disequilibrium with nearby MHC loci, and future studies of this region will need to address the potential confounding impact of LD with MHC variants.
Genes with insufficient data to assess.
A number of genes and polymorphisms have been analyzed in single studies with very small sample sizes or using a control population that is not in HWE for the SNP of interest. Due to the limited amount of data on these genes, it is difficult to assess these findings with any certainty. These genes include procollagen IIIα1 (COL3A) (one study with 26 patients and 14 controls) (155), heat shock protein A1A (HSPA1A [formerly HSP70-1]) (one study with 49 patients and 38 controls) (235), transporter associated with antigen processing 2 (TAP2) (one study with 57 patients and 40 controls) (236), beta-2 glycoprotein I (APOH) (one study with 113 patients and 113 controls [control group not in HWE for SNP with disease association]) (45), and cytotoxic-T-lymphocyte-associated antigen (CTLA4) (one study with 26 patients and 14 controls) (155). In addition, there are a small number of genes for which single studies suggested no effect. For example, DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin), encoding a C-type lectin that binds to pathogens displaying surface carbohydrate moieties (16, 107, 318), is a promising candidate gene because of functional studies demonstrating this receptor's ability to bind M. leprae (23) as well as other studies showing an enrichment of DC-SIGN+ macrophages and an absence of CD1b+ DCs in lepromatous compared to tuberculoid lesions (164). However, DC-SIGN polymorphisms were recently investigated in 194 leprosy cases (109 lepromatous and 85 tuberculoid cases) and 78 controls in a Pakistani population, and no associations with leprosy overall or with polar disease were detected (23). Other genes with a reported lack of association include BTNL2, a candidate costimulatory molecule (one study with 72 families and 208 affected individuals; association was attributed to LD with the HLA-DR region) (150), IL-12 receptor β1 (IL12RB1) (one study with 93 patients and 94 controls) (173), and gamma interferon receptor 1 (IFNGR1) (one study with 93 patients and 94 controls) (173).
GENOME-WIDE ASSOCIATION STUDIES
Genome-wide association studies (GWASs), to date, have been uncommon in infectious diseases (86, 125, 141, 153, 188, 245). These studies typically include thousands of unrelated individuals and genotype hundreds of thousands of polymorphisms spaced across the genome to look for associations with disease using commercially available platforms. Typically, a “discovery sample set” is genotyped, and areas of association are then more finely mapped with specific polymorphisms covering candidate genes in regions with the greatest association in the discovery set analysis. The advantage of such studies is the ability to combine the breadth of unbiased discovery previously unique to family-based genetic linkage studies with the resolution of associations down to the SNP level. In addition, GWASs apply to large populations, rather than family pedigrees or small case-control studies, and so are designed to capture gene effects that may be entirely missed by the narrow focus and smaller sample sizes of candidate gene studies. The disadvantage of these studies is the need for large sample sizes, driven by the need to correct for multiple comparisons for hundreds of thousands of SNPs.
Recently, the first leprosy GWAS was reported for a population in China (348). This large, seminal study relied on a discovery set of 706 patients, 1,225 controls, and three independent replication sets that together comprised 3,254 patients and 5,955 controls (348). For genotyping, Zhang and coinvestigators used an Illumina Human 610-Quad BeadChip containing 500,000 SNPs. The initial discovery analysis found strong associations within the MHC region on chromosome 6p21 at both the HLA-B/HLA-C locus (MHC class I) and the HLA-DR-DQ locus (MHC class II) that were independently significant; additional associations were also found for chromosome 16q21 and chromosome 13q14. Ninety-three SNPs from the 60 top non-MHC regions of association (defined as having a P value of <5.0 × 10−4) were then genotyped in the replication sets.
Interestingly, the replication studies failed to show an association with the MHC class I region but strongly confirmed the association for an SNP in the HLA-DR-DQ locus, rs602875 (OR, 0.67; P = 5.35 × 10−27, combined analysis). Among the non-MHC genes, nine SNPs in five different genes were replicated. Two polymorphisms (rs42490 and rs40457) in RIPK2 (receptor-interacting serine-threonine kinase 2), a gene in the NOD2 signaling pathway, were associated with protection from leprosy (OR, 0.76 [P = 1.38 × 10−16] and OR, 0.77 [P = 1.34 ×10−12], respectively [combined analysis]). SNP rs42490 was more strongly associated with MB leprosy than PB leprosy. Two SNPs in the NOD2 region (rs9302752 and rs7194886) lying in the intergenic region between NOD2 and its 5′ upstream neighbor, SNX20, were both associated with elevated leprosy risk (OR of 1.59 [P = 3.77 × 10−40] and OR of 1.63 [P = 1.77 × 10−30], respectively [combined analysis]). These two SNPs were more strongly associated with leprosy than two other SNPs within the NOD2 gene, rs8057341 (P = 5.22 × 10−2) and rs3135499 (P = 9.21 × 10−2). The effect of SNP rs9302752 was also stronger for MB than PB leprosy (348). In addition, three polymorphisms (rs4574921, rs10114470, and rs6478108) in TNFSF15 (tumor necrosis factor [ligand] superfamily member 15), two SNPS (rs3764147 and rs10507522) in C13orf31 (chromosome 13, open reading frame 31), and two SNPs (rs9533634 and rs3088362) in CCDC122 (coiled-coil domain containing 122) were associated with increased or decreased leprosy susceptibility (348). Those investigators also analyzed the haplotype blocks surrounding the risk-associated variants and analyzed any gene associated with this block. For all genes except for those in the MHC, only one gene was found per haplotype block.
This GWAS confirmed the previous long-standing association of the MHC class II region with leprosy risk but failed to validate the association of leprosy with the PARK2/PACRG (196, 197) and LTA (8) genes or the association of PB leprosy with chromosome 10p13 region (282). Notably, although those authors investigated 13 SNPs in the PARK2/PACRG regulatory region, they did not evaluate PARK2_e01(−2599) and rs1040079, the two SNPs found to capture all of the associations of the chromosome 6q25 region with leprosy risk (196). Similarly, eight SNPs in the LTA gene were examined, but not LTA+80, the SNP identified as the causative variant for LTA's association with leprosy (8). In addition, the mean age at the onset of diagnosis of all participants in this study was 23.3 years. In the previous linkage study reporting an association of the LTA gene with leprosy, the risk was highest for patients under the age of 16 years or between the ages of 16 and 25 years (8). In the 10p13 region, more than 10 SNPs were investigated, none of which had an association with paucibacillary leprosy in the GWAS. The identity of the causative variant at this locus remains elusive. In addition, two new candidate genes of unknown function were identified: C13orf31 and CCDC122 (348).
Following the study by Zhang et al. in China, another group reported a second genome-wide association study for leprosy that was carried out in India and Mali (336, 337). That study used a gene-centric 50,000-SNP microarray to assess associations for >2,000 genes across the genome (10-fold-lower SNP density than the GWAS performed in China). A primary-association analysis was performed with 258 leprosy cases and 300 controls in New Delhi, India. SNPs with significant associations (P < 1 × 10−4) in this initial screen were investigated in two or three different replication studies: a separate case-control study in Bengal, India (220 cases and 162 controls); a TDT study in Tamil Nadu, India (161 families); and a case-control study in Mali (336, 337). In the HLA-DRB1/DQA1 region, two SNPs with strong associations with leprosy susceptibility were identified (rs1071630 and rs9270650 [P = 3.1 × 10−11 and 4.9 × 10−14, respectively, combined allelic analysis]) (336). In addition, the TLR1 I602S (T1805G) SNP was strongly associated with protection from leprosy in the New Delhi and Bengal populations (OR, 0.27 to 0.40; P = 1.3 × 10−4 to 0.02, allelic analysis) but was borderline in the TDT study (OR, 0.61; P = 0.09). Nonetheless, this SNP association remained highly significant in the combined analysis (P = 5.7 × 10−8) (336). This GWAS also confirmed the association of two other SNPs identified in the Chinese GWAS, rs3764147 (C13orf31) and rs9533634 (CCDC122), with leprosy susceptibility (OR, 1.59 [95% confidence interval {CI}, 1.34 to 1.89]; P = 6.11 × 10−8, combined analysis) and leprosy resistance (OR, 0.70 [95% CI, 0.59 to 0.82]; P = 1.12 × 10−5, combined analysis), respectively (337).
The finding of an association of the TLR1 SNP T1805G (I602S) replicates previous reports of an association of this SNP with protection from leprosy or type 1 reactions (149, 198). All three studies also showed the same direction of effect, strongly supporting a role for TLR1 in leprosy immunopathogenesis. TLR1 has widely varying frequencies across different ethnic groups (121, 149, 336, 338). The lack of an association of TLR1 with leprosy in the China GWAS likely stems from the very low frequency of the 602S allele in this population (1.7%) (336) and the resultant lack of power to detect associations at this locus.
For NOD2, in contrast to the GWAS findings in China (348), no associations with leprosy were found in the second leprosy GWAS (337). As mentioned above, we recently found associations of NOD2 polymorphisms with leprosy in Nepal (27), consistent with data from the China study. Although the polymorphisms examined in our study and the study by Zhang et al. were mostly nonoverlapping and there is no known functional mechanism for any of these variants, these two studies support a role for the NOD2 pathway in leprosy pathogenesis (27). The reason for the lack of an association in the Indian and Malian populations is unclear but could reflect different underlying LD (haplotype) structures in these gene regions or the presence of alternate susceptibility loci that mask or reduce the effect of NOD2 alleles. Other associations that were not replicated in the second leprosy GWAS by Wong et al. included LTA, NOD2, PARK2 and PACRG, RIPK2, SLC11A1, TLR2, TLR4, TNFSF15, and VDR (337).
CONCLUSIONS
In this review, we have presented a detailed view of the field of leprosy genetics and commented on the quality of evidence that exists for each gene or susceptibility locus. Of the many reports of genes associated with leprosy, relatively few have been replicated in additional study populations. Among those that have been validated, genes with a potential or demonstrated biological mechanism form an even smaller subset. There are several reasons for this rather modest track record. In the first place, many of the candidate gene studies are simply underpowered to detect risk alleles of modest effect. Sufficient power to detect modest effects (OR of ∼1.5) requires large sample sizes (thousands) and/or polymorphisms present at high frequencies. Linkage studies require even greater numbers to have sufficient power to uncover modest effects in complex diseases (251) and are best at detecting rare alleles with large effects (296). The effect of risk alleles could also be modified by the preponderance of certain clinical forms of leprosy in one geographic region compared to another (195, 215). If 10p13 is associated with PB leprosy exclusively, the effect may be more difficult to demonstrate in a region where MB disease predominates, for example. Second, there is likely to be a heterogeneity of genetic effects across populations. The 10p13 region is a risk factor for leprosy in India (282) but not in Brazil (195), and conversely, the HLA region is a risk factor for leprosy in Brazil (277) but not in India (282). This phenomenon can arise from alterations in the linkage structures of alleles between populations (population-specific linkage disequilibrium) (11), differences in the polymorphisms or genes that confer disease risk in different ethnic groups, variability in penetrance, or epistatic interactions. Third, differences in leprosy case ascertainment or categorization of leprosy type could also exist. Finally, failure to correct for population stratification between cases and controls may also confound study results.
Traditionally, common infectious diseases, such as leprosy, are thought to have a complex or polygenic pattern of inheritance. In this model, the disease trait arises out of the additive effect of multiple genes, each with a modest effect on the phenotype (one disease and many genes). Conversely, the inheritance pattern of primary immunodeficiency syndromes, such as SCID (severe combined immunodeficiency) or Bruton's agammaglobulinemia, are monogenic. Primary immunodeficiency syndromes in this traditional sense are the consequence of the Mendelian transmission of single mutations producing a severe phenotype, often with multiple infections of different microbial etiologies (one highly penetrant gene and many infections) (54, 217, 234). This view has recently been challenged by some experts in the field, who point to the narrow impact of some Mendelian mutations that unexpectedly alter susceptibility to single rather than multiple pathogens (7, 234). For example, herpes simplex encephalitis was shown to be due to rare, highly penetrant Mendelian mutations in TLR3 or Unc93B (7, 55). Could some common infectious diseases be determined by monogenic or oligogenic inheritance as well? In the case of leprosy, data from genome-wide linkage studies might suggest that rather than being a disease of complex or polygenic inheritance, susceptibility to leprosy or forms of leprosy may be controlled mostly by a few major genes (8, 196, 197, 234). However, even for this well-studied disease, some large effects from linkage studies have not been replicated in alternate populations, while candidate gene association studies have uncovered modest-risk genes, such as NOD2, that have not been detected by linkage studies. Further studies are needed to assess whether leprosy susceptibility is governed by oligogenic or polygenic inheritance patterns.
Which genomic regions and which genes have the best data to support a role in leprosy susceptibility? The strongest evidence from linkage analysis exists for chromosome 10p13 (two separate linkage studies, one in India and one in Vietnam) (197, 282), the PARK2/PACRG promoter region (three populations [two from Vietnam {linkage} and one from Brazil {case-control SNP association study}]) (196, 197), and chromosome 6p21 and the LTA gene (two populations [one from Vietnam {linkage} and one from India {case-control}]) (8). Each of these studies involved large numbers of families, sibling pairs, and/or unrelated cases and controls and contained an internal replication set. Reasonable evidence also exists for the linkage of SLC11A1 (NRAMP) with the Mitsuda reaction (2, 10, 241, 242). The putative disease-associated SNPs have been identified for PARK2/PACRG [PARK2_e01(−2599) and rs1040079] and for chromosome 6p21 (LTA+80) but not for 10p13. Subsequent studies of these PARK2/PACRG SNPs in alternate ethnic groups have not validated these specific variants. The PARK2 gene encodes an E3 ubiquitin ligase, which facilities the proteosomal degradation of proteins, but was also recently reported to regulate cyclin E (319, 320). Cyclin E is a tumor suppressor protein that is normally suppressed by PARK2. The release of this negative control causes neuronal apoptosis (319), which could be a mechanism by which M. leprae causes nerve damage, although this is speculative.
Among the candidate gene studies, the best-quality evidence exists for TLR1 SNP T1805G or A743G (four studies, including a GWAS) (149, 198, 271, 336), the NOD2 gene (two positive studies, including data from a GWAS, with associations in nonoverlapping SNPs) (27, 337, 348), and the IL10 promoter SNP C−819T (five studies, including one meta-analysis) (93, 96, 185, 206, 225). In addition, the first leprosy GWAS (348) identified SNPs in two genes of unknown function, CCDC122 (rs9533634) and C13orf31 (rs3764147) (348), which were validated in a second GWAS (337). Functionally, the TLR1 variant T1805G has been shown to regulate the surface expression of TLR1 (149), which is a major receptor for M. leprae, and to regulate the recognition of M. leprae by human monocytes (198). The IL10 promoter SNP −819T has been associated with decreased IL-10 levels (225), although it remains unclear if this mechanism is related to the SNP's variable association with leprosy type. The mechanisms underlying NOD2 polymorphisms are not known.
