Abstract
The 37/67 kDa human laminin receptor (LamR) is a cell surface receptor for laminin, prion protein, and a variety of viruses. Because of its wide range of ligands, LamR plays a role in numerous pathologies. LamR overexpression correlates with a highly invasive cell phenotype and increased metastatic ability, mediated by interactions between LamR and laminin. In addition, the specific targeting of LamR with small-interfering RNAs (siRNAs), blocking antibodies, and Sindbis viral vectors confers anti-tumor effects. We adopted a structure-based approach to map a laminin binding site on human LamR, by comparing the sequences and crystal structures of LamR and A. fulgidus S2p, a non-laminin binding ortholog. Here, we identify a laminin binding site on LamR, comprising residues Phe32, Glu35, and Arg155, which are conserved among mammalian species. Mutation of these residues results in a significant loss of laminin binding. Further, recombinant wild-type LamR is able to act as a soluble decoy to inhibit cellular migration toward laminin. Mutation of this laminin binding site results in loss of migration inhibition, which demonstrates the physiological role of Phe32, Glu35, and Arg155 for laminin binding activity. Mapping of the LamR binding site should contribute to the development of therapeutics that inhibit LamR interactions with laminin, and may aid in the prevention of tumor growth and metastasis.
The 37/67 kDa human laminin receptor (LamR), was originally identified as a non-integrin cell surface receptor for the extracellular matrix molecule laminin1;2;3. Laminins, other glycoproteins, collagen IV and proteoglycans constitute a tight network to form the basement membrane. Laminin-1, a 900 kDa glycoprotein, contains many bioactive domains involved in binding both integrin and nonintegrin receptors and is vital for basement membrane assembly4. Interactions between LamR and laminin play a major role in mediating changes in the cellular environment that affect cell adhesion5, neurite outgrowth4, and tumor growth and metastasis6.
Overexpression of LamR has been shown in many cancers, including lung7, breast8, gastric9, colon10;11, ovarian12;13, uterine14, thyroid15, prostate16, liver17, and melanoma18;19. LamR expression correlates with a highly invasive cell phenotype and increased metastatic ability, mediated by the high-affinity interactions between LamR and laminin20;21;22;23;24. Interactions between LamR and laminin contribute to tumor cell attachment to the basement membrane. These properties render LamR a prognostic factor in determining the degree of malignancy of human cancer patients20. Understanding how LamR and laminin interact may provide insight into tumor invasion and metastasis.
LamR has been observed to act as the cell surface receptor for pathogenic prion protein25 and a variety of viruses, including Sindbis virus26;27;28. LamR has also been reported to bind EGCG, a polyphenol extract found in green tea29. Interestingly, both human LamR and the p40 ribosomal protein are encoded by the same gene, 37LRP/p4030. Thus, human LamR precursor protein has a dual function as a component of the translational machinery and as a cell surface receptor for laminin and other molecules. Intracellular LamR is found on the 40S ribosome and polysomes31;32 and in the nucleus33, which suggests that LamR is a multifunctional protein. Silencing LamR expression in mammalian cells in vitro induces G1 cell cycle arrest, protein translation shutdown, and loss of LamR association with the polysomes32. Sequence conservation of 37LRP/p40 genes across species has been demonstrated, with evolution of the C-terminal tail convergent with vertebrates30. At the cell surface, LamR, which is targeted to the membrane via fatty-acid acylation34, exists as both a monomer (37 kDa) and a dimer (67 kDa)25. Although the homo- or hetero-dimeric state of LamR has not been fully resolved, both 37 kDa and 67 kDa LamR bind laminin35;36.
Characterization of LamR interactions with laminin has not been fully conclusive. Several LamR segments, including residues 161–180 (peptide G)37, residues 205–22938, and TEDWS-containing C-terminal repeats39 have been implicated as binding epitopes for laminin. We previously determined a high-resolution crystal structure of the majority of LamR, residues 1 through 220 (abbreviated LamR220)36 (PDB code 3BCH). LamR220 binds laminin with similar affinity as full-length LamR and inhibits Sindbis virus infection in vitro36. It has also been shown that some of the phenotypic effects of silencing endogenous LamR can be rescued by exogenous full-length LamR and LamR22040. Based on analyses of sequence conservation and of the crystal structures of human LamR and A. fulgidus S2 ribosomal protein, we introduced a number of point mutations into LamR220 to probe LamR binding to laminin. In the current study, we have mapped a laminin binding site on LamR, comprising residues Phe32, Glu35, and Arg155.
Expression and characterization of LamR mutants
We previously solved the crystal structure of a biologically active domain of human LamR, LamR22036. LamR220 binds laminin with similar affinity to full-length LamR (LamR295)36, demonstrating that LamR220 is sufficient for laminin binding. For these studies, LamR220 was used for mutagenesis due to its higher level of expression and purity compared to LamR295. Despite the implication in an earlier study37 of residues 161–180 in laminin binding, the laminin binding site has remained elusive since the crystal structure of human LamR revealed that this domain is not solvent accessible36. To better understand LamR interactions with laminin, we designed LamR mutants based upon analysis of sequence conservation and the crystal structures of human LamR22036 (PDB code 3BCH) and its non-laminin binding ortholog, Archaeoglobus fulgidus S2 ribosomal protein41 (PDB code 1VI6).
