Abstract
Summary: FtsZ, a bacterial homolog of tubulin, is well established as forming the cytoskeletal framework for the cytokinetic ring. Recent work has shown that purified FtsZ, in the absence of any other division proteins, can assemble Z rings when incorporated inside tubular liposomes. Moreover, these artificial Z rings can generate a constriction force, demonstrating that FtsZ is its own force generator. Here we review light microscope observations of how Z rings assemble in bacteria. Assembly begins with long-pitch helices that condense into the Z ring. Once formed, the Z ring can transition to short-pitch helices that are suggestive of its structure. FtsZ assembles in vitro into short protofilaments that are ∼30 subunits long. We present models for how these protofilaments might be further assembled into the Z ring. We discuss recent experiments on assembly dynamics of FtsZ in vitro, with particular attention to how two regulatory proteins, SulA and MinC, inhibit assembly. Recent efforts to develop antibacterial drugs that target FtsZ are reviewed. Finally, we discuss evidence of how FtsZ generates a constriction force: by protofilament bending into a curved conformation.
INTRODUCTION
FtsZ is the major cytoskeletal protein in the bacterial cytokinesis machine. It forms a ring (the Z ring) under the membrane at the center of the cell, and this Z ring constricts to initiate division of the cell. In addition to FtsZ, there are a dozen accessory proteins that are essential for cell division in Escherichia coli. These are mostly transmembrane proteins that are involved in remodeling the cell wall, and they will not be discussed here. The present article focuses on FtsZ. For more comprehensive reviews of bacterial cell division, including the downstream proteins, see references 36, 59, 69, 111, 195, and 198. A recent review by Adams and Errington (2) focuses on the proteins that interact directly with FtsZ and may regulate its assembly. The evolutionary relationships of FtsZ to tubulin and of MreB to actin have been discussed in two articles, each providing some different perspectives (46, 102).
LIGHT MICROSCOPY OF Z RINGS IN BACTERIA
Constriction of the Z Ring and Reassembly of New Z Rings
The pioneering immunoelectron microscopy of Bi and Lutkenhaus (22) provided the first evidence that FtsZ is a cytoskeletal protein localized in a ring at the center of the cell. They found that in a cell just starting septation, antibody-coated gold beads localized near the membrane at the site of constriction. Subsequently, Levin and Losick (97) used immunofluorescence at the light microscope level and made the surprising discovery that Z rings were present at the centers of almost all cells, not just dividing ones. Wang and Lutkenhaus (197) independently developed immunofluorescence technology and discovered that Z rings were present in 50% of the cells in a culture of the archaeon Haloferax volcanii. The fraction of cells with Z rings is 85 to 95% in rapidly dividing Bacillus subtilis and E. coli (3, 97). This means that FtsZ assembles in the daughter cells very soon after division and remains assembled in the cell for most of the cell cycle.
Green fluorescent protein (GFP) labeling of FtsZ was introduced by Ma et al. (108), confirming the localization seen by immunofluorescence. That study also found that FtsA-GFP had a localization very similar to that of FtsZ. The powerful advantage of GFP labeling is that the localization can be followed over time in living bacteria. The Margolin lab later used FtsZ-GFP to observe the dynamics of the Z ring throughout the cell cycle and during constriction (186, 191). In those studies FtsZ-GFP was used as a dilute label in the presence of wild-type FtsZ expressed from the genome. As long as the level of FtsZ-GFP is less than ca. one-third of that of the wild-type FtsZ, it labels the Z ring without introducing obvious defects in division. Our lab has recently derived an E. coli strain that can use FtsZ-YFP as the sole source of FtsZ (138). This strain has a second-site mutation, whose nature is not known, somewhere in the genome. This strain should be useful for future studies, but for most observations, including those shown in Fig. 1, we have used FtsZ as a dilute label.
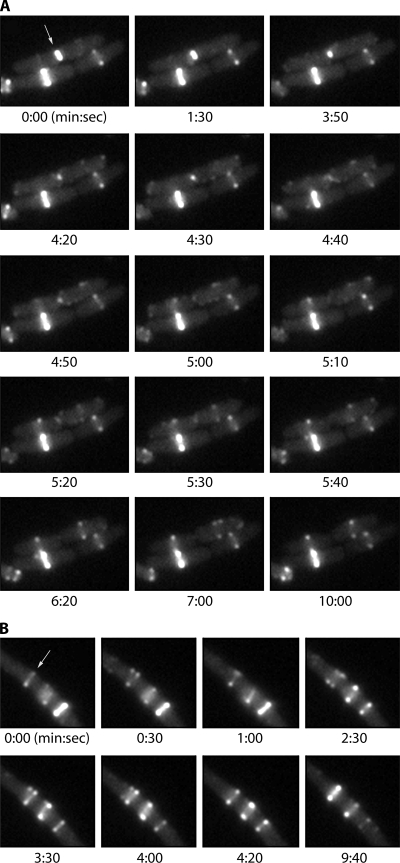
FIG. 1.
Time-lapse observation of Z rings in E. coli, using FtsZ-GFP as a dilute label, expressed at about one-third the level of genomic FtsZ. The cells were induced to produce FtsZ-GFP for about 1 h and then immobilized on an agar pad for time-lapse observation at 37°C. (A) Three cells. We will ignore the two on the bottom (with a bright Z ring [left] and a dim Z ring [right]) and focus on the upper one (arrow), which is undergoing division. The constriction of the Z ring, its concurrent disassembly, and the assembly of new Z rings in the daughter cells are described in the text. (B) A cell with three Z rings (perhaps induced by excessive production of FtsZ-GFP). The upper Z ring (arrow) alternately opens into a short-pitch helix and collapses into an apparent circle. Frames from both panels A and B are taken from Movie S1 in the supplemental material.
Figure 1A shows our own use of FtsZ-GFP to follow the Z ring through a division event. The typical appearance of the Z ring is a pair of bright dots on either side of the cell, as seen in the lower right Z ring in Fig. 1A. The Z ring is actually a continuous circular filament of fairly uniform density. The bright dots on the edges are due to the ring being more intense when viewed in vertical projection. Some very bright rings do not show the two-dot structure due to saturation of the image, and in highly constricted cells the dots are too close to resolve.
We will look first at the constriction of the Z ring. In Fig. 1A, the panel at 0:00 (min:s) shows the mother cell (arrow) with a bright Z ring in the initial stage of constriction. This Z ring is about half the diameter of the other two Z rings in the field, so it began constriction at some undetermined earlier time. Complete constriction of the mother Z ring took about another 4 min; the total time for constriction was probably 8 to 10 min (about half of the 20-min cell cycle). As the Z ring constricted, it disassembled completely. Several studies have noted that the Z ring disassembles as it constricts (40, 120, 186). A recent study reported that the density of FtsZ in the Z ring actually increased during constriction (91). However, the single example presented there measured FtsZ over a constriction of only 200 nm. This is less than the 250-nm resolution of the light microscope and much less than the 1,000-nm diameter of an E. coli cell. In our time-lapse movies, the Z ring decreased in brightness during constriction and disappeared completely at the end (Fig. 1A).
The mechanism by which the Z ring disassembles as it constricts is not known. As discussed below, FtsZ rapidly cycles between the Z ring and the cytoplasmic pool. The cycling continues at the same rate when rings begin constricting (183). However, it appears that something may block the return of FtsZ to the Z ring, while still permitting its loss. In a temperature shift experiment with FtsZ84, Addinall et al. noted that Z rings rapidly disappeared when cells were shifted to 42°C and rapidly reformed when cells were shifted back to 30°C (4). There was one exception: sites with a visible constriction did not reform a Z ring but rather assembled Z rings at the one- and three-quarter positions, where the Z rings would assemble in the daughter cells. This is consistent with some mechanism that blocks return of FtsZ to the Z-ring site once constriction has begun.
Faint Z rings are already visible in the daughter cells in the 0:00 frame (Fig. 1). This confirms the observation of Sun and Margolin (186) that the Z rings are able to initially assemble in daughter cells before constriction of the mother cell is complete. However, this initial assembly is transient and seems to have disappeared at 1:30 and 3:50. From 4:20 to 5:20 the FtsZ in the daughter cells appears to form foci scattered throughout the cell (discussed below). At 5:10 in the left-hand cell and 5:30 in the right-hand cell, the peripheral foci disappear and Z rings are formed. The characteristic two-dot structure of the Z ring is clearly seen in the right-hand cell at 5:30, and it becomes brighter at 6:20 and 10:00.
Aarsman et al. (1) studied the assembly and maturation of the Z ring over the course of the cell cycle. In LMC500 cells grown with a doubling time of 40 min, the Z ring appeared after 15% of the cell cycle. Proteins downstream of FtsK appeared after a substantial lag, 49% of the cell cycle, and visible constrictions appeared almost immediately after this. When the cell cycle time was increased (slower growth), the Z ring appeared later in the cycle and there was a further delay before the downstream proteins assembled. It is likely that the Z ring generates a constriction force as soon as it is assembles, but constriction can begin only when all of the downstream proteins are assembled to remodel the cell wall.
A number of studies agree that the constriction phase occupies approximately the last half of the cell cycle. den Blaauwen and colleagues (1, 40) found that constriction began at ∼50% into the 40-min cell cycle. Reshes et al. (158) used sophisticated analysis of phase-contrast images to follow the course of the constriction. Their analysis concluded that constriction began about half way through the 20-min cell cycle. Inoue et al. (80) created a strain in which FtsZ-GFP was expressed from the genome at a level ∼10% that of wild-type FtsZ, giving a steady level of fluorescence with no apparent aberration in cell division. For a 100-min cell cycle they found 8 to 12% of cells with a visible constriction, implying a constriction time of 8 to 12 min. This was a much shorter time for constriction than was determined in other studies. The timing of when constriction begins can be complicated by the different techniques used.
In a recent study, Strömqvist et al. (184) used fluorescence recovery after photobleaching (FRAP) to measure diffusion of GFP across dividing E. coli cells. From the rate of diffusion, they calculated the radius of the constriction. Their analysis was valid mostly for the later stages of constriction, which they concluded proceeded at a linear rate on average.
Two Kinds of Z-Ring Helices
Ben-Yehuda and Losick (13) reported that when B. subtilis was entering into sporulation, the single central Z ring spun off helical structures in both directions, which then condensed into two polar Z rings. Helical intermediates were also found to precede the formation of sporulation Z rings in Streptomyces coelicolor (62). Thanedar and Margolin (191) and Peters et al. (143) analyzed time-lapse images of FtsZ-GFP in E. coli and B. subtilis and reported localization to moving, membrane-bound spots throughout the cell, especially prior to formation of the Z ring. They used deconvolution microscopy to make a convincing case that these spots were mostly projection views of helices. The helices eventually collapsed toward the center of the cell and condensed into the Z ring. Thanedar and Margolin (191) noted two types of dynamic movement of these helices. They changed position on a rapid time scale of a few seconds, and on a longer time scale they oscillated from one side to the other in elongated cells. This movement resembled the oscillation of MinD, and indeed it was dependent on an operational MinCDE system.
We will refer to this first type of helix as cytoplasmic or long-pitch helices. Figure 1A shows a cell completing division and assembling new Z rings in the daughter cells. The Z ring of the mother cell constricts and disappears at 4:50 to 5:00. From 4:20 to 5:20 small patches of FtsZ are seen in the daughter cells. These patches are similar to those resolved as long-pitch helices by deconvolution microscopy (143, 191). These helices then condense to form Z rings. From 5:40 each daughter cell has a single Z ring, and the cytoplasmic patches/helices have largely disappeared. It seems that the cytoplasmic helices are the preferred FtsZ structure when there is no Z ring, but they are incorporated into the Z ring once it forms; however, occasional peripheral foci continue to appear and disappear even in the presence of the Z ring (see Movie S1 in the supplemental material). The transition from diffuse helices to condensed Z rings is also demonstrated in a recent study by Monahan et al. (Fig. 6 in reference 120).
A second type of helical structure emerges from the Z ring once it is formed. At 6:20 in the right-hand daughter cell, the Z ring shows the characteristic structure of one dot on the upper membrane and one on the lower. At 7:00 the upper dot has separated into two, suggesting that the Z ring has split into a helix. At 10:00 it has collapsed back into a ring. The separation of the Z ring into helices is seen in more detail in Fig. 1B, which shows a segment of a cell that has elongated and assembled three Z rings. The Z ring at the upper left appears as a closed ring (a line or two bright dots on the sides of the cell) at 0:00, 4:20, and 9:40. At 0:30, 2:30, 3:30, and 4:00 the dots separate into two, suggesting that this Z ring is actually a helix of ∼2 turns. This suggests that the Z ring is not a closed circle but a helix. Most of the time the gyres are too close to be resolved in the light microscope (<250 nm), but they occasionally separate to ∼350 nm apart. We refer to this as a short-pitch helix.
The two types of helices (diffuse and long-pitch helices over the cytoplasm and short-pitch helices emerging from the Z ring) point to a common feature of FtsZ assembly in the cell: its tendency to form very long filamentous structures. If a long filament is tethered to the membrane, its generic form will be a helix. The shape of filaments tethered to the membrane and generating bending forces in various directions was analyzed in detail by Andrews and Arkin (9).
Assembly Dynamics of FtsZ In Vivo
As seen in Fig. 1A, Z-ring constriction and disassembly takes place over several minutes, and condensation of the long-pitch helices into definitive Z rings takes about 30 to 60 s. The fluctuations between a ring and a short-pitch helix are on a similar 30- to 60-s time scale (Fig. 1B). On a scale shorter than ∼30 s, the Z ring appears to be static. However, FRAP (fluorescence recovery after photobleaching) analysis showed that the Z ring is much more dynamic. Recovery of a bleached spot on the Z ring occurs with a half time of 8 to 9 s, which means that subunits in the Z ring are exchanging with those in the cytoplasm on this time scale (7). The turnover was very similar in E. coli and B. subtilis, suggesting that it is a common feature in bacteria. Turnover was only modestly altered by null mutations in regulatory proteins MinCD, ZapA, and EzrA (7). A subsequent study of Mycobacterium smegmatis, which has a slower cell cycle, gave an average turnover half time of 34 s, with a broad spread from 10 to 70 s (27).
In an independent study of E. coli, Geissler et al. (55) found a turnover half time of 11 s for FtsZ, which is very similar to our value. They also measured the turnover of FtsA. This was 16 s for FtsA-GFP, which was rather toxic when expressed with the genomic FtsA. The turnover half time was 12 s for their interesting mutant FtsA*-GFP, which was much less toxic. An earlier study showed that ZipA, a transmembrane protein with a cytoplasmic domain that binds FtsZ (121), was turning over at a rate very close to that of FtsZ (183).
Niu and Yu studied FtsZ dynamics by tracking single molecules of FtsZ-GFP in E. coli (126). They were able to track for only 1 to 2 s, but this was sufficient to define two classes of molecules. FtsZ molecules in the center of the cell, presumably those in the Z ring, were stationary over the 1 to 2 s. (Had they been able to track molecules for 20 s, they would presumably have seen the ∼8-s turnover measured by FRAP.) FtsZ molecules outside the Z ring were highly mobile, with a diffusion coefficient similar to that of monomeric membrane proteins. This mobility is much faster than the movement of patches and long-pitch helices (143, 191), an apparent contradiction that remains to be resolved. Single-molecule tracking seems to be a promising technique that should see important future applications to the Z ring.
Still unresolved is the oligomeric state of the FtsZ outside the Z ring. For cells with a total concentration of 4 μM FtsZ, if 30% is in the Z ring, the cytoplasmic concentration should be 2.8 μM. If the critical concentration is 1 μM, one would expect 1.8 μM FtsZ to be assembled into protofilaments. One might expect these protofilaments to be tethered to the membrane, but surprisingly, after the Z ring has formed, the cytoplasmic FtsZ appears to be diffuse and not localized to the membrane (Fig. 1). One possible explanation for the lack of membrane attachment could be that all of the FtsA and ZipA are sequestered into the Z ring. Rueda et al. (162) determined for one strain of E. coli that the total amount of FtsA plus ZipA was about half the total amount of FtsZ, so it is possible that these membrane-tethering molecules are mostly sequestered in the Z ring. Unfortunately, there is presently no measure for what fraction of FtsA and ZipA are in and outside the Z ring.