Over the last decade there has been an enormous expansion in both the methodology and affordable technology available to perform sophisticated genetic studies of disease associations. It is now financially and technically possible to scrutinize over 1 million SNPs in thousands of samples and identify allelic variants associated with disease. Our understanding of the genetic risk factors for specific infections is quickly expanding, to cover more populations and more genes, and deepening, to discover more SNPs per gene and better understand how risk alleles might be modified, appear, or disappear in different populations. A small but growing number of genome-wide association studies have been carried out for infectious diseases, including malaria (141), HIV (86, 125), Creutzfeldt-Jakob disease (188), hepatitis B and C (153, 245), and now leprosy (336, 337, 348). Information on new types of genetic variation, including copy number variants, is just emerging. These new tools promise an era of rapid acquisition of data describing genetic variation in diverse populations and the elaboration of fresh theories of infectious pathogenesis, novel means of immunomodulation, and improvements in drug design. In the case of leprosy and tuberculosis, diseases for which our pharmaceutical armamentarium is underdeveloped, such new insights from the human genome could make large contributions to global health.
Acknowledgments
This work was supported by National Institutes of Health grants K08 AI080952 (W.R.B.), K08 AI076604 (E.A.M.), and F32AR057293-01 (J.C.V.). Other grant support includes the Burroughs Wellcome Fund (T.R.H.), the Heiser Foundation (T.R.H. and W.R.B.), the American Skin Association (J.C.V.), and the Dermatology Foundation (J.C.V.).
REFERENCES
- 1.Abel, L., and F. Demenais. 1988. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Desirade island. Am. J. Hum. Genet. 42:256-266. [PMC free article] [PubMed] [Google Scholar]
- 2.Abel, L., F. O. Sanchez, J. Oberti, N. V. Thuc, L. V. Hoa, V. D. Lap, E. Skamene, P. H. Lagrange, and E. Schurr. 1998. Susceptibility to leprosy is linked to the human NRAMP1 gene. J. Infect. Dis. 177:133-145. [DOI] [PubMed] [Google Scholar]
- 3.Abel, L., D. L. Vu, J. Oberti, V. T. Nguyen, V. C. Van, M. Guilloud-Bataille, E. Schurr, and P. H. Lagrange. 1995. Complex segregation analysis of leprosy in southern Vietnam. Genet. Epidemiol. 12:63-82. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal, D. P., H. G. Benkmann, H. W. Goedde, R. Rohde, H. Delbruck, and A. Rougemont. 1974. Levels of serum beta 1C-beta 1A globulin (C3) and its polymorphism in leprosy patients and healthy controls from Ethiopia and Mali. Humangenetik 21:355-359. [DOI] [PubMed] [Google Scholar]
- 5.Agnese, D. M., J. E. Calvano, S. J. Hahm, S. M. Coyle, S. A. Corbett, S. E. Calvano, and S. F. Lowry. 2002. Human Toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of Gram-negative infections. J. Infect. Dis. 186:1522-1525. [DOI] [PubMed] [Google Scholar]
- 6.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 7.Alcais, A., L. Abel, and J.-L. Casanova. 2009. Human genetics of infectious diseases: between proof of principle and paradigm. J. Clin. Invest. 119:2506-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcaïs, A., A. Alter, G. Antoni, M. Orlova, N. V. Thuc, M. Singh, P. R. Vanderborght, K. Katoch, M. T. Mira, V. H. Thai, N. T. Huong, N. N. Ba, M. Moraes, N. Mehra, E. Schurr, and L. Abel. 2007. Stepwise replication identifies a low-producing lymphotoxin-[alpha] allele as a major risk factor for early-onset leprosy. Nat. Genet. 39:517-522. [DOI] [PubMed] [Google Scholar]
- 9.Alcaïs, A., M. Mira, J.-L. Casanova, E. Schurr, and L. Abel. 2005. Genetic dissection of immunity in leprosy. Curr. Opin. Immunol. 17:44-48. [DOI] [PubMed] [Google Scholar]
- 10.Alcais, A., F. O. Sanchez, N. V. Thuc, V. D. Lap, J. Oberti, P. H. Lagrange, E. Schurr, and L. Abel. 2000. Granulomatous reaction to intradermal injection of lepromin (Mitsuda reaction) is linked to the human NRAMP1 gene in Vietnamese leprosy sibships. J. Infect. Dis. 181:302-308. [DOI] [PubMed] [Google Scholar]
- 11.Alter, A., A. Alcaïs, L. Abel, and E. Schurr. 2008. Leprosy as a genetic model for susceptibility to common infectious diseases. Hum. Genet. 123:227-235. [DOI] [PubMed] [Google Scholar]
- 12.Alter, A., L. de Leseleuc, N. V. Thuc, V. H. Thai, N. T. Huong, N. N. Ba, C. C. Cardoso, A. V. Grant, L. Abel, M. O. Moraes, A. Alcais, and E. Schurr. 2010. Genetic and functional analysis of common MRC1 exon 7 polymorphisms in leprosy susceptibility. Hum. Genet. 127:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altmuller, J., L. J. Palmer, G. Fischer, H. Scherb, and M. Wjst. 2001. Genomewide scans of complex human diseases: true linkage is hard to find. Am. J. Hum. Genet. 69:936-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarado-Navarro, A., M. Montoya-Buelna, J. F. Muñoz-Valle, R. I. López-Roa, C. Guillén-Vargas, and M. Fafutis-Morris. 2008. The 3′UTR 1188 A/C polymorphism in the interleukin-12p40 gene (IL-12B) is associated with lepromatous leprosy in the west of Mexico. Immunol. Lett. 118:148-151. [DOI] [PubMed] [Google Scholar]
- 15.Ananthakrishnan, R., H. Walter, G. Kellermann, and T. Matznetter. 1973. Further studies on associations between leprosy and genetic markers in human serum. Humangenetik 19:183-192. [DOI] [PubMed] [Google Scholar]
- 16.Appelmelk, B. J., I. van Die, S. J. van Vliet, C. M. J. E. Vandenbroucke-Grauls, T. B. H. Geijtenbeek, and Y. van Kooyk. 2003. carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J. Immunol. 170:1635-1639. [DOI] [PubMed] [Google Scholar]
- 17.Arbour, N. C., E. Lorenz, B. C. Schutte, J. Zabner, J. N. Kline, M. Jones, K. Frees, J. L. Watt, and D. A. Schwartz. 2000. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 25:187-191. [DOI] [PubMed] [Google Scholar]
- 18.Ardlie, K. G., L. Kruglyak, and M. Seielstad. 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3:299-309. [DOI] [PubMed] [Google Scholar]
- 19.Austin, C. M., X. Ma, and E. A. Graviss. 2008. Common nonsynonymous polymorphisms in the NOD2 gene are associated with resistance or susceptibility to tuberculosis disease in African Americans. J. Infect. Dis. 197:1713-1716. [DOI] [PubMed] [Google Scholar]
- 20.Bafica, A., C. A. Scanga, C. G. Feng, C. Leifer, A. Cheever, and A. Sher. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 202:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakker, M. I., M. Hatta, A. Kwenang, P. Van Mosseveld, W. R. Faber, P. R. Klatser, and L. Oskam. 2006. Risk factors for developing leprosy—a population-based cohort study in Indonesia. Lepr. Rev. 77:48-61. [PubMed] [Google Scholar]
- 22.Barreiro, L. B., M. Ben-Ali, H. Quach, G. Laval, E. Patin, J. K. Pickrell, C. Bouchier, M. Tichit, O. Neyrolles, B. Gicquel, J. R. Kidd, K. K. Kidd, A. Alcais, J. Ragimbeau, S. Pellegrini, L. Abel, J. L. Casanova, and L. Quintana-Murci. 2009. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5:e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barreiro, L. B., H. Quach, J. Krahenbuhl, S. Khaliq, A. Mohyuddin, S. Q. Mehdi, B. Gicquel, O. Neyrolles, and L. Quintana-Murci. 2006. DC-SIGN interacts with Mycobacterium leprae but sequence variation in this lectin is not associated with leprosy in the Pakistani population. Hum. Immunol. 67:102-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayley, J. P., T. H. Ottenhoff, and C. L. Verweij. 2004. Is there a future for TNF promoter polymorphisms? Genes Immun. 5:315-329. [DOI] [PubMed] [Google Scholar]
- 25.Bellamy, R., C. Ruwende, T. Corrah, K. P. McAdam, H. C. Whittle, and A. V. Hill. 1998. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N. Engl. J. Med. 338:640-644. [DOI] [PubMed] [Google Scholar]
- 26.Berrington, W. R., and T. R. Hawn. 2007. Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter? Immunol. Rev. 219:167-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berrington, W. R., M. Macdonald, S. Khadge, B. R. Sapkota, M. Janer, D. A. Hagge, G. Kaplan, and T. R. Hawn. 2010. Common polymorphisms in the NOD2 gene region are associated with leprosy and its reactive states. J. Infect. Dis. 201:1422-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beutler, B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430:257-263. [DOI] [PubMed] [Google Scholar]
- 29.Bhalla, A. K., E. P. Amento, and S. M. Krane. 1986. Differential effects of 1,25-dihydroxyvitamin D3 on human lymphocytes and monocyte/macrophages: inhibition of interleukin-2 and augmentation of interleukin-1 production. Cell. Immunol. 98:311-322. [DOI] [PubMed] [Google Scholar]
- 30.Blackwell, J. M. 1998. Genetics of host resistance and susceptibility to intramacrophage pathogens: a study of multicase families of tuberculosis, leprosy and leishmaniasis in north-eastern Brazil. Int. J. Parasitol. 28:21-28. [DOI] [PubMed] [Google Scholar]
- 31.Blackwell, J. M., G. F. Black, C. S. Peacock, E. N. Miller, D. Sibthorpe, D. Gnananandha, J. J. Shaw, F. Silveira, Z. Lins-Lainson, F. Ramos, A. Collins, and M. A. Shaw. 1997. Immunogenetics of leishmanial and mycobacterial infections: the Belem Family Study. Philos. Trans. R. Soc. Lond. B Biol. Sci. 352:1331-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwell, J. M., and S. Searle. 1999. Genetic regulation of macrophage activation: understanding the function of Nramp1 (=Ity/Lsh/Bcg). Immunol. Lett. 65:73-80. [DOI] [PubMed] [Google Scholar]
- 33.Blake, L. A., C. W. Burton, C. H. Lary, and J. R. Todd IV. 1987. Environmental nonhuman sources of leprosy. Rev. Infect. Dis. 9:562-577. [DOI] [PubMed] [Google Scholar]
- 34.Bochud, P. Y., T. R. Hawn, and A. Aderem. 2003. A Toll-like receptor 2 polymorphism that is associated with lepromatous leprosy is unable to mediate mycobacterial signaling. J. Immunol. 170:3451-3454. [DOI] [PubMed] [Google Scholar]
- 35.Bochud, P. Y., T. R. Hawn, M. R. Siddiqui, P. Saunderson, S. Britton, I. Abraham, A. T. Argaw, M. Janer, L. P. Zhao, G. Kaplan, and A. Aderem. 2008. Toll-like receptor 2 (TLR2) polymorphisms are associated with reversal reaction in leprosy. J. Infect. Dis. 197:253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochud, P. Y., D. Sinsimer, A. Aderem, M. R. Siddiqui, P. Saunderson, S. Britton, I. Abraham, A. T. Argaw, M. Janer, T. R. Hawn, and G. Kaplan. 2009. Polymorphisms in Toll-like receptor 4 (TLR4) are associated with protection against leprosy. Eur. J. Clin. Microbiol. Infect. Dis. 28:1055-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boddingius, J. 1974. The occurrence of Mycobacterium leprae within axons of peripheral nerves. Acta Neuropathol. 27:257-270. [DOI] [PubMed] [Google Scholar]
- 38.Bopst, M., I. Garcia, R. Guler, M. L. Olleros, T. Rulicke, M. Muller, S. Wyss, K. Frei, M. Le Hir, and H. P. Eugster. 2001. Differential effects of TNF and LTα in the host defense against M. bovis BCG. Eur. J. Immunol. 31:1935-1943. [DOI] [PubMed] [Google Scholar]
- 39.Borras, S. G., C. Cotorruelo, L. Racca, M. Recarte, C. Garcias, C. Biondi, O. Bottasso, and A. Racca. 2008. Association of leprosy with HLA-DRB1 in an Argentinean population. Ann. Clin. Biochem. 45:96-98. [DOI] [PubMed] [Google Scholar]
- 40.Bouma, G., J. B. Crusius, M. O. Pool, J. J. Kolkman, B. M. von Blomberg, P. J. Kostense, M. J. Giphart, G. M. Schreuder, S. G. Meuwissen, and A. S. Pena. 1996. Secretion of tumour necrosis factor alpha and lymphotoxin alpha in relation to polymorphisms in the TNF genes and HLA-DR alleles. Relevance for inflammatory bowel disease. Scand. J. Immunol. 43:456-463. [DOI] [PubMed] [Google Scholar]
- 41.Bradley, D. J. 1974. Genetic control of natural resistance to Leishmania donovani. Nature 250:353-354. [DOI] [PubMed] [Google Scholar]
- 42.Bradley, D. J. 1977. Regulation of Leishmania populations within the host. II. Genetic control of acute susceptibility of mice to Leishmania donovani infection. Clin. Exp. Immunol. 30:130-140. [PMC free article] [PubMed] [Google Scholar]
- 43.Brinkman, B. M., D. Zuijdeest, E. L. Kaijzel, F. C. Breedveld, and C. L. Verweij. 1995. Relevance of the tumor necrosis factor alpha (TNF alpha) −308 promoter polymorphism in TNF alpha gene regulation. J. Inflamm. 46:32-41. [PubMed] [Google Scholar]
- 44.Britton, W. J., and D. N. Lockwood. 2004. Leprosy. Lancet 363:1209-1219. [DOI] [PubMed] [Google Scholar]
- 45.Brochado, M. J., J. F. Figueiredo, C. T. Mendes-Junior, P. Louzada-Junior, O. M. Kim, and A. M. Roselino. 2010. Correlation between beta-2-glycoprotein I gene polymorphism and anti-beta-2 glycoprotein I antibodies in patients with multibacillary leprosy. Arch. Dermatol. Res. 302:583-591. [DOI] [PubMed] [Google Scholar]
- 46.Brown, I. N., A. A. Glynn, and J. Plant. 1982. Inbred mouse strain resistance to Mycobacterium lepraemurium follows the Ity/Lsh pattern. Immunology 47:149-156. [PMC free article] [PubMed] [Google Scholar]
- 47.Brown, J. A., and M. M. Stone. 1958. Tuberculoid leprosy in identical twins. Lepr. Rev. 29:53-55. [DOI] [PubMed] [Google Scholar]
- 48.Browne, S. G. 1985. The history of leprosy, p. 1-14. In J. C. Hastings (ed.), Leprosy. Churchill-Livingstone, Edinburgh, United Kingdom.
