Abstract
Previously, we showed that surface protective antigen (Spa) proteins of Erysipelothrix rhusiopathiae can be classified into three molecular species—SpaA, SpaB, and SpaC—and that SpaC is the most broadly cross-protective antigen among the three Spa proteins. In this study, we examined the ability of the α-helical domain, which comprises the N-terminal half of SpaC, to elicit cross-protective immunity in mice and pigs. Mice actively immunized with the full-length protein (rSpaC664) or the α-helical domain (rSpaC427), but not the C-terminal domain (rSpaC253), were protected against challenge with E. rhusiopathiae serovars 1a, 2, 6, 19, and 18 expressing heterologous (SpaA or SpaB) and homologous (SpaC) Spas. The α-helical domain seemed to provide better protection than rSpaC664, although the differences did not reach statistical significance. Similarly, mice passively immunized with rabbit anti-rSpaC664 or anti-rSpaC427 sera, but not anti-rSpaC253 serum, were protected from challenge with various serovars. Pigs immunized with SpaC427 also developed specific antibodies against Spa proteins and were protected from challenge with the highly virulent heterologous E. rhusiopathiae strain Fujisawa (serovar 1a). Taken together, these results demonstrate for the first time the striking protective efficacy of the α-helical domain-mediated immunization in both mice and pigs, thereby highlighting its utility as the most promising candidate for the development of a safe and effective vaccine against erysipelas.
Erysipelothrix rhusiopathiae is a small Gram-positive rod bacterium that causes erysipelas in swine and a variety of diseases in other animals, as well as erysipeloid, a skin disease of humans. Swine erysipelas, a disease causing enormous economic losses in pig production, can occur as an acute septicemia or chronic polyarthritis, lymphadenitis, and endocarditis (25). The genus Erysipelothrix contains two main species, Erysipelothrix rhusiopathiae (including serovars 1a, 1b, 2a, 2b, 4, 5, 6, 8, 9, 11, 12, 15, 16, 17, 19, and 21 and type N) and Erysipelothrix tonsillarum (including serovars 3, 7, 10, 14, 20, 22, and 23), and two unclassified species (including serovars 13 and 18) (19).
In erysipelas, antibodies against a cell surface component(s) of the organism have been known to play an important role in protection (4). It has been reported that a 66- to 64-kDa protein in a Triton X-100 extract of cell surface antigen is a protective molecule (3). Recently, a gene encoding a 69.9-kDa surface protective antigen (spaA) was cloned from Tama-96 (serovar 2) (9) and Fujisawa strains (serovar 1a) (6, 16), and its nucleotide sequence was determined. The genetic region responsible for protective immunity in SpaA molecule has also been suggested.
Our previous study revealed that the surface protective antigen (Spa) proteins of E. rhusiopathiae could be classified into three molecular species, named SpaA (produced by serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17, and N), SpaB (produced by serovars 4, 6, 11, 19, and 21), and SpaC (produced by serovar 18), and also indicated that SpaC is the most broadly cross-protective antigen among the three Spa proteins in murine model. Sequence analysis showed that the amino acid sequence similarities within each Spa of various serovar strains are 96 to 99% in SpaA and 96 to 99% in SpaB. In contrast, the similarities between different Spas are 61 to 64% (between SpaA and SpaB), 63 to 65% (between SpaA and SpaC), and 66 to 67% (between SpaB and SpaC). It has been demonstrated that the signal and repetitive amino acid regions are highly conserved among Spa proteins (100% and 83 to 88% identity, respectively), and the α-helical coding region is highly variable among Spas (∼50% identity) (21).
Like other surface proteins of Gram-positive bacteria (1, 5, 28), the SpaC protein is also made up of three major amino acid sequence regions. The C-terminal 20-amino-acid repeat region attaches the protein to the cell surface, which is conserved among the Spa proteins. Upstream of this region is the proline-rich region consisting of five tandem repeats of 6 amino acids. The N-terminal half of SpaC is the α-helical and amphipathic structures, which is exposed on the bacterial surface. This region is hypervariable in both size and sequence among Spa proteins and is shown to play a role in immunoprotection against E. rhusiopathiae infection. In the present study, we evaluated whether the α-helical domain of SpaC induces cross-protective immunity against challenge with various serovars in mice, as well as pigs, a target host of this vaccine candidate.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. rhusiopathiae strains used in the present study were Fujisawa (serovar 1a), ATCC 19414T (serovar 2), Dolphin E-1 (serovar 6), IV 12/8 (serovar 11), 2017 (serovar 19), and 715 (serovar 18). E. rhusiopathiae strains were grown in tryptose phosphate broth supplemented with 1% proteose peptone no. 3 (Difco Laboratories, Detroit, MI) and 0.1% Tween 80 (pH 7.8).