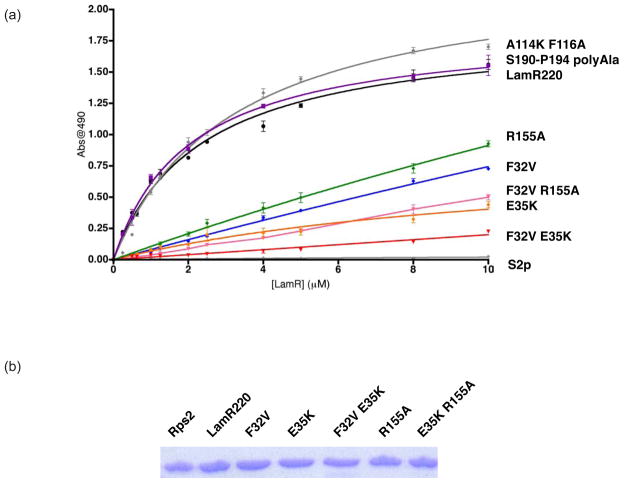
In total, 14 mutants were analyzed individually (Table 1, Fig. 1a). Protein expression and purification of each mutant, including by gel filtration, was similar to that of wild-type LamR220, which indicates that the mutations did not interfere with protein folding or stability. The purity of the proteins was monitored by SDS-PAGE (Fig. 2b). All mutants were examined for their ability to bind immobilized laminin. Purified recombinant LamR220 (greater than 95% purity) was incubated in serial dilution on pre-coated laminin plates, and binding was detected by anti-His HRP. As a control for binding specificity, we tested laminin binding for A. fulgidus S2p ribosomal protein, which should not interact with laminin. A. fulgidus S2p does not exhibit laminin-binding activity, as expected (Fig. 2a). These data support our hypothesis that differences between human LamR and A. fulgidus S2p are important for laminin binding.
Table 1.
Effect of mutations of LamR on laminin binding activity. Half-maximal binding (Kd) was generated using nonlinear regression of binding curves in GraphPad Prism. Relative binding refers to the percentage ratio of LamR220 wild-type Kd/mutant Kd. NB indicates mutations that result in loss of laminin binding activity.
| Mutant | Half-maximal binding [μM] | Relative binding (%age) |
|---|---|---|
| WT LamR220 | 2.3 +/− 0.16 | 100 |
| L16Q | 2.8 +/− 0.15 | 82 |
| R53A E56A K57A | 1.9 +/− 0.21 | 121 |
| R102A | 2.7 +/− 0.16 | 85 |
| A114K F116A | 3.5 +/− 0.31 | 66 |
| H131A | 5.4 +/− 0.28 | 43 |
| Y139A | 6.2 +/− 0.9 | 37 |
| N141A | 2.2 +/− 0.13 | 105 |
| K166A | 4.3 +/− 0.92 | 53 |
| S190-P194 polyAla | 2.0 +/− 0.17 | 115 |
| F32V | 97.0 +/− 41.8 | 2 |
| E35K | 18.5 +/− 2.9 | 12 |
| R155A | 52.8 +/− 20.8 | 4 |
| F32V E35K | NB | N/A |
| F32V R155A | NB | N/A |
Figure 1.
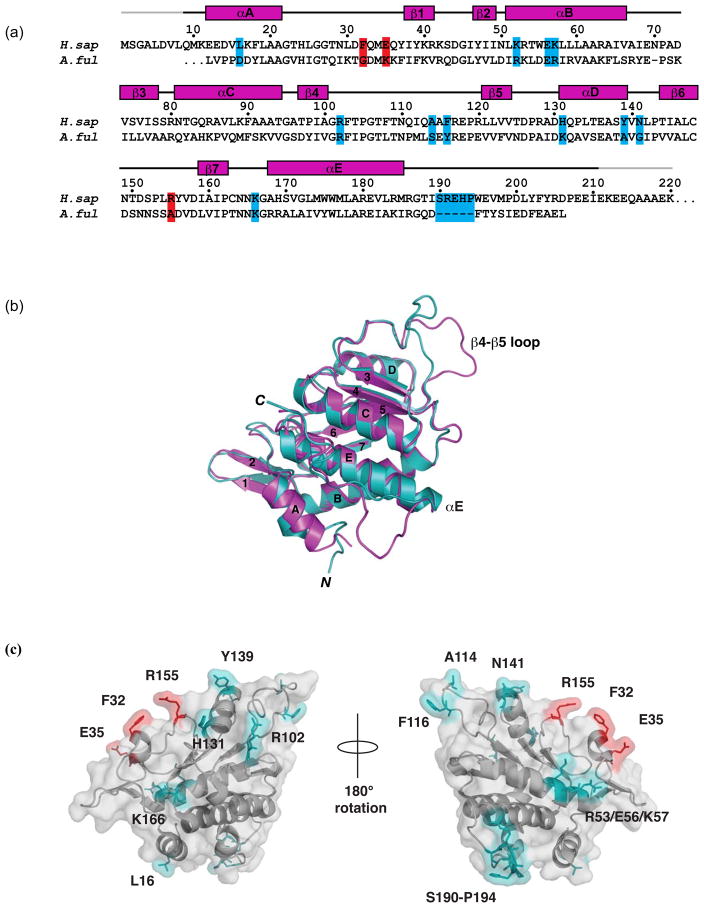
Analysis of LamR structural and sequence features. (a) Sequence alignment between 37LRP/p40 orthologs H. sapiens LamR (residues 1–220) and A. fulgidus S2p ribosomal protein (residues 1–198). Residue numbering is for human LamR. Mutated residues that cause a significant change in laminin binding are in red. Mutated residues that cause a slight or no reduction in affinity are in cyan. Secondary structure (α helices and β strands, magenta) for LamR220 is shown according to PROCHECK49. Gray lines indicate the N- and C-terminal portions of the structure that are disordered. (b) Superimposition of human LamR220 (magenta) and A. fulgidus S2p (cyan) in ribbon format. α helices and β strands are labeled according to (a). The regions of structural divergence, between the β4-β5 loop (residues 111–118) and after αE (residues 190–194), are labeled. (c) Surface representation of LamR220 mutations. LamR220 (grey) with Phe32, Glu35, and Arg155 labeled (red) and shown in stick, mutations that do not affect laminin binding (cyan) are also labeled and shown in stick.
Figure 2.
Analysis of LamR binding affinity for laminin. (a) Binding of LamR and mutants to laminin. LamR220 (black), A. fulgidus S2p (light grey), LamR220 A114K/F116A (dark grey), LamR220 S190-P194 polyAla (purple), LamR220 Phe32Val (blue), LamR220 R155A (green), LamR220 E35K (orange), LamR220 F32V/R155A (pink), and LamR220 F32V/E35K (red). n = 3 +/− SEM. (b) Coomassie blue staining of A. fulgidus S2p, LamR220 and mutants that affect binding for experimental loading control. Residues 1–220 of human 37 kDa LamR precursor protein (LamR220) and LamR220 mutants were expressed and purified as