- 49.Bulut, Y., E. Faure, L. Thomas, O. Equils, and M. Arditi. 2001. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167:987-994. [DOI] [PubMed] [Google Scholar]
- 50.Burman, K. D., A. Rijall, S. Agrawal, A. Agarwalla, and K. K. Verma. 2003. Childhood leprosy in eastern Nepal: a hospital-based study. Indian J. Lepr. 75:47-52. [PubMed] [Google Scholar]
- 51.Cabrera, M., M. A. Shaw, C. Sharples, H. Williams, M. Castes, J. Convit, and J. M. Blackwell. 1995. Polymorphism in tumor necrosis factor genes associated with mucocutaneous leishmaniasis. J. Exp. Med. 182:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calvano, J. E., D. J. Bowers, S. M. Coyle, M. Macor, M. T. Reddell, A. Kumar, S. E. Calvano, and S. F. Lowry. 2006. Response to systemic endotoxemia among humans bearing polymorphisms of the Toll-like receptor 4 (hTLR4). Clin. Immunol. 121:186-190. [DOI] [PubMed] [Google Scholar]
- 53.Casanova, J.-L., and L. Abel. 2002. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20:581-620. [DOI] [PubMed] [Google Scholar]
- 54.Casanova, J. L., and L. Abel. 2007. Human genetics of infectious diseases: a unified theory. EMBO J. 26:915-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casrouge, A., S.-Y. Zhang, C. Eidenschenk, E. Jouanguy, A. Puel, K. Yang, A. Alcais, C. Picard, N. Mahfoufi, N. Nicolas, L. Lorenzo, S. Plancoulaine, B. Senechal, F. Geissmann, K. Tabeta, K. Hoebe, X. Du, R. L. Miller, B. Heron, C. Mignot, T. B. de Villemeur, P. Lebon, O. Dulac, F. Rozenberg, B. Beutler, M. Tardieu, L. Abel, and J.-L. Casanova. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314:308-312. [DOI] [PubMed] [Google Scholar]
- 56.Chackravartti, M. R., and F. Vogel. 1973. A twin study on leprosy. Georg Thieme Publishers, Stuttgart, Germany.
- 57.Chen, M., M. Divangahi, H. Gan, D. S. Shin, S. Hong, D. M. Lee, C. N. Serhan, S. M. Behar, and H. G. Remold. 2008. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med. 205:2791-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho, S. N., R. V. Cellona, L. G. Villahermosa, T. T. Fajardo, Jr., M. V. Balagon, R. M. Abalos, E. V. Tan, G. P. Walsh, J. D. Kim, and P. J. Brennan. 2001. Detection of phenolic glycolipid I of Mycobacterium leprae in sera from leprosy patients before and after start of multidrug therapy. Clin. Diagn. Lab. Immunol. 8:138-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clark, B. M., C. K. Murray, L. L. Horvath, G. A. Deye, M. S. Rasnake, and R. N. Longfield. 2008. Case-control study of armadillo contact and Hansen's disease. Am. J. Trop. Med. Hyg. 78:962-967. [PubMed] [Google Scholar]
- 60.Clark, K. A., S. H. Kim, L. F. Boening, M. J. Taylor, T. G. Betz, and F. V. McCasland. 1987. Leprosy in armadillos (Dasypus novemcinctus) from Texas. J. Wildl. Dis. 23:220-224. [DOI] [PubMed] [Google Scholar]
- 61.Convit, J., P. G. Smith, M. Zuniga, C. Sampson, M. Ulrich, J. A. Plata, J. Silva, J. Molina, and A. Salgado. 1993. BCG vaccination protects against leprosy in Venezuela: a case-control study. Int. J. Lepr. Other Mycobact. Dis. 61:185-191. [PubMed] [Google Scholar]
- 62.Cooke, G. S., S. J. Campbell, J. Sillah, P. Gustafson, B. Bah, G. Sirugo, S. Bennett, K. P. W. J. McAdam, O. Sow, C. Lienhardt, and A. V. S. Hill. 2006. Polymorphism within the interferon-γ/receptor complex is associated with pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 174:339-343. [DOI] [PubMed] [Google Scholar]
- 63.Cooper, A. M., D. K. Dalton, T. A. Stewart, J. P. Griffin, D. G. Russell, and I. M. Orme. 1993. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178:2243-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coulombe, F., M. Divangahi, F. Veyrier, L. de Leseleuc, J. L. Gleason, Y. Yang, M. A. Kelliher, A. K. Pandey, C. M. Sassetti, M. B. Reed, and M. A. Behr. 2009. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 206:1709-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cunha, S. S., L. C. Rodrigues, V. Pedrosa, I. M. Dourado, M. L. Barreto, and S. M. Pereira. 2004. Neonatal BCG protection against leprosy: a study in Manaus, Brazilian Amazon. Lepr. Rev. 75:357-366. [PubMed] [Google Scholar]
- 66.D'Ambrosio, D., M. Cippitelli, M. G. Cocciolo, D. Mazzeo, P. Di Lucia, R. Lang, F. Sinigaglia, and P. Panina-Bordignon. 1998. Inhibition of IL-12 production by 1,25-dihydroxyvitamin D3. Involvement of NF-kappaB downregulation in transcriptional repression of the p40 gene. J. Clin. Invest. 101:252-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davey, T. F., and R. J. Rees. 1974. The nasal discharge in leprosy: clinical and bacteriological aspects. Lepr. Rev. 45:121-134. [DOI] [PubMed] [Google Scholar]
- 68.Davila, S., M. L. Hibberd, R. H. Dass, H. E. Wong, E. Sahiratmadja, C. Bonnard, B. Alisjahbana, J. S. Szeszko, Y. Balabanova, F. Drobniewski, R. van Crevel, E. van de Vosse, S. Nejentsev, T. H. Ottenhoff, and M. Seielstad. 2008. Genetic association and expression studies indicate a role of Toll-like receptor 8 in pulmonary tuberculosis. PLoS Genet. 4:e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Messias, I. J., J. Santamaria, M. Brenden, A. Reis, and G. Mauff. 1993. Association of C4B deficiency (C4B*Q0) with erythema nodosum in leprosy. Clin. Exp. Immunol. 92:284-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Messias-Reason, I., P. G. Kremsner, and F. J. K. Jurgen. 2009. Functional haplotypes that produce normal ficolin-2 levels protect against clinical leprosy. J. Infect. Dis. 199:801-804. [DOI] [PubMed] [Google Scholar]
- 71.de Messias-Reason, I. J., A. B. Boldt, A. C. Moraes Braga, E. Von Rosen Seeling Stahlke, L. Dornelles, L. Pereira-Ferrari, P. G. Kremsner, and J. F. Kun. 2007. The association between mannan-binding lectin gene polymorphism and clinical leprosy: new insight into an old paradigm. J. Infect. Dis. 196:1379-1385. [DOI] [PubMed] [Google Scholar]
- 72.Dennehy, K. M., G. Ferwerda, I. Faro-Trindade, E. Pyz, J. A. Willment, P. R. Taylor, A. Kerrigan, S. V. Tsoni, S. Gordon, F. Meyer-Wentrup, G. J. Adema, B. J. Kullberg, E. Schweighoffer, V. Tybulewicz, H. M. Mora-Montes, N. A. Gow, D. L. Williams, M. G. Netea, and G. D. Brown. 2008. Syk kinase is required for collaborative cytokine production induced through dectin-1 and Toll-like receptors. Eur. J. Immunol. 38:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deps, P. D., B. L. Alves, C. G. Gripp, R. L. Aragao, B. Guedes, J. B. Filho, M. K. Andreatta, R. S. Marcari, I. Prates, and L. C. Rodrigues. 2008. Contact with armadillos increases the risk of leprosy in Brazil: a case control study. Indian J. Dermatol. Venereol. Leprol. 74:338-342. [DOI] [PubMed] [Google Scholar]
- 74.Divangahi, M., M. Chen, H. Gan, D. Desjardins, T. T. Hickman, D. M. Lee, S. Fortune, S. M. Behar, and H. G. Remold. 2009. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol. 10:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Divangahi, M., S. Mostowy, F. Coulombe, R. Kozak, L. Guillot, F. Veyrier, K. S. Kobayashi, R. A. Flavell, P. Gros, and M. A. Behr. 2008. NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. J. Immunol. 181:7157-7165. [DOI] [PubMed] [Google Scholar]
- 76.Dornelles, L. N., L. Pereira-Ferrari, and I. Messias-Reason. 2006. Mannan-binding lectin plasma levels in leprosy: deficiency confers protection against the lepromatous but not the tuberculoid forms. Clin. Exp. Immunol. 145:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eichbaum, Q., P. Clerc, G. Bruns, F. McKeon, and R. A. B. Ezekowitz. 1994. Assignment of the human macrophage mannose receptor gene (MRC1) to 10p13 by in situ hybridization and PCR-based somatic cell hybrid mapping. Genomics 22:656-658. [DOI] [PubMed] [Google Scholar]
- 78.Eisen, D. P., and R. M. Minchinton. 2003. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin. Infect. Dis. 37:1496-1505. [DOI] [PubMed] [Google Scholar]
- 79.Enard, D., F. Depaulis, and H. R. Crollius. 2010. Human and non-human primate genomes share hotspots of positive selection. PLoS Genet. 6:e1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erridge, C., J. Stewart, and I. R. Poxton. 2003. Monocytes heterozygous for the Asp299Gly and Thr399Ile mutations in the Toll-like receptor 4 gene show no deficit in lipopolysaccharide signalling. J. Exp. Med. 197:1787-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Faber, W. R., A. J. Jensema, and W. F. Goldschmidt. 2006. Treatment of recurrent erythema nodosum leprosum with infliximab. N. Engl. J. Med. 355:739. [DOI] [PubMed] [Google Scholar]
- 82.Fageras Bottcher, M., M. Hmani-Aifa, A. Lindstrom, M. C. Jenmalm, X.-M. Mai, L. Nilsson, H. A. Zdolsek, B. Bjorksten, P. Soderkvist, and O. Vaarala. 2004. A TLR4 polymorphism is associated with asthma and reduced lipopolysaccharide-induced interleukin-12(p70) responses in Swedish children. J. Allergy Clin. Immunol. 114:561-567. [DOI] [PubMed] [Google Scholar]
- 83.Farhud, D. D., R. Ananthakrishnan, and H. Walter. 1972. Association between C′3 phenotypes and various diseases. Humangenetik 17:57-60. [PubMed] [Google Scholar]
- 84.Feitosa, M. F., I. Borecki, H. Krieger, B. Beiguelman, and D. C. Rao. 1995. The genetic epidemiology of leprosy in a Brazilian population. Am. J. Hum. Genet. 56:1179-1185. [PMC free article] [PubMed] [Google Scholar]
- 85.Felix, J. S. V., S. C. G. Salazar, R. C. Velazquez, J. G. R. Maldonado, and H. R. Villalobos. 2009. Association between the TaqI polymorphism of vitamin D receptor gene and lepromatous leprosy in a Mexican population sample. Salud Publica Mex. 51:59-61. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 86.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fernandes, H., B. Koneru, N. Fernandes, M. Hameed, M. C. Cohen, E. Raveche, and S. Cohen. 2002. Investigation of promoter polymorphisms in the tumor necrosis factor-alpha and interleukin-10 genes in liver transplant patients. Transplantation 73:1886-1891. [DOI] [PubMed] [Google Scholar]
- 88.Fernando, S. L., and W. J. Britton. 2006. Genetic susceptibility to mycobacterial disease in humans. Immunol. Cell Biol. 84:125-137. [DOI] [PubMed] [Google Scholar]
- 89.Ferreira, F. R., L. R. Goulart, H. D. Silva, and I. M. Goulart. 2004. Susceptibility to leprosy may be conditioned by an interaction between the NRAMP1 promoter polymorphisms and the lepromin response. Int. J. Lepr. Other Mycobact. Dis. 72:457-467. [DOI] [PubMed] [Google Scholar]
- 90.Ferwerda, G., S. E. Girardin, B. J. Kullberg, L. Le Bourhis, D. J. de Jong, D. M. Langenberg, R. van Crevel, G. J. Adema, T. H. Ottenhoff, J. W. Van der Meer, and M. G. Netea. 2005. NOD2 and Toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 1:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferwerda, G., B. J. Kullberg, D. J. de Jong, S. E. Girardin, D. M. Langenberg, R. van Crevel, T. H. Ottenhoff, J. W. Van Der Meer, and M. G. Netea. 2007. Mycobacterium paratuberculosis is recognized by Toll-like receptors and NOD2. J. Leukoc. Biol. 82:1011-1018. [DOI] [PubMed] [Google Scholar]
- 92.Fiorentino, D. F., A. Zlotnik, P. Vieira, T. R. Mosmann, M. Howard, K. W. Moore, and A. O'Garra. 1991. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J. Immunol. 146:3444-3451. [PubMed] [Google Scholar]
- 93.Fitness, J., S. Floyd, D. K. Warndorff, L. Sichali, L. Mwaungulu, A. C. Crampin, P. E. Fine, and A. V. Hill. 2004. Large-scale candidate gene study of leprosy susceptibility in the Karonga district of northern Malawi. Am. J. Trop. Med. Hyg. 71:330-340. [PubMed] [Google Scholar]
- 94.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 95.Flynn, J. L., J. Chan, K. J. Triebold, D. K. Dalton, T. A. Stewart, and B. R. Bloom. 1993. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Franceschi, D. S., P. S. Mazini, C. C. Rudnick, A. M. Sell, L. T. Tsuneto, M. L. Ribas, P. R. Peixoto, and J. E. Visentainer. 2009. Influence of TNF and IL10 gene polymorphisms in the immunopathogenesis of leprosy in the south of Brazil. Int. J. Infect. Dis. 13:493-498. [DOI] [PubMed] [Google Scholar]
- 97.Franceschi, D. S. A., P. S. Mazini, C. C. C. Rudnick, A. M. Sell, L. T. Tsuneto, F. C. de Melo, M. A. Braga, P. R. F. Peixoto, and J. E. L. Visentainer. 2008. Association between killer-cell immunoglobulin-like receptor genotypes and leprosy in Brazil. Tissue Antigens 72:478-482. [DOI] [PubMed] [Google Scholar]
- 98.Franchi, L., N. Warner, K. Viani, and G. Nunez. 2009. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 227:106-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fraser, D. A., L. Bulat-Kardum, J. Knezevic, P. Babarovic, N. Matakovic-Mileusnic, J. Dellacasagrande, D. Matanic, J. Pavelic, Z. Beg-Zec, and Z. Dembic. 2003. Interferon-gamma receptor-1 gene polymorphism in tuberculosis patients from Croatia. Scand. J. Immunol. 57:480-484. [DOI] [PubMed] [Google Scholar]
- 100.Gagnon, L., L. G. Filion, C. Dubois, and M. Rola-Pleszczynski. 1989. Leukotrienes and macrophage activation: augmented cytotoxic activity and enhanced interleukin 1, tumor necrosis factor and hydrogen peroxide production. Agents Actions 26:141-147. [DOI] [PubMed] [Google Scholar]
- 101.Gantner, B. N., R. M. Simmons, S. J. Canavera, S. Akira, and D. M. Underhill. 2003. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197:1107-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garcia, V. E., K. Uyemura, P. A. Sieling, M. T. Ochoa, C. T. Morita, H. Okamura, M. Kurimoto, T. H. Rea, and R. L. Modlin. 1999. IL-18 promotes type 1 cytokine production from NK cells and T cells in human intracellular infection. J. Immunol. 162:6114-6121. [PubMed] [Google Scholar]
- 103.Reference deleted.