PCR amplification, cloning, and expression of recombinant fusion proteins.
PCR products containing the nucleotide sequences of interest (the full-length or truncated DNA fragments of SpaC) were amplified from genomic DNA of E. rhusiopathiae strain 715 (serovar 18). Oligonucleotide primers used in the PCR amplification experiments were obtained from a commercial source (Sawady Technology Co., Ltd., Tokyo, Japan) and are listed in Table 1. The PCR was performed as previously described (23). The PCR products were ligated into plasmid pGEM-T Easy (Promega, Madison, WI), and both strands of the cloned DNA were sequenced to verify that no changes have occurred during the PCR process. Then, BamHI-SalI fragments containing sequences coding for the full-length SpaC (residues 1 to 664, 664 amino acids, SpaC664) and the N-terminal half of SpaC (residues 1 to 427, 427 amino acids, SpaC427) were ligated into BamHI/SalI-digested pQE30 (Qiagen, Santa Clarita, CA). The BamHI-XhoI fragment containing sequence coding for the C-terminal domain of SpaC (residues 412 to 664, 253 amino acids, SpaC253) was ligated into BamHI/XhoI-digested pET21a (Novagen, Madison, WI). The ligated DNAs were transformed into Escherichia coli XL1-Blue or E. coli BL21(DE3) by electroporation as described elsewhere (21, 29).
TABLE 1.
Primers used for PCR amplificationa
| Target fragment of spaC | Primerb | Sequence (5′-3′) |
|---|---|---|
| Fragment encoding the full length | Er-1F | AGGATCCATGAAAAAGAAAAAACACCTATTTCCGAAAGTA |
| Er-2R | GAAGCTTCTATTTTAAACTTCCATCGTTCTTAAATGCATA | |
| Fragment encoding the N-terminal end | Er-1F | AGGATCCATGAAAAAGAAAAAACACCTATTTCCGAAAGTA |
| Er-3R | CGGTTTCTTTTGATCACCCGGT | |
| Fragment encoding the C-terminal end | Er-4F | TGTTGGATCCCCTGAAAGTCCTATTAAAGTA |
| Er-5R | CTGTCTCGAGTTTTAAACTTCCATCGTTCTT |
All PCRs were started by initial denaturing at 94°C for 3 min, followed by 25 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 5 min. Primers were designed on the basis of spaC nucleotide sequence of E. rhusiopathiae strain 715 (serovar 18; GenBank accession number AB238210).
F, forward; R, reverse.
The expression of recombinant histidine-tagged proteins in E. coli was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM. The recombinant proteins were purified by column chromatography with Ni-nitrilotriacetic acid resin (Qiagen) according to the recommendations of the manufacturer.
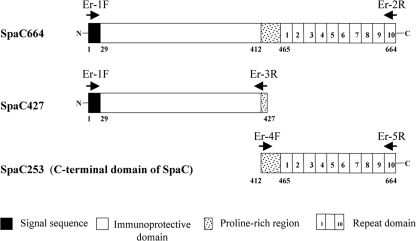
The SpaC664 full-length or truncated gene products, SpaC427 and SpaC253, generated in the present study are illustrated in Fig. 1.
FIG. 1.
Schematic representation of the full-length and truncated gene products (SpaC427, the N-terminal-half domain; and SpaC253, the C-terminal domain). The numbers indicate the first and last amino acid (aa) residues of each construct. The 29-amino-acid signal sequence, immunoprotective domain (the α-helical domain), proline-rich region, and 20-amino-acid repeat domain are indicated by solid, open, stippled, and numbered boxes, respectively. Arrows indicate the position and orientation of primer sites. F, forward; R, reverse.