Another possible explanation is that the critical concentration in the cytoplasm may be higher than the 1 μM measured in dilute solution in vitro, perhaps due to negative regulatory proteins. In this case the cytoplasmic FtsZ may not be assembled into protofilaments. Monomeric FtsZ may not bind efficiently to the membrane through the FtsA and ZipA tethers.
SUBSTRUCTURE OF THE Z RING
Structure of the FtsZ Subunit and Protofilaments
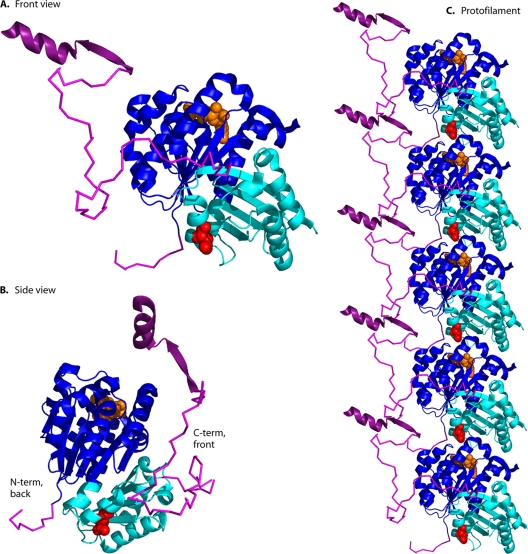
Figure 2 shows a cartoon image of an FtsZ protein subunit, based on the crystal structure of FtsZ from Pseudomonas aeruginosa (32), which is 67% identical to that of E. coli over the globular domain. We use the amino acid numbering from E. coli for this discussion. Figure 2A shows the view that we call “front,” since it corresponds to the view of a tubulin subunit from the outside of a microtubule. Figure 2B shows the FtsZ subunit rotated 90 degrees and viewed from the left side.
FIG. 2.
(A) Structure of the FtsZ subunit. The globular domain, shown in cartoon format, comprises two subdomains colored blue (N-terminal) and cyan (C-terminal). This is from the X-ray structure of P. aeruginosa FtsZ, PDB 1OFU (32). The GDP is shown in orange space fill, and the synergy loop amino acid D212 (E. coli numbering) is in red. This view corresponds to that of a tubulin subunit seen from the outside of a microtubule, and is designated the “front view.” A 10-amino-acid segment on the N terminus and a 50-amino-acid segment on the C terminus are shown in magenta, each modeled as flexible peptides. Shown in dark purple are the extended beta strand and alpha helix formed by the C-terminal 17-amino-acid peptide when bound to ZipA (from PDB 1F47 [121]). The model was constructed using the program PyMol (39). (B) The FtsZ subunit viewed from the side. This shows that the C-terminal peptide emerges from the front face and the N-terminal peptide from the back face, ∼180 degrees away. (C) A protofilament is assembled by stacking subunits on top of each other so that the D212 of the upper subunit is just above the GDP of the one below.
The globular domain comprises two subdomains, which can be expressed separately and are independently folding (133, 139). The N-terminal subdomain, colored dark blue, has the structure of a Rossman fold and contains all the amino acids of the GTP-binding site and the entire lower side of the interface of the longitudinal protofilament bond. The C-terminal domain, colored cyan, contains all of the amino acids of the upper side of the interface, including the “synergy” (T7) loop (discussed under “GTP Hydrolysis” below). The border between the two subdomains is not completely clear. Oliva et al. (133) terminated the N-terminal domain at amino acid 179 of the E. coli sequence, placing the H7 helix entirely in the C-terminal domain. Osawa and Erickson (139) suggested terminating the N-terminal domain at amino acid 195, putting the first half of helix H7 (which contains amino acids that contact the GTP and make contact across the protofilament interface) in the N-terminal domain.
The C-terminal globular domain terminates at amino acid G316. The following ∼50 amino acids are highly divergent in sequence across bacterial species, and this segment is invisible in crystal structures. It is generally considered to be an unstructured peptide that can act as a flexible linker. In the absence of an extension force, the peptide behaves as a worm-like chain and collapses to an average end-to-end distance of 5 nm (130). This segment is represented here as an irregular ribbon (magenta) of alpha carbons. The N-terminal 10 amino acids are also a disordered segment and are similarly represented. When exerting a force upon the membrane, this 50-amino acid linker could be extended to a maximum of 17 nm, its contour length.
The final ∼17 amino acids are highly conserved across bacteria; this peptide binds to FtsA, ZipA, and several other proteins (discussed below). It is shown in Fig. 2 in darker purple as the extended beta strand and alpha helix that it adopts when it binds ZipA (121).
FtsZ subunits assemble into protofilaments by stacking vertically (Fig. 2C). This results in the GTP (shown in orange space fill on the top) being sandwiched between its binding site and the subunit above. The subunit above the GTP has three highly conserved amino acids (N207, D209, and D212) that play a critical role in GTP hydrolysis (45, 157, 168). Thus, FtsZ is considered to act as its own GTPase-activating protein (GAP), and hydrolysis occurs only after subunits come into contact in the protofilament. The catalytic amino acid D212 is shown in red space fill on the bottom of the model.
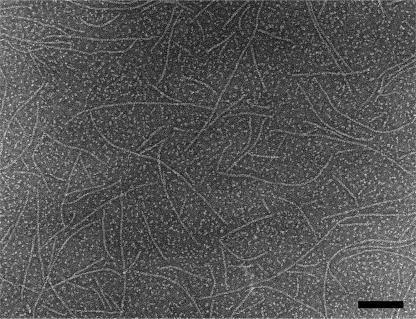
Figure 3 shows a negatively stained electron microscopy (EM) image of FtsZ protofilaments assembled in vitro. These protofilaments are one subunit thick and show little or no tendency to associate laterally (78, 161).
FIG. 3.
Electron micrograph of negatively stained protofilaments assembled in vitro from E. coli FtsZ (1 μM FtsZ, 50 mM MES [morpholineethanesulfonic acid] [pH 6.5], 100 mM KAc, 5 mM MgAc, 1 mM GTP). The bar is 100 nm. This specimen was prepared on a carbon film treated with UV light and ozone to render it hydrophilic (24). With these carbon films we obtain protofilaments at lower FtsZ concentrations, and they are longer than those previously reported. The protofilaments here are mostly straight, but some show a tendency to curve.
The Z Ring Modeled as Short, Overlapping Protofilaments
FtsZ assembles in vitro into protofilaments that are one subunit thick and average 120 to 200 nm long (30 to 50 subunits) (30, 78, 79, 161) (Fig. 2C and 3). Under some conditions the protofilaments associate further into paired filaments or larger bundles, but in dilute physiological buffers (100 to 300 mM potassium acetate [KAc], 5 mM Mg, pH 7.7) single protofilaments are the predominant form for E. coli FtsZ. It is generally assumed that these protofilaments are the basic structural unit and that they are somehow assembled further to make the Z ring.
To propose a structure of the Z ring, we need to know how much FtsZ it contains. A number of studies have used quantitative Western blotting to determine the number of molecules per cell. These values, and in some cases the number of FtsA molecules, are collected in Table 1. Most strains seem to have 5,000 to 7,000 FtsZ molecules per cell, although some have up to 15,000. An E. coli cell measuring 0.96 μm in diameter and 3.6 μm long has a volume of 2.5 μm3 (89, 158). Six thousand molecules per 2.5 μm3 gives a concentration of 4 μM; 15,000 molecules in the same volume would be 10 μM. These numbers are well above the ∼1 μM critical concentration, suggesting that most FtsZ in the cell is assembled into protofilaments.
TABLE 1.
Quantitation of FtsZ and FtsA in cells
| Species and strain | No. of molecules/cell |
Reference | |
|---|---|---|---|
| FtsZ | FtsA | ||
| E. coli | |||
| BL21 (B/rA) | 15,000 | 106 | |
| B/rK | 4,000a | 740 | 162 |
| W3110 | 10-14,000 | 157 | |
| 6,500a | 149 | ||
| 7,000a | 37 | ||
| MC4100 | 4,800 | 119 | |
| B. subtilis | 6,250a | 1,000 | 52 |
| 5,500a | 970 | 81 | |
| S. pneumoniae | 3.750a | 2,200 | 94 |
| C. crescentus | 9,500a | 153 | |
| M. tuberculosis | 30,000 | 44 | |
| M. smegmatis | 12,000 | 44 | |
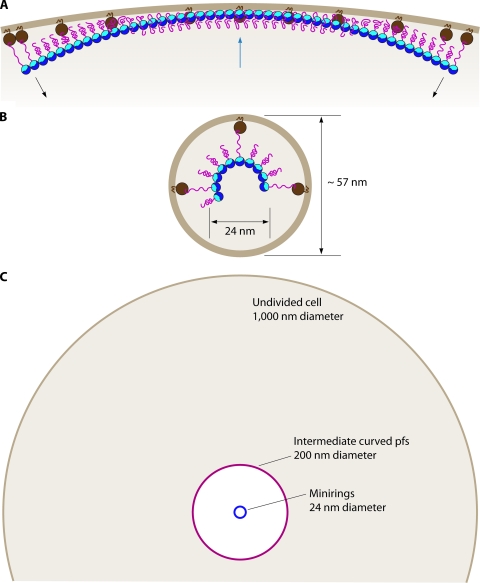
We found by quantitative fluorescence imaging that only 30% of the total FtsZ was in the Z ring in both E. coli and B. subtilis; the remaining 70% was cytoplasmic (7). Geissler et al. (55) found that 40% of FtsZ was in the Z ring for their strain of E. coli. For a cell with 6,000 FtsZ molecules, 2,100 FtsZ molecules in the Z ring would be sufficient to make a total protofilament length of 8,400 nm (at 4 nm per subunit). This would encircle a 1-μm-diameter cell two and a half times.
Figure 4 A shows a model for how protofilaments might be further assembled to make the Z ring. Since the protofilaments are much shorter than the circumference of the bacterium, they must be assembled with a staggered overlap. Support for this kind of model came from a study of Caulobacter by cryo-EM tomography (98). These images showed short filaments scattered around the circumference of the cell a short distance from the membrane. Control experiments, in which FtsZ was depleted or overexpressed, provided evidence that the filaments were in fact FtsZ.
FIG. 4.
(A) A model for how short protofilaments might be arranged to make the Z ring. The average 125-nm length is much shorter than the 3,000-nm circumference, so protofilaments would be arranged in a staggered overlap. (B) An alternative structure where the short protofilaments are proposed to anneal into one or a few long protofilaments.
An alternative model, discussed below, proposes that the short protofilaments might anneal into one or a few longer protofilaments (Fig. 4B).
Tethering FtsZ to the Membrane
One question raised by either model is how the protofilaments are attached to the membrane. This question has been resolved by Pichoff and Lutkenhaus. They first showed that FtsZ could assemble a Z ring if the cell had either FtsA or ZipA, but not in the absence of both (147). ZipA is a transmembrane protein whose cytoplasmic domain is known to bind the C-terminal peptide of FtsZ. Nuclear magnetic resonance (NMR) and X-ray crystal structures show this peptide forming a helix and a beta strand along a hydrophobic groove of the cytoplasmic globular domain of ZipA (121, 122). This binding therefore provides a tether of FtsZ to the membrane. The same C-terminal peptide of FtsZ is known to bind FtsA (41, 109), and the binding site has been localized to a patch on subdomain 2B (145). FtsA has long been considered a membrane-associated protein, but the detailed mechanism for its membrane binding was only recently discovered. Pichoff and Lutkenhaus (146) showed that the C-terminal peptide of FtsA forms an amphipathic helix that inserts into the lipid bilayer and anchors FtsA to the membrane. Thus, FtsZ is also tethered to the membrane by FtsA. Since ZipA is found only in gammaproteobacteria and a gain-of-function point mutation in FtsA can render ZipA nonessential in E. coli (14), we will consider FtsA the primary tether to the membrane.
The C-terminal peptide that binds FtsA and ZipA is highly conserved across bacteria, including some that have neither FtsA nor ZipA. In B. subtilis, which has no ZipA, FtsA can be eliminated and the cells are still viable, although with defects in division variably described as severe or moderate (10, 85). What tethers FtsZ to the membrane in the absence of FtsA and ZipA?
In Mycobacterium tuberculosis FtsW binds this peptide, but this probably does not occur in other species, because the binding site on FtsW is a C-terminal extension that is unique to mycobacteria (38). Also, even in mycobacteria, FtsW is apparently a late recruit to the Z ring (54, 155), so something else must provide the primary membrane tether. Several other proteins that regulate the Z ring (reviewed in reference 2) bind to this C-terminal peptide of FtsZ: EzrA (175), SepF (176), ClpX (25, 185), and the C-terminal domain of MinC (170). EzrA is probably the best candidate for a tether, because it is a transmembrane protein with a topology like that of ZipA (96). Also EzrA localizes to the Z ring early, along with FtsA and ZapA (54). On the other hand EzrA is best characterized as a negative regulator of Z rings (66, 96), so its role as a tether is not intuitive.
The C-terminal peptide is conserved in bacteria such as Mycoplasma, which have no candidate protein for binding it (194). We considered the possibility that these peptides might bind the membrane directly, but several sequences that we examined showed no features of an amphipathic helix. It is attractive to think that this C-terminal peptide provides the membrane tether in all species, but some additional binding partners or mechanisms are waiting to be discovered.
Lateral Bonds: Do They Exist?
The Z ring appears by light microscopy as a very long filament of mostly uniform density. The model in Fig. 4A, where the Z ring is made from overlapping short protofilaments, raises the question of how the protofilaments are associated with each other. One possibility is lateral bonds, which would involve specific contacts between subunits in adjacent protofilaments. This is the way that protofilaments are assembled to make the microtubule wall.
If FtsZ formed lateral bonds like tubulin, protofilaments should assemble into a two-dimensional (2-D) array with semicrystalline regularity. Sheets of protofilaments have been seen under certain conditions for FtsZ from Methanococcus jannaschii (103, 104, 134) and from Thermotoga maritima (106). FtsZ from M. tuberculosis forms long, two-stranded filaments, in which the two strands must be connected by some type of regular lateral bond (27, 199). FtsZ from most species, however, assembles one-stranded protofilaments that show no consistent lateral association in physiological buffer.
In 10 mM Mg, protofilaments of E. coli FtsZ associate into long, thin bundles that are several protofilaments thick (29, 35, 124). One study reported similar bundles in a more physiological buffer (5 mM Mg) (116); however, other labs have reported only one-stranded protofilaments under these conditions. Ca at 10 mM produces bundles that are somewhat thicker and quite long (202). When examined in cross section, the Ca-induced bundles showed irregular profiles of protofilaments but no regular protofilament lattice (105). Ruthenium red generated bundles similar to those in Ca (164).
The bacterial cytoplasm is a crowded environment due to the high concentration of proteins and nucleic acids. Agents such as Ficoll, polyvinyl alcohol, and methylcellulose, which are thought to mimic the physical chemistry of crowding, have a dramatic effect on FtsZ assembly, producing very large bundles, ∼100 nm in diameter and many that are micrometers long (151). In the original study of FtsZ assembly in Ficoll, the polymers were described as ribbons that were one subunit thick (60). However, we have found by embedding and sectioning that the bundles formed in a variety of crowding agents are round (D.E.A. and H.P.E., unpublished observations). This is also the interpretation from negatively stained specimens (151). Under some conditions the bundles were straight, and under others they curved into toroids or spirals. Importantly, diffraction patterns of these bundles showed very poor order. A diffuse equatorial spot indicated an average spacing of protofilaments of 6.8 nm, which is substantially larger than the width of an FtsZ protofilament (maximum width of 4.5 to 5 nm). The diffuseness of this spot and the lack of any second-order reflection suggested that the protofilaments are not spaced on a lattice, as they would be if held by regular lateral contacts between subunits, but are packed together with a liquid-crystalline order.