previously described36. Residues 1–208 of A. fulgidus S2p were cloned, expressed and purified utilizing the same conditions as human LamR220. The LamR220 construct described above was used as the backbone for generating LamR220 point mutants in the LamR coding region with the QuikChange site-directed mutagenesis kit XL II (Stratagene). Complementary forward and reverse primers were designed to mutate LamR220 residues. PCR-based point mutations were made using thermal cycling according to the Stratagene protocol, the PCR product was digested with DpnI restriction enzyme (NE Biolabs) and transformed into E. coli strain XLII Blue (Stratagene). LamR220 vectors positive for mutations were verified by automated DNA sequencing. Wild-type and mutant LamR220 binding affinity for laminin were tested in vitro. Protein concentration was determined by Protein Dc Assay (Biorad). Clear polystyrene ELISA 96-well microplates, precoated with murine laminin (NEBiolabs), were blocked for 1 hr at 37° C with blocking buffer (2% FCS, 1mg/ml BSA, 0.1% sodium azide in PBS). Wells were incubated with increasing concentrations of purified wild-type or mutant LamR220, which all contain a 6XHis-tag, for 1 h at 37° C. Each well was washed three times with wash buffer (0.5% Tween in PBS). Penta-His HRP conjugate (1:500) (Qiagen) was incubated for 2 h at RT. After washing, substrate solution was added and fluorescent absorbance was detected at 490 nm on an ELISA plate reader (ELX800, Biotek Instruments, Inc.). A binding curve, Kd, and standard error for Kd were generated for wild-type and mutant LamR220 using GraphPad Prism software. The Kd was calculated using a one-site binding hyperbola and the equation . Each group was tested in triplicate and binding affinity was determined by normalizing to background fluorescence.
Mapping of the laminin-binding region of LamR by site-directed mutagenesis
Structural and sequence analyses afforded selection of solvent-exposed residues that could contribute to laminin binding. Our studies examined the role of three categories of mutations: structurally divergent regions between human LamR and A. fulgidus S2p, residues previously implicated in laminin binding, and non-conserved residues between human LamR and A. fulgidus (Fig. 1a). Human LamR and its ortholog A. fulgidus S2p share 32% sequence identity. Superimposition of the structures of LamR220 and A. fulgidus S2p revealed two areas in which the structures are divergent (Fig. 1b): a segment between β4 and β5 (residues 111–118 in LamR) and a segment after the last α helix (αE) (residues 190–194 in LamR), in which LamR contains a five-residue insertion relative to A. fulgidus S2p. In A. fulgidus S2p, the segment between β4 and β5 (residues 111–118 in LamR) is stabilized in a folded-back conformation via a salt bridge between Arg113 and Asp93. These residues, which correspond to Arg117 and Thr97 in LamR (Fig. 1a and 1b), project away from the domain in LamR220 and instead pack against a symmetry (two-fold)-related molecule in the crystal structure. Within this crystallographic dimer, Ala114 packs into a tight pocket in the symmetry-related molecule and Phe116 is in van der Waals contact with Tyr139. We mutated LamR Ala114 to Lys and Phe116 to Ala (abbreviated A114F/K116A), the corresponding residues in the S. cerevisiae S2 ribosomal protein ortholog. Wild-type LamR220 bound laminin with an Kd of 2.3 μM, and A. fulgidus S2p showed no binding to laminin (Table 1, Fig. 2a). A114F/K116A had no effect on laminin binding (Table 1, Fig. 2a).