Preparation of polyclonal antisera.
For preparation of rabbit anti-rSpaC664, anti-rSpaC427, and anti-rSpaC253 antisera, the purified rSpaC664, rSpaC427 and rSpaC253 proteins were run and excised from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The gel slices were washed three times (20 min each) and then ground in distilled water. The antigen mixture was emulsified with an equal volume of Freund complete adjuvant just before immunization. Groups of two rabbits were injected subcutaneously (s.c.) with antigen preparation (250 μg of the purified rSpaC664, rSpaC427, or rSpaC253 protein in 1 ml of emulsion of the antigen mixture and Freund complete adjuvant). Booster injections were given thrice in weeks 3, 5, and 7 as described above, except that Freund incomplete adjuvant was used. Blood samples were collected before immunization and at week 9 to determine serum antibody titers by Western blotting. Rabbits were exsanguinated at week 10 to collect the sera.
To prepare mouse polyclonal antisera against E. rhusiopathiae strains expressing SpaA, SpaB, and SpaC, groups of five mice aged 5 weeks (Japan SLC, Shizuoka, Japan) were immunized s.c. twice at 2-week intervals with 0.5 ml of formalinized whole-cell vaccines (approximately 5 × 108 cells) of Fujisawa (serovar 1a, SpaA), Dolphin E-1 (serovar 6, SpaB), or 715 (serovar 18, SpaC) in alum adjuvant. Two weeks after immunization, mice were challenged s.c. with approximately 2.2 × 102 CFU of the strain Fujisawa, 3.6 × 104 CFU of the strain Dolphin E-1, or 6.1 × 102 CFU of the strain 715. Two weeks after the challenge, mice were euthanized and bled, and sera were filtered and stored at −20°C.
Preparation of native SpaA, SpaB, and SpaC.
Native Spa proteins of E. rhusiopathiae strains Fujisawa (serovar 1a), Dolphin E-1 (serovar 6), and 715 (serovar 18) were extracted by alkaline treatment as described previously (4). Briefly, smooth colonies of the three strains were grown in a medium at 37°C overnight. Cells harvested and washed with phosphate-buffered saline (PBS) were suspended in 10 mM NaOH at 0.05 g (wet weight)/ml and incubated with constant stirring for 18 h at 4°C. After neutralization, the suspension was centrifuged, concentrated, filter sterilized, and stored at −70°C.
SDS-PAGE and Western blotting.
SDS-PAGE was performed as previously described (7). Antigenic proteins were probed with polyclonal rabbit and mouse antisera and then visualized with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse immunoglobulins (Zymed Laboratories, Inc.) and a substrate solution containing 4-chloro-1-naphthol and hydrogen peroxide.
Mouse immunization and challenge.
Specific-pathogen-free (SPF) ddY mice aged 4 weeks were obtained from commercial vendor (Japan SLC, Shizuoka, Japan) and quarantined for 1 week before use. For passive immunization experiments, mice were injected intraperitoneally (i.p.) with 0.20 ml of anti-rSpaC664, anti-rSpaC427, or anti-rSpaC253 rabbit antisera. Four hours after injection of the antiserum, control and immunized mice (ten per group) were challenged s.c. with 1.8 × 101 CFU of the strain Fujisawa (serovar 1a), 3.1 × 101 CFU of the strain 2017 (serovar 19), or 4.2 × 101 CFU of the strain 715 (serovar 18) and were observed for death for 14 days. For active immunization experiments, mice were immunized s.c. twice at 3-week intervals with approximately 40 μg of the purified His-tagged recombinant proteins rSpaC664, rSpaC427, or rSpaC253 emulsified with an equal volume of an oil-based adjuvant, Montanide IMS 1113 (SEPPIC, Paris, France). Nonimmunized mice were added to experimental group as negative controls. At 2 weeks after boosting, the control and immunized mice were subdivided into groups of 10 each, and each group was challenged s.c. with 3.3 × 102 CFU of the strain Fujisawa (serovar 1a), 3.5 × 102 CFU of the strain ATCC 19414T (serovar 2), 4.6 × 104 CFU of the strain Dolphin E-1 (serovar 6), 3.2 × 102 CFU of the strain 2017 (serovar 19), or 7.1 × 102 CFU of the strain 715 (serovar 18). Mouse mortality was monitored daily for the following 14 days. To examine specific antibody responses to recombinant SpaA, SpaB, SpaC664, SpaC427, and SpaC253 proteins, the control and immunized mice (ten per group) were bled 14 days after the boosting, and sera were stored at −20°C.