Long filamentous bundles and toroids of FtsZ were also formed when FtsZ-GFP was expressed in yeast cytoplasm, which is a natural crowded environment (181). FRAP showed that both the bundles and toroids were turning over subunits with a half time of 11 s, similar to the 8-s turnover of the Z ring in bacteria (7) and the 3.5 to 7 s for turnover of protofilaments in vitro (29). This is additional evidence that the protofilaments in the bundles are not associated by specific lateral contacts, since lateral bonds would be expected to slow down exchange. Protofilaments in the centers of the bundles apparently are also turning over, which means that they have access to subunits in solution.
Assembly in 1 M sodium glutamate produced very large bundles that resembled the ones made under crowding conditions (17). The GTPase was reduced only by about half in 1 M glutamate, suggesting that FtsZ, even in the interiors of the bundles, is rapidly exchanging with solution. This suggests a very loose structure similar to that of the bundles generated in the yeast cytoplasm. Bundles induced by Ca and Mg have substantially lower GTPase (29 202; our unpublished observations), suggesting that they may involve lateral contacts that inhibit subunit exchange.
Indirect evidence for bundling came from study of a temperature-sensitive B. subtilis FtsZ mutant (120). At the nonpermissive temperature, the mutant formed long-pitch helices in the cytoplasm but seemed to be incapable of collapsing them to make a Z ring. That study found that wild-type FtsZ assembled into toroid bundles in vitro at both 35 and 22°C. The mutant FtsZ formed toroids at 22°C, but at 35°C it formed thin, one-stranded protofilaments, suggesting that it was defective in bundle formation at the higher temperature. Another line of evidence for the bundling potential of FtsZ was its ability to form elastic gels in vitro (35, 51). These were formed in buffers with a low salt concentration, pH 6.5, and 10 mM Mg, so the gel formation may be less under the physiological conditions of the cytoplasm.
Several division proteins interact with FtsZ and cause it to form bundles or sheets. One of the first discovered was ZipA, which bundles most efficiently at a pH of <6 (67, 156). ZapA, a protein that enhances Z-ring stability, generates a variety of bundled forms (65, 101, 119, 178). ZapB, a small coiled-coil protein that stimulates Z-ring assembly, induces FtsZ bundles with a striking ∼10-nm banding pattern. FtsA*, a gain-of-function mutant of FtsA, caused FtsZ to assemble sheets of protofilaments that appeared to adopt the ∼200-nm-diameter intermediate curvature (see below) (18). SlmA produced curled sheets of FtsZ with sharp edges and a seeming regular structure (15). It is surprising that SlmA induced polymers of FtsZ, because SlmA was identified as a nucleoid occlusion factor and was thought to inhibit Z-ring formation over the nucleoid. The bonds between FtsZ and the bundling proteins may involve electrostatic interactions (15) but are mostly unknown.
Large round bundles of protofilaments are probably not relevant inside the bacterium. With only 5,000 to 7,000 molecules of FtsZ in the typical bacterial cell (Table 1), there is only enough FtsZ to make ∼7 protofilaments 3 μm long. In addition, because FtsZ protofilaments in the Z ring are tethered to the membrane, they should be limited to a 2-D ribbon or sheet. Expression of an FtsZ-GFP fusion with the C terminus of FtsZ deleted did lead to formation of rods in the central cytoplasm (108). When the C terminus is present, however, tethering to the membrane and subsequent Z-ring assembly apparently trump the tendency to form cytoplasmic bundles. Also, the effects of crowding on FtsZ (or other) assembly may have been overestimated. McGuffee and Elcock (112) have recently shown that the steric effects of crowding, which are the main ones considered in previous analyses, can be largely canceled by the favorable ionic and hydrophobic interactions of (FtsZ) subunits with the multiple other proteins in the cytoplasm. That study suggests that polymerization of FtsZ in the bacterial cytoplasm might be more similar to polymerization in dilute buffer than to that in Ficoll and polyvinyl alcohol. Expression of FtsZ in yeast did produce linear bundles and toroids, similar to the structures in polyvinyl alcohol, but this might have involved a high level of expression.
Hamon et al. (68) have obtained superb images of FtsZ protofilaments by atomic force microscopy (AFM). They found that adsorption to mica enhances the effective concentration and can support protofilament assembly from solution concentrations well below the critical concentration. Remarkably, that study found that tubulin also formed short, curved single protofilaments when adsorbed onto mica from dilute solution. In solution, tubulin protofilaments are unstable and exist only when stabilized by lateral bonds to neighboring protofilaments (49). Adsorption to the 2-D mica surface appears to provide the stabilization needed to observe these small intermediates. This technology has great promise for observing polymer assembly reactions, including intermediates that are too weak to be observed in bulk solution.
At higher FtsZ concentrations, protofilaments become very crowded on mica and tend to associate in parallel sheets and spiral structures (71-73, 141). The packing appears in places to be just the result of the density of protofilaments, but the authors have presented mathematical models in which lateral bonds play an important role in determining the packing. The lateral interactions between adjacent protofilaments are not regular lateral bonds like those in the microtubule wall, because the protofilaments show irregular packing with variable spaces. Instead the authors proposed a Lennard-Jones type interaction, which is maximal when the protofilaments are about 13 nm apart and becomes repulsive at closer distances (discussed also in reference 48). If the Lennard-Jones attraction was independent of rotation around the protofilament axis, it might be related to the round bundles formed under crowding conditions. Still lacking, however, is a chemical basis for the proposed lateral attractions that could explain the apparent two-dimensional packing on mica and three-dimensional packing in round bundles in solution.
In summary, lateral bonds between FtsZ protofilaments probably do not exist as the kind of repetitive protein-protein contacts that produce the microtubule wall. The association of protofilaments into bundles seems to involve irregular packing into a liquid-crystalline array. The protofilaments are not in physical contact with each other but may interact through electrostatic forces involving the ions between the protofilaments, as proposed for packing of actin bundles (150, 152, 190), or through other solvent effects.
Alternatives to Lateral Bonds
How might the long FtsZ filament, either the ring or the extended helices, be constructed apart from lateral bonds? Shlomovitz and Gov (173) have proposed a mechanism based on forces transmitted across the membrane. In their model, bending Z rings exert a constriction force that distorts the membrane, and these membrane distortions interact when Z rings approach each other. Their model predicts that adjacent Z rings will undergo an initial attraction that would cause them to slide together. The attraction would bring them to a certain distance that is semistable, but eventually they would move together and coalesce. This prediction fits the observed movement of Z rings in tubular liposomes (137). This attraction was mediated solely by the distortions of the membrane generated by the two adjacent rings. The model assumed that the Z ring was a uniform circle generating an inward constriction force. It will be interesting to see if a future model can be applied to scattered, short protofilaments. In particular, could short protofilaments with a preferential bend communicate through membrane distortions to generate the uniform distribution of filaments around the Z ring? An attractive feature of this model is its agreement with the cryo-EM tomography (98), which showed the FtsZ as short filaments not making contact with each other.
Another possibility is that the protofilaments do not remain as the short, ∼30-subunit structures that assemble in solution but that they anneal into much longer filaments (Fig. 4B). Annealing has been visualized directly by AFM imaging of FtsZ protofilaments adsorbed to mica (117). The protofilaments were able to diffuse on the 2-D mica surface, and examples of fragmentation and annealing were shown. In a recent study, Chen and Erickson (29) reported indirect evidence for annealing of protofilaments in bulk solution.
In the cell the protofilaments are tethered to the membrane and are probably restricted to the center of the cell by the Min and nucleoid occlusion systems. Surovtsev et al. (187) proposed that FtsZ tethered to the membrane might be confined to a distance 8 nm from the surface (a reasonable estimate of the flexibility provided by the 50-amino-acid tail of FtsZ [Fig. 2]) and to an axial zone 100 nm wide (a reasonable estimate for the width of the Z ring). The volume of this shell would be only 1/1,000 of the total cytoplasmic volume. If the total cytoplasmic FtsZ is ∼4 μM, and 30% of it is in the Z ring, the effective concentration of FtsZ in this restricted volume would be 1,200 μM. More importantly, the concentration of filament ends would be 40 μM, assuming protofilaments 30 subunits long. Annealing should therefore be much more favorable than adding single subunits from the 1 μM cytoplasmic pool. This simple numerical argument suggests that annealing could play a major role in determining the structure of the Z ring. Of course, the high concentration of protofilaments would also enhance any lateral interactions. A diagram of the Z ring as a single long filament is shown in Fig. 4B.
One problem with an annealing model is to reconcile the very long protofilaments with the rapid turnover of subunits in the Z ring. In the 8-s half time for turnover (discussed below), about 45 subunits could be added to a protofilament, given the measured kinetics of assembly (7). This turnover could be explained for a Z ring made from short protofilaments averaging 30 to 50 subunits (Fig. 4A). However, if the Z ring was one long, continuous protofilament, this exchange would have to occur from multiple interior sites, which would involve frequent breakage and dissociation of subunits, followed by reassembly and reannealing. The long protofilament produced by annealing would then seem to be constantly breaking and reannealing. This frequent breakage should also occur for a filament of short protofilaments connected by lateral bonds. The rapid subunit exchange in the Z ring is thus difficult to reconcile with the evidence that the basic architecture of the Z ring is a long, continuous filament, whether produced by annealing or by lateral bonds.
The observation that the Z ring can separate into a short-pitch, two-turn helix (Fig. 1B) is quite consistent with its structure being a long, continuous filament. If the Z ring consisted of scattered short protofilaments, as depicted in Fig. 4A, lateral drift should produce only a diffuse Z ring, not separate gyres of a helix.
ASSEMBLY OF FtsZ IN VITRO
Buffer Conditions for In Vitro Studies
Early in vitro studies of FtsZ assembly were done at pH 6.5, which appeared to give more robust assembly than a higher pH (50, 124). However, the pH of E. coli cytoplasm varies from 7.4 to 7.8 depending on the external pH (177). Recent measurements showed a cytoplasmic pH of 7.7 for an external pH of 7.5 (200). Potassium is the major cytoplasmic cation, and it varies from 0.14 to 0.76 M depending on external osmolarity (26). The major anions in the cytoplasm are nucleic acids. The major monomeric anion is glutamate, but its concentration is only about one-fifth that of potassium (26). We generally use potassium acetate as the salt, but we have found no effect on FtsZ assembly or GTPase activity when the anion was changed to chloride. (While the nature of the anion is probably not important, that of the cation is. Substituting sodium for potassium increased the critical concentration from 1 to 20 μM [189].) A KAc concentration of 350 mM is probably the best approximation to physiological salt, but for some assays, such as negative-stain EM, 100 mM KAc gives better results. There was only a small difference in the GTPase of E. coli FtsZ between 100 and 350 mM KAc (29). The use of physiological buffer for in vitro studies has been addressed previously (60, 106).
Magnesium is an important variable affecting FtsZ assembly and dynamics. Various in vitro studies have used Mg concentrations of from 2.5 to 10 mM. Earlier work showed that at 2.5 to 5 mM Mg, FtsZ assembles into protofilaments that are one subunit thick (78, 161). These studies were done at pH 6.5, but we have repeatedly observed single protofilaments at pH 7.7 with these low Mg concentrations. With 10 mM Mg the protofilaments associate into long, thin bundles, several protofilaments thick, and the rates of GTP hydrolysis and subunit exchange are reduced 3-fold (29, 35). Mohammadi et al. (119) recently undertook a comprehensive study of how pH and Mg affect the interaction of FtsZ and ZapA. They found striking differences between 5 and 10 mM Mg, as well as between pH 6.5 and 7.5.
The total cellular Mg has been measured to be 20 to 80 mM, but it has long been known that most of this is bound by RNA and other polyanions. How much Mg is unbound in the cytoplasm? The question was resolved recently using the fluorescent dye mag-fura to measure the concentration of free cytoplasmic Mg: it is 0.9 mM (53). This suggests that FtsZ assembly in vitro should be studied at ∼1 mM free Mg. In selecting an Mg concentration for in vitro assembly experiments, it should be kept in mind that GTP will chelate an equivalent amount of Mg. If the reaction mixture contains 1 mM GTP, the buffer should contain 2 to 2.5 mM Mg. We should also note that we have not found any difference in assembly between 2.5 and 5 mM Mg.
GTP Hydrolysis
G proteins bind GTP in their active state, and when GTP is hydrolyzed to GDP, they switch to an inactive state. The level of intrinsic GTPase activity of G proteins is very low, and hydrolysis is generated by a separate GTPase-activating protein (GAP), which binds to the G protein at the GTP-binding pocket. The GAP provides catalytic side chains that contact the GTP and initiate hydrolysis.
FtsZ, like tubulin, is its own GAP. Monomeric FtsZ hydrolyzes GTP very slowly, if at all. When assembled into a protofilament, the bottom interface of one FtsZ makes contact with the GTP pocket of the subunit below (Fig. 2). A key player is the synergy or T7 loop, which contains the sequence 207NxDFAD212 (E. coli sequence numbers). This sequence is highly conserved in all FtsZ proteins and is NxDxxE in all α-tubulins. Mutation of the conserved amino acids severely cripples the GTPase activity, leading to the proposal that the synergy loop acts as a GAP to activate hydrolysis of the GTP in the subunit below (45, 127, 157, 168). Details of the mechanism have been imaged in a crystal structure (133). The two aspartate residues are in good hydrogen-bonding positions to polarize a water molecule that attacks the gamma phosphate of the GTP. The role of cations and pH has been further explored by molecular dynamics (113).
FtsZ polymers contain a substantial amount of GTP, which suggests that hydrolysis occurs with some lag following assembly. Romberg and Mitchison measured the nucleotide content of protofilaments at steady state and found the GDP/GTP ratio to be 20:80 (160). This is quite different from the case for microtubules, where the nucleotide is almost entirely GDP, with only a small cap of GTP at each end. Chen and Erickson (29) repeated this assay for FtsZ and found that the fraction of GDP increased as the external concentration of GTP was increased from 20 to 120 μM. Above 100 μM GTP, the GDP/GTP ratio plateaued at 50:50. Total assembly and the rate of GTP hydrolysis at steady state also increased at the higher external GTP concentrations. The half maximum for these reactions (increasing GDP in polymer, total assembly, and GTPase) occurred at ∼50 μM GTP. This is far above the binding affinity for GTP and GDP nucleotides, which Huecas et al. (79) have found to be in the 10 to 100 nM range (that reference should be consulted for a comprehensive study of the thermodynamics and kinetics of nucleotide binding to M. jannaschii apo-FtsZ). In an earlier study, treating the GTPase reaction as a classical Michaelis-Menten reaction, Sossong et al. (180) determined a Km of 82 μM. From these studies it seems clear that some aspect of FtsZ assembly is modulated by GTP concentrations in the range of ∼50 to 100 μM. It is not clear what mechanism is involved.
Earlier studies suggested that unlike microtubules, FtsZ protofilaments could exchange their nucleotide for GTP in solution (60, 116, 133, 189). Oliva et al. (133) supported this interpretation with a crystal of M. jannaschii FtsZ that showed dimers that were similar in structure to the αβ-tubulin dimer. These dimers showed a gap from which the GTP could exit, and indeed the nucleotide pocket of the dimers could be filled by soaking the crystals in GMPCPP.
Experimental evidence clarifying the exchange of nucleotide into protofilaments was provided by Huecas et al. (79). They showed that apo-FtsZ of M. jannaschii could assemble into protofilaments and that GDP or mant-GTP could associate into these polymers. However, when FtsZ was assembled with nucleotide, the bound nucleotide was very slow to dissociate. They concluded that the slow dissociation of bound nucleotide presented a kinetic block to the exchange of nucleotide into polymer. The data of Chen and Erickson agreed with this and suggested that previous indications for nucleotide exchange into polymer were probably due to the pool of subunits that exchange without GTP hydrolysis (29). These studies now agree that nucleotide exchange occurs only when the GTP-binding pocket is exposed to the solvent, at the plus end of a protofilament or on free monomers.
It is known that FtsZ-GDP can assemble but that it does so much more weakly than FtsZ-GTP (79, 159, 180). However, it is not known what happens following GTP hydrolysis in a protofilament. One possibility is that the protofilament fragments at the site of the GDP. This fragmentation would have to follow a lag comparable to the lag for hydrolysis in order to account for the 50:50 GDP/GTP ratio. Another possibility is that fragmentation is minimal, and subunits dissociate primarily from the ends of protofilaments. In this model the end subunit would dissociate much faster if it was bound by a GDP interface.