The second region of structural divergence between human LamR and A. fulgidus S2p is a 5-residue insertion in human LamR, comprising residues Ser190 through Pro194 (Fig. 1b). Mutation of all five residues (Ser190-Pro194) to Ala resulted in no change in laminin binding compared to wild-type LamR220 (Table 1, Fig. 2a). In addition, we examined mutation of a previously identified putative laminin binding site, peptide G. Peptide G, residues 161–180, was implicated as a binding epitope in assays that utilized peptide segments of LamR to bind laminin37. Of this segment, only residues Lys166 and His169 are solvent accessible in the LamR220 crystal structure36. Mutation of Lys166 to Ala does not affect laminin binding (Table 1, Fig. 1a), which suggests that Lys166 is not essential for the laminin-binding function of LamR.
Lastly, we selected LamR residues Leu16, Phe32, Glu35, Arg53, Glu56, Lys57, Arg102, His131, Tyr139, Asn141, and Arg155 (Fig. 1a and 1c), which are solvent-exposed in the LamR220 structure and show sequence conservation among laminin-binding species30, but are not conserved in the sequence of the non-binding A. fulgidus S2p. These mutations include changes in charge (for Glu35, Arg53, Glu56, Lys57, Arg102, His131, and Arg155) and in hydrophobicity (for Leu16, Phe32, Tyr139, and N141). Only mutations Phe32, Glu35, and Arg155 resulted in an appreciable loss of binding affinity (Table 1, Fig. 2a). Single point mutation of LamR Phe32 to Val (the corresponding residue in S. cerevisiae S2 ribosomal protein), or Glu35 to Lys, or Arg155 to Ala (the corresponding residues in A. fulgidus S2 ribosomal protein) resulted in a decrease in laminin binding (8- to 42-fold increase in Kd) (Table 1, Fig. 2a). Double point mutation of Phe32 and Glu35 (abbreviated F32V/E35K) or Phe32 and Arg155 (abbreviated F32V/R155A) resulted in a greater loss of laminin binding activity than the single mutants (Table 1, Fig. 2a). Mutation of His131, Tyr139, and Asn141, which are in close proximity to Arg155, Phe32, and Glu35, did not affect laminin binding (Table 1, Fig. 1c). These data demonstrate that Phe32, Glu35, and Arg155 (Fig. 1c) comprise a primary laminin binding site.
Physiological significance of LamR mutants
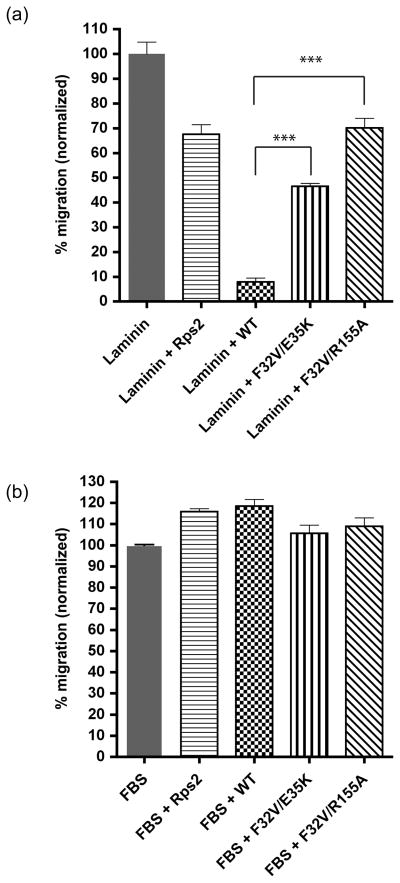
To assess the physiological significance of residues Phe32, Glu35, and Arg155 in laminin binding, we developed a cellular assay to test wild-type and mutant LamR function. Previously, it has been demonstrated that LamR localizes to both the cell membrane and cytoplasm32;42. Silencing LamR expression in HT-1080 cells results in protein translation and cell cycle arrest40. Although transfection of FLAG-tagged LamR cDNA into cells is able to rescue the effects of silencing endogenous LamR40, transfected LamR does not localize to the cell membrane (D.M., unpublished data). These results render studying the role of mutations in LamR at the cell surface difficult. As a result, we utilized purified recombinant LamR as a soluble decoy to interfere with endogenous LamR cell surface interactions with laminin in a cell migration assay. Cell migration to laminin has been previously observed43;44. We examined the ability of recombinant wild-type or mutant LamR220 to inhibit HT-1080 cell migration towards laminin in a Boyden chamber in which cells were added to the top chamber and laminin or laminin with recombinant protein were added to the bottom chamber. When recombinant wild-type LamR220 is added with laminin, HT-1080 cell migration towards laminin is 8% of the migration observed relative to migration to laminin alone (Fig. 3a, WT). These data suggest that recombinant LamR220 interacts with laminin in solution, inhibiting cellular migration towards laminin. Cellular migration to recombinant protein alone is not observed. Protein purity and