Pig immunization and challenge.
Nine SPF pigs at the age of 4 weeks obtained from a farm free from erysipelas where no pigs were vaccinated against the disease were divided into two groups. Six pigs were immunized intramuscularly with 100 μg of the purified His-tagged recombinant protein rSpaC427 emulsified with an equal volume of an oil-based adjuvant Montanide IMS 1113 twice at a 3-week interval. The remaining three pigs served as nonimmunized controls. The immunized pigs were daily observed for adverse reactions following injections of vaccine. Two weeks after the second immunization, the pigs were challenged intradermally with 1.0 × 106 CFU of serovar 1a strain Fujisawa. After the challenge, clinical signs of erysipelas, including increased body temperature, depression of activity, skin lesions, and death, were monitored and recorded. Dead pigs were autopsied on the day of death, and pigs that survived were euthanized and autopsied 1 week after challenge. Organs (heart, lung, liver, spleen, kidney, lymph nodes, and tonsils), synovial fluid of hock joints, and skin erythema lesions of pigs were examined by bacterial isolation. Blood samples collected immediately before vaccination and before challenge were used for the determination of anti-Spa antibodies by enzyme-linked immunosorbent assay (ELISA).
All of the animals used in the present study were cared for in accordance with the guidelines for animal treatment of Nippon Institute for Biological Science, which conform to the standard principles of laboratory animal care.
ELISA.
The procedures used to determine the titers of Spa antibodies in mouse and pig sera were similar to those described previously (22), with minor modifications. Briefly, polystyrene microtiter plates (96-well assay plate; Becton Dickinson, Franklin Lakes, NJ) were coated overnight at 4°C with 50 μl per well of the purified His-tagged recombinant proteins rSpaA, rSpaB, rSpaC664, rSpaC427, and rSpaC253 at concentrations of 5 μg/ml in bicarbonate coating buffer. Various dilutions of sera in PBS containing 0.1% Tween 80 and 1% gelatin were tested in duplicate. The secondary antibodies used were horseradish peroxidase-conjugated goat anti-mouse immunoglobulin IgG (Zymed Laboratories, Inc.) and horseradish peroxidase-conjugated anti-swine immunoglobulin IgG (heavy and light chains; Rockland). Optical densities were read at 492 nm (OD492) by using a SpectraMax 250 automated microplate reader (Molecular Devices, Sunnyvale, CA) with SoftMax Pro 2.0 software. The serum dilution for which an absorbance reading of 0.1 was recorded after background subtraction was considered the titer of this serum.
Statistical methods.
Comparisons of live-versus-dead numbers in mouse experiments were compared by the Fisher exact test using SAS software (SAS Institute, Inc., Cary, NC) (13).
RESULTS
Expression and purification of recombinant and native Spas.
The sequence analysis of the amplified DNA fragments revealed that the sizes of DNA fragments coding for SpaC664, SpaC427, and SpaC253 were 1,995, 1,281, and 759 nucleotides, respectively (encoding 664, 427, and 253 amino acids, respectively). The predicted molecular masses of the three proteins were 76.9, 49.4, and 29.3 kDa, respectively. These DNA fragments were then excised from the cloning vector pGEM-T Easy and ligated to the expression vectors pQE30 and pET21a, which had also been digested with the same enzymes. The histidine-tagged fusion proteins were expressed in E. coli and were purified by column chromatography with Ni-nitrilotriacetic acid resin.