Finally, we note that GTP hydrolysis is not required for assembly. Assembly occurs with GTP plus EDTA, which chelates Mg, but hydrolysis is completely blocked (28, 124). Assembly is well supported by the GTP analog GMPCPP, which is hydrolyzed 50 times slower than GTP (179). When aluminum fluoride is added to FtsZ-GDP, the AlF binds to the GDP and mimics the gamma phosphate, producing a nonhydrolyzable GTP analog. FtsZ assembled efficiently but with slower kinetics in GDP-AlF (117). Two analogs that do not work are GMPPNP and GTPγS (169). GMPPNP bound FtsZ very weakly and could not compete GTP. GTPγS bound tightly, but it could not support polymerization unless mixed with GTP.
GTP binding is not even required for assembly. Huecas and Andreu showed that apo-FtsZ from M. jannaschii assembled as well as FtsZ-GTP, with a similar critical concentration and polymer morphology (77). GDP strongly destabilized the assembly. The role of GTP therefore appears to be to provide a means for destabilizing the polymer following hydrolysis, leading to a constant recycling of FtsZ subunits.
Assembly Dynamics of FtsZ In Vitro
Several years ago we discovered a tryptophan mutant of FtsZ that exhibited a 2.5-fold increase in fluorescence emission upon assembly (28). This provided an important tool to assay assembly, since the fluorescence was obtained in real time and was directly proportional to the number of subunits forming interfaces in protofilaments. This assay was used to measure the kinetics of assembly initiated by adding GTP. The data were interpreted with a model of cooperative assembly with a weak dimer nucleus. The structural nature of this dimer is still not known.
An alternative fluorescence assay was developed based on fluorescence resonance energy transfer (FRET) (30). This assay provided a way to measure the exchange of subunits between protofilaments at steady state. Protofilaments were preassembled from donor- and acceptor-labeled FtsZ and then mixed. Initially there was no FRET, since the donor and acceptor labels were on separate protofilaments. A FRET signal developed as the protofilaments disassembled and the subunits reassembled into mixed protofilaments. The half time for subunit exchange was 7 s in the original study at pH 6.5 and 100 mM KAc (30). At pH 7.5 the half time for exchange was 3.5 s with 100 mM KAc and 17 s with 350 mM KAc (29).
This FRET assay was used to measure the protofilament turnover in a variety of different buffers and to compare the turnover with the time required for hydrolysis of GTP (29). The half time for subunit exchange varied from 3.5 to 35 s for the different buffers. The time required for a subunit to hydrolyze a GTP varied from 13 to 100 s. Over this range, the time for nearly full subunit exchange (two times the half time) was generally twice as fast as the time to hydrolyze a GTP. This suggested that there are two mechanisms for subunit exchange from protofilaments. One is coupled to GTP hydrolysis and likely involves exchange of one FtsZ for every hydrolysis event. The second mechanism involves exchange of FtsZ-GTP subunits at the ends of protofilaments, without GTP hydrolysis. These two mechanisms contribute roughly equally to the overall exchange.
Conditions resulting in slower exchange times and slower GTP hydrolysis correlated with association of protofilaments into bundles. (These bundles were similar to the Ca/Mg bundles, rather than the large bundles of loose-packed protofilaments assembled in crowding agents, which maintain a rapid exchange.) In the simplest model, once a subunit has hydrolyzed its GTP, it must dissociate to exchange GDP for GTP before it can repolymerize and hydrolyze another. Although it is possible that reagents such as calcium act to inhibit GTPase, and this leads to bundling, we believe the opposite: that bundling is the primary action and reduced GTPase is the secondary effect.
Two classes of dynamics are known for the eukaryotic cytoskeleton. Actin filaments undergo treadmilling at steady state, where the filaments grow at one end and shorten at the other. Microtubules undergo dynamic instability, where each end switches between extended phases of growth and shortening. (Dynamic instability can be accompanied by treadmilling.) Analysis of FtsZ cap mutants, which block either the top or bottom assembly interface, gave evidence for treadmilling of FtsZ, with subunits adding preferentially to the bottom end (away from the GTP) (157).
Dynamic instability remains a possibility for FtsZ. Scheffers et al. investigated the nonhydrolyzable analog GTPγS and discovered an important clue to the mechanism of dynamics (169). GTPγS could bind to FtsZ with an affinity similar to that of GTP but could not support assembly on its own. When FtsZ assembly was initiated with a limiting amount of GTP (20 μM) and no GTPγS, turbidity reached a peak and then fell as the GTP was hydrolyzed (this assembly was in a pH 6.5 buffer with 10 mM Ca, which produces bundles and strong turbidity). However, if 50 to 200 μM GTPγS was included in the reaction, the assembly was stabilized at the peak. This result is remarkably similar to the ability of nonhydrolyzable analogs to stabilize microtubules assembled in limited GTP (129). It is likely that the stabilization involves GTPγS coassembled with GTP throughout the protofilaments, but the ratio and distribution are not known. Further exploration may shed light on the mechanisms of dynamics.
Inhibition of FtsZ Assembly by SulA
A number of proteins, called negative regulators, that appear to inhibit Z-ring assembly have been identified. The best studied are SulA and MinC. The mechanisms by which these proteins inhibit FtsZ are beginning to shed light on how FtsZ assembles into protofilaments and the Z ring.
SulA is expressed during the SOS response to DNA damage. It blocks cell division until the DNA is repaired, and SulA then is degraded. Overexpression of SulA in E. coli was shown to block Z-ring assembly (21, 37, 123). Two studies in 1998 showed that SulA inhibited both the GTPase activity and assembly of FtsZ (123, 192). A recent study by Dajkovic et al. (37) has made several important advances. An assay for the critical concentration provided good evidence for a simple mechanism. In the absence of SulA the critical concentration was 0.9 μM, and this increased to 4.3 and 5.9 μM, respectively, when 3.5 and 4.0 μM SulA were present. The reaction behaved as if the concentration of active FtsZ was equal to the total FtsZ minus the concentration of SulA. This suggests that SulA acts by binding free subunits of FtsZ and blocking their assembly. A plot of GTPase versus FtsZ concentration was similarly shifted, as if an amount of FtsZ equivalent to that of SulA was sequestered from the reaction. An important extension of this interpretation is that the affinity of SulA for FtsZ must be higher than that of FtsZ for binding the end of a protofilament. This has not yet been demonstrated directly.
This simple sequestration mechanism was predicted earlier from the crystal structure of a SulA-FtsZ (32). SulA bound to the bottom of FtsZ, near the T7/synergy loop. Bound SulA would sterically block the formation of the protofilament interface, and all FtsZ subunits with a bound SulA would be essentially dead for the assembly reaction. Dajkovic et al. (37) also found that Z rings disappeared in vivo at a SulA concentration of around half that of FtsZ. The substoichiometric activity probably reflects the critical concentration (one needs to sequester only the FtsZ above the critical concentration to block FtsZ assembly) and the fact that only 30 to 40% of the cell's FtsZ is in the Z ring.
The sequestration mechanism of SulA action would appear to resolve this issue, were it not for a truly remarkable discovery. Dajkovic et al. (37) found that FtsZ was no longer sensitive to SulA when assembly was induced by GMPCPP, GTP plus EDTA, or GDP plus AlF. Each of these nonhydrolyzable analogs eliminates the cycling of subunits coupled to GTP hydrolysis. There is still an equilibrium exchange of subunits from the ends of protofilaments (29), but it seems that the more rapid cycling of subunits coupled to GTP hydrolysis is needed for SulA to inhibit assembly. This is not consistent with a simple sequestration mechanism.
Dajkovic et al. (37) suggested an explanation based on high- and low-affinity conformations for FtsZ, which has been proposed previously by three groups to explain cooperative assembly (35, 79, 90, 118; see also reference 48). In the Dajkovic model SulA was proposed to bind only the high-affinity conformation. A second requirement was that the off rate of FtsZ from stabilized protofilaments (assembled in GMPCPP, GDP-AlF, or EDTA) must be 1,000-fold lower than the off rate from SulA and much lower than the off rate from Mg-GTP protofilaments. There is, however, contradictory evidence for both points. First, there is no candidate for a high-affinity conformation among the many crystal structures of FtsZ; they all have the same conformation (135). Most important, in the crystal structure of FtsZ bound to SulA (32), the conformation of FtsZ was the same as in the other crystals. This conformation would presumably be the abundant, low-affinity conformation. Second, the half time for dilution-induced disassembly was 7 s with EDTA and 1.4 s with Mg (29). The off rate from the EDTA protofilaments is at most 5 times lower than that from Mg protofilaments. Although this particular explanation appears to have problems, the mechanism of SulA inhibition provides an important direction for new research. This phenomenon may be related to the mechanism giving the appearance of cooperative assembly (see below), and its study may help unravel this enigma.
Inhibition of FtsZ Assembly by MinC
The MinCDE system helps localize the Z ring to the cell center by inhibiting its assembly at the poles, where MinCD has its highest average concentration (107). If the Min system is deleted, division near the poles produces minicells. Although most cells still have central septa, their placement was less precise in cells lacking MinC (64). MinD and MinE operate to localize MinC. MinC is the protein that interacts with FtsZ to inhibit Z-ring assembly. Assembly of Z rings is blocked by overexpression of MinCD in E. coli (75, 144).
In vitro analysis showed that MinC inhibited FtsZ polymerization, as measured by a centrifugation assay, at approximately a 1:1 stoichiometry (75). Remarkably, however, the GTPase was not inhibited even by a 2.5-fold excess of MinC. This suggests that MinC does not inhibit the assembly of protofilaments but blocks their association into bundles. This is also suggested by the specifics of the centrifugation assay used to monitor polymers. The assembly was done in a pH 6.5 buffer with 10 mM Mg, conditions that enhance the formation of long, thin protofilament bundles. These bundles are pelleted much more readily than short, one-stranded protofilaments. This suggests that MinC inhibits Z-ring assembly not by affecting the assembly of protofilaments but by inhibiting their association into bundles.
Dajkovic et al. (35) supported this interpretation by use of two different assays. They used a FRET assay (30) to show that total polymer was not inhibited by MinC. They then explored an assay measuring the elasticity of gels formed by FtsZ at high concentrations (25 μM). The elasticity is a measure of association of protofilaments to form a three-dimensional gel. Addition of MinC to a preformed FtsZ gel, at a 1:2 or 1:1 MinC/FtsZ ratio, resulted in loss of elasticity over 10 to 20 min. At 2:1, the elasticity was lost immediately. Most in vitro studies of MinC have used the E. coli system, but Scheffers has recently studied the interaction of FtsZ and MinC from B. subtilis (167). He used a centrifugation assay that included DEAE-dextran, which was needed to obtain FtsZ polymers large enough to pellet. MinC inhibition was found to be pH sensitive. It was minimal at pH 6.5, but at pH 7.5 MinC inhibited assembly 80% (although this required a 50% excess of MinC over FtsZ). As in the E. coli system, MinC had no effect on GTP hydrolysis, suggesting that it did not inhibit assembly of protofilaments but blocked their association into DEAE-dextran bundles.
An intriguing observation in the study by Dajkovic et al. (35) was that MinC was completely inactive against FtsZ polymers assembled in GMPCPP or in GDP-AlF. This is remarkably similar to the case with SulA, which inhibited assembly only when GTP hydrolysis contributed to cycling of FtsZ subunits. This is surprising, however, because SulA and MinC are supposed to be operating by very different mechanisms (SulA inhibits protofilament assembly, and MinC inhibits protofilament bundling). Since MinC does not inhibit either the assembly of protofilaments or GTP hydrolysis, it is difficult to see how cycling of subunits would matter to the inhibition. Nevertheless, as with SulA, this observation seems of exceptional importance and may lead to new understanding of FtsZ assembly dynamics and cooperativity.
The essential role of GTP hydrolysis for sensitivity to MinC, suggested by the in vitro experiments referenced above, is also supported by in vivo observations. Three point mutants of FtsZ, when expressed as the sole source of FtsZ, showed resistance to overexpression of MinC: ftsZ2 (D212G, on the bottom interface), ftsZ9 (an insert of VG between V18 and G19, buried near the GTP in the Rossman fold), and ftsZ100 (A70T on the upper interface) (Fig. 5 in reference 144). The locations of these mutations are scattered and do not suggest a binding site. The mutants do, however, have one thing in common: their GTPase is greatly reduced or not detectable (34). The resistance of these FtsZ mutants to MinC may be due not to their inability to bind MinC but to their reduced GTPase-dependent assembly dynamics.
FIG. 5.
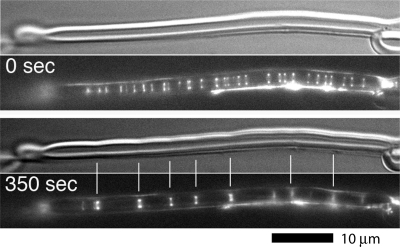
Z rings assembled in tubular liposomes from purified, membrane-targeted FtsZ. The upper frame at the two times uses Nomarski optics to show the profile of the liposome. The lower panel shows FtsZ localized by yellow fluorescent protein (YFP) fluorescence. At time zero (actually about 10 min after specimen preparation), there are many dim Z rings. After 350 s, the dim Z rings have coalesced into fewer bright ones, and these are generating constrictions in the liposome. The full sequence may be seen in Movie S2 in the supplemental material. (This figure is reprinted and Movie S2 is reproduced from reference 137 with permission from AAAS.)
Recent studies have suggested that the Min system in B. subtilis may function other than by blocking Z-ring assembly at the poles. Gregory et al. (63) expressed a functional MinC-GFP from the chromosomal minCD locus, which should produce a normal cytoplasmic level of the fusion protein. They found that MinC-GFP localized prominently to the Z ring late in divisome assembly and prior to septation. MinC-GFP remained transiently at the poles following septation, where it appeared to destabilize newly forming Z rings and block their maturation into division competency. The prominent polar localization observed in previous studies was attributed to an overexpression artifact. Bramkamp and colleagues (23, 193) also found that MinC localized to Z rings late in the cycle, and they concluded that “the main function of the Min system is to prevent minicell formation adjacent to recently completed division sites by promoting the disassembly of the cytokinetic ring” during constriction. These observations question the simple paradigm that MinCD acts simply to block FtsZ-ring assembly at the poles and thereby localize it to the cell center. These studies have so far been done only with B. subtilis, so it remains to be seen if similar phenomena are found in E. coli.
MinC has two domains. The N-terminal domain, MinC-N, which is considered the primary inhibitory domain, blocked Z-ring assembly when overexpressed in vivo, and it inhibited FtsZ polymers in vitro (74, 171). The C-terminal domain, MinC-C, mediates dimerization of MinC and its binding to MinD (31, 74). Binding of MinC to MinD serves to localize it to the membrane, and also enhances its association to the Z ring and its disassembly activity. Shiomi and Margolin (172) found that MinC-C could also inhibit Z-ring assembly, but this required coexpression of MinD. A similar activation of MinC is produced by its complex with DicB, a prophage protein whose expression results in MinC-mediated destruction of Z rings (86). There appear to be two steps in the activation of MinC. Johnson et al. (87) showed that tethering MinC to the membrane (the tether was a truncated ZipA comprising the transmembrane domain and the ∼160-amino-acid flexible P/Q segment) rendered it toxic at a 9-fold-lower concentration than when MinC was expressed as a soluble protein. The inhibitory activity of tethered MinC was further increased by expression of MinD, which also resulted in the MinC localizing to Z rings. A simple summary of these results is that MinC itself can inhibit Z-ring assembly at sufficiently high concentration; its activity is enhanced by tethering it to the membrane and further still by targeting it directly to Z rings. These last two steps may be explained by an increase in effective concentration of MinC.