- 104.Garcia-Gonzalez, M. A., M. A. Aisa, M. Strunk, R. Benito, E. Piazuelo, P. Jimenez, F. Sopena, and A. Lanas. 2009. Relevance of IL-1 and TNF gene polymorphisms on interleukin-1β and tumor necrosis factor-α gastric mucosal production. Hum. Immunol. 70:935-945. [DOI] [PubMed] [Google Scholar]
- 105.Garred, P., M. Harboe, T. Oettinger, C. Koch, and A. Svejgaard. 1994. Dual role of mannan-binding protein in infections: another case of heterosis? Eur. J. Immunogenet. 21:125-131. [DOI] [PubMed] [Google Scholar]
- 106.Garred, P., F. Larsen, J. Seyfarth, R. Fujita, and H. O. Madsen. 2006. Mannose-binding lectin and its genetic variants. Genes Immun. 7:85-94. [DOI] [PubMed] [Google Scholar]
- 107.Geijtenbeek, T. B. H., R. Torensma, S. J. van Vliet, G. C. F. van Duijnhoven, G. J. Adema, Y. van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 108.Geluk, A., and T. H. Ottenhoff. 2006. HLA and leprosy in the pre and postgenomic eras. Hum. Immunol. 67:439-445. [DOI] [PubMed] [Google Scholar]
- 109.Genc, M. R., S. Vardhana, M. L. Delaney, A. Onderdonk, R. Tuomala, E. Norwitz, and S. S. Witkin. 2004. Relationship between a Toll-like receptor-4 gene polymorphism, bacterial vaginosis-related flora and vaginal cytokine responses in pregnant women. Eur. J. Obstet. Gynecol. Reprod. Biol. 116:152-156. [DOI] [PubMed] [Google Scholar]
- 110.Gibson, A. W., J. C. Edberg, J. Wu, R. G. Westendorp, T. W. Huizinga, and R. P. Kimberly. 2001. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J. Immunol. 166:3915-3922. [DOI] [PubMed] [Google Scholar]
- 111.Goldman, G., R. Welbourn, L. Kobzik, C. R. Valeri, D. Shepro, and H. B. Hechtman. 1993. Lavage with leukotriene B4 induces lung generation of tumor necrosis factor-alpha that in turn mediates neutrophil diapedesis. Surgery 113:297-303. [PubMed] [Google Scholar]
- 112.Gomes, G. I., E. P. Nahn, Jr., R. K. Santos, W. D. Da Silva, and T. L. Kipnis. 2008. The functional state of the complement system in leprosy. Am. J. Trop. Med. Hyg. 78:605-610. [PubMed] [Google Scholar]
- 113.Goulart, L. R., F. R. Ferreira, and I. M. Goulart. 2006. Interaction of TaqI polymorphism at exon 9 of the vitamin D receptor gene with the negative lepromin response may favor the occurrence of leprosy. FEMS Immunol. Med. Microbiol. 48:91-98. [DOI] [PubMed] [Google Scholar]
- 114.Greiner, J., F. J. Weber, G. Mauff, and M. Baur. 1980. Genetic polymorphisms of properdin factor B (Bf), the second component (C2), and the fourth component (C4) of complement in leprosy patients and healthy controls from Thailand. Immunobiology 158:134-138. [DOI] [PubMed] [Google Scholar]
- 115.Hagge, D. A., S. O. Robinson, D. Scollard, G. McCormick, and D. L. Williams. 2002. A new model for studying the effects of Mycobacterium leprae on Schwann cell and neuron interactions. J. Infect. Dis. 186:1283-1296. [DOI] [PubMed] [Google Scholar]
- 116.Hagge, D. A., B. M. Saunders, G. J. Ebenezer, N. A. Ray, V. T. Marks, W. J. Britton, J. L. Krahenbuhl, and L. B. Adams. 2009. Lymphotoxin-alpha and TNF have essential but independent roles in the evolution of the granulomatous response in experimental leprosy. Am. J. Pathol. 174:1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haile, R. W., L. Iselius, P. E. Fine, and N. E. Morton. 1985. Segregation and linkage analyses of 72 leprosy pedigrees. Hum. Hered. 35:43-52. [DOI] [PubMed] [Google Scholar]
- 118.Hansen, G. H. A. 1875. On the etiology of leprosy. Chir. Rev. 55:459-489. [PMC free article] [PubMed] [Google Scholar]
- 119.Harboe, M. 1973. Armauer Hansen—the man and his work. Int. J. Lepr. Other Mycobact. Dis. 41:417-424. [PubMed] [Google Scholar]
- 120.Hatagima, A., D. V. Opromolla, S. Ura, M. F. Feitosa, B. Beiguelman, and H. Krieger. 2001. No evidence of linkage between Mitsuda reaction and the NRAMP1 locus. Int. J. Lepr. Other Mycobact. Dis. 69:99-103. [PubMed] [Google Scholar]
- 121.Hawn, T. R., E. A. Misch, S. J. Dunstan, G. E. Thwaites, N. T. Lan, H. T. Quy, T. T. Chau, S. Rodrigues, A. Nachman, M. Janer, T. T. Hien, J. J. Farrar, and A. Aderem. 2007. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur. J. Immunol. 37:2280-2289. [DOI] [PubMed] [Google Scholar]
- 122.Hawn, T. R., A. Verbon, M. Janer, L. P. Zhao, B. Beutler, and A. Aderem. 2005. Toll-like receptor 4 polymorphisms are associated with resistance to Legionnaires' disease. Proc. Natl. Acad. Sci. U. S. A. 102:2487-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Heldwein, K. A., and M. J. Fenton. 2002. The role of Toll-like receptors in immunity against mycobacterial infection. Microbes Infect. 4:937-944. [DOI] [PubMed] [Google Scholar]
- 124.Helgadottir, A., A. Manolescu, A. Helgason, G. Thorleifsson, U. Thorsteinsdottir, D. F. Gudbjartsson, S. Gretarsdottir, K. P. Magnusson, G. Gudmundsson, A. Hicks, T. Jonsson, S. F. A. Grant, J. Sainz, S. J. O'Brien, S. Sveinbjornsdottir, E. M. Valdimarsson, S. E. Matthiasson, A. I. Levey, J. L. Abramson, M. P. Reilly, V. Vaccarino, M. L. Wolfe, V. Gudnason, A. A. Quyyumi, E. J. Topol, D. J. Rader, G. Thorgeirsson, J. R. Gulcher, H. Hakonarson, A. Kong, and K. Stefansson. 2006. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat. Genet. 38:68-74. [DOI] [PubMed] [Google Scholar]
- 125.Herbeck, J. T., G. S. Gottlieb, C. A. Winkler, G. W. Nelson, P. An, B. S. Maust, K. G. Wong, J. L. Troyer, J. J. Goedert, B. D. Kessing, R. Detels, S. M. Wolinsky, J. Martinson, S. Buchbinder, G. D. Kirk, L. P. Jacobson, J. B. Margolick, R. A. Kaslow, S. J. O'Brien, and J. I. Mullins. 2010. Multistage genomewide association study identifies a locus at 1q41 associated with rate of HIV disease progression to clinical AIDS. J. Infect. Dis. 201:618-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Herpers, B. L., M.-M. Immink, B. A. W. de Jong, H. van Velzen-Blad, B. M. de Jongh, and E. J. van Hannen. 2006. Coding and non-coding polymorphisms in the lectin pathway activator L-ficolin gene in 188 Dutch blood bank donors. Mol. Immunol. 43:851-855. [DOI] [PubMed] [Google Scholar]
- 127.Hill, A. V. S. 2006. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 40:469. [DOI] [PubMed] [Google Scholar]
- 128.Hoeven, T. A., E. A. Fischer, D. Pahan, and J. H. Richardus. 2008. Social distance and spatial distance are not the same, observations on the use of GIS in leprosy epidemiology. Epidemiol. Infect. 136:1624-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsieh, C. S., A. B. Heimberger, J. S. Gold, A. O'Garra, and K. M. Murphy. 1992. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an alpha beta T-cell-receptor transgenic system. Proc. Natl. Acad. Sci. U. S. A. 89:6065-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hsieh, C. S., S. E. Macatonia, C. S. Tripp, S. F. Wolf, A. O'Garra, and K. M. Murphy. 1993. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260:547-549. [DOI] [PubMed] [Google Scholar]
- 131.Hugot, J. P., M. Chamaillard, H. Zouali, S. Lesage, J. P. Cezard, J. Belaiche, S. Almer, C. Tysk, C. A. O'Morain, M. Gassull, V. Binder, Y. Finkel, A. Cortot, R. Modigliani, P. Laurent-Puig, C. Gower-Rousseau, J. Macry, J. F. Colombel, M. Sahbatou, and G. Thomas. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411:599-603. [DOI] [PubMed] [Google Scholar]
- 132.Hummelshoj, T., L. Munthe-Fog, H. O. Madsen, T. Fujita, M. Matsushita, and P. Garred. 2005. Polymorphisms in the FCN2 gene determine serum variation and function of ficolin-2. Hum. Mol. Genet. 14:1651-1658. [DOI] [PubMed] [Google Scholar]
- 133.Hummelshoj, T., N. M. Thielens, H. O. Madsen, G. J. Arlaud, R. B. Sim, and P. Garred. 2007. Molecular organization of human ficolin-2. Mol. Immunol. 44:401-411. [DOI] [PubMed] [Google Scholar]
- 134.Ilangumaran, S., S. Ramanathan, N. Shankernarayan, G. Ramu, and V. Muthukkarauppan. 1996. Immunological profiles of leprosy patients and healthy family contacts toward M. leprae antigens. Int. J. Lepr. Other Mycobact. Dis. 64:6-14. [PubMed] [Google Scholar]
- 135.Ingles, S. A., R. W. Haile, B. E. Henderson, L. N. Kolonel, G. Nakaichi, C. Y. Shi, M. C. Yu, R. K. Ross, and G. A. Coetzee. 1997. Strength of linkage disequilibrium between two vitamin D receptor markers in five ethnic groups: implications for association studies. Cancer Epidemiol. Biomarkers Prev. 6:93-98. [PubMed] [Google Scholar]
- 136.Ip, W. K., K. Takahashi, R. A. Ezekowitz, and L. M. Stuart. 2009. Mannose-binding lectin and innate immunity. Immunol. Rev. 230:9-21. [DOI] [PubMed] [Google Scholar]
- 137.Ip, W. K., K. Takahashi, K. J. Moore, L. M. Stuart, and R. A. Ezekowitz. 2008. Mannose-binding lectin enhances Toll-like receptors 2 and 6 signaling from the phagosome. J. Exp. Med. 205:169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ishida, H., R. Hastings, L. Thompson-Snipes, and M. Howard. 1993. Modified immunological status of anti-IL-10 treated mice. Cell. Immunol. 148:371-384. [DOI] [PubMed] [Google Scholar]
- 139.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 140.Jacobson, R. R., and J. L. Krahenbuhl. 1999. Leprosy. Lancet 353:655-660. [DOI] [PubMed] [Google Scholar]
- 141.Jallow, M., Y. Y. Teo, K. S. Small, K. A. Rockett, P. Deloukas, T. G. Clark, K. Kivinen, K. A. Bojang, D. J. Conway, M. Pinder, G. Sirugo, F. Sisay-Joof, S. Usen, S. Auburn, S. J. Bumpstead, S. Campino, A. Coffey, A. Dunham, A. E. Fry, A. Green, R. Gwilliam, S. E. Hunt, M. Inouye, A. E. Jeffreys, A. Mendy, A. Palotie, S. Potter, J. Ragoussis, J. Rogers, K. Rowlands, E. Somaskantharajah, P. Whittaker, C. Widden, P. Donnelly, B. Howie, J. Marchini, A. Morris, M. Sanjoaquin, E. A. Achidi, T. Agbenyega, A. Allen, O. Amodu, P. Corran, A. Djimde, A. Dolo, O. K. Doumbo, C. Drakeley, S. Dunstan, J. Evans, J. Farrar, D. Fernando, T. T. Hien, R. D. Horstmann, M. Ibrahim, N. Karunaweera, G. Kokwaro, K. A. Koram, M. Lemnge, J. Makani, K. Marsh, P. Michon, D. Modiano, M. E. Molyneux, I. Mueller, M. Parker, N. Peshu, C. V. Plowe, O. Puijalon, J. Reeder, H. Reyburn, E. M. Riley, A. Sakuntabhai, P. Singhasivanon, S. Sirima, A. Tall, T. E. Taylor, M. Thera, M. Troye-Blomberg, T. N. Williams, M. Wilson, and D. P. Kwiatkowski. 2009. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat. Genet. 41:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jamieson, S. E., E. N. Miller, G. F. Black, C. S. Peacock, H. J. Cordell, J. M. Howson, M. A. Shaw, D. Burgner, W. Xu, Z. Lins-Lainson, J. J. Shaw, F. Ramos, F. Silveira, and J. M. Blackwell. 2004. Evidence for a cluster of genes on chromosome 17q11-q21 controlling susceptibility to tuberculosis and leprosy in Brazilians. Genes Immun. 5:46-57. [DOI] [PubMed] [Google Scholar]
- 143.Jindal, N., V. Shanker, G. R. Tegta, M. Gupta, and G. K. Verma. 2009. Clinico-epidemiological trends of leprosy in Himachal Pradesh: a five year study. Indian J. Lepr. 81:173-179. [PubMed] [Google Scholar]
- 144.Jo, E. K. 2008. Mycobacterial interaction with innate receptors: TLRs, C-type lectins, and NLRs. Curr. Opin. Infect. Dis. 21:279-286. [DOI] [PubMed] [Google Scholar]
- 145.Job, C. K. 1980. Genetics and leprosy. Lepr. India 52:353-358. [PubMed] [Google Scholar]
- 146.Job, C. K. 1989. Nerve damage in leprosy. Int. J. Lepr. Other Mycobact. Dis. 57:532-539. [PubMed] [Google Scholar]
- 147.Job, C. K. 1971. Pathology of peripheral nerve lesions in lepromatous leprosy—a light and electron microscopic study. Int. J. Lepr. Other Mycobact. Dis. 39:251-268. [PubMed] [Google Scholar]
- 148.Job, C. K., J. Jayakumar, M. Kearney, and T. P. Gillis. 2008. Transmission of leprosy: a study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am. J. Trop. Med. Hyg. 78:518-521. [PubMed] [Google Scholar]
- 149.Johnson, C. M., E. A. Lyle, K. O. Omueti, V. A. Stepensky, O. Yegin, E. Alpsoy, L. Hamann, R. R. Schumann, and R. I. Tapping. 2007. A common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J. Immunol. 178:7520-7524. [DOI] [PubMed] [Google Scholar]
- 150.Johnson, C. M., J. A. Traherne, S. E. Jamieson, M. Tremelling, S. Bingham, M. Parkes, J. M. Blackwell, and J. Trowsdale. 2007. Analysis of the BTNL2 truncating splice site mutation in tuberculosis, leprosy and Crohn's disease. Tissue Antigens 69:236-241. [DOI] [PubMed] [Google Scholar]
- 151.Jullien, D., P. Sieling, K. Uyemura, N. Mar, T. Rea, and R. Modlin. 1997. IL-15, an immunomodulator of T cell responses in intracellular infection. J. Immunol. 158:800-806. [PubMed] [Google Scholar]