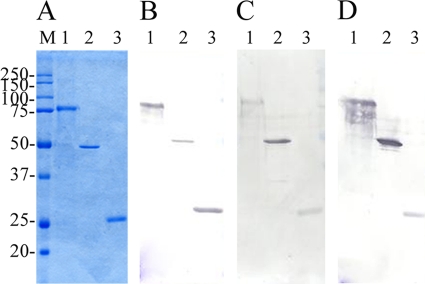
The purified full-length protein rSpaC664 gave a prominent 77-kDa band on SDS-PAGE gels. The purified truncated proteins, rSpaC427 and rSpA253, gave the prominent bands at ca. 49.4 and 29.3 kDa, respectively (Fig. 2A). In Escherichia coli, production level for the truncated rSpaC427 protein is higher than that for rSpaC664 (data not shown). In Western blotting analysis, anti-rSpaC664 rabbit antisera reacted with the bands of the rSpaC664, rSpaC427, and rSpaC253 proteins, and anti-rSpaC427 rabbit antisera reacted with the bands of rSpaC664 and rSpaC427 proteins but did not react with that of the rSpaC253 protein. In addition, anti-rSpaC253 rabbit antisera reacted only with the bands of rSpaC664 and rSpaC253 proteins (data not shown).
FIG. 2.
(A to D) Coomassie blue-stained SDS-12% PAGE gels (A) and immunoblot analysis of purified recombinant full-length and truncated SpaC proteins probed with polyclonal antisera raised against E. rhusiopathiae strain Fujisawa expressing SpaA (B), Dolphin E-1 expressing SpaB (C), and 715 expressing SpaC (D). Lane 1, rSpaC664 protein; lane 2, rSpaC427 protein; lane 3, rSpaC253 protein. Lane M, molecular masses in kilodaltons.
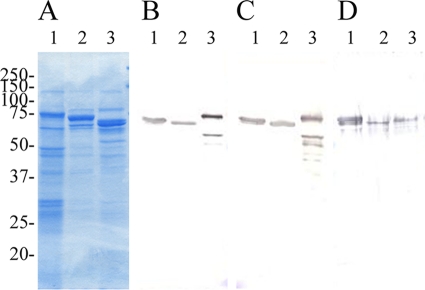
SDS-PAGE profiles of alkaline extracts of E. rhusiopathiae strains, which contained native Spas, are shown in Fig. 3A. The SDS-PAGE profiles of the extracts found in the present study were very similar to those reported previously (21), in which it was suggested that the alkaline extracts of E. rhusiopathiae strains contained mainly native Spa proteins.
FIG. 3.
(A to D) Coomassie blue-stained SDS-12% PAGE gels (A) and immunoblot analysis of alkaline extracts of E. rhusiopathiae strains (native Spa proteins) probed with polyclonal antisera from mice immunized with rSpaC664 protein (B), rSpaC427 protein (C), and rSpaC253 protein (D). The alkaline extracts of E. rhusiopathiae strains Fujisawa expressing SpaA (lane 1), Dolphin E-1 expressing SpaB (lane 2), and 715 expressing SpaC (lane 3). Numbers on the left are molecular masses in kilodaltons.
Reactivity of native and recombinant Spas with their polyclonal mouse antisera.
To examine the cross-reactivity of native Spas (SpaA, SpaB, and SpaC) and rSpaC proteins (rSpaC664, rSpaC427, and rSpaC253) with their mouse antisera, the specificity of antibodies produced against the native or recombinant Spa proteins was determined by Western blotting.
The polyclonal mouse antisera raised against strains Fujisawa (expressing SpaA), Dolphin E-1 (expressing SpaB), or 715 (expressing SpaC) reacted strongly with the three recombinant proteins rSpaC664, rSpaC427, and rSpaC253 (Fig. 2B, C, and D). The anti-rSpaC664, rSpaC427, and rSpaC253 mouse antisera also reacted strongly with the native Spa proteins SpaA, SpaB, and SpaC from alkaline extracts (Fig. 3B, C, and D) or from whole-cell lysates prepared from strains Fujisawa, Dolphin E-1, and 715, respectively (data not shown). The results indicated that the animals immunized to the native Spas also made antibodies reactive with both native and recombinant Spas, and vice versa.
Active immunization experiments.
To determine the region mainly responsible for cross-protective activity of SpaC protein, mice were actively immunized with the truncated and full-length SpaC proteins and then challenged s.c. with strains Fujisawa (serovar 1a) and ATCC 19414T (serovar 2), which express SpaA; Dolphin E-1 (serovars 6) and 2017 (serovars 19), which express SpaB; and 715 (serovars 18), which expresses SpaC (Table 2).