Shen and Lutkenhaus used random mutagenesis to identify the sites on FtsZ that bind MinC-C and MinC-N (170, 171). They found four mutations in FtsZ that rendered it insensitive to MinC-C. These were all in the C-terminal peptide that binds FtsA and ZipA, identifying this peptide as the binding site for MinC-C. The same strategy was then used to find mutations of FtsZ that would render it insensitive to MinC-N (171). Four amino acid changes in FtsZ were found, and they clustered into a small patch (a maximum of 14 Å apart) that probably corresponds to the binding site for MinC-N. This patch shows substantial overlap with the binding site for SulA, with two of the amino acids (R271 and L205) being buried in the FtsZ-SulA interface (32). In addition, all four correspond to amino acids that are buried in the longitudinal interface of the αβ-tubulin protofilament (128). This suggests that the MinC-N binding site is buried for all FtsZ subunits in the middle of protofilaments. Its binding site will be exposed only on the terminal subunit at the minus end of a protofilament. It has been suggested that MinC-N might act by weakening the longitudinal interfaces in the protofilament (35), but this could only happen if the interface were opened by bending. Even the 23-degree bend of the highly curved miniring conformation is probably not enough to permit access of the globular MinC-N.
The affinity of MinC for binding to FtsZ has been measured with a plasmon resonance assay, with a KD (equilibrium dissociation constant) reported to be 6 μM (171). This is surprisingly weak compared to the 1 μM critical concentration for FtsZ assembly, suggesting that MinC would show minimal binding to the pool of free FtsZ monomers in the cytoplasm. It should also be noted that the concentration of MinC in E. coli is only 400 molecules per cell (188). The weak binding affinity might be compensated for by tethering MinC to the membrane via MinD. However, the low stoichiometry relative to FtsZ would seem to pose problems for models developed from in vitro experiments at an equal or higher stoichiometry.
FtsZ as a Target for Drugs
FtsZ has long been recognized as an attractive target for potential antibiotic drugs. A number of natural compounds with antibacterial activity have been shown to inhibit FtsZ (19, 42, 43, 76, 84, 154). Most of these studies are still preliminary, and it is possible that some of the compounds target pathways other than FtsZ. Two labs have studied GTP analogs for their potential to inhibit FtsZ polymerization in vitro. C8 MeOGTP inhibited polymerization of M. jannaschii FtsZ competitively, with a 50% inhibitory concentration (IC50) of 15 μM (in 60 μM GTP) (93). Compound 14 in another study showed a similar inhibition in vitro and also showed substantial inhibition of Staphylococcus aureus growth in an agar diffusion assay (142). In contrast to these agents, which act by blocking FtsZ polymer, the compound OTBA appeared to act by stabilizing FtsZ polymer and causing its association into large bundles (20). This is similar to the mechanism of PC190723, which is discussed below.
Several groups have used in vitro assays to screen libraries of hundreds of thousands of compounds for inhibition of FtsZ. One study used an assay involving assembly of fluorescent FtsZ with DEAE-dextran to screen extracts of microbial fermentation broths and plants (196). They identified a viriditoxin, a previously characterized metabolite of Aspergillus, as a promising lead candidate. Another study screened large chemical libraries for compounds that inhibited the GTPase activity of FtsZ, and those authors identified a half dozen compounds that they called “zantrins” (110). The zantrins operated by two different mechanisms. Some inhibited the formation of protofilaments, and others caused the protofilaments to associate into bundles. Either mechanism could block the function of FtsZ for cell division. The zantrins blocked Z-ring assembly in E. coli and killed a range of bacterial species. Another study used a cellular screening system to identify inhibitors of chromosome partitioning in E. coli (82). Reasoning that some of these might function by blocking FtsZ, this group screened 138 of the most active compounds for the ability to block assembly and GTPase of FtsZ. They identified A189 as an attractive lead compound.
Stokes et al. (182) developed a cell-based assay to screen specifically for inhibitors of cell division. Their assay used a reporter specific for the asymmetric cell division during sporulation of B. subtilis. Out of 105,000 compounds screened, one hit was selected for further study. The screen could have picked up inhibitors of any of the seven essential cell division proteins, but the selected hit compound was shown to target FtsZ. It blocked assembly of the Z ring in cells, and the compound inhibited pelleting of FtsZ and GTP hydrolysis in vitro. Several point mutants of FtsZ that were resistant to the compound were isolated.
Probably the most attractive compound targeting FtsZ is PC190723. Its discovery originated with the compound 3-methoxybenzamide, which was earlier shown to have anti-FtsZ activity but only at high concentrations (132). Haydon et al. (70) undertook a medicinal chemistry program and tested 500 modifications of the simple 3-methoxybenzamide. They identified one compound, PC190723, with vastly improved potency. PC190723 was bactericidal against B. subtilis and all staphylococcal strains tested, including methicillin-resistant S. aureus. It was inactive toward Gram-negative bacteria, several Gram-positive bacteria, and yeast. The compound was effective in mice, providing 100% protection from a lethal dose of S. aureus. Several spontaneous mutations that caused resistance to the compound arose in S. aureus. Each mutant had a single amino acid change in FtsZ, at one of six positions. These positions formed a cluster that was also identified as a binding site for PC190723 by a docking program. The binding site was equivalent to that for taxanes on β-tubulin.
When B. subtilis was exposed to PC190723 the FtsZ-GFP was mislocalized to discrete foci along the elongated cells, rather than being completely disassembled as with some other inhibitors. Andreu et al. have studied the interaction of PC190723 with B. subtilis FtsZ in vitro, and they found that it stabilized FtsZ protofilaments and caused them to associate into protofilament bundles, toroids, and helical bundles (8). It thus seems to block Z-ring function by sequestering FtsZ into these inactive condensates.
Cooperative Assembly and Treadmilling of FtsZ
An enduring enigma is how FtsZ, which assembles into protofilaments that are one subunit thick, can exhibit features of cooperative assembly. Three groups have suggested that the appearance of cooperativity can be generated by postulating a conformational change that favors assembly and is itself favored by the act of assembly (35, 79, 90, 118). However, no one has yet suggested a possible model, at the level of atomic structure, that could generate this enhancement.
An intriguing result with FtsZ interface mutants may be relevant to cooperative assembly. Our lab prepared and characterized a number of single-amino-acid mutants that targeted the interface connecting subunits into the protofilament (157). The original goal was to produce what we thought would be “cap mutants” that would block either the top or bottom interface. We speculated that a “bottom cap” mutant, with a crippling defect on its bottom surface, would be able to bind to the bottom of a growing protofilament and block further assembly at that end.
We first tested the mutants in vivo. We found nine bottom and eight top mutants that were unable to complement an FtsZ null mutant. We then tested for dominant-negative effects, expecting that some mutants might poison the wild-type FtsZ substoichiometrically. Surprisingly, none of the mutants showed dominant effects until they were expressed at levels 3 to 5 times that of the genomic wild-type FtsZ. If we assume an FtsZ concentration of 4 μM (discussed above) and a critical concentration of 1 μM, the mutant FtsZ needs to be present at 12 to 20 times the 1 μM concentration of soluble wild-type FtsZ. This indicates a surprisingly weak interaction.
Nine of 10 mutants on the bottom interface were dominant negative at these high expression levels (157). A related study examined various truncated FtsZ proteins (139). Truncation after amino acid 195 permits the N-terminal domain to fold and form a functional top interface, while the C-terminal domain (bottom interface) is nonfunctional. All such truncations, with only the top interface functional, were dominant negative, acting like bottom cap mutants (139).
The top mutants were even more surprising. Seven of eight mutants showed no dominant-negative effects at the highest levels tested. This suggests a directional effect equivalent to treadmilling, where subunits assemble preferentially onto the bottom of the protofilament and disassemble preferentially from the top. When examined in vitro, bottom cap mutants inhibited the GTPase of wild-type FtsZ, but top cap mutants did not (157).
Expression of wild-type FtsZ at comparable levels (3 to 5 times the genomic FtsZ) produced a 1.5-fold increase in the length of cells growing in solution (139) and no reduction in colony size (157). Although Dai and Lutkenhaus (33) reported that cell division was inhibited by 3-fold overexpression of FtsZ, they noted that “the filamentation … became less severe after several passages.” Our observations on cell length were made after growth to log phase in liquid culture from the original colony. Filamentation due to excess FtsZ may depend on strain or growth conditions.
One suggestion from the cap mutant study (157) is that a point mutation on either the top or bottom interface surface substantially weakens the ability of that FtsZ subunit to bind to protofilaments with its supposedly good interface. This means that the binding of a subunit to the end of a protofilament is very weak unless it is itself able to bind the next subunit. This may be related to the mechanism of cooperative assembly. In particular, Miraldi et al. (118) pointed out that in order for their conformational change to produce cooperativity, they had to postulate that both the top and bottom surfaces had to switch simultaneously to the high-affinity form. Our observation that crippling either interface cripples the binding affinity of the other may be related to this, but it has not yet been incorporated into any theory.
Martin-Galiano et al. (111a) have recently identified several hinge points that would permit rotational movements of subdomains of FtsZ. These bending mutations may be related to the conformational changes needed for the high-affinity cooperative binding (111a). The most important next step to advance the conformational-change hypothesis will be to suggest specific contact amino acids that might be involved in the proposed conformational change and then to test them by mutagenesis.
FtsZ AS A FORCE GENERATOR: BENDING PROTOFILAMENTS
The Z-Centric Hypothesis and Reconstitution of Z Rings in Liposomes
In 1997 a “Z-centric hypothesis” proposed that FtsZ forms the cytoskeletal framework of the Z ring and may also generate the constriction force, all by itself (47). The rationale for this second role was that (i) no motor molecules had been identified in bacteria, (ii) some bacteria lacked any homologs of the dozen accessory division proteins identified in E. coli, and (iii) FtsZ protofilaments underwent a conformational change from a straight to a curved conformation that would be capable of generating a force. The curved conformation will be discussed in the next section.
A step toward confirming this hypothesis was the observation that divergent FtsZ from foreign bacteria could replace E. coli FtsZ and function in cell division in E. coli (138). We argued that, because of the extensive sequence divergence, the FtsZ of B. subtilis would not be able to bind any E. coli division protein. We found that B. subtilis FtsZ could function in division in E. coli, but only if we gave it the E. coli C-terminal peptide so it could bind ZipA and FtsA (which recruits the downstream proteins). We also needed to generate a suppressor mutation somewhere in the E. coli genome; these suppressor mutations have not been identified, but they probably facilitate division by a partly crippled FtsZ. We concluded that FtsZ needed to bind only itself and FtsA to function for cell division. This would exclude any role for a motor molecule.
The Z-centric hypothesis was confirmed when we succeeded in reconstituting Z rings in liposomes (137). For this we spliced an amphipathic helix onto the C terminus of FtsZ so it could tether itself to the membrane, eliminating the need for FtsA, which normally provides the tether (146). This membrane-targeted FtsZ, when incorporated inside tubular multilamellar liposomes, assembled multiple Z rings. Figure 5 shows an example where there were initially many dim Z rings, and they generated no visible constrictions on the membrane. Over 6 min the Z rings slid back and forth, collided, and coalesced to make brighter Z rings. The bright Z rings coincided with visible constrictions in the wall of the liposome. Other examples showed deeper constrictions. If the system was allowed to run out of GTP, the constrictions suddenly relaxed (137). FtsZ can therefore assemble a Z ring and generate a constriction force without any other protein. It needs only to associate with itself and be tethered to the membrane.
This reconstitution required two fortuitous events. First was the formation of tubular liposomes. Initially the multilamellar liposomes were mostly spherical, and they aggregated in clumps. Applying the coverslip caused some of the liposomes to transform into a tubular shape, probably by shearing forces. Importantly, many had inside diameters of ∼1 μm, the same as for E. coli. This diameter seems to be optimal for Z-ring assembly. The cylindrical geometry is probably also essential, since we have failed to generate Z rings inside spherical liposomes. The second fortuitous event was that the FtsZ, which was initially on the outside, became incorporated inside many of the liposomes. We have found that about half of the tubular liposomes are permeable or leaky to proteins, and this permits FtsZ and GTP to equilibrate inside. These and other details of the liposome preparation have been described in detail (140).
Two Different Curved Conformations of FtsZ Protofilaments
The early rationale that FtsZ might generate a constriction force all by itself was based on the observation that FtsZ protofilaments had two conformations: straight and highly curved. The highly curved protofilaments formed mini-rings or helical tubes 24 nm in diameter (45a, 105). The miniring contained 16 subunits with a 23-degree bend at each interface. The straight conformation was thought to be favored by bound GTP and the curved conformation by GDP (105). Thus, the hydrolysis of GTP could power the conformational change and generate a bending force.
Evidence has accumulated recently that there is a second curved conformation of FtsZ protofilaments. These have an intermediate curvature, corresponding to a 2.5-degree bend between subunits and producing a curvature of ∼200 nm diameter. Mingorance et al. (117) obtained AFM images of FtsZ protofilaments adsorbed to mica, showing a mixture of straight and curved protofilaments. Hamon et al. (68) obtained AFM images of protofilaments assembled on mica from very dilute FtsZ solutions. The filaments were mostly straight when assembled for only 5 s, but after 30 s they were mostly curved with an ∼160-nm diameter. At later times they formed multiturn spiral structures with an outer diameter of ∼160 nm and inner spirals as small as 80 nm.
Protofilaments with this intermediate curvature have been seen by conventional negative-stain EM (28, 61, 78, 117). These images show protofilaments with an extended contour of uniform curvature, about 200 nm in diameter, mixed with straight protofilaments. Sometimes the curved protofilaments form closed circles. Under some conditions, especially with crowding agents, the curved protofilaments form loose spiral bundles or toroids with a diameter of ∼200 nm (150, 151). Similar toroids were assembled from B. subtilis FtsZ in the presence of calcium (114) or in a low-ionic-strength buffer (120). The antibiotic candidate PC190273 caused FtsZ from B. subtilis and S. aureus FtsZ to associate into very similar toroids and helical bundles (8). Beuria et al. (18) found the intermediate curved conformation when they mixed E. coli FtsZ with a mutant of FtsA (FtsA*, which has enhanced activity). Assembly of this mixture produced protofilaments or sheets with a striking ∼200-nm-diameter curvature.
The intermediate curved conformation seems not to require GTP hydrolysis. It was documented by both negative-stain and atomic force microscopy for assembly supported by GDP-AlF, a nonhydrolyzable analog (117). Circles were also formed by the mutant L68W assembled in EDTA, which blocks GTP hydrolysis (28), and we have found circles assembled by wild-type FtsZ in EDTA (Y. Chen and H. P. Erickson, unpublished observations).
It has been noted that the FtsZ mutant D212G, which hydrolyzes GTP at less than 1% the rate of the wild type, can achieve cell division (36, 125). This requires generation of a second-site mutation somewhere in the E. coli genome (138), but the fact that the cells can divide with a GTPase-dead FtsZ strongly suggests that the energy of GTP hydrolysis is not needed to generate the constriction force (36). Two studies showed that FtsZ-D212G plus DEAE-dextran assembled tubes (125, 192). Since these tubes are a manifestation of the highly curved miniring conformation (105), this suggests that this conformation is not strictly determined by GTP hydrolysis. It may be that the tilt giving this curved conformation can be achieved by removing either the gamma phosphate of GTP or the side chain of the D212 that makes contact with the GTP.
An important question is whether the intermediate curved conformation represents a preferred and rigid conformation or represents the bending by thermal forces of protofilaments that are straight but very flexible. Two studies have assumed the latter scenario and estimated a persistence length (Lp) of 0.18 μm (35) or 0.054 nm (78). Lan et al. (91) noted that these persistence lengths would mean a very flexible protofilament, not strong enough to generate any significant bending force on a membrane. Hörger et al. (72), in contrast, assumed the first scenario and measured the curvature of protofilaments adsorbed on mica, without a prior assumption that the preferred conformation was straight. These filaments were predominantly curved, with a diameter ranging from 80 to 500 nm, with an average of 200 nm. Importantly, these authors measured the deviation from this average curvature and determined a persistence length of ∼4 μm.
In a classic study of the flexural rigidity of protein polymers, Gittes and Howard (57) measured the thermal bending of actin and microtubules. They concluded that both of these polymers had a Young's modulus equivalent to that of hard plastic (Plexiglas). By extension this Young's modulus would apply to globular proteins in general and their polymers. Mickey and Howard (115) estimated the flexural rigidity for a single protofilament of a microtubule to be 1.2 × 10−26 nm2, which corresponds to a persistence length of 2.9 μm. This is remarkably close to the value determined experimentally by Hörger et al. (72).