equal loading was examined by SDS-PAGE. A. fulgidus S2p, which does not exhibit laminin binding activity in vitro, was utilized as a negative control for migration inhibition (Fig. 3a, Rps2). LamR220 mutants F32V/E35K and F32V/R155A, which exhibited loss of laminin binding in vitro, were examined for loss of inhibition and restoration of cell migration to laminin. Both LamR220 mutants F32V/E35K and F32V/R155A demonstrate a significant loss of inhibition of cell migration to laminin (Fig. 3a, F32V/E35K and F32V/R155A). LamR220 F32V/R155A behaves similarly to A. fulgidus S2p (negative control), with 70% migration observed compared to migration to laminin alone. LamR220 F32V/E35K results in 46% of migration compared to migration to laminin alone. Cell migration towards 10% FBS is not inhibited by wild-type LamR220, mutant LamR220, or A. fulgidus S2p, which suggests that inhibition of cell migration to laminin by wild-type LamR220 is specific to interactions between LamR and laminin (Fig. 3b). Together, these data demonstrate the physiological role of Phe32, Glu35, and Arg155 in laminin binding.
Figure 3.
Recombinant wild-type LamR inhibits HT-1080 cell migration towards laminin. (a) Inhibition of cell migration to laminin by wild-type LamR, but not LamR mutants. Laminin alone (grey), laminin + A. fulgidus S2p (horizontal stripes), laminin + wild-type LamR220 (checked), laminin + LamR220 F32V/E35K (vertical stripes), laminin + LamR220 F32V/R155A (diagonal stripes). n = 3 +/− SEM. The differences between wild-type LamR220 inhibition and either LamR220 F32V/R155A or F32V/E35K are both statistically significant (P<0.0001). (b) HT-1080 cell migration towards FBS is not inhibited by recombinant wild-type or mutant LamR. FBS alone (grey), FBS + A. fulgidus S2p (horizontal stripes), FBS + wild-type LamR220 (checked), FBS + LamR220 F32V/E35K (vertical stripes), FBS + LamR220 F32V/R155A (diagonal stripes). n = 3 +/− SEM. HT-1080 cells were tested for their ability to migrate to 10 ug/mL purified laminin (Invitrogen) or 10% FBS using the CytoSelectTM 24-Well Cell Migration Assay (8μm, Colorimetric Format) (Cell Biolabs, Inc.). HT-1080 cells were obtained from the American Type Culture Collection (Mannasas, VA). HT-1080 cells were maintained in DMEM 4.5 g/ml glucose with 10% FCS. All basal media was supplemented with 100 μg/ml of penicillin-streptomycin and 0.5 μg/ml amphotericin B (all from Mediatech). Briefly, 1.5×105 cells in unsupplemented media were added to the upper insert chamber of each well. For (a), the reservoir below contained 500μl of either unsupplemented media, purified laminin, or purified laminin and recombinant wild-type LamR, mutant LamR, or A. fulgidus S2p. For (b), the reservoir below contained 500μl of either unsupplemented media, media supplemented with 10% FBS, or 10% FBS and recombinant wild-type LamR, mutant LamR, or A. fulgidus S2p. All protein was at 1 μM final concentration. Protein concentration was determined by Protein Dc Assay (Biorad). After the incubation at 37°C for 5 hours the upper membrane of each insert was thoroughly washed with dH2O 3X to remove non-migratory cells and then incubated in cell stain solution for 20 minutes at room temperature. Inserts were washed again in dH2O and then air-dried. After agitation for 10 minutes at RT in extraction solution, 100μl of each sample was transferred to an ELISA plate and read at 630nm. To calculate migration, background migration to unsupplemented media was subtracted. Migration to laminin or FBS in the presence of either wild-type LamR, mutant LamR, or A. fulgidus S2p was normalized to migration to laminin or FBS alone. Assays were performed in triplicate. Results were plotted using GraphPad Prism software and statistical significance was calculated using a standard Student’s t-test. (P<.0001).