TABLE 2.
Active immunization of mice against challenge with various serovar strains of E. rhusiopathiae
| Immunizationa | Challenge strain (serovar, Spa protein expressed)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 715 (18, SpaA) |
2017 (19, SpaB) |
Dolphin E (6, SpaB) |
ATCC 19414T (2, SpaA) |
Fujisawa (1a, SpaA) |
||||||
| No. alive/ total no. | P | No. alive/ total no. | P | No. alive/ total no. | P | No. alive/ total no. | P | No. alive/ total no. | P | |
| rSpaC664 | 8/10 | 0.0004 | 9/10 | 0.0001 | 7/10 | 0.0015 | 8/10 | 0.0004 | 7/10 | 0.0015 |
| rSpaC427 | 10/10 | 0.0000 | 10/10 | 0.0000 | 9/10 | 0.0001 | 10/10 | 0.0000 | 10/10 | 0.0000 |
| rSpaC253 | 0/10 | 1.0000 | 0/10 | 1.0000 | 1/10 | 0.3049 | 0/10 | 1.0000 | 0/10 | 1.0000 |
| Control | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | |||||
Mice were immunized and boosted 3 weeks apart with 40 μg and challenged 2 weeks after the boost.
Data are presented as the number of mice alive/the total number of animals tested. The survival of mice was monitored for 14 days after challenge with strains of E. rhusiopathiae. P values are based on a comparison of alive/dead ratios for immunized mice with those for control mice, as calculated by the Fisher exact test.
After challenge, all rSpaC253-immunized and control mice showed clinical symptoms such as depression and anorexia; and died within 3 to 6 days, whereas 7 to 9 of 10 mice and 9 to 10 of 10 mice in groups immunized with rSpaC664 and SpaC427, respectively, survived until day 14 when the experiment was terminated. The mice immunized with rSpaC664 and rSpaC427 were also protected against challenge with not only the homologous strains (715 expressing SpaC) but also the heterologous strains (Fujisawa and ATCC 19414T expressing SpaA; Dolphin E-1 and 2017 expressing SpaB). The observed protection in the mice immunized with rSpaC664 and rSpaC427 was shown using the Fisher exact test to be significantly different than the control mice (Table 2). The number of rSpaC427-immunized mice that survived the homologous and heterologous challenges was higher than the number of survivors in the groups of mice immunized with rSpaC664, although the differences did not reach statistical significance. Taken together, these results indicate that protection elicited by the rSpaC427 is much more complete, per gram, than that elicited by rSpaC664, suggesting that the protection-eliciting epitopes of SpaC would be mainly located within the α-helical domain. These results also indicate that the C-terminal domain alone cannot function as a vaccine to provide protection against challenge exposure with E. rhusiopathiae strains.
Passive immunization experiments.
We then sought to determine whether anti-rSpaC664, anti-rSpaC427, or anti-rSpaC253 sera can confer cross-protection against challenge with various serovars. At 4 h after i.p. injection of 0.2 ml of the antiserum, mice were challenged s.c. with three E. rhusiopathiae strains, Fujisawa (serovar 1a), 2017 (serovar 19), and 715 (serovar 18) expressing SpaA, SpaB, and SpaC, respectively, and observed for clinical symptoms and death for 14 days.
After challenge exposure, 9 to 10 (90 to 100%) of anti-rSpaC253 serum-treated and control mice died within 4 to 6 days, whereas 9 to 10 (90 to 100%) of anti-rSpaC664 or anti-rSpaC427 serum-treated mice survived until day 14 (Table 3). Our findings indicate that anti-rSpaC664 and anti-rSpaC427 sera could provide passive protection against challenge with various E. rhusiopathiae serovar strains and also suggest that antibodies directed against the α-helical domain of SpaC could confer broad cross-protection.
TABLE 3.