- 152.Kahawita, I. P., and D. N. Lockwood. 2008. Towards understanding the pathology of erythema nodosum leprosum. Trans. R. Soc. Trop. Med. Hyg. 102:329-337. [DOI] [PubMed] [Google Scholar]
- 153.Kamatani, Y., S. Wattanapokayakit, H. Ochi, T. Kawaguchi, A. Takahashi, N. Hosono, M. Kubo, T. Tsunoda, N. Kamatani, H. Kumada, A. Puseenam, T. Sura, Y. Daigo, K. Chayama, W. Chantratita, Y. Nakamura, and K. Matsuda. 2009. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 41:591-595. [DOI] [PubMed] [Google Scholar]
- 154.Kang, T. J., and G. T. Chae. 2001. Detection of Toll-like receptor 2 (TLR2) mutation in the lepromatous leprosy patients. FEMS Immunol. Med. Microbiol. 31:53-58. [DOI] [PubMed] [Google Scholar]
- 155.Kaur, G., G. Sachdeva, L. K. Bhutani, and R. Bamezai. 1997. Association of polymorphism at COL3A and CTLA4 loci on chromosome 2q31-33 with the clinical phenotype and in-vitro CMI status in healthy and leprosy subjects: a preliminary study. Hum. Genet. 100:43-50. [DOI] [PubMed] [Google Scholar]
- 156.Kerr-Pontes, L. R., M. L. Barreto, C. M. Evangelista, L. C. Rodrigues, J. Heukelbach, and H. Feldmeier. 2006. Socioeconomic, environmental, and behavioural risk factors for leprosy in North-east Brazil: results of a case-control study. Int. J. Epidemiol. 35:994-1000. [DOI] [PubMed] [Google Scholar]
- 157.Kéry, V., J. J. F. Krepinsky, C. D. Warren, P. Capek, and P. D. Stahl. 1992. Ligand recognition by purified human mannose receptor. Arch. Biochem. Biophys. 298:49-55. [DOI] [PubMed] [Google Scholar]
- 158.Knight, J. C., B. J. Keating, and D. P. Kwiatkowski. 2004. Allele-specific repression of lymphotoxin-alpha by activated B cell factor-1. Nat. Genet. 36:394-399. [DOI] [PubMed] [Google Scholar]
- 159.Knight, J. C., B. J. Keating, K. A. Rockett, and D. P. Kwiatkowski. 2003. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat. Genet. 33:469-475. [DOI] [PubMed] [Google Scholar]
- 160.Krahenbuhl, J. L., L. D. Sibley, and G.-T. Chae. 1990. Gamma interferon in experimental leprosy. Diagn. Microbiol. Infect. Dis. 13:405-409. [DOI] [PubMed] [Google Scholar]
- 161.Krarup, A., S. Thiel, A. Hansen, T. Fujita, and J. C. Jensenius. 2004. L-ficolin is a pattern recognition molecule specific for acetyl groups. J. Biol. Chem. 279:47513-47519. [DOI] [PubMed] [Google Scholar]
- 162.Kroeger, K. M., K. S. Carville, and L. J. Abraham. 1997. The −308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol. Immunol. 34:391-399. [DOI] [PubMed] [Google Scholar]
- 163.Krutzik, S. R., M. T. Ochoa, P. A. Sieling, S. Uematsu, Y. W. Ng, A. Legaspi, P. T. Liu, S. T. Cole, P. J. Godowski, Y. Maeda, E. N. Sarno, M. V. Norgard, P. J. Brennan, S. Akira, T. H. Rea, and R. L. Modlin. 2003. Activation and regulation of Toll-like receptors 2 and 1 in human leprosy. Nat. Med. 9:525-532. [DOI] [PubMed] [Google Scholar]
- 164.Krutzik, S. R., B. Tan, H. Li, M. T. Ochoa, P. T. Liu, S. E. Sharfstein, T. G. Graeber, P. A. Sieling, Y. J. Liu, T. H. Rea, B. R. Bloom, and R. L. Modlin. 2005. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat. Med. 11:653-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kubota, T., D. M. McNamara, J. J. Wang, M. Trost, C. F. McTiernan, D. L. Mann, and A. M. Feldman. 1998. Effects of tumor necrosis factor gene polymorphisms on patients with congestive heart failure. VEST Investigators for TNF Genotype Analysis. Vesnarinone Survival Trial. Circulation 97:2499-2501. [DOI] [PubMed] [Google Scholar]
- 166.Kuhn, R., J. Lohler, D. Rennick, K. Rajewsky, and W. Muller. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75:263-274. [DOI] [PubMed] [Google Scholar]
- 167.Lahiri, R., and J. L. Krahenbuhl. 2008. The role of free-living pathogenic amoeba in the transmission of leprosy: a proof of principle. Lepr. Rev. 79:401-409. [PubMed] [Google Scholar]
- 168.Lander, E., and L. Kruglyak. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11:241-247. [DOI] [PubMed] [Google Scholar]
- 169.Lane, J. E., W. M. Meyers, and D. S. Walsh. 2009. Armadillos as a source of leprosy infection in the Southeast. South. Med. J. 102:113-114. [DOI] [PubMed] [Google Scholar]
- 170.Lavania, M., K. Katoch, V. M. Katoch, A. K. Gupta, D. S. Chauhan, R. Sharma, R. Gandhi, V. Chauhan, G. Bansal, P. Sachan, S. Sachan, V. S. Yadav, and R. Jadhav. 2008. Detection of viable Mycobacterium leprae in soil samples: insights into possible sources of transmission of leprosy. Infect. Genet. Evol. 8:627-631. [DOI] [PubMed] [Google Scholar]
- 171.Lazaro, F. P., R. I. Werneck, C. C. Mackert, A. Cobat, F. C. Prevedello, R. P. Pimentel, G. M. Macedo, M. A. Eleueterio, G. Vilar, L. Abel, M. B. Xavier, A. Alcais, and M. T. Mira. 2010. A major gene controls leprosy susceptibility in a hyperendemic isolated population from North of Brazil. J. Infect. Dis. 201:1598-1605. [DOI] [PubMed] [Google Scholar]
- 172.Lee, H., E. K. Jo, S. Y. Choi, S. B. Oh, K. Park, J. S. Kim, and S. J. Lee. 2006. Necrotic neuronal cells induce inflammatory Schwann cell activation via TLR2 and TLR3: implication in Wallerian degeneration. Biochem. Biophys. Res. Commun. 350:742-747. [DOI] [PubMed] [Google Scholar]
- 173.Lee, S. B., B. C. Kim, S. H. Jin, Y. G. Park, S. K. Kim, T. J. Kang, and G. T. Chae. 2003. Missense mutations of the interleukin-12 receptor beta 1 (IL12RB1) and interferon-gamma receptor 1 (IFNGR1) genes are not associated with susceptibility to lepromatous leprosy in Korea. Immunogenetics 55:177-181. [DOI] [PubMed] [Google Scholar]
- 174.Lemire, J. M., J. S. Adams, R. Sakai, and S. C. Jordan. 1984. 1 alpha,25-dihydroxyvitamin D3 suppresses proliferation and immunoglobulin production by normal human peripheral blood mononuclear cells. J. Clin. Invest. 74:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Levee, G., J. Liu, B. Gicquel, S. Chanteau, and E. Schurr. 1994. Genetic control of susceptibility to leprosy in French Polynesia; no evidence for linkage with markers on telomeric human chromosome 2. Int. J. Lepr. Other Mycobact. Dis. 62:499-511. [PubMed] [Google Scholar]
- 176.Levee, G., E. Schurr, and J. P. Pandey. 1997. Tumor necrosis factor-alpha, interleukin-1-beta and immunoglobulin (GM and KM) polymorphisms in leprosy. A linkage study. Exp. Clin. Immunogenet. 14:160-165. [PubMed] [Google Scholar]
- 177.Lima, M. C., G. M. Pereira, F. D. Rumjanek, H. M. Gomes, N. Duppre, E. P. Sampaio, I. M. Alvim, J. A. Nery, E. N. Sarno, and M. C. Pessolani. 2000. Immunological cytokine correlates of protective immunity and pathogenesis in leprosy. Scand. J. Immunol. 51:419-428. [DOI] [PubMed] [Google Scholar]
- 178.Little, D., S. Khanolkar-Young, A. Coulthart, S. Suneetha, and D. N. Lockwood. 2001. Immunohistochemical analysis of cellular infiltrate and gamma interferon, interleukin-12, and inducible nitric oxide synthase expression in leprosy type 1 (reversal) reactions before and during prednisolone treatment. Infect. Immun. 69:3413-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu, P. T., S. Stenger, H. Li, L. Wenzel, B. H. Tan, S. R. Krutzik, M. T. Ochoa, J. Schauber, K. Wu, C. Meinken, D. L. Kamen, M. Wagner, R. Bals, A. Steinmeyer, U. Zugel, R. L. Gallo, D. Eisenberg, M. Hewison, B. W. Hollis, J. S. Adams, B. R. Bloom, and R. L. Modlin. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311:1770-1773. [DOI] [PubMed] [Google Scholar]
- 180.Lockwood, D. N., S. Vinayakumar, J. N. Stanley, K. P. McAdam, and M. J. Colston. 1993. Clinical features and outcome of reversal (type 1) reactions in Hyderabad, India. Int. J. Lepr. Other Mycobact. Dis. 61:8-15. [PubMed] [Google Scholar]
- 181.Louis, E., D. Franchimont, A. Piron, Y. Gevaert, N. Schaaf-Lafontaine, S. Roland, P. Mahieu, M. Malaise, D. De Groote, R. Louis, and J. Belaiche. 1998. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin. Exp. Immunol. 113:401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lwin, K., T. Sundaresan, M. M. Gyi, L. M. Bechelli, C. Tamondong, P. G. Garbajosa, H. Sansarricq, and S. K. Noordeen. 1985. BCG vaccination of children against leprosy: fourteen-year findings of the trial in Burma. Bull. World Health Organ. 63:1069-1078. [PMC free article] [PubMed] [Google Scholar]
- 183.Mahapatra, S., D. C. Crick, M. R. McNeil, and P. J. Brennan. 2008. Unique structural features of the peptidoglycan of Mycobacterium leprae. J. Bacteriol. 190:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Malhotra, D., K. Darvishi, M. Lohra, H. Kumar, C. Grover, S. Sood, B. S. Reddy, and R. N. Bamezai. 2006. Association study of major risk single nucleotide polymorphisms in the common regulatory region of PARK2 and PACRG genes with leprosy in an Indian population. Eur. J. Hum. Genet. 14:438-442. [DOI] [PubMed] [Google Scholar]
- 185.Malhotra, D., K. Darvishi, S. Sood, S. Sharma, C. Grover, V. Relhan, B. S. Reddy, and R. N. Bamezai. 2005. IL-10 promoter single nucleotide polymorphisms are significantly associated with resistance to leprosy. Hum. Genet. 118:295-300. [DOI] [PubMed] [Google Scholar]
- 186.Malhotra, D., V. Relhan, B. S. N. Reddy, and R. Bamezai. 2005. TLR2 Arg677Trp polymorphism in leprosy: revisited. Hum. Genet. 116:413-415. [DOI] [PubMed] [Google Scholar]
- 187.McGuire, W., A. V. Hill, C. E. Allsopp, B. M. Greenwood, and D. Kwiatkowski. 1994. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature 371:508-510. [DOI] [PubMed] [Google Scholar]
- 188.Mead, S., M. Poulter, J. Uphill, J. Beck, J. Whitfield, T. E. F. Webb, T. Campbell, G. Adamson, P. Deriziotis, S. J. Tabrizi, H. Hummerich, C. Verzilli, M. P. Alpers, J. C. Whittaker, and J. Collinge. 2009. Genetic risk factors for variant Creutzfeldt-Jakob disease: a genome-wide association study. Lancet Neurol. 8:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Means, T. K., B. W. Jones, A. B. Schromm, B. A. Shurtleff, J. A. Smith, J. Keane, D. T. Golenbock, S. N. Vogel, and M. J. Fenton. 2001. Differential effects of a Toll-like receptor antagonist on Mycobacterium tuberculosis-induced macrophage responses. J. Immunol. 166:4074-4082. [DOI] [PubMed] [Google Scholar]
- 190.Means, T. K., S. Wang, E. Lien, A. Yoshimura, D. T. Golenbock, and M. J. Fenton. 1999. Human Toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J. Immunol. 163:3920-3927. [PubMed] [Google Scholar]
- 191.Meisner, S. J., S. Mucklow, G. Warner, S. O. Sow, C. Lienhardt, and A. V. Hill. 2001. Association of NRAMP1 polymorphism with leprosy type but not susceptibility to leprosy per se in West Africans. Am. J. Trop. Med. Hyg. 65:733-735. [DOI] [PubMed] [Google Scholar]
- 192.Miceli-Richard, C., S. Lesage, M. Rybojad, A. M. Prieur, S. Manouvrier-Hanu, R. Hafner, M. Chamaillard, H. Zouali, G. Thomas, and J. P. Hugot. 2001. CARD15 mutations in Blau syndrome. Nat. Genet. 29:19-20. [DOI] [PubMed] [Google Scholar]
- 193.Michel, O., T. D. LeVan, D. Stern, M. Dentener, J. Thorn, D. Gnat, M. L. Beijer, P. Cochaux, P. G. Holt, F. D. Martinez, and R. Rylander. 2003. Systemic responsiveness to lipopolysaccharide and polymorphisms in the Toll-like receptor 4 gene in human beings. J. Allergy Clin. Immunol. 112:923-929. [DOI] [PubMed] [Google Scholar]
- 194.Mikita, N., N. Kanazawa, M. Ozaki, M. Kosaka, N. Ishii, H. Nishimura, and F. Furukawa. 2009. No involvement of non-synonymous TLR2 polymorphisms in Japanese leprosy patients. J. Dermatol. Sci. 54:48-49. [DOI] [PubMed] [Google Scholar]
- 195.Miller, E. N., S. E. Jamieson, C. Joberty, M. Fakiola, D. Hudson, C. S. Peacock, H. J. Cordell, M. A. Shaw, Z. Lins-Lainson, J. J. Shaw, F. Ramos, F. Silveira, and J. M. Blackwell. 2004. Genome-wide scans for leprosy and tuberculosis susceptibility genes in Brazilians. Genes Immun. 5:63-67. [DOI] [PubMed] [Google Scholar]
- 196.Mira, M. T., A. Alcais, N. V. Thuc, M. O. Moraes, C. Di Flumeri, V. H. Thai, M. C. Phuong, N. T. Huong, N. N. Ba, P. X. Khoa, E. N. Sarno, A. Alter, A. Montpetit, M. E. Moraes, J. R. Moraes, C. Dore, C. J. Gallant, P. Lepage, A. Verner, E. van de Vosse, T. J. Hudson, L. Abel, and E. Schurr. 2004. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427:636-640. [DOI] [PubMed] [Google Scholar]
- 197.Mira, M. T., A. Alcais, N. V. Thuc, V. H. Thai, N. T. Huong, N. N. Ba, A. Verner, T. J. Hudson, L. Abel, and E. Schurr. 2003. Chromosome 6q25 is linked to susceptibility to leprosy in a Vietnamese population. Nat. Genet. 33:412-415. [DOI] [PubMed] [Google Scholar]