We have used the small-angle approximation of the beam equation for a cantilevered beam (a rod with one end fixed) to estimate the force that could be generated by a bending protofilament. Based on this approximation, the force needed to bend the free end of the rod a distance y is given by F = (3EI/L3)·y, where E is the Young's modulus, I is the second moment of inertia, and L is the length of the rod. To get an idea of the forces required to bend a 130-nm protofilament into (or out of) the ∼200-nm-diameter intermediate curved conformation, we can consider the protofilament to be fixed in the middle; thus, each half can be modeled as a 65-nm cantilevered beam (Fig. 6). Using 1.4 GPa for the Young's modulus (115), 8.4 × 10−36 m4 for the second moment (6), 65 nm for the length, and 20 nm for the deflection of the end, we estimate a force on the end of each half-protofilament of approximately 2.6 pN. This is a significant amount of force for just one protofilament and is in general agreement with estimations by others that forces on the order of several pN would be required to deform the bacterial membrane (92). This simple calculation is a rough approximation. A more complete model, which would distribute the force to multiple attachment points along the length of protofilament, is in progress.
FIG. 6.
Scale model of protofilament bending. A 130-nm-long protofilament (solid line) is shown fixed at the center (dot) in the straight and the intermediate curved conformations. The two 65-nm halves behave as individual cantilevered beams as the protofilament bends to the radius of the intermediate curved conformation. Arrows show the direction of force (lighter central arrows represent the opposite force that would necessarily push back against the membrane if the protofilament were free).
Allard and Cytrynbaum (6) have formulated a model for generation of a constriction force by protofilament bending, using the flexural rigidity of Plexiglas and assuming that the preferred curvature for FtsZ-GDP subunits was the 24-nm diameter of the miniring. They concluded that protofilament bending could generate a substantial inward-directed constriction force. Our simple analysis in Fig. 6 supports this interpretation. Readers with a background in physics will be interested in two recent papers that examine how protofilaments with an intrinsic curvature are affected by applied bending forces, including the thermal forces acting on a worm-like chain (12, 56).
Evidence That the Constriction Force Is Generated by Bending Protofilaments
There are two general classes of models for how the Z ring generates a constriction force (reviewed in reference 48). One class is based on lateral bonds and proposes that if protofilaments can slide and increase the number of lateral bonds, this would decrease the circumference and constrict the Z ring. The second proposes that protofilaments exert a bending force on the membrane as they are induced to a curved conformation. Recent experiments with liposomes support the bending model (136).
In those experiments, membrane-targeted FtsZ was applied to the outside of liposomes, where it bound and produced visible distortions. Initially the spherical liposomes present a convex surface on the outside. The membrane-targeted FtsZ distorted this surface into multiple concave depressions (Fig. 7 A). The constriction force generating these concave depressions is in the same direction as that of the Z rings inside the liposomes, which bind to a concave surface and constrict to make it more concave.
FIG. 7.
FtsZ on the outside of liposomes produces distortions. (A) Normal membrane-targeted FtsZ, with the membrane tether on the C terminus, forms concave depressions. (B) When the tether is switched to the N terminus, it forms convex protrusions. (C) Diagram showing the curvature and two sides for attaching the membrane targeting sequence (MTS). (Reprinted from reference 136 with permission from Macmillan Publishers Ltd.)
A very informative experiment was to switch the point of attachment of the membrane-targeting amphipathic helix. Normally the FtsA-binding peptide, which we replaced with an amphipathic helix, is at the C terminus of FtsZ. This is at the end of the 50-amino-acid linker (Fig. 2A), which is attached to the globular domain in the center of the “front face,” equivalent to the outside of a microtubule. The N terminus of FtsZ is attached to the opposite face of the subunit, approximately 180 degrees away. We switched the amphipathic helix (and linker) from the C to the N terminus and tested it on the outside of liposomes. This construct formed convex protrusions on the outside of liposomes (Fig. 7B), a bending opposite to the concave depressions formed by the normal C-terminal attachment (136). This is exactly what we would expect if the FtsZ protofilaments have a fixed curvature, with the normal, C-terminal attachment on the outside of the curve. When the membrane attachment was switched to the inside of the curve, it would produce convex protrusions. We do need to remind the reader that the direction of curvature proposed from the FtsZ experiments, where the front face (C terminus) is on the outside of the curve, is the opposite of that proposed for tubulin, where the front face is on the inside of the tubulin rings (see reference 136 for references).
Constriction force models based on the other model, sliding protofilaments, are difficult to reconcile with another aspect of the liposome experiments. In order for sliding filaments to generate a force on the membrane, they would need to be anchored. One possible anchor would be to have the Z ring form a complete circle, with the sliding generated where the ends overlap. Another would be for shorter filaments to be anchored to the rigid peptidoglycan wall. However, the concave and convex distortions of liposomes are generated by partial Z rings, and the fluid lipid bilayer would provide no anchor. Protofilament bending seems the most attractive mechanism for producing the concave depressions and convex protrusions on the outside of liposomes and therefore the constriction force of the Z ring.
Incomplete FtsZ Rings Can Generate Constriction
The initial assembly of FtsZ in newborn daughter cells produces wide-pitch helices, discussed above, but these helices then collapse into a ring. The Z ring usually appears to be a closed circle but occasionally separates into a short-pitch helix (Fig. 1), showing perhaps its true structure. The density of the ring is relatively uniform: if it is bright, it is bright all over; if it is dim, it is dim all over. This applies to Z rings both in bacteria (Fig. 1) and in liposomes (Fig. 5). This raises the question of whether FtsZ needs to form a complete circle in order to generate a constriction.
An early indication that closed circular rings are not essential was the study of the ftsZ26 mutant (5). This mutant forms normal-looking Z rings about 50% of the time, but the other 50% are distinctly spirals. When examined by scanning EM, many bacteria showed spiral constrictions, suggesting that the spiral Z rings were capable of generating a constriction that matched their shape. Similar twisted septa have been shown recently for the B. subtilis mutant TS1 (120). The earlier study also showed some examples of partial Z rings or arcs in cells that had a mutation causing them to grow as spheres. A recent study of MreBCD depletion mutants growing as large spheres provides convincing evidence for asymmetric constriction, with the constriction located at an arc of FtsZ (11).
A dramatic example of asymmetric septation is seen in the division of the cyanelle, a primitive plastid of the protist Cyanophora paradoxa (165, 166). The Z ring extends from one-fourth to one-half way around the cell and is located over a very asymmetrically forming septum (Fig. 8).
FIG. 8.
Asymmetric division of isolated cyanelle plastids. (A) Three cyanelles imaged by scanning EM, showing furrows progressing from shallow (arrow) to deep (double arrowhead). (B to D) Increasingly deep furrows correspond to increasing length of the FtsZ arc. Row 1 shows Nomarski images, row 2 shows the cytoplasm imaged by autofluorescence of chlorophyll and phycobilin, row 3 shows immunofluorescence staining of FtsZ, and row 4 shows overlap of rows 2 and 3. (Reprinted from reference 165 with kind permission from Springer Science+Business Media.)
In all of these examples, including the cyanelle, there is a peptidoglycan cell wall, and the generation of a constriction force could involve coupling of the Z ring to the rigid wall. However, the concave depressions on the outside of liposomes demonstrate that peptidoglycan is not necessary for generating force on the membrane. This simple system shows that an incomplete arc of protofilaments is capable of generating a bending force on a fluid membrane.
Finally, we should note one major problem for a simple model of bending protofilaments. These are thought to be tethered to the membrane by attaching to FtsA (Fig. 9 A and B) and ZipA (not shown). In both cases the C-terminal peptide that binds FtsA and ZipA is at the end of a 50-amino-acid segment that is thought to be a flexible linker (Fig. 2A). This has a contour length of 17 nm and a relaxed end-to-end distance of 5 nm. ZipA has an additional 150-amino-acid tether between the FtsZ-binding domain and the membrane (131). If these tethers are flexible, then why would a short protofilament, with a preferred curvature, not simply roll 90 degrees and curve in the plane of the membrane? Some additional structural feature is needed to limit the rolling of the protofilament and maintain its plane of curvature perpendicular to the membrane.
FIG. 9.
Bending protofilaments can only constrict so far. (A) An FtsZ protofilament is shown tethered to the membrane of an undivided cell. The blue FtsZ subunits are connected into a protofilament that has a preferred bend. About one out of five FtsZ proteins are shown tethered to an FtsA, indicated by an orange circle. An amphipathic helix extends from FtsA into the membrane bilayer. The FtsZ tether is shown extended to ∼10 nm at the ends of the protofilament, where the bending toward the cell center is maximal. Near the middle of the protofilament it is pushing outward on the membrane, and the tethers are compressed. (B) When fully in the curved conformation, the 24-nm miniring plus an ∼10-nm tether plus FtsA could constrict the membrane to about a 57-nm outside diameter. (C) The diameters of the undivided cell, the intermediate curved conformation, and the miniring are compared.
Finishing Division: Membrane Scission without FtsZ
It is now thought that the FtsZ protofilament has three preferred conformations: straight, intermediate curved (200-nm diameter, 2.5-degree bend per subunit) and highly curved (24-nm diameter, 23-degree bend per subunit). Although we initially focused on the highly curved (miniring) conformation, which seemed to be coupled to GTP hydrolysis (50, 105), there is accumulating evidence that the intermediate curved conformation may play the most important role. However, neither curved conformation can pull the membrane to a complete scission. We suggest here that the scission might be completed by membrane fluctuations.
The membrane of an undivided cell has a diameter of 1,000 nm. A protofilament attached to this slightly curved membrane would need a bend of 0.5 degrees per FtsZ, which is close to the straight conformation. A straight protofilament would tend to align along the axis of the cell, but if it had any inherent curvature it would tend to align circumferentially (9). Initially the protofilaments would not be able to bend because they are tethered to the approximately straight membrane (Fig. 9A). However, if their preferred conformation is curved, the protofilaments will generate a bending force on the membrane. As long as the membrane is attached to the rigid peptidoglycan wall, the force will not result in constriction. However, as the Z ring recruits transmembrane proteins that remodel the wall, it will be able to follow the constricting membrane, initiating septation.
The highly curved conformation could pull the septum to a diameter of ∼50 nm (24 nm for the outside of the ring plus ∼8 to 12 nm on each side for the 50-amino-acid tether plus 5 nm for FtsA [Fig. 9B]). If the intermediate curved conformation is producing the force, it could pull the septum to a diameter of ∼250 nm. In neither case would the curved FtsZ protofilaments be able to pull the membrane to complete closure and scission. How is the division process completed?
One possibility is that the remodeling of the cell wall, which initially followed the membrane invagination produced by FtsZ, eventually becomes a positive force pushing the septum toward closure. Favoring this possibility, Joseleau-Petit et al. (88) reported that unstable L forms of E. coli, which they could generate by overnight growth in cefsulodin, retained about 7% of the wild-type level of peptidoglycan. Moreover, they required peptidoglycan synthesis to grow and divide. This suggested that the remodeling of the peptidoglycan might be powering division, at least partly, by pushing from without. Another possibility is that the constriction of the Z ring is regulated by feedback from the transmembrane proteins of peptidoglycan synthesis.
In contrast to these L forms, Leaver et al. (95) created L forms of B. subtilis and provided convincing evidence that peptidoglycan was completely absent. One might have hoped to use these L forms to observe how FtsZ could divide cells having only a plasma membrane. However, Leaver et al. went on to show that FtsZ was not used for division and was completely dispensable for growth of the L forms. The cells sometimes appeared to divide by extruding membrane buds from the large spherical mother cell. Another mechanism, probably related, involved extrusion of a pseudopod-like extension, which then fragmented into small spherical bodies, with at least some presumably containing a complete nucleoid. The authors suggested that extrusion likely involved a force produced by a cytoskeletal element; an active chromosome segregation system was one possibility. This force would form the extended pseudopod, and resolution might then be achieved by collapse and resealing of membranes.
Perhaps related to the mechanism of division of this L form is the existence of some bacteria that can divide without FtsZ. The entire phylum of Crenarcheota has no FtsZ, but these archaea are now known to use a completely different system, which is related to the eukaryotic ESCRT system (99, 163). However, there are no ESCRT proteins in eubacteria. The bacterial phyla Chlamydiaceae and Planctomycetes have no FtsZ. Pilhofer et al. (148) have noted that several genes of the dcw cluster are present in various species of these phyla, and they suggested that these organisms descended from an ancestor that had FtsZ and have lost it. At least two Mycoplasma species, Ureaplasma urealyticum (58) and Mycoplasma mobile (83), have no FtsZ. A recent study showed that Mycoplasma genitalium can grow and divide when its ftsZ is deleted (48a, 100). These organisms grow as gliding cells attached to a substrate and apparently divide by “traction-mediated cytofission,” where a cell develops two gliding organelles that move in opposite directions and pull the cell apart. Mycoplasma species without ftsZ probably use the same mechanism.
If these bacteria and the B. subtilis L form can divide without FtsZ, perhaps the same mechanism might apply to the normal process of completing scission. More specifically, we suggest that membrane collapse and scission may occur as natural fluctuations of the membrane, independent of force from cytoskeleton or motor molecules (48a). In support of this, Bendezú and de Boer (11) reported that defects in the mre or mrd pathways caused cells to lose their rod shape and become round, and the round cells accumulated membrane-enclosed vesicles inside. The large round cells had an excess of lipid bilayer, and the formation and excision of vesicles might occur to accommodate the excess. We suggest that related membrane fluctuations may drive the final steps of septum closure and scission. FtsZ protofilament curvature would then be responsible for bringing the septum to a 200- or 50-nm diameter, with the final step of scission completed by membrane fluctuations.
In a theoretical study, Zhang and Robinson (204) concluded that differential tension and pressure of the membrane at the poles and center of the cell can drive constriction in the complete absence of a contractile machine. In an in vitro system, Yanagisawa et al. (201) showed that liposomes prepared with lipids with different melting temperatures could be induced to form different shapes in response to osmotic pressure. Phase separation following a temperature downshift could cause them to bud spherical vesicles. In one example, a long tubular liposome resolved into a necklace of beaded domains, resembling somewhat the resolving pseudopod of the B. subtilis L form (95). Finally, we should note the progress of Szostak and colleagues in creating artificial “protocells” (205). Fatty acid vesicles fed with micelles developed an elongated tubular morphology. Then, upon mild shear, the tubules divided into small spherical vesicles. The growth and division could be repeated, mimicking the growth and division of cells. These in vitro results show that membrane growth and fluctuations can generate effects related to cytokinesis without the involvement of proteins. Similar mechanisms may play a role in cytokinesis in cells, especially the final scission event.
FUTURE DIRECTIONS
The field needs a more definitive model for the substructure of the Z ring. At present there is support for a model of overlapping short protofilaments (Fig. 4A) but also some indication that protofilaments may anneal into much longer filaments (Fig. 4B). A promising new approach is superresolution light microscopy, especially photoactivation localization microscopy (PALM) (16, 174). PALM can achieve a resolution of 20 to 30 nm, which might be sufficient to resolve the protofilaments in the Z ring. A particular advantage of PALM is that the protofilaments are colored red by the fused fluorophore, and there is no noise from unrelated cytoplasmic structures. There are many technical hurdles, but the study of the structure of the Z ring would seem to be one of the most attractive applications for PALM.
An enduring enigma from in vitro studies is how the one-subunit-thick protofilaments can exhibit cooperative assembly. The proposals for a conformational change seem attractive, but we now need suggestions for specific amino acids or domains that might be involved. The inhibition of assembly by SulA and MinC may be related to the mechanism of cooperative assembly. SulA and MinC now bring their own enigma, i.e., that they inhibit assembly only when subunits are hydrolyzing GTP and undergoing the GTPase-dependent cycling. An important step in understanding the mechanisms will be to determine the association and kinetic constants for FtsZ binding to these inhibitors.
We know very little about the mechanism of GTP hydrolysis and related subunit exchange. Does hydrolysis in the middle of a protofilament lead to fragmentation, or do GDP subunits dissociate only when they are at the end? What are the time course and molecular mechanism for GTP hydrolysis after a subunit enters a protofilament? Do protofilament dynamics involve a mechanism like dynamic instability, where the protofilament has excursions of growth and disassembly? The mechanism of microtubule dynamic instability is thought to be explained by a GTP cap, but we really do not know what determines the dynamics inside the cap. There is some hope that understanding the dynamics of FtsZ may explain the GTP cap of microtubules.