Passive immunization of mice against challenge with various serovar strains of E. rhusiopathiae
| Immunizationa | Challenge strain (serovar, Spa protein expressed)b |
|||||
|---|---|---|---|---|---|---|
| 715 (18, SpaC) |
2017 (19, SpaB) |
Fujisawa (1a, SpaA) |
||||
| No. alive/ total no. | P | No. alive/ total no. | P | No. alive/ total no. | P | |
| rSpaC664 | 10/10 | 0.0000 | 10/10 | 0.0000 | 9/10 | 0.0001 |
| rSpaC427 | 10/10 | 0.0000 | 10/10 | 0.0000 | 9/10 | 0.0001 |
| rSpaC253 | 1/10 | 0.3049 | 1/10 | 0.3049 | 0/10 | 1.0000 |
| Control | 0/10 | 0/10 | 0/10 | |||
At 4 h after i.p. injection of 0.2 ml of serum, mice were challenged s.c. with strains of E. rhusiopathiae.
Data are presented as the number of mice alive/the total number of animals tested. The survival of mice was monitored for 14 days after challenge with strains of E. rhusiopathiae. P values are based on a comparison of alive/dead ratios for immunized mice with those for control mice, as calculated by the Fisher exact test.
Antibody response in mice immunized with rSpaC664, rSpaC427, and rSpaC253.
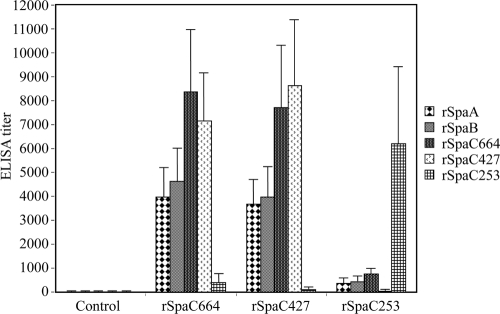
Humoral immune response in mice immunized with the recombinant proteins (rSpaC664, rSpaC427, or rSpaC253) was evaluated by using ELISA with the rSpaA, rSpaB, rSpaC664, rSpaC427, and rSpaC253 proteins as coating antigens (Fig. 4). Immunization with either rSpaC664 or rSpaC427 led to significant rises in antibody titers to the homologous and heterologous rSpa proteins. The titers obtained with the homologous antigens (rSpaC664 and rSpaC427) were significantly higher than those observed with the heterologous antigens (rSpaA and rSpaB), but the differences between anti-rSpaC664 and anti-rSpaC427 titers were not statistically significant.
FIG. 4.
ELISA detection of rSpaA, rSpaB, rSpaC664, rSpaC427, and rSpaC253 antibody titers in sera of mice vaccinated with rSpaC664, rSpaC427, or rSpaC253 and of the nonimmunized control mice. Mice were immunized and boosted 3 weeks apart with 40 μg of rSpas. Serum samples were collected at 14 days after boosting. Antibody titers are expressed as the average titer ± the standard deviation of the average of 10 mice each.
Pig protection study.
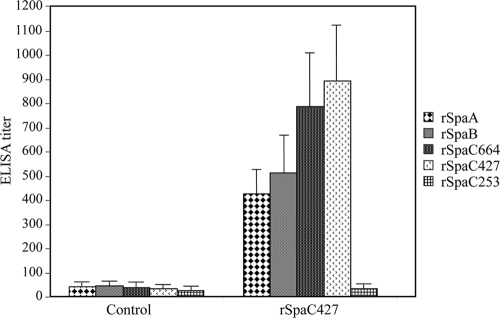
We did not detect abnormal clinical signs or local reactions in all pigs following the vaccination with purified rSpaC427. All pigs were seronegative before the first vaccination. Vaccinated pigs were seropositive 14 days after the second vaccination, whereas control pigs remained seronegative throughout the study. Pigs immunized with the purified rSpaC427 generated high levels of antibodies to the homologous and heterologous Spa proteins, as detected by ELISA (Fig. 5). Six pigs immunized with rSpaC427 did not show any clinical symptoms and remained healthy for 7 days after challenge with virulent strain Fujisawa expressing SpaA. The bacteria were not isolated from any organs examined. The control pigs showed typical clinical responses of E. rhusiopathiae infection, i.e., depression, anorexia, pyrexia (above 41°C), lameness due to arthritis, and generalized skin erythema. The control animals died within 4 days after challenge, and numerous E. rhusiopathiae bacteria were isolated from all body organs (Table 4). In our preliminary infection experiments, three E. rhusiopathiae strains—Dolphin E-1 (serovar 6) and IV 12/8 (serovar 11) expressing SpaB and 715 (serovar 18) expressing SpaC—could not induce any clinical sign of erysipelas in swine; therefore, these strains were not used for challenging pigs.