- 198.Misch, E. A., M. Macdonald, C. Ranjit, B. R. Sapkota, R. D. Wells, M. R. Siddiqui, G. Kaplan, and T. R. Hawn. 2008. Human TLR1 deficiency is associated with impaired mycobacterial signaling and protection from leprosy reversal reaction. PLoS Negl. Trop. Dis. 2:e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Misra, N., M. Selvakumar, S. Singh, M. Bharadwaj, V. Ramesh, R. S. Misra, and I. Nath. 1995. Monocyte derived IL 10 and PGE2 are associated with the absence of Th 1 cells and in vitro T cell suppression in lepromatous leprosy. Immunol. Lett. 48:123-128. [DOI] [PubMed] [Google Scholar]
- 200.Mocharla, H., A. W. Butch, A. A. Pappas, J. T. Flick, R. S. Weinstein, P. De Togni, R. L. Jilka, P. K. Roberson, A. M. Parfitt, and S. C. Manolagas. 1997. Quantification of vitamin D receptor mRNA by competitive polymerase chain reaction in PBMC: lack of correspondence with common allelic variants. J. Bone Miner. Res. 12:726-733. [DOI] [PubMed] [Google Scholar]
- 201.Mock, B., J. Blackwell, J. Hilgers, M. Potter, and C. Nacy. 1993. Genetic control of Leishmania major infection in congenic, recombinant inbred and F2 populations of mice. Eur. J. Immunogenet. 20:335-348. [DOI] [PubMed] [Google Scholar]
- 202.Modlin, R. L. 2010. The innate immune response in leprosy. Curr. Opin. Immunol. 22:48-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Moet, F. J., A. Meima, L. Oskam, and J. H. Richardus. 2004. Risk factors for the development of clinical leprosy among contacts, and their relevance for targeted interventions. Lepr. Rev. 75:310-326. [PubMed] [Google Scholar]
- 204.Moet, F. J., D. Pahan, R. P. Schuring, L. Oskam, and J. H. Richardus. 2006. Physical distance, genetic relationship, age, and leprosy classification are independent risk factors for leprosy in contacts of patients with leprosy. J. Infect. Dis. 193:346-353. [DOI] [PubMed] [Google Scholar]
- 205.Moore, K. W., A. O'Garra, R. de Waal Malefyt, P. Vieira, and T. R. Mosmann. 1993. Interleukin-10. Annu. Rev. Immunol. 11:165-190. [DOI] [PubMed] [Google Scholar]
- 206.Moraes, M. O., A. G. Pacheco, J. J. M. Schonkeren, P. R. Vanderborght, J. A. C. Nery, A. R. Santos, M. E. Moraes, J. R. Moraes, T. H. M. Ottenhoff, E. P. Sampaio, T. W. J. Huizinga, and E. N. Sarno. 2004. Interleukin-10 promoter single-nucleotide polymorphisms as markers for disease susceptibility and disease severity in leprosy. Genes Immun. 5:592-595. [DOI] [PubMed] [Google Scholar]
- 207.Morahan, G., G. Kaur, M. Singh, C. C. Rapthap, N. Kumar, K. Katoch, N. K. Mehra, and D. Huang. 2007. Association of variants in the IL-12B gene with leprosy and tuberculosis. Tissue Antigens. 69:234-236. [DOI] [PubMed] [Google Scholar]
- 208.Moro, J. R., M. Iwata, and U. H. von Andriano. 2008. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat. Rev. Immunol. 8:685-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Morrison, N. A., J. C. Qi, A. Tokita, P. J. Kelly, L. Crofts, T. V. Nguyen, P. N. Sambrook, and J. A. Eisman. 1994. Prediction of bone density from vitamin D receptor alleles. Nature 367:284-287. [DOI] [PubMed] [Google Scholar]
- 210.Munthe-Fog, L., T. Hummelshøj, B. E. Hansen, C. Koch, H. O. Madsen, K. Skjødt, and P. Garred. 2007. The impact of FCN2 polymorphisms and haplotypes on the ficolin-2 serum levels. Scand. J. Immunol. 65:383-392. [DOI] [PubMed] [Google Scholar]
- 211.Murray, P. J., L. Wang, C. Onufryk, R. I. Tepper, and R. A. Young. 1997. T cell-derived IL-10 antagonizes macrophage function in mycobacterial infection. J. Immunol. 158:315-321. [PubMed] [Google Scholar]
- 212.Narayan, N. P., G. Ramu, K. V. Desikan, and R. S. Vallishayee. 2001. Correlation of clinical, histological and immunological features across the leprosy spectrum. Indian J. Lepr. 73:329-342. [PubMed] [Google Scholar]
- 213.Nedwin, G. E., S. L. Naylor, A. Y. Sakaguchi, D. Smith, J. Jarrett-Nedwin, D. Pennica, D. V. Goeddel, and P. W. Gray. 1985. Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic Acids Res. 13:6361-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ng, V., G. Zanazzi, R. Timpl, J. F. Talts, J. L. Salzer, P. J. Brennan, and A. Rambukkana. 2000. Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of Mycobacterium leprae. Cell 103:511-524. [DOI] [PubMed] [Google Scholar]
- 215.Nordeen, S. K. 1985. The epidemiology of leprosy, p. 15-30. In J. C. Hastings (ed.), Leprosy. Churchill-Livingstone, Edinburgh, United Kingdom.
- 216.Nordeen, S. K. 1994. The epidemiology of leprosy, p. 29-48. In J. C. Hastings (ed.), Leprosy, 2nd ed. Churchill-Livingstone, Edinburgh, United Kingdom.
- 217.Notarangelo, L. D., A. Fischer, R. S. Geha, J.-L. Casanova, H. Chapel, M. E. Conley, C. Cunningham-Rundles, A. Etzioni, L. Hammartrom, S. Nonoyama, H. D. Ochs, J. Puck, C. Roifman, R. Seger, and J. Wedgwood. 2009. Primary immunodeficiencies: 2009 update. J. Allergy Clin. Immunol. 124:1161-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ohyama, H., K. Ogata, K. Takeuchi, M. Namisato, Y. Fukutomi, F. Nishimura, H. Naruishi, T. Ohira, K. Hashimoto, T. Liu, M. Suzuki, Y. Uemura, and S. Matsushita. 2005. Polymorphism of the 5′ flanking region of the IL-12 receptor β2 gene partially determines the clinical types of leprosy through impaired transcriptional activity. J. Clin. Pathol. 58:740-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Oliveira, R. B., M. T. Ochoa, P. A. Sieling, T. H. Rea, A. Rambukkana, E. N. Sarno, and R. L. Modlin. 2003. Expression of Toll-like receptor 2 on human Schwann cells: a mechanism of nerve damage in leprosy. Infect. Immun. 71:1427-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pandey, A. K., Y. Yang, Z. Jiang, S. M. Fortune, F. Coulombe, M. A. Behr, K. A. Fitzgerald, C. M. Sassetti, and M. A. Kelliher. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 5:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Patrocinio, L. G., I. M. B. Goulart, L. R. Goulart, J. A. Patrocinio, F. R. Ferreira, and R. N. Fleury. 2005. Detection of Mycobacterium leprae in nasal mucosa biopsies by the polymerase chain reaction. FEMS Immunol. Med. Microbiol. 44:311-316. [DOI] [PubMed] [Google Scholar]
- 222.Paulus, S. C., A. F. Hirschfeld, R. E. Victor, J. Brunstein, E. Thomas, and S. E. Turvey. 2007. Common human Toll-like receptor 4 polymorphisms—role in susceptibility to respiratory syncytial virus infection and functional immunological relevance. Clin. Immunol. 123:252-257. [DOI] [PubMed] [Google Scholar]
- 223.Pedley, J. C. 1973. The nasal mucus in leprosy. Lepr. Rev. 44:33-35. [DOI] [PubMed] [Google Scholar]
- 224.Penna, G., and L. Adorini. 2000. 1α,25-dihydroxyvitamin D3 inhibits differentiation, maturation, activation, and survival of dendritic cells leading to impaired alloreactive T cell activation. J. Immunol. 164:2405-2411. [DOI] [PubMed] [Google Scholar]
- 225.Pereira, A. C., V. N. Brito-de-Souza, C. C. Cardoso, I. M. F. Dias-Baptista, F. P. C. Parelli, J. Venturini, F. R. Villani-Moreno, A. G. Pacheco, and M. O. Moraes. 2008. Genetic, epidemiological and biological analysis of interleukin-10 promoter single-nucleotide polymorphisms suggests a definitive role for −819C/T in leprosy susceptibility. Genes Immun. 10:174-180. [DOI] [PubMed] [Google Scholar]
- 226.Plant, J., and A. A. Glynn. 1976. Genetics of resistance to infection with Salmonella typhimurium in mice. J. Infect. Dis. 133:72-78. [DOI] [PubMed] [Google Scholar]
- 227.Plant, J., and A. A. Glynn. 1974. Natural resistance to Salmonella infection, delayed hypersensitivity and Ir genes in different strains of mice. Nature 248:345-347. [DOI] [PubMed] [Google Scholar]
- 228.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 229.Porter, R. R. 1985. The complement components coded in the major histocompatibility complexes and their biological activities. Immunol. Rev. 87:7-17. [DOI] [PubMed] [Google Scholar]
- 230.Prado-Montes de Oca, E., J. S. Velarde-Félix, J. J. Ríos-Tostado, V. J. Picos-Cárdenas, and L. E. Figuera. 2009. SNP 668C (−44) alters a NF-kappaB1 putative binding site in non-coding strand of human [beta]-defensin 1 (DEFB1) and is associated with lepromatous leprosy. Infect. Genet. Evol. 9:617-625. [DOI] [PubMed] [Google Scholar]
- 231.Pravica, V., A. Asderakis, C. Perrey, A. Hajeer, P. J. Sinnott, and I. V. Hutchinson. 1999. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur. J. Immunogenet. 26:1-3. [DOI] [PubMed] [Google Scholar]
- 232.Propping, P., and F. Vogel. 1976. Twin studies in medical genetics. Acta Genet. Med. Gemellol. (Roma) 25:249-258. [DOI] [PubMed] [Google Scholar]
- 233.Quesniaux, V., C. Fremond, M. Jacobs, S. Parida, D. Nicolle, V. Yeremeev, F. Bihl, F. Erard, T. Botha, M. Drennan, M. N. Soler, M. Le Bert, B. Schnyder, and B. Ryffel. 2004. Toll-like receptor pathways in the immune responses to mycobacteria. Microbes Infect. 6:946-959. [DOI] [PubMed] [Google Scholar]
- 234.Quintana-Murci, L., A. Alcais, L. Abel, and J.-L. Casanova. 2007. Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases. Nat. Immunol. 8:1165-1171. [DOI] [PubMed] [Google Scholar]
- 235.Rajalingam, R., N. K. Mehra, and D. P. Singal. 2000. Polymorphism in heat-shock protein 70-1 (HSP70-1) gene promoter region and susceptibility to tuberculoid leprosy and pulmonary tuberculosis in Asian Indians. Indian J. Exp. Biol. 38:658-662. [PubMed] [Google Scholar]
- 236.Rajalingam, R., D. P. Singal, and N. K. Mehra. 1997. Transporter associated with antigen-processing (TAP) genes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis. Tissue Antigens 49:168-172. [DOI] [PubMed] [Google Scholar]
- 237.Ralston, S. H., and B. de Crombrugghe. 2006. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 20:2492-2506. [DOI] [PubMed] [Google Scholar]
- 238.Rambukkana, A. 2001. Molecular basis for the peripheral nerve predilection of Mycobacterium leprae. Curr. Opin. Microbiol. 4:21-27. [DOI] [PubMed] [Google Scholar]
- 239.Rambukkana, A., J. L. Salzer, P. D. Yurchenco, and E. I. Tuomanen. 1997. Neural targeting of Mycobacterium leprae mediated by the G domain of the laminin-alpha2 chain. Cell 88:811-821. [DOI] [PubMed] [Google Scholar]
- 240.Rambukkana, A., H. Yamada, G. Zanazzi, T. Mathus, J. L. Salzer, P. D. Yurchenco, K. P. Campbell, and V. A. Fischetti. 1998. Role of alpha-dystroglycan as a Schwann cell receptor for Mycobacterium leprae. Science 282:2076-2079. [DOI] [PubMed] [Google Scholar]
- 241.Ranque, B., A. Alcais, N. V. Thuc, S. Woynard, V. H. Thai, N. T. Huong, N. N. Ba, P. X. Khoa, E. Schurr, and L. Abel. 2005. A recessive major gene controls the Mitsuda reaction in a region endemic for leprosy. J. Infect. Dis. 192:1475-1482. [DOI] [PubMed] [Google Scholar]
- 242.Ranque, B., A. Alter, M. Mira, N. V. Thuc, V. H. Thai, N. T. Huong, N. N. Ba, P. X. Khoa, E. Schurr, L. Abel, and A. Alcais. 2007. Genomewide linkage analysis of the granulomatous Mitsuda reaction implicates chromosomal regions 2q35 and 17q21. J. Infect. Dis. 196:1248-1252. [DOI] [PubMed] [Google Scholar]
- 243.Ranque, B., V. T. Nguyen, H. T. Vu, T. H. Nguyen, N. B. Nguyen, X. K. Pham, E. Schurr, L. Abel, and A. Alcais. 2007. Age is an important risk factor for onset and sequelae of reversal reactions in Vietnamese patients with leprosy. Clin. Infect. Dis. 44:33-40. [DOI] [PubMed] [Google Scholar]
- 244.Rao, N. L., P. J. Dunford, X. Xue, X. Jiang, K. A. Lundeen, F. Coles, J. P. Riley, K. N. Williams, C. A. Grice, J. P. Edwards, L. Karlsson, and A. M. Fourie. 2007. Anti-inflammatory activity of a potent, selective leukotriene A4 hydrolase inhibitor in comparison with the 5-lipoxygenase inhibitor zileuton. J. Pharmacol. Exp. Ther. 321:1154-1160. [DOI] [PubMed] [Google Scholar]
- 245.Rauch, A., Z. Kutalik, P. Descombes, T. Cai, J. Di Iulio, T. Mueller, M. Bochud, M. Battegay, E. Bernasconi, J. Borovicka, S. Colombo, A. Cerny, J.-F. Dufour, H. Furrer, H. F. Gunthard, M. Heim, B. Hirschel, R. Malinverni, D. Moradpour, B. Mullhaupt, A. Witteck, J. S. Beckmann, T. Berg, S. Bergmann, F. Negro, A. Telenti, and P.-Y. Bochud. 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138:1338.e7-1345.e7. [DOI] [PubMed] [Google Scholar]
- 246.Reichel, H., H. P. Koeffler, A. Tobler, and A. W. Norman. 1987. 1 alpha,25-dihydroxyvitamin D3 inhibits gamma-interferon synthesis by normal human peripheral blood lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 84:3385-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Remus, N., A. Alcais, and L. Abel. 2003. Human genetics of common mycobacterial infections. Immunol. Res. 28:109-129. [DOI] [PubMed] [Google Scholar]