Single-molecule fluorescence microscopy of FtsZ protofilaments may be a useful approach, but the small size of the protofilaments, which is less than the resolution of the light microscope, will pose formidable challenges.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM66014.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Aarsman, M. E., A. Piette, C. Fraipont, T. M. Vinkenvleugel, M. Nguyen-Disteche, and T. den Blaauwen. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55:1631-1645. [DOI] [PubMed] [Google Scholar]
- 2.Adams, D. W., and J. Errington. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642-653. [DOI] [PubMed] [Google Scholar]
- 3.Addinall, S. G., E. F. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. Temperature shift experiments with ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 179:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 6.Allard, J. F., and E. N. Cytrynbaum. 2009. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc. Natl. Acad. Sci. U. S. A. 106:145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson, D. E., F. J. Gueiros-Filho, and H. P. Erickson. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreu, J. M., C. Schaffner-Barbero, S. Huecas, D. Alonso, M. L. Lopez-Rodriguez, L. B. Ruiz-Avila, R. Nunez-Ramirez, O. Llorca, and A. J. Martin-Galiano. 2010. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J. Biol. Chem. 285:14239-14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews, S. S., and A. P. Arkin. 2007. A mechanical explanation for cytoskeletal rings and helices in bacteria. Biophys. J. 93:1872-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beall, B., and J. Lutkenhaus. 1992. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J. Bacteriol. 174:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendezu, F. O., C. A. Hale, T. G. Bernhardt, and P. A. de Boer. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28:193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benetatos, P., and E. M. Terentjev. 2010. Stretching weakly bending filaments with spontaneous curvature in two dimensions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 81:031802. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 14.Bernard, C. S., M. Sadasivam, D. Shiomi, and W. Margolin. 2007. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 64:1289-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernhardt, T. G., and P. A. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betzig, E., G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz, and H. F. Hess. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642-1645. [DOI] [PubMed] [Google Scholar]
- 17.Beuria, T. K., S. S. Krishnakumar, S. Sahar, N. Singh, K. Gupta, M. Meshram, and D. Panda. 2003. Glutamate-induced assembly of bacterial cell division protein FtsZ. J. Biol. Chem. 278:3735-3741. [DOI] [PubMed] [Google Scholar]
- 18.Beuria, T. K., S. Mullapudi, E. Mileykovskaya, M. Sadasivam, W. Dowhan, and W. Margolin. 2009. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J. Biol. Chem. 284:14079-14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beuria, T. K., M. K. Santra, and D. Panda. 2005. Sanguinarine blocks cytokinesis in bacteria by inhibiting FtsZ assembly and bundling. Biochemistry 44:16584-16593. [DOI] [PubMed] [Google Scholar]
- 20.Beuria, T. K., P. Singh, A. Surolia, and D. Panda. 2009. Promoting assembly and bundling of FtsZ as a strategy to inhibit bacterial cell division: a new approach for developing novel antibacterial drugs. Biochem. J. 423:61-69. [DOI] [PubMed] [Google Scholar]
- 21.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 23.Bramkamp, M., R. Emmins, L. Weston, C. Donovan, R. A. Daniel, and J. Errington. 2008. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556-1569. [DOI] [PubMed] [Google Scholar]
- 24.Burgess, S. A., M. L. Walker, K. Thirumurugan, J. Trinick, and P. J. Knight. 2004. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 147:247-258. [DOI] [PubMed] [Google Scholar]
- 25.Camberg, J. L., J. R. Hoskins, and S. Wickner. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 106:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cayley, S., B. A. Lewis, H. J. Guttman, and M. T. Record Jr. 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity: implications for protein-DNA interactions in vivo. J. Mol. Biol. 222:281-300. [DOI] [PubMed] [Google Scholar]
- 27.Chen, Y., D. E. Anderson, M. Rajagopalan, and H. P. Erickson. 2007. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 282:27736-27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, Y., K. Bjornson, S. D. Redick, and H. P. Erickson. 2005. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 88:505-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, Y., and H. P. Erickson. 2009. FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry 48:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, Y., and H. P. Erickson. 2005. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 280:22549-22554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordell, S. C., R. E. Anderson, and J. Löwe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordell, S. C., E. J. Robinson, and J. Lowe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. U. S. A. 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai, K., A. Mukherjee, Y. Xu, and J. Lutkenhaus. 1994. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J. Bacteriol. 176:130-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dajkovic, A., G. Lan, S. X. Sun, D. Wirtz, and J. Lutkenhaus. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18:235-244. [DOI] [PubMed] [Google Scholar]
- 36.Dajkovic, A., and J. Lutkenhaus. 2006. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 11:140-151. [DOI] [PubMed] [Google Scholar]
- 37.Dajkovic, A., A. Mukherjee, and J. Lutkenhaus. 2008. Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J. Bacteriol. 190:2513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta, P., A. Dasgupta, S. Bhakta, and J. Basu. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 277:24983-24987. [DOI] [PubMed] [Google Scholar]
- 39.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 40.den Blaauwen, T., N. Buddelmeijer, M. E. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ Assembly in Escherichia coli. J. Bacteriol. 181:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Din, N., E. M. Quardokus, M. J. Sackett, and Y. V. Brun. 1998. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 27:1051-1063. [DOI] [PubMed] [Google Scholar]
- 42.Domadia, P., S. Swarup, A. Bhunia, J. Sivaraman, and D. Dasgupta. 2007. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 74:831-840. [DOI] [PubMed] [Google Scholar]
- 43.Domadia, P. N., A. Bhunia, J. Sivaraman, S. Swarup, and D. Dasgupta. 2008. Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47:3225-3234. [DOI] [PubMed] [Google Scholar]
- 44.Dziadek, J., M. V. Madiraju, S. A. Rutherford, M. A. Atkinson, and M. Rajagopalan. 2002. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology 148:961-971. [DOI] [PubMed] [Google Scholar]
- 45.Erickson, H. P. 1998. Atomic structures of tubulin and FtsZ. Trends Cell Biol. 8:133-137. [DOI] [PubMed] [Google Scholar]
- 45a.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U.S.A. 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erickson, H. P. 2007. Evolution of the cytoskeleton. Bioessays 29:668-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erickson, H. P. 1997. FtsZ, a tubulin homolog, in prokaryote cell division. Trends Cell Biol. 7:362-367. [DOI] [PubMed] [Google Scholar]
- 48.Erickson, H. P. 2009. Modeling the physics of FtsZ assembly and force generation. Proc. Natl. Acad. Sci. U. S. A. 106:9238-9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Erickson, H. P., and M. Osawa. 2010. Cell division without FtsZ—a variety of redundant mechanisms. Mol. Microbiol. 78:267-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erickson, H. P., and D. Pantaloni. 1981. The role of subunit entropy in cooperative assembly. Nucleation of microtubules and other two-dimensional polymers. Biophys.J. 34:293-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U. S. A. 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esue, O., Y. Tseng, and D. Wirtz. 2005. The rapid onset of elasticity during the assembly of the bacterial cell-division protein FtsZ. Biochem. Biophys. Res. Commun. 333:508-516. [DOI] [PubMed] [Google Scholar]
- 52.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 53.Froschauer, E. M., M. Kolisek, F. Dieterich, M. Schweigel, and R. J. Schweyen. 2004. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol. Lett. 237:49-55. [DOI] [PubMed] [Google Scholar]
- 54.Gamba, P., J. W. Veening, N. J. Saunders, L. W. Hamoen, and R. A. Daniel. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 191:4186-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geissler, B., D. Shiomi, and W. Margolin. 2007. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh, S. K., K. Singh, and A. Sain. 2009. Effect of intrinsic curvature on semiflexible polymers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 80:051904. [DOI] [PubMed] [Google Scholar]
- 57.Gittes, F., B. Mickey, J. Nettleton, and J. Howard. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 59.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez, J. M., M. Jimenez, M. Velez, J. Mingorance, J. M. Andreu, M. Vicente, and G. Rivas. 2003. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 278:37664-37671. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez, J. M., M. Velez, M. Jimenez, C. Alfonso, P. Schuck, J. Mingorance, M. Vicente, A. P. Minton, and G. Rivas. 2005. Cooperative behavior of Escherichia coli cell-division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc. Natl. Acad. Sci. U. S. A. 102:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grantcharova, N., U. Lustig, and K. Flardh. 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 187:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregory, J. A., E. C. Becker, and K. Pogliano. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22:3475-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guberman, J. M., A. Fay, J. Dworkin, N. S. Wingreen, and Z. Gitai. 2008. PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput. Biol. 4:e1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haeusser, D. P., R. L. Schwartz, A. M. Smith, M. E. Oates, and P. A. Levin. 2004. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52:801-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hale, C. A., A. C. Rhee, and P. A. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamon, L., D. Panda, P. Savarin, V. Joshi, J. Bernhard, E. Mucher, A. Mechulam, P. A. Curmi, and D. Pastre. 2009. Mica surface promotes the assembly of cytoskeletal proteins. Langmuir 25:3331-3335. [DOI] [PubMed] [Google Scholar]
- 69.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precison. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 70.Haydon, D. J., N. R. Stokes, R. Ure, G. Galbraith, J. M. Bennett, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington, and L. G. Czaplewski. 2008. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321:1673-1675. [DOI] [PubMed] [Google Scholar]
- 71.Horger, I., F. Campelo, A. Hernandez-Machado, and P. Tarazona. 2010. Constricting force of filamentary protein rings evaluated from experimental results. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 81:031922. [DOI] [PubMed] [Google Scholar]
- 72.Horger, I., E. Velasco, J. Mingorance, G. Rivas, P. Tarazona, and M. Velez. 2008. Langevin computer simulations of bacterial protein filaments and the force-generating mechanism during cell division. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 77:011902. [DOI] [PubMed] [Google Scholar]
- 73.Horger, I., E. Velasco, G. Rivas, M. Velez, and P. Tarazona. 2008. FtsZ bacterial cytoskeletal polymers on curved surfaces: the importance of lateral interactions. Biophys. J. 94:L81-L83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu, Z. L., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu, Z. L., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. U. S. A. 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang, Q., F. Kirikae, T. Kirikae, A. Pepe, A. Amin, L. Respicio, R. A. Slayden, P. J. Tonge, and I. Ojima. 2006. Targeting FtsZ for antituberculosis drug discovery: noncytotoxic taxanes as novel antituberculosis agents. J. Med. Chem. 49:463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huecas, S., and J. M. Andreu. 2004. Polymerization of nucleotide-free, GDP- and GTP-bound cell division protein FtsZ: GDP makes the difference. FEBS Lett. 569:43-48. [DOI] [PubMed] [Google Scholar]
- 78.Huecas, S., O. Llorca, J. Boskovic, J. Martin-Benito, J. M. Valpuesta, and J. M. Andreu. 2008. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys. J. 94:1796-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huecas, S., C. Schaffner-Barbero, W. Garcia, H. Yebenes, J. M. Palacios, J. F. Diaz, M. Menendez, and J. M. Andreu. 2007. The interactions of cell division protein FtsZ with guanine nucleotides. J. Biol. Chem. 282:37515-37528. [DOI] [PubMed] [Google Scholar]
- 80.Inoue, I., R. Ino, and A. Nishimura. 2009. New model for assembly dynamics of bacterial tubulin in relation to the stages of DNA replication. Genes Cells 14:435-444. [DOI] [PubMed] [Google Scholar]
- 81.Ishikawa, S., Y. Kawai, K. Hiramatsu, M. Kuwano, and N. Ogasawara. 2006. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60:1364-1380. [DOI] [PubMed] [Google Scholar]
- 82.Ito, H., A. Ura, Y. Oyamada, A. Tanitame, H. Yoshida, S. Yamada, M. Wachi, and J. Yamagishi. 2006. A 4-aminofurazan derivative—A189—inhibits assembly of bacterial cell division protein FtsZ in vitro and in vivo. Microbiol. Immunol. 50:759-764. [DOI] [PubMed] [Google Scholar]
- 83.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaiswal, R., T. K. Beuria, R. Mohan, S. K. Mahajan, and D. Panda. 2007. Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry 46:4211-4220. [DOI] [PubMed] [Google Scholar]
- 85.Jensen, S. O., L. S. Thompson, and E. J. Harry. 2005. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-ring assembly. J. Bacteriol. 187:6536-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson, J. E., L. L. Lackner, C. A. Hale, and P. A. de Boer. 2004. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J. Bacteriol. 186:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joseleau-Petit, D., J. C. Liebart, J. A. Ayala, and R. D'Ari. 2007. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J. Bacteriol. 189:6512-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kubitschek, H. E. 1986. Increase of cell mass during the division cycle of Escherichia coli B/rA. J. Bacteriol. 168:613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lan, G., A. Dajkovic, D. Wirtz, and S. X. Sun. 2008. Polymerization and bundling kinetics of FtsZ filaments. Biophys. J. 95:4045-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lan, G., B. R. Daniels, T. M. Dobrowsky, D. Wirtz, and S. X. Sun. 2009. Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 106:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lan, G., C. W. Wolgemuth, and S. X. Sun. 2007. Z-ring force and cell shape during division in rod-like bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:16110-16115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lappchen, T., V. A. Pinas, A. F. Hartog, G. J. Koomen, C. Schaffner-Barbero, J. M. Andreu, D. Trambaiolo, J. Lowe, A. Juhem, A. V. Popov, and T. den Blaauwen. 2008. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem. Biol. 15:189-199. [DOI] [PubMed] [Google Scholar]
- 94.Lara, B., A. I. Rico, S. Petruzzelli, A. Santona, J. Dumas, J. Biton, M. Vicente, J. Mingorance, and O. Massidda. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55:699-711. [DOI] [PubMed] [Google Scholar]
- 95.Leaver, M., P. Dominguez-Cuevas, J. M. Coxhead, R. A. Daniel, and J. Errington. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849-853. [DOI] [PubMed] [Google Scholar]
- 96.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levin, P. A., and R. Losick. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10:478-488. [DOI] [PubMed] [Google Scholar]
- 98.Li, Z., M. J. Trimble, Y. V. Brun, and G. J. Jensen. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lindas, A. C., E. A. Karlsson, M. T. Lindgren, T. J. Ettema, and R. Bernander. 2008. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U. S. A. 105:18942-18946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lluch-Senar, M., E. Querol, and J. Pinol. 2010. Cell division in a minimal bacterium in the absence of ftsZ. Mol. Microbiol. 78:278-289. [DOI] [PubMed] [Google Scholar]
- 101.Low, H. H., M. C. Moncrieffe, and J. Lowe. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341:839-852. [DOI] [PubMed] [Google Scholar]
- 102.Lowe, J., and L. A. Amos. 2009. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int. J. Biochem. Cell Biol. 41:323-329. [DOI] [PubMed] [Google Scholar]
- 103.Löwe, J., and L. A. Amos. 2000. Helical tubes of FtsZ from Methanococcus jannaschii. Biol. Chem. 381:993-999. [DOI] [PubMed] [Google Scholar]
- 104.Löwe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu, C., M. Reedy, and H. P. Erickson. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskel. 40:71-86. [DOI] [PubMed] [Google Scholar]
- 107.Lutkenhaus, J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539-562. [DOI] [PubMed] [Google Scholar]