Summary
The interaction between LamR and laminin mediates changes in the cell environment that affect cell adhesion and tumor growth and metastasis5;6;22;42. We utilized sequence and structure analyses to guide LamR mutagenesis to map a laminin binding site on LamR. Both the mutagenesis data in the context of the LamR crystal structure and cell migration data indicate a primary laminin binding site comprising residues Phe32, Glu35, and Arg155 (Fig. 2). These residues are conserved among mammalian laminin-binding species30, but not in A. fulgidus S2 ribosomal protein ortholog, which does not bind laminin or inhibit cell migration to laminin. Interestingly, mutation of two regions of structural divergence between human LamR and A. fulgidus S2p, the β4-β5 linker and a five-residue insertion after αE, did not affect laminin binding, suggesting that they contribute to other LamR functions. In addition, mutation of Lys166 to Ala, which was previously implicated in laminin binding37, does not affect binding in vitro.
In the setting of cancer, interactions between LamR and laminin contribute to modifications of the extracellular matrix structure that affect cancer cell growth and proliferation as well as tumor invasion and metastasis, and activate proteolytic enzymes and their regulators6;45;46. The specific inhibition of LamR interactions with laminin has been shown to elicit anti-tumor effects32;42;47;48. Targeting LamR in the tumor environment may be possible due to a larger percentage of unoccupied LamR sites compared to normal cells, on which laminin binding is saturated22;23;24. Mapping the LamR binding site for laminin may contribute to the development of therapeutics that inhibit LamR interactions with laminin and aid in the prevention of tumor growth and metastasis.
Acknowledgments
U.S. Public Health Service grants CA100687 (to D.M.) and CA68498 (to D.M.) from the National Cancer Institute, National Institutes of Health, and Department of Health and Human Services supported this study. S.R.H. is a recipient of an Irma T. Hirschl-Monique Weill-Caulier Career Scientist Award. Funding was also provided by the Litwin Foundation. We thank Dr. Christine Pampeno and Vincent DiGiacomo for critical reading of this manuscript and helpful discussions.
Footnotes
Competing Interests Statement
Some of the authors have competing interests. Specifically, the contents of this paper are being utilized for a patent. According to the rules and regulations of New York University School of Medicine, if this patent is licensed by a third party, some of the authors (D.M., S.R.H., K.V.J.) may receive benefits in the form of royalties or equity participation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rao NC, Barsky SH, Terranova VP, Liotta LA. Isolation of a tumor cell laminin receptor. Biochem Biophys Res Commun. 1983;111:804–8. doi: 10.1016/0006-291x(83)91370-0. [DOI] [PubMed] [Google Scholar]
- 2.Terranova VP, Rao CN, Kalebic T, Margulies IM, Liotta LA. Laminin receptor on human breast carcinoma cells. Proc Natl Acad Sci U S A. 1983;80:444–8. doi: 10.1073/pnas.80.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malinoff HL, Wicha MS. Isolation of a cell surface receptor protein for laminin from murine fibrosarcoma cells. J Cell Biol. 1983;96:1475–9. doi: 10.1083/jcb.96.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mecham RP. Receptors for laminin on mammalian cells. Faseb J. 1991;5:2538–46. doi: 10.1096/fasebj.5.11.1651264. [DOI] [PubMed] [Google Scholar]
- 5.Graf J, Ogle RC, Robey FA, Sasaki M, Martin GR, Yamada Y, Kleinman HK. A pentapeptide from the laminin B1 chain mediates cell adhesion and binds the 67,000 laminin receptor. Biochemistry. 1987;26:6896–900. doi: 10.1021/bi00396a004. [DOI] [PubMed] [Google Scholar]
- 6.Narumi K, Inoue A, Tanaka M, Isemura M, Shimo-Oka T, Abe T, Nukiwa T, Satoh K. Inhibition of experimental metastasis of human fibrosarcoma cells by anti-recombinant 37-kDa laminin binding protein antibody. Jpn J Cancer Res. 1999;90:425–31. doi: 10.1111/j.1349-7006.1999.tb00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh K, Narumi K, Isemura M, Sakai T, Abe T, Matsushima K, Okuda K, Motomiya M. Increased expression of the 67kDa-laminin receptor gene in human small cell lung cancer. Biochem Biophys Res Commun. 1992;182:746–52. doi: 10.1016/0006-291x(92)91795-r. [DOI] [PubMed] [Google Scholar]
- 8.Martignone S, Menard S, Bufalino R, Cascinelli N, Pellegrini R, Tagliabue E, Andreola S, Rilke F, Colnaghi MI. Prognostic significance of the 67-kilodalton laminin receptor expression in human breast carcinomas. J Natl Cancer Inst. 1993;85:398–402. doi: 10.1093/jnci/85.5.398. [DOI] [PubMed] [Google Scholar]