- 248.Reynard, M. P., D. Turner, A. P. Junqueira-Kipnis, M. Ramos de Souza, C. Moreno, and C. V. Navarrete. 2003. Allele frequencies for an interferon-gamma microsatellite in a population of Brazilian leprosy patients. Eur. J. Immunogenet. 30:149-151. [DOI] [PubMed] [Google Scholar]
- 249.Ridley, D. S., and W. H. Jopling. 1966. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 34:255-273. [PubMed] [Google Scholar]
- 250.Rigby, W. F., T. Stacy, and M. W. Fanger. 1984. Inhibition of T lymphocyte mitogenesis by 1,25-dihydroxyvitamin D3 (calcitriol). J. Clin. Invest. 74:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Risch, N., and K. Merikangas. 1996. The future of genetic studies of complex human diseases. Science 273:1516-1517. [DOI] [PubMed] [Google Scholar]
- 252.Rittner, C., and J. Bertrams. 1981. On the significance of C2, C4, and factor B polymorphisms in disease. Hum. Genet. 56:235-247. [DOI] [PubMed] [Google Scholar]
- 253.Roberts, M., B. A. Mock, and J. M. Blackwell. 1993. Mapping of genes controlling Leishmania major infection in CXS recombinant inbred mice. Eur. J. Immunogenet. 20:349-362. [DOI] [PubMed] [Google Scholar]
- 254.Roger, M., G. Levee, S. Chanteau, B. Gicquel, and E. Schurr. 1997. No evidence for linkage between leprosy susceptibility and the human natural resistance-associated macrophage protein 1 (NRAMP1) gene in French Polynesia. Int. J. Lepr. Other Mycobact. Dis. 65:197-202. [PubMed] [Google Scholar]
- 255.Rothfuchs, A. G., A. Bafica, C. G. Feng, J. G. Egen, D. L. Williams, G. D. Brown, and A. Sher. 2007. Dectin-1 interaction with Mycobacterium tuberculosis leads to enhanced IL-12p40 production by splenic dendritic cells. J. Immunol. 179:3463-3471. [DOI] [PubMed] [Google Scholar]
- 256.Roy, S., A. Frodsham, B. Saha, S. K. Hazra, C. G. Mascie-Taylor, and A. V. Hill. 1999. Association of vitamin D receptor genotype with leprosy type. J. Infect. Dis. 179:187-191. [DOI] [PubMed] [Google Scholar]
- 257.Roy, S., W. McGuire, C. G. N. Mascie-Taylor, B. Saha, S. K. Hazra, A. V. S. Hill, and D. Kwiatkowski. 1997. Tumor necrosis factor promoter polymorphism and susceptibility to lepromatous leprosy. J. Infect. Dis. 176:530-532. [DOI] [PubMed] [Google Scholar]
- 258.Salgame, P., M. Yamamura, B. R. Bloom, and R. L. Modlin. 1992. Evidence for functional subsets of CD4+ and CD8+ T cells in human disease: lymphokine patterns in leprosy. Chem. Immunol. 54:44-59. [PubMed] [Google Scholar]
- 259.Samuelsson, B., and C. D. Funk. 1989. Enzymes involved in the biosynthesis of leukotriene B4. J. Biol. Chem. 264:19469-19472. [PubMed] [Google Scholar]
- 260.Sansonetti, P., and P. H. Lagrange. 1981. The immunology of leprosy: speculations on the leprosy spectrum. Rev. Infect. Dis. 3:422-469. [DOI] [PubMed] [Google Scholar]
- 261.Santos, A. R., A. S. Almeida, P. N. Suffys, M. O. Moraes, V. F. Filho, H. J. Mattos, J. A. Nery, P. H. Cabello, E. P. Sampaio, and E. N. Sarno. 2000. Tumor necrosis factor promoter polymorphism (TNF2) seems to protect against development of severe forms of leprosy in a pilot study in Brazilian patients. Int. J. Lepr. Other Mycobact. Dis. 68:325-327. [PubMed] [Google Scholar]
- 262.Santos, A. R., P. N. Suffys, P. R. Vanderborght, M. O. Moraes, L. M. Vieira, P. H. Cabello, A. M. Bakker, H. J. Matos, T. W. Huizinga, T. H. Ottenhoff, E. P. Sampaio, and E. N. Sarno. 2002. Role of tumor necrosis factor-alpha and interleukin-10 promoter gene polymorphisms in leprosy. J. Infect. Dis. 186:1687-1691. [DOI] [PubMed] [Google Scholar]
- 263.Santos, A. S., D. S. Castro, and A. Falqueto. 2008. Risk factors for leprosy transmission. Rev. Bras. Enferm. 61(Spec. No.):738-743. (In Portugese.) [DOI] [PubMed] [Google Scholar]
- 264.Sarno, E. N., G. E. Grau, L. M. Vieira, and J. A. Nery. 1991. Serum levels of tumour necrosis factor-alpha and interleukin-1 beta during leprosy reactional states. Clin. Exp. Immunol. 84:103-108. [PMC free article] [PubMed] [Google Scholar]
- 265.Schippers, E. F., C. van't Veer, S. van Voorden, C. A. E. Martina, T. W. J. Huizinga, S. le Cessie, and J. T. van Dissel. 2005. IL-10 and Toll-like receptor-4 polymorphisms and the in vivo and ex vivo response to endotoxin. Cytokine 29:215-228. [DOI] [PubMed] [Google Scholar]
- 266.Schlesinger, L., and M. Horwitz. 1991. Phagocytosis of Mycobacterium leprae by human monocyte-derived macrophages is mediated by complement receptors CR1 (CD35), CR3 (CD11b/CD18), and CR4 (CD11c/CD18) and IFN-gamma activation inhibits complement receptor function and phagocytosis of this bacterium. J. Immunol. 147:1983-1994. [PubMed] [Google Scholar]
- 267.Schlesinger, L. S., and M. A. Horwitz. 1991. Phenolic glycolipid-1 of Mycobacterium leprae binds complement component C3 in serum and mediates phagocytosis by human monocytes. J. Exp. Med. 174:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Schlesinger, L. S., T. M. Kaufman, S. Iyer, S. R. Hull, and L. K. Marchiando. 1996. Differences in mannose receptor-mediated uptake of lipoarabinomannan from virulent and attenuated strains of Mycobacterium tuberculosis by human macrophages. J. Immunol. 157:4568-4575. [PubMed] [Google Scholar]
- 269.Schoenen, H., B. Bodendorfer, K. Hitchens, S. Manzanero, K. Werninghaus, F. Nimmerjahn, E. M. Agger, S. Stenger, P. Andersen, J. Ruland, G. D. Brown, C. Wells, and R. Lang. 2010. Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J. Immunol. 184:2756-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Schroder, N. W., and R. R. Schumann. 2005. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect. Dis. 5:156-164. [DOI] [PubMed] [Google Scholar]
- 271.Schuring, R. P., L. Hamann, W. R. Faber, D. Pahan, J. H. Richardus, R. R. Schumann, and L. Oskam. 2009. Polymorphism N248S in the human Toll-like receptor 1 gene is related to leprosy and leprosy reactions. J. Infect. Dis. 199:1816-1819. [DOI] [PubMed] [Google Scholar]
- 272.Scollard, D. M. 2008. The biology of nerve injury in leprosy. Lepr. Rev. 79:242-253. [PubMed] [Google Scholar]
- 273.Scollard, D. M., L. B. Adams, T. P. Gillis, J. L. Krahenbuhl, R. W. Truman, and D. L. Williams. 2006. The continuing challenges of leprosy. Clin. Microbiol. Rev. 19:338-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Scollard, D. M., M. P. Joyce, and T. P. Gillis. 2006. Development of leprosy and type 1 leprosy reactions after treatment with infliximab: a report of 2 cases. Clin. Infect. Dis. 43:e19-e22. [DOI] [PubMed] [Google Scholar]
- 275.Serjeantson, S., S. R. Wilson, and B. J. Keats. 1979. The genetics of leprosy. Ann. Hum. Biol. 6:375-393. [DOI] [PubMed] [Google Scholar]
- 276.Shaw, M. A., S. Atkinson, H. Dockrell, R. Hussain, Z. Lins-Lainson, J. Shaw, F. Ramos, F. Silveira, S. Q. Mehdi, F. Kaukab, et al. 1993. An RFLP map for 2q33-q37 from multicase mycobacterial and leishmanial disease families: no evidence for an Lsh/Ity/Bcg gene homologue influencing susceptibility to leprosy. Ann. Hum. Genet. 57:251-271. [DOI] [PubMed] [Google Scholar]
- 277.Shaw, M. A., I. J. Donaldson, A. Collins, C. S. Peacock, Z. Lins-Lainson, J. J. Shaw, F. Ramos, F. Silveira, and J. M. Blackwell. 2001. Association and linkage of leprosy phenotypes with HLA class II and tumour necrosis factor genes. Genes Immun. 2:196-204. [DOI] [PubMed] [Google Scholar]
- 278.Shepard, C. C. 1960. Acid-fast bacilli in nasal excretions in leprosy, and results of inoculation of mice. Am. J. Hyg. 71:147-157. [DOI] [PubMed] [Google Scholar]
- 279.Shepard, C. C. 1962. The nasal excretion of Mycobacterium leprae in leprosy. Int. J. Lepr. 30:10-18. [PubMed] [Google Scholar]
- 280.Shields, E. D., D. A. Russell, and M. A. Pericak-Vance. 1987. Genetic epidemiology of the susceptibility to leprosy. J. Clin. Invest. 79:1139-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Shimoji, Y., V. Ng, K. Matsumura, V. A. Fischetti, and A. Rambukkana. 1999. A 21-kDa surface protein of Mycobacterium leprae binds peripheral nerve laminin-2 and mediates Schwann cell invasion. Proc. Natl. Acad. Sci. U. S. A. 96:9857-9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Siddiqui, M. R., S. Meisner, K. Tosh, K. Balakrishnan, S. Ghei, S. E. Fisher, M. Golding, N. P. S. Narayan, T. Sitaraman, U. Sengupta, R. Pitchappan, and A. V. S. Hill. 2001. A major susceptibility locus for leprosy in India maps to chromosome 10p13. Nat. Genet. 27:439-441. [DOI] [PubMed] [Google Scholar]
- 283.Sieling, P. A., X. H. Wang, M. K. Gately, J. L. Oliveros, T. McHugh, P. F. Barnes, S. F. Wolf, L. Golkar, M. Yamamura, Y. Yogi, et al. 1994. IL-12 regulates T helper type 1 cytokine responses in human infectious disease. J. Immunol. 153:3639-3647. [PubMed] [Google Scholar]
- 284.Singh, P. K., Ratna, P. K. Jain, and V. P. Mital. 1984. Immunological studies in leprosy. III. Cutaneous response to antigens in spectrum of leprosy. Indian J. Lepr. 56:257-263. [PubMed] [Google Scholar]
- 285.Skamene, E. 1994. The Bcg gene story. Immunobiology 191:451-460. [DOI] [PubMed] [Google Scholar]
- 286.Skamene, E., P. Gros, A. Forget, P. A. Kongshavn, C. St. Charles, and B. A. Taylor. 1982. Genetic regulation of resistance to intracellular pathogens. Nature 297:506-509. [DOI] [PubMed] [Google Scholar]
- 287.Skamene, E., P. Gros, A. Forget, P. J. Patel, and M. N. Nesbitt. 1984. Regulation of resistance to leprosy by chromosome 1 locus in the mouse. Immunogenetics 19:117-124. [DOI] [PubMed] [Google Scholar]
- 288.Skamene, M., E. Schurr, and P. Gros. 1998. Infection genomics: Nramp1 as a major determinant of natural resistance to intracellular infections. Annu. Rev. Med. 49:275-287. [DOI] [PubMed] [Google Scholar]
- 289.Smith, D. G. 1979. The genetic hypothesis for susceptibility to lepromatous leprosy. Hum. Genet. 50:163-177. [DOI] [PubMed] [Google Scholar]
- 290.Søborg, C., H. O. Madsen, A. B. Andersen, T. Lillebaek, A. Kok-Jensen, and P. Garred. 2003. Mannose-binding lectin polymorphisms in clinical tuberculosis. J. Infect. Dis. 188:777-782. [DOI] [PubMed] [Google Scholar]
- 291.Somoskovi, A., G. Zissel, U. Seitzer, J. Gerdes, M. Schlaak, and J. M. Quernheim. 1999. Polymorphisms at position −308 in the promoter region of the TNF-alpha and in the first intron of the TNF-beta genes and spontaneous and lipopolysaccharide-induced TNF-alpha release in sarcoidosis. Cytokine 11:882-887. [DOI] [PubMed] [Google Scholar]
- 292.Spierings, E., T. de Boer, B. Wieles, L. B. Adams, E. Marani, and T. H. Ottenhoff. 2001. Mycobacterium leprae-specific, HLA class II-restricted killing of human Schwann cells by CD4+ Th1 cells: a novel immunopathogenic mechanism of nerve damage in leprosy. J. Immunol. 166:5883-5888. [DOI] [PubMed] [Google Scholar]
- 293.Spierings, E., T. De Boer, L. Zulianello, and T. H. Ottenhoff. 2000. Novel mechanisms in the immunopathogenesis of leprosy nerve damage: the role of Schwann cells, T cells and Mycobacterium leprae. Immunol. Cell Biol. 78:349-355. [DOI] [PubMed] [Google Scholar]
- 294.Srivastava, L. M., D. P. Agarwal, H. G. Benkmann, and H. W. Goedde. 1975. Biochemical, immunological and genetic studies in leprosy. III. Genetic polymorphism of C3 and immunoglobulin profile in leprosy patients, healthy family members and controls. Tropenmed. Parasitol. 26:426-430. [PubMed] [Google Scholar]
- 295.Stanley, S. J., C. Howland, M. M. Stone, and I. Sutherland. 1981. BCG vaccination of children against leprosy in Uganda: final results. J. Hyg. (Lond.) 87:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Stein, C. 2010. Identifying genes underlying human inherited disease. In Encyclopedia of life sciences. John Wiley & Sons, Ltd., Chichester, United Kingdom. doi: 10.1002/9780470015902.a0022395. [DOI]
- 297.Stoner, G. L. 1979. Importance of the neural predilection of Mycobacterium leprae in leprosy. Lancet ii:994-996. [DOI] [PubMed] [Google Scholar]
- 298.Super, M., S. Thiel, J. Lu, R. J. Levinsky, and M. W. Turner. 1989. Association of low levels of mannan-binding protein with a common defect of opsonisation. Lancet ii:1236-1239. [DOI] [PubMed] [Google Scholar]
- 299.Tailleux, L., N. Pham-Thi, A. Bergeron-Lafaurie, J. L. Herrmann, P. Charles, O. Schwartz, P. Scheinmann, P. H. Lagrange, J. de Blic, A. Tazi, B. Gicquel, and O. Neyrolles. 2005. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med. 2:e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Tailleux, L., O. Schwartz, J. L. Herrmann, E. Pivert, M. Jackson, A. Amara, L. Legres, D. Dreher, L. P. Nicod, J. C. Gluckman, P. H. Lagrange, B. Gicquel, and O. Neyrolles. 2003. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J. Exp. Med. 197:121-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Tang, G. J., S. L. Huang, H. W. Yien, W. S. Chen, C. W. Chi, C. W. Wu, W. Y. Lui, J. H. Chiu, and T. Y. Lee. 2000. Tumor necrosis factor gene polymorphism and septic shock in surgical infection. Crit. Care Med. 28:2733-2736. [DOI] [PubMed] [Google Scholar]