- 108.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Margalit, D. N., L. Romberg, R. B. Mets, A. M. Hebert, T. J. Mitchison, M. W. Kirschner, and D. RayChaudhuri. 2004. Targeting cell division: small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc. Natl. Acad. Sci. U. S. A. 101:11821-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111a.Martin-Galiano, A. J., R. M. Buey, M. Cabezas, and J. M. Andreu. 2010. Mapping flexibility and the assembly switch of cell division protein FtsZ by computational and mutational approaches. J. Biol. Chem. 285: 22554-22565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McGuffee, S. R., and A. H. Elcock. 2010. Diffusion, crowding and protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 6:e1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mendieta, J., A. I. Rico, E. Lopez-Vinas, M. Vicente, J. Mingorance, and P. Gomez-Puertas. 2009. Structural and functional model for ionic (K(+)/Na(+)) and pH dependence of GTPase activity and polymerization of FtsZ, the prokaryotic ortholog of tubulin. J. Mol. Biol. 390:17-25. [DOI] [PubMed] [Google Scholar]
- 114.Michie, K. A., L. G. Monahan, P. L. Beech, and E. J. Harry. 2006. Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J. Bacteriol. 188:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mickey, B., and J. Howard. 1995. Rigidity of microtubules is increased by stabilizing agents. J. Cell Biol. 130:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mingorance, J., S. Rueda, P. Gomez-Puertas, A. Valencia, and M. Vicente. 2001. Escherichia coli FtsZ polymers contain mostly GTP and have a high nucleotide turnover. Mol. Microbiol. 41:83-91. [DOI] [PubMed] [Google Scholar]
- 117.Mingorance, J., M. Tadros, M. Vicente, J. M. Gonzalez, G. Rivas, and M. Velez. 2005. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280:20909-20914. [DOI] [PubMed] [Google Scholar]
- 118.Miraldi, E. R., P. J. Thomas, and L. Romberg. 2008. Allosteric models for cooperative polymerization of linear polymers. Biophys. J. 95:2470-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mohammadi, T., G. E. Ploeger, J. Verheul, A. D. Comvalius, A. Martos, C. Alfonso, J. van Marle, G. Rivas, and T. den Blaauwen. 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48:11056-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Monahan, L. G., A. Robinson, and E. J. Harry. 2009. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol. Microbiol. 74:1004-1017. [DOI] [PubMed] [Google Scholar]
- 121.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moy, F. J., E. Glasfeld, L. Mosyak, and R. Powers. 2000. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry 39:9146-9156. [DOI] [PubMed] [Google Scholar]
- 123.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mukherjee, A., and J. Lutkenhaus. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17:462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mukherjee, A., C. Saez, and J. Lutkenhaus. 2001. Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J. Bacteriol. 183:7190-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Niu, L., and J. Yu. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys. J. 95:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nogales, E., K. H. Downing, L. A. Amos, and J. Lowe. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5:451-458. [DOI] [PubMed] [Google Scholar]
- 128.Nogales, E., M. Whittaker, R. A. Milligan, and K. H. Downing. 1999. High-resolution model of the microtubule. Cell 96:79-88. [DOI] [PubMed] [Google Scholar]
- 129.O'Brien, E. T., and H. P. Erickson. 1989. Assembly of pure tubulin in the absence of free GTP: effect of magnesium, glycerol, ATP, and the nonhydrolyzable GTP analogues. Biochemistry 28:1413-1422. [DOI] [PubMed] [Google Scholar]
- 130.Ohashi, T., S. D. Galiacy, G. Briscoe, and H. P. Erickson. 2007. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 16:1429-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ohashi, T., C. A. Hale, P. A. De Boer, and H. P. Erickson. 2002. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J. Bacteriol. 184:4313-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ohashi, Y., Y. Chijiiwa, K. Suzuki, K. Takahashi, H. Nanamiya, T. Sato, Y. Hosoya, K. Ochi, and F. Kawamura. 1999. The lethal effect of a benzamide derivative, 3-methoxybenzamide, can be suppressed by mutations within a cell division gene, ftsZ, in Bacillus subtilis. J. Bacteriol. 181:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oliva, M. A., S. C. Cordell, and J. Lowe. 2004. Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11:1243-1250. [DOI] [PubMed] [Google Scholar]
- 134.Oliva, M. A., S. Huecas, J. M. Palacios, J. Martin-Benito, J. M. Valpuesta, and J. M. Andreu. 2003. Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J. Biol. Chem. 278:33562-33570. [DOI] [PubMed] [Google Scholar]
- 135.Oliva, M. A., D. Trambaiolo, and J. Lowe. 2007. Structural insights into the conformational variability of FtsZ. J. Mol. Biol. 373:1229-1242. [DOI] [PubMed] [Google Scholar]
- 136.Osawa, M., D. E. Anderson, and H. P. Erickson. 2009. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28:3476-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Osawa, M., D. E. Anderson, and H. P. Erickson. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Osawa, M., and H. P. Erickson. 2006. FtsZ from divergent foreign bacteria can function for cell division in Escherichia coli. J. Bacteriol. 188:7132-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Osawa, M., and H. P. Erickson. 2005. Probing the domain structure of FtsZ by random truncation and insertion of GFP. Microbiology 151:4033-4043. [DOI] [PubMed] [Google Scholar]
- 140.Osawa, M., and H. P. Erickson. 2009. Tubular liposomes with variable permeability for reconstitution of FtsZ rings. Methods Enzymol. 464:3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paez, A., P. Mateos-Gil, I. Horger, J. Mingorance, G. Rivas, M. Vicente, M. Velez, and P. Tarazona. 2009. Simple modeling of FtsZ polymers on flat and curved surfaces: correlation with experimental in vitro observations. PMC Biophys. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paradis-Bleau, C., M. Beaumont, F. Sanschagrin, N. Voyer, and R. C. Levesque. 2007. Parallel solid synthesis of inhibitors of the essential cell division FtsZ enzyme as a new potential class of antibacterials. Bioorg. Med. Chem. 15:1330-1340. [DOI] [PubMed] [Google Scholar]
- 143.Peters, P. C., M. D. Migocki, C. Thoni, and E. J. Harry. 2007. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol. Microbiol. 64:487-499. [DOI] [PubMed] [Google Scholar]
- 144.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pichoff, S., and J. Lutkenhaus. 2007. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol. 64:1129-1138. [DOI] [PubMed] [Google Scholar]
- 146.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 147.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pilhofer, M., K. Rappl, C. Eckl, A. P. Bauer, W. Ludwig, K. H. Schleifer, and G. Petroni. 2008. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J. Bacteriol. 190:3192-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pla, J., M. Sanchez, P. Palacios, M. Vicente, and M. Aldea. 1991. Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol.Microbiol. 5:1681-1686. [DOI] [PubMed] [Google Scholar]
- 150.Popp, D., M. Iwasa, H. P. Erickson, A. Narita, Y. Maeda, and R. C. Robinson. 2010. Suprastructures and dynamic properties of mycobacterium tuberculosis FtsZ. J. Biol. Chem. 285:11281-11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Popp, D., M. Iwasa, A. Narita, H. P. Erickson, and Y. Maeda. 2009. FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91:340-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Popp, D., A. Narita, M. Iwasa, Y. Maeda, and R. C. Robinson. 2010. Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem. Biophys. Res. Commun. 391:1598-1603. [DOI] [PubMed] [Google Scholar]
- 153.Quardokus, E. M., N. Din, and Y. V. Brun. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39:949-959. [DOI] [PubMed] [Google Scholar]
- 154.Rai, D., J. K. Singh, N. Roy, and D. Panda. 2008. Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem. J. 410:147-155. [DOI] [PubMed] [Google Scholar]
- 155.Rajagopalan, M., E. Maloney, J. Dziadek, M. Poplawska, H. Lofton, A. Chauhan, and M. V. Madiraju. 2005. Genetic evidence that mycobacterial FtsZ and FtsW proteins interact, and colocalize to the division site in Mycobacterium smegmatis. FEMS Microbiol. Lett. 250:9-17. [DOI] [PubMed] [Google Scholar]
- 156.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Redick, S. D., J. Stricker, G. Briscoe, and H. P. Erickson. 2005. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Reshes, G., S. Vanounou, I. Fishov, and M. Feingold. 2008. Cell shape dynamics in Escherichia coli. Biophys. J. 94:251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rivas, G., A. Lopez, J. Mingorance, M. J. Ferrandiz, S. Zorrilla, A. P. Minton, M. Vicente, and J. M. Andreu. 2000. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer—the primary steps for FtsZ assembly. J. Biol. Chem. 275:11740-11749. [DOI] [PubMed] [Google Scholar]
- 160.Romberg, L., and T. J. Mitchison. 2004. Rate-limiting guanosine 5′-triphosphate hydrolysis during nucleotide turnover by FtsZ, a prokaryotic tubulin homologue involved in bacterial cell division. Biochemistry 43:282-288. [DOI] [PubMed] [Google Scholar]
- 161.Romberg, L., M. Simon, and H. P. Erickson. 2001. Polymerization of FtsZ, a bacterial homolog of tubulin. Is assembly cooperative? J. Biol. Chem. 276:11743-11753. [DOI] [PubMed] [Google Scholar]
- 162.Rueda, S., M. Vicente, and J. Mingorance. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Samson, R. Y., T. Obita, S. M. Freund, R. L. Williams, and S. D. Bell. 2008. A role for the ESCRT system in cell division in archaea. Science 322:1710-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Santra, M. K., T. K. Beuria, A. Banerjee, and D. Panda. 2004. Ruthenium red-induced bundling of bacterial cell division protein, FtsZ. J. Biol. Chem. 279:25959-25965. [DOI] [PubMed] [Google Scholar]
- 165.Sato, M., Y. Mogi, T. Nishikawa, S. Miyamura, T. Nagumo, and S. Kawano. 2009. The dynamic surface of dividing cyanelles and ultrastructure of the region directly below the surface in Cyanophora paradoxa. Planta 229:781-791. [DOI] [PubMed] [Google Scholar]
- 166.Sato, M., T. Nishikawa, H. Kajitani, and S. Kawano. 2007. Conserved relationship between FtsZ and peptidoglycan in the cyanelles of Cyanophora paradoxa similar to that in bacterial cell division. Planta 227:177-187. [DOI] [PubMed] [Google Scholar]
- 167.Scheffers, D. J. 2008. The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett. 582:2601-2608. [DOI] [PubMed] [Google Scholar]
- 168.Scheffers, D. J., J. G. de Wit, T. den Blaauwen, and A. J. Driessen. 2002. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41:521-529. [DOI] [PubMed] [Google Scholar]
- 169.Scheffers, D. J., T. Den Blaauwen, and A. J. M. Driessen. 2000. Non-hydrolysable GTP-gamma-S stabilizes the FtsZ polymer in a GDP-bound state. Mol. Microbiol. 35:1211-1219. [DOI] [PubMed] [Google Scholar]
- 170.Shen, B., and J. Lutkenhaus. 2009. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol. Microbiol. 72:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shen, B., and J. Lutkenhaus. 2010. Examination of the interaction between FtsZ and MinC in E. coli suggests how MinC disrupts Z rings. Mol. Microbiol. 75:1285-1298. [DOI] [PubMed] [Google Scholar]
- 172.Shiomi, D., and W. Margolin. 2007. The C-terminal domain of MinC inhibits assembly of the Z ring in Escherichia coli. J. Bacteriol. 189:236-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shlomovitz, R., and N. S. Gov. 2009. Membrane-mediated interactions drive the condensation and coalescence of FtsZ rings. Phys Biol. 6:46017. [DOI] [PubMed] [Google Scholar]
- 174.Shroff, H., C. G. Galbraith, J. A. Galbraith, and E. Betzig. 2008. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Singh, J. K., R. D. Makde, V. Kumar, and D. Panda. 2007. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ. Biochemistry 46:11013-11022. [DOI] [PubMed] [Google Scholar]
- 176.Singh, J. K., R. D. Makde, V. Kumar, and D. Panda. 2008. SepF increases the assembly and bundling of FtsZ polymers and stabilizes FtsZ protofilaments by binding along its length. J. Biol. Chem. 283:31116-31124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. U. S. A. 78:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Small, E., R. Marrington, A. Rodger, D. J. Scott, K. Sloan, D. Roper, T. R. Dafforn, and S. G. Addinall. 2007. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J. Mol. Biol. 369:210-221. [DOI] [PubMed] [Google Scholar]
- 179.Sontag, C. A., H. Sage, and H. P. Erickson. 2009. BtubA-BtubB heterodimer is an essential intermediate in protofilament assembly. PLoS One 4:e7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sossong, T. M., M. R. Brigham-Burke, P. Hensley, and K. H. Pearce. 1999. Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry 38:14843-14850. [DOI] [PubMed] [Google Scholar]
- 181.Srinivasan, R., M. Mishra, L. Wu, Z. Yin, and M. K. Balasubramanian. 2008. The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 22:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Stokes, N. R., J. Sievers, S. Barker, J. M. Bennett, D. R. Brown, I. Collins, V. M. Errington, D. Foulger, M. Hall, R. Halsey, H. Johnson, V. Rose, H. B. Thomaides, D. J. Haydon, L. G. Czaplewski, and J. Errington. 2005. Novel inhibitors of bacterial cytokinesis identified by a cell-based antibiotic screening assay. J. Biol. Chem. 280:39709-39715. [DOI] [PubMed] [Google Scholar]
- 183.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U. S. A. 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Strömqvist, J., K. Skoog, D. O. Daley, J. Widengren, and G. von Heijne. 2010. Estimating Z-ring radius and contraction in dividing Escherichia coli. Mol. Microbiol. 76:151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sugimoto, S., K. Yamanaka, S. Nishikori, A. Miyagi, T. Ando, and T. Ogura. 2010. AAA+ chaperone ClpX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J. Biol. Chem. 285:6648-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sun, Q., and W. Margolin. 1998. FtsZ dynamics during the division cycle of live Escherichia coli cells. J. Bacteriol. 180:2050-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Surovtsev, I. V., J. J. Morgan, and P. A. Lindahl. 2008. Kinetic modeling of the assembly, dynamic steady state, and contraction of the FtsZ ring in prokaryotic cytokinesis. PLoS Comput. Biol. 4:e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tadros, M., J. M. Gonzalez, G. Rivas, M. Vicente, and J. Mingorance. 2006. Activation of the Escherichia coli cell division protein FtsZ by a low-affinity interaction with monovalent cations. FEBS Lett. 580:4941-4946. [DOI] [PubMed] [Google Scholar]
- 190.Tang, J. X., and P. A. Janmey. 1996. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J. Biol. Chem. 271:8556-8563. [DOI] [PubMed] [Google Scholar]
- 191.Thanedar, S., and W. Margolin. 2004. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Trusca, D., S. Scott, C. Thompson, and D. Bramhill. 1998. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 180:3946-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.van Baarle, S., and M. Bramkamp. 2010. The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One 5:e9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vaughan, S., B. Wickstead, K. Gull, and S. G. Addinall. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58:19-29. [DOI] [PubMed] [Google Scholar]
- 195.Vicente, M., A. I. Rico, R. Martinez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang, J., A. Galgoci, S. Kodali, K. B. Herath, H. Jayasuriya, K. Dorso, F. Vicente, A. Gonzalez, D. Cully, D. Bramhill, and S. Singh. 2003. Discovery of a small molecule that inhibits cell division by blocking FtsZ, a novel therapeutic target of antibiotics. J. Biol. Chem. 278:44424-44428. [DOI] [PubMed] [Google Scholar]
- 197.Wang, X., and J. Lutkenhaus. 1996. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria. Mol.Microbiol. 21:313-319. [DOI] [PubMed] [Google Scholar]
- 198.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 199.White, E. L., L. J. Ross, R. C. Reynolds, L. E. Seitz, G. D. Moore, and D. W. Borhani. 2000. Slow polymerization of Mycobacterium tuberculosis FtsZ. J. Bacteriol. 182:4028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wilks, J. C., and J. L. Slonczewski. 2007. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189:5601-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yanagisawa, M., M. Imai, and T. Taniguchi. 2008. Shape deformation of ternary vesicles coupled with phase separation. Phys. Rev. Lett. 100:148102. [DOI] [PubMed] [Google Scholar]
- 202.Yu, X. C., and W. Margolin. 1997. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 16:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yu, X. C., and W. Margolin. 1998. Inhibition of assembly of bacterial cell division protein FtsZ by the hydrophobic dye 5,5′-bis-(8-anilino-1-naphthalenesulfonate). J. Biol. Chem. 273:10216-10222. [DOI] [PubMed] [Google Scholar]
- 204.Zhang, W., and D. N. Robinson. 2005. Balance of actively generated contractile and resistive forces controls cytokinesis dynamics. Proc. Natl. Acad. Sci. U. S. A. 102:7186-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhu, T. F., and J. W. Szostak. 2009. Coupled growth and division of model protocell membranes. J. Am. Chem. Soc. 131:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.