- 9.de Manzoni G, Guglielmi A, Verlato G, Tomezzoli A, Pelosi G, Schiavon I, Cordiano C. Prognostic significance of 67-kDa laminin receptor expression in advanced gastric cancer. Oncology. 1998;55:456–60. doi: 10.1159/000011895. [DOI] [PubMed] [Google Scholar]
- 10.Cioce V, Castronovo V, Shmookler BM, Garbisa S, Grigioni WF, Liotta LA, Sobel ME. Increased expression of the laminin receptor in human colon cancer. J Natl Cancer Inst. 1991;83:29–36. doi: 10.1093/jnci/83.1.29. [DOI] [PubMed] [Google Scholar]
- 11.Sanjuan X, Fernandez PL, Miquel R, Munoz J, Castronovo V, Menard S, Palacin A, Cardesa A, Campo E. Overexpression of the 67-kD laminin receptor correlates with tumour progression in human colorectal carcinoma. J Pathol. 1996;179:376–80. doi: 10.1002/(SICI)1096-9896(199608)179:4<376::AID-PATH591>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 12.Liebman JM, Burbelo PD, Yamada Y, Fridman R, Kleinman HK. Altered expression of basement-membrane components and collagenases in ascitic xenografts of OVCAR-3 ovarian cancer cells. Int J Cancer. 1993;55:102–9. doi: 10.1002/ijc.2910550119. [DOI] [PubMed] [Google Scholar]
- 13.van den Brule FA, Castronovo V, Menard S, Giavazzi R, Marzola M, Belotti D, Taraboletti G. Expression of the 67 kD laminin receptor in human ovarian carcinomas as defined by a monoclonal antibody, MLuC5. Eur J Cancer. 1996;32A:1598–602. doi: 10.1016/0959-8049(96)00119-0. [DOI] [PubMed] [Google Scholar]
- 14.van den Brule FA, Buicu C, Berchuck A, Bast RC, Deprez M, Liu FT, Cooper DN, Pieters C, Sobel ME, Castronovo V. Expression of the 67-kD laminin receptor, galectin-1, and galectin-3 in advanced human uterine adenocarcinoma. Hum Pathol. 1996;27:1185–91. doi: 10.1016/s0046-8177(96)90313-5. [DOI] [PubMed] [Google Scholar]
- 15.Montuori N, Muller F, De Riu S, Fenzi G, Sobel ME, Rossi G, Vitale M. Laminin receptors in differentiated thyroid tumors: restricted expression of the 67-kilodalton laminin receptor in follicular carcinoma cells. J Clin Endocrinol Metab. 1999;84:2086–92. doi: 10.1210/jcem.84.6.5721. [DOI] [PubMed] [Google Scholar]
- 16.Waltregny D, de Leval L, Menard S, de Leval J, Castronovo V. Independent prognostic value of the 67-kd laminin receptor in human prostate cancer. J Natl Cancer Inst. 1997;89:1224–7. doi: 10.1093/jnci/89.16.1224. [DOI] [PubMed] [Google Scholar]
- 17.Ozaki I, Yamamoto K, Mizuta T, Kajihara S, Fukushima N, Setoguchi Y, Morito F, Sakai T. Differential expression of laminin receptors in human hepatocellular carcinoma. Gut. 1998;43:837–42. doi: 10.1136/gut.43.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vacca A, Ribatti D, Roncali L, Lospalluti M, Serio G, Carrel S, Dammacco F. Melanocyte tumor progression is associated with changes in angiogenesis and expression of the 67-kilodalton laminin receptor. Cancer. 1993;72:455–61. doi: 10.1002/1097-0142(19930715)72:2<455::aid-cncr2820720222>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Taraboletti G, Belotti D, Giavazzi R, Sobel ME, Castronovo V. Enhancement of metastatic potential of murine and human melanoma cells by laminin receptor peptide G: attachment of cancer cells to subendothelial matrix as a pathway for hematogenous metastasis. J Natl Cancer Inst. 1993;85:235–40. doi: 10.1093/jnci/85.3.235. [DOI] [PubMed] [Google Scholar]
- 20.Menard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Res Treat. 1998;52:137–45. doi: 10.1023/a:1006171403765. [DOI] [PubMed] [Google Scholar]
- 21.Castronovo V. Laminin receptors and laminin-binding proteins during tumor invasion and metastasis. Invasion Metastasis. 1993;13:1–30. [PubMed] [Google Scholar]
- 22.Liotta LA, Rao NC, Barsky SH, Bryant G. The laminin receptor and basement membrane dissolution: role in tumour metastasis. Ciba Found Symp. 1984;108:146–62. doi: 10.1002/9780470720899.ch10. [DOI] [PubMed] [Google Scholar]
- 23.Aznavoorian S, Murphy AN, Stetler-Stevenson WG, Liotta LA. Molecular aspects of tumor cell invasion and metastasis. Cancer. 1993;71:1368–83. doi: 10.1002/1097-0142(19930215)71:4<1368::aid-cncr2820710432>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 24.Liotta LA. Tumor Invasion and Metastases Role of the Extracellular-Matrix - Rhoads Memorial Award Lecture. Cancer Research. 1986;46:1–7. [PubMed] [Google Scholar]
- 25.Gauczynski S, Peyrin JM, Haik S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, Lasmezas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. Embo J. 2001;20:5863–75. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig GV, Kondig JP, Smith JF. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J Virol. 1996;70:5592–9. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80:9831–6. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tachibana H, Koga K, Fujimura Y, Yamada K. A receptor for green tea polyphenol EGCG. Nat Struct Mol Biol. 2004;11:380–1. doi: 10.1038/nsmb743. [DOI] [PubMed] [Google Scholar]