- 302.Tapping, R. I., K. O. Omueti, and C. M. Johnson. 2007. Genetic polymorphisms within the human Toll-like receptor 2 subfamily. Biochem. Soc. Trans. 35:1445-1448. [DOI] [PubMed] [Google Scholar]
- 303.Tarkowski, E., A. M. Liljeroth, A. Nilsson, A. Ricksten, P. Davidsson, L. Minthon, and K. Blennow. 2000. TNF gene polymorphism and its relation to intracerebral production of TNFα and TNFβ in AD. Neurology 54:2077-2081. [DOI] [PubMed] [Google Scholar]
- 304.Tobin, D. M., J. C. Vary, Jr., J. P. Ray, G. S. Walsh, S. J. Dunstan, N. D. Bang, D. A. Hagge, S. Khadge, M. C. King, T. R. Hawn, C. B. Moens, and L. Ramakrishnan. 2010. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140:717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Todd, J. R., B. C. West, and J. C. McDonald. 1990. Human leukocyte antigen and leprosy: study in northern Louisiana and review. Rev. Infect. Dis. 12:63-74. [DOI] [PubMed] [Google Scholar]
- 306.Tosh, K., S. Meisner, M. R. Siddiqui, K. Balakrishnan, S. Ghei, M. Golding, U. Sengupta, R. M. Pitchappan, and A. V. Hill. 2002. A region of chromosome 20 is linked to leprosy susceptibility in a South Indian population. J. Infect. Dis. 186:1190-1193. [DOI] [PubMed] [Google Scholar]
- 307.Tosh, K., M. Ravikumar, J. T. Bell, S. Meisner, A. V. S. Hill, and R. Pitchappan. 2006. Variation in MICA and MICB genes and enhanced susceptibility to paucibacillary leprosy in South India. Hum. Mol. Genet. 15:2880-2887. [DOI] [PubMed] [Google Scholar]
- 308.Towers, T. L., and L. P. Freedman. 1998. Granulocyte-macrophage colony-stimulating factor gene transcription is directly repressed by the vitamin D3 receptor. Implications for allosteric influences on nuclear receptor structure and function by a DNA element. J. Biol. Chem. 273:10338-10348. [DOI] [PubMed] [Google Scholar]
- 309.Triccas, J. A., P. W. Roche, N. Winter, C. G. Feng, C. R. Butlin, and W. J. Britton. 1996. A 35-kilodalton protein is a major target of the human immune response to Mycobacterium leprae. Infect. Immun. 64:5171-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Trinchieri, G. 1993. Interleukin-12 and its role in the generation of TH1 cells. Immunol. Today 14:335-338. [DOI] [PubMed] [Google Scholar]
- 311.Truman, R. 2008. Armadillos as a source of infection for leprosy. South. Med. J. 101:581-582 doi: 10.1097/SMJ.0b013e318172dd6c. [DOI] [PubMed] [Google Scholar]
- 312.Turner, D. M., D. M. Williams, D. Sankaran, M. Lazarus, P. J. Sinnott, and I. V. Hutchinson. 1997. An investigation of polymorphism in the interleukin-10 gene promoter. Eur. J. Immunogenet. 24:1-8. [DOI] [PubMed] [Google Scholar]
- 313.van der Graaf, C., B. J. Kullberg, L. Joosten, T. Verver-Jansen, L. Jacobs, J. W. M. Van der Meer, and M. G. Netea. 2005. Functional consequences of the Asp299Gly Toll-like receptor-4 polymorphism. Cytokine 30:264-268. [DOI] [PubMed] [Google Scholar]
- 314.van de Veerdonk, F. L., A. C. Teirlinck, J. Kleinnijenhuis, B. J. Kullberg, R. van Crevel, J. W. M. van der Meer, L. A. B. Joosten, and M. G. Netea. 18 March 2010, posting date. Mycobacterium tuberculosis induces IL-17A responses through TLR4 and dectin-1 and is critically dependent on endogenous IL-1. J. Leukoc. Biol. [Epub ahead of print.] doi: 10.1189/jlb.0809550. [DOI] [PubMed]
- 315.van Eden, W., and R. R. de Vries. 1984. HLA and leprosy: a re-evaluation. Lepr. Rev. 55:89-104. [DOI] [PubMed] [Google Scholar]
- 316.van Eden, W., R. R. de Vries, N. K. Mehra, M. C. Vaidya, J. D'Amaro, and J. J. van Rood. 1980. HLA segregation of tuberculoid leprosy: confirmation of the DR2 marker. J. Infect. Dis. 141:693-701. [DOI] [PubMed] [Google Scholar]
- 317.van Eden, W., N. M. Gonzalez, R. R. de Vries, J. Convit, and J. J. van Rood. 1985. HLA-linked control of predisposition to lepromatous leprosy. J. Infect. Dis. 151:9-14. [DOI] [PubMed] [Google Scholar]
- 318.van Kooyk, Y., and T. B. H. Geijtenbeek. 2003. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 319.Veeriah, S., L. G. Morris, D. Solit, and T. A. Chan. 2010. The familial Parkinson disease gene PARK2 is a multisite tumor suppressor on chromosome 6q25.2-27 that regulates cyclin E. Cell Cycle 9:1451-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Veeriah, S., B. S. Taylor, S. Meng, F. Fang, E. Yilmaz, I. Vivanco, M. Janakiraman, N. Schultz, A. J. Hanrahan, W. Pao, M. Ladanyi, C. Sander, A. Heguy, E. C. Holland, P. B. Paty, P. S. Mischel, L. Liau, T. F. Cloughesy, I. K. Mellinghoff, D. B. Solit, and T. A. Chan. 2010. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet. 42:77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Vejbaesya, S., P. Mahaisavariya, P. Luangtrakool, and C. Sermduangprateep. 2007. TNF alpha and NRAMP1 polymorphisms in leprosy. J. Med. Assoc. Thai. 90:1188-1192. [PubMed] [Google Scholar]
- 322.Veltkamp, M., P. A. H. M. Wijnen, C. H. M. van Moorsel, G. T. Rijkers, H. J. T. Ruven, M. Heron, O. Bekers, A. M. E. Claessen, M. Drent, J. M. M. van den Bosch, and J. C. Grutters. 2007. Linkage between Toll-like receptor (TLR) 2 promotor and intron polymorphisms: functional effects and relevance to sarcoidosis. Clin. Exp. Immunol. 149:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Verbeek, W., A. F. Gombart, M. Shiohara, M. Campbell, and H. P. Koeffler. 1997. Vitamin D receptor: no evidence for allele-specific mRNA stability in cells which are heterozygous for the TaqI restriction enzyme polymorphism. Biochem. Biophys. Res. Commun. 238:77-80. [DOI] [PubMed] [Google Scholar]
- 324.Verdu, P., L. B. Barreiro, E. Patin, A. Gessain, O. Cassar, J. R. Kidd, K. K. Kidd, D. M. Behar, A. Froment, E. Heyer, L. Sica, J. L. Casanova, L. Abel, and L. Quintana-Murci. 2006. Evolutionary insights into the high worldwide prevalence of MBL2 deficiency alleles. Hum. Mol. Genet. 15:2650-2658. [DOI] [PubMed] [Google Scholar]
- 325.Vidyarani, M., P. Selvaraj, S. P. Anand, M. S. Jawahar, A. R. Adhilakshmi, and P. R. Narayanan. 2006. Interferon gamma (IFNγ) & interleukin-4 (IL-4) gene variants & cytokine levels in pulmonary tuberculosis. Indian J. Med. Res. 124:403-410. [PubMed] [Google Scholar]
- 326.von Aulock, S., N. W. J. Schroder, K. Gueinzius, S. Traub, S. Hoffmann, K. Graf, S. Dimmeler, T. Hartung, R. R. Schumann, and C. Hermann. 2003. Heterozygous Toll-like receptor 4 polymorphism does not influence lipopolysaccharide-induced cytokine release in human whole blood. J. Infect. Dis. 188:938-943. [DOI] [PubMed] [Google Scholar]
- 327.Wagener, D. K., V. Schauf, K. E. Nelson, D. Scollard, A. Brown, and T. Smith. 1988. Segregation analysis of leprosy in families of northern Thailand. Genet. Epidemiol. 5:95-105. [DOI] [PubMed] [Google Scholar]
- 328.Walker, S. L., and D. N. Lockwood. 2007. Leprosy. Clin. Dermatol. 25:165-172. [DOI] [PubMed] [Google Scholar]
- 329.Walker, S. L., and D. N. Lockwood. 2008. Leprosy type 1 (reversal) reactions and their management. Lepr. Rev. 79:372-386. [PubMed] [Google Scholar]
- 330.Wallace, C., J. Fitness, B. Hennig, L. Sichali, L. Mwaungulu, J. M. Ponnighaus, D. K. Warndorff, D. Clayton, P. E. Fine, and A. V. Hill. 2004. Linkage analysis of susceptibility to leprosy type using an IBD regression method. Genes Immun. 5:221-225. [DOI] [PubMed] [Google Scholar]
- 331.Wang, L. M., A. Kimura, M. Satoh, and S. Mineshita. 1999. HLA linked with leprosy in southern China: HLA-linked resistance alleles to leprosy. Int. J. Lepr. Other Mycobact. Dis. 67:403-408. [PubMed] [Google Scholar]
- 332.WHO. 1998. Expert Committee on Leprosy, seventh report, vol. 874. WHO, Geneva, Switzerland.
- 333.WHO. 2009. Global leprosy situation. Wkly. Epidemiol. Rec. 84:333-340. [PubMed] [Google Scholar]
- 334.Wibawa, T., H. Soebono, and M. Matsuo. 2002. Association of a missense mutation of the laminin alpha2 gene with tuberculoid type of leprosy in Indonesian patients. Trop. Med. Int. Health 7:631-636. [DOI] [PubMed] [Google Scholar]
- 335.Wilson, A. G., J. A. Symons, T. L. McDowell, H. O. McDevitt, and G. W. Duff. 1997. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 94:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Wong, S. H., S. Gochhait, D. Malhotra, F. H. Pettersson, Y. Y. Teo, C. C. Khor, A. Rautanen, S. J. Chapman, T. C. Mills, A. Srivastava, A. Rudko, M. B. Freidin, V. P. Puzyrev, S. Ali, S. Aggarwal, R. Chopra, B. S. N. Reddy, V. K. Garg, S. Roy, S. Meisner, S. K. Hazra, B. Saha, S. Floyd, B. J. Keating, C. Kim, B. P. Fairfax, J. C. Knight, P. C. Hill, R. A. Adegbola, H. Hakonarson, P. E. M. Fine, R. M. Pitchappan, R. N. K. Bamezai, A. V. S. Hill, and F. O. Vannberg. 2010. Leprosy and the adaptation of human Toll-like receptor 1. PLoS Pathog. 6:e1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Wong, S. H., A. V. Hill, and F. O. Vannberg. 2010. Genomewide association study of leprosy. N. Engl. J. Med. 362:1446-1447. [DOI] [PubMed] [Google Scholar]
- 338.Wurfel, M. M., A. C. Gordon, T. D. Holden, F. Radella, J. Strout, O. Kajikawa, J. T. Ruzinski, G. Rona, R. A. Black, S. Stratton, G. P. Jarvik, A. M. Hajjar, D. A. Nickerson, M. Rieder, J. Sevransky, J. P. Maloney, M. Moss, G. Martin, C. Shanholtz, J. G. Garcia, L. Gao, R. Brower, K. C. Barnes, K. R. Walley, J. A. Russell, and T. R. Martin. 2008. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am. J. Respir. Crit. Care Med. 178:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Xu, K. Y., R. R. de Vries, H. M. Fei, A. van Leeuwen, R. B. Chen, and G. Y. Ye. 1985. HLA-linked control of predisposition to lepromatous leprosy. Int. J. Lepr. Other Mycobact. Dis. 53:56-63. [PubMed] [Google Scholar]
- 340.Yamamura, M. 1992. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science 255:12. [DOI] [PubMed] [Google Scholar]
- 341.Yamamura, M., K. Uyemura, R. J. Deans, K. Weinberg, T. H. Rea, B. R. Bloom, and R. L. Modlin. 1991. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science 254:277-279. [DOI] [PubMed] [Google Scholar]
- 342.Yamamura, M., X. H. Wang, J. D. Ohmen, K. Uyemura, T. H. Rea, B. R. Bloom, and R. L. Modlin. 1992. Cytokine patterns of immunologically mediated tissue damage. J. Immunol. 149:1470-1475. [PubMed] [Google Scholar]
- 343.Yamazaki, K., M. Takazoe, T. Tanaka, T. Kazumori, and Y. Nakamura. 2002. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn's disease. J. Hum. Genet. 47:469-472. [DOI] [PubMed] [Google Scholar]
- 344.Yang, Y., C. Yin, A. Pandey, D. Abbott, C. Sassetti, and M. A. Kelliher. 2007. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J. Biol. Chem. 282:36223-36229. [DOI] [PubMed] [Google Scholar]
- 345.Yim, J. J., H. W. Lee, H. S. Lee, Y. W. Kim, S. K. Han, Y. S. Shim, and S. M. Holland. 2006. The association between microsatellite polymorphisms in intron II of the human Toll-like receptor 2 gene and tuberculosis among Koreans. Genes Immun. 7:150-155. [DOI] [PubMed] [Google Scholar]
- 346.Zenaro, E., M. Donini, and S. Dusi. 2009. Induction of Th1/Th17 immune response by Mycobacterium tuberculosis: role of dectin-1, mannose receptor, and DC-SIGN. J. Leukoc. Biol. 86:1393-1401. [DOI] [PubMed] [Google Scholar]
- 347.Zerva, L., B. Cizman, N. Mehra, S. Alahari, R. Murali, C. Zmijewski, M. Kamoun, and D. Monos. 1996. Arginine at positions 13 or 70-71 in pocket 4 of HLA-DRB1 alleles is associated with susceptibility to tuberculoid leprosy. J. Exp. Med. 183:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Zhang, F. R., W. Huang, S. M. Chen, L. D. Sun, H. Liu, Y. Li, Y. Cui, X. X. Yan, H. T. Yang, R. D. Yang, T. S. Chu, C. Zhang, L. Zhang, J. W. Han, G. Q. Yu, C. Quan, Y. X. Yu, Z. Zhang, B. Q. Shi, L. H. Zhang, H. Cheng, C. Y. Wang, Y. Lin, H. F. Zheng, X. A. Fu, X. B. Zuo, Q. Wang, H. Long, Y. P. Sun, Y. L. Cheng, H. Q. Tian, F. S. Zhou, H. X. Liu, W. S. Lu, S. M. He, W. L. Du, M. Shen, Q. Y. Jin, Y. Wang, H. Q. Low, T. Erwin, N. H. Yang, J. Y. Li, X. Zhao, Y. L. Jiao, L. G. Mao, G. Yin, Z. X. Jiang, X. D. Wang, J. P. Yu, Z. H. Hu, C. H. Gong, Y. Q. Liu, R. Y. Liu, D. M. Wang, D. Wei, J. X. Liu, W. K. Cao, H. Z. Cao, Y. P. Li, W. G. Yan, S. Y. Wei, K. J. Wang, M. L. Hibberd, S. Yang, X. J. Zhang, and J. J. Liu. 2009. Genomewide association study of leprosy. N. Engl. J. Med. 361:2609-2618. [DOI] [PubMed] [Google Scholar]