- 30.Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME, Colnaghi MI, Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol Biol Evol. 1998;15:1017–25. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- 31.Auth D, Brawerman G. A 33-kDa polypeptide with homology to the laminin receptor: component of translation machinery. Proc Natl Acad Sci U S A. 1992;89:4368–72. doi: 10.1073/pnas.89.10.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheiman J, Tseng JC, Zheng Y, Meruelo D. Multiple functions of the 37/67-kd laminin receptor make it a suitable target for novel cancer gene therapy. Mol Ther. 2010;18:63–74. doi: 10.1038/mt.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato M, Kinoshita K, Kaneda Y, Saeki Y, Iwamatsu A, Tanaka K. Analysis of nuclear localization of laminin binding protein precursor p40 (LBP/p40) Biochem Biophys Res Commun. 1996;229:896–901. doi: 10.1006/bbrc.1996.1899. [DOI] [PubMed] [Google Scholar]
- 34.Buto S, Tagliabue E, Ardini E, Magnifico A, Ghirelli C, van den Brule F, Castronovo V, Colnaghi MI, Sobel ME, Menard S. Formation of the 67-kDa laminin receptor by acylation of the precursor. J Cell Biochem. 1998;69:244–51. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 35.Fatehullah A, Doherty C, Pivato G, Allen G, Devine L, Nelson J, Timson DJ. Interactions of the 67 kDa laminin receptor and its precursor with laminin. Biosci Rep. 2010;30:73–9. doi: 10.1042/BSR20090023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamieson KV, Wu J, Hubbard SR, Meruelo D. Crystal structure of the human laminin receptor precursor. J Biol Chem. 2008;283:3002–5. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- 37.Castronovo V, Taraboletti G, Sobel ME. Functional domains of the 67-kDa laminin receptor precursor. J Biol Chem. 1991;266:20440–6. [PubMed] [Google Scholar]
- 38.Landowski TH, Uthayakumar S, Starkey JR. Control pathways of the 67 kDa laminin binding protein: surface expression and activity of a new ligand binding domain. Clin Exp Metastasis. 1995;13:357–72. doi: 10.1007/BF00121912. [DOI] [PubMed] [Google Scholar]
- 39.Kazmin DA, Hoyt TR, Taubner L, Teintze M, Starkey JR. Phage display mapping for peptide 11 sensitive sequences binding to laminin-1. J Mol Biol. 2000;298:431–45. doi: 10.1006/jmbi.2000.3680. [DOI] [PubMed] [Google Scholar]
- 40.Scheiman J, Jamieson KV, Ziello J, Tseng JC, Meruelo D. Extraribosomal functions associated with the C terminus of the 37/67 kDa laminin receptor are required for maintaining cell viability. Cell Death and Disease. 2010;1 doi: 10.1038/cddis.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–96. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 42.Zuber C, Knackmuss S, Zemora G, Reusch U, Vlasova E, Diehl D, Mick V, Hoffmann K, Nikles D, Frohlich T, Arnold GJ, Brenig B, Wolf E, Lahm H, Little M, Weiss S. Invasion of tumorigenic HT1080 cells is impeded by blocking or downregulating the 37-kDa/67-kDa laminin receptor. J Mol Biol. 2008;378:530–9. doi: 10.1016/j.jmb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy JB, Furcht LT. Laminin and fibronectin promote the haptotactic migration of B16 mouse melanoma cells in vitro. J Cell Biol. 1984;98:1474–80. doi: 10.1083/jcb.98.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy JB, Palm SL, Furcht LT. Migration by haptotaxis of a Schwann cell tumor line to the basement membrane glycoprotein laminin. J Cell Biol. 1983;97:772–7. doi: 10.1083/jcb.97.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pupa SM, Menard S, Forti S, Tagliabue E. New insights into the role of extracellular matrix during tumor onset and progression. J Cell Physiol. 2002;192:259–67. doi: 10.1002/jcp.10142. [DOI] [PubMed] [Google Scholar]
- 46.Graf J, Iwamoto Y, Sasaki M, Martin GR, Kleinman HK, Robey FA, Yamada Y. Identification of an amino acid sequence in laminin mediating cell attachment, chemotaxis, and receptor binding. Cell. 1987;48:989–96. doi: 10.1016/0092-8674(87)90707-0. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka M, Narumi K, Isemura M, Abe M, Sato Y, Abe T, Saijo Y, Nukiwa T, Satoh K. Expression of the 37-kDa laminin binding protein in murine lung tumor cell correlates with tumor angiogenesis. Cancer Lett. 2000;153:161–8. doi: 10.1016/s0304-3835(00)00365-7. [DOI] [PubMed] [Google Scholar]
- 48.Tseng JC, Levin B, Hurtado A, Yee H, Perez de Castro I, Jimenez M, Shamamian P, Jin R, Novick RP, Pellicer A, Meruelo D. Systemic tumor targeting and killing by Sindbis viral vectors. Nat Biotechnol. 2004;22:70–7. doi: 10.1038/nbt917. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the sterochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]