FIG. 5.
ELISA detection of rSpaA, rSpaB, rSpaC664, rSpaC427, and rSpaC253 antibody titers in the sera of pigs vaccinated with SpaC427 (n = 6) and of the nonimmunized control pigs (n = 3). Antibody titers are expressed as the average titer ± the standard deviation of the average of six or three pigs. Pigs were immunized and boosted 3 weeks apart with 100 μg of rSpac427. Serum samples were collected just before challenge.
TABLE 4.
Protection of pigs against erysipelas after immunization with truncated rSpaC427
| Immunizationa | Pig no. | Presence of clinical sign after challenge |
||||
|---|---|---|---|---|---|---|
| Tempb | Erythemac | Depression | Death | Isolation of E. rhusiopathiaed | ||
| rSpaC427 | 115 | − | + | No | No | No |
| 116 | + | + | No | No | No | |
| 117 | + | + | No | No | No | |
| 118 | + | + | No | No | No | |
| 119 | − | + | No | No | No | |
| 120 | + | + | No | No | No | |
| Control | 121 | ++ | +++ | Yes | Yes | Yes |
| 122 | +++ | +++ | Yes | Yes | Yes | |
| 123 | +++ | +++ | Yes | Yes | Yes | |
Pigs were immunized and boosted 3 weeks apart with 100 μg of rSpaC427 and challenged 2 weeks after the boost.
Maximum measured rectal temperature. -, <40°C; +, >40 to 41°C; ++, 41 to 42°C; +++, >42°C.
Skin lesion. +, urticarial lesion at the injection site; +++, systemic erythema.
Isolation results from tonsils, spleens, hearts, livers, lungs, kidneys, lymph nodes, and synovial fluid of hock joints.
DISCUSSION
Currently available live attenuated or killed vaccines have been used for the control of swine erysipelas for many years (14, 25). However, it has been suggested that the vaccines do not prevent the chronic form of the disease and that vaccination may cause an increase in arthritis lesions by hypersensitizing the animal to subsequent contact with the bacteria (2, 6, 11, 16, 24). Thus, the development of safer and more effective vaccines against erysipelas is highly desired. Our previous study revealed that (i) the Spa proteins could be divided into three molecular species, named SpaA, SpaB, and SpaC, and (ii) the rSpaC protein is the most potential cross-protective antigen in the murine lethal challenge model (21). In the present study, we evaluated a panel of truncated and full-length rSpaC proteins in the animal (mouse and pig) lethal challenge model and found that the α-helical domain (the N-terminal half) of the SpaC protein elicited cross-protective responses against challenge with different E. rhusiopathiae serovars.
The Spa proteins of E. rhusiopathiae have been proposed as good vaccine candidates, although Spa has been reported to be variable among strains (6, 9, 21). To be a potential vaccine candidate, the protein should induce antibodies that are able to cross-react and cross-protect. Our results revealed that active immunization with rSpaC427 (which corresponds to amino acid residues 1 to 427 of SpaC protein of strain 715 [serovar 18]) or passive administration of anti-rSpaC427 rabbit antisera could confer broadly cross-protective immunity against challenge with different E. rhusiopathiae strains in mice as well as pigs, the target species of this vaccine candidate. The present study also provided suggestive evidence that the proline-rich hydrophobic and C-terminal 20-amino-acid repeat region is not necessary for protection.
Our observation that the protection-eliciting epitopes of SpaC are located within the α-helical domain of the protein but not the proline-rich and 20-amino-acid repeat region is consistent with data from previous reports by Shimoji et al. (16) and Imada et al. (6). Shimoji et al. found that (i) in active immunization, mice immunized with rSpaA protein (from strain Fujisawa [serovar 1a]), but not the C-terminal amino acid repeat region, were protected against challenge with strain Fujisawa and that (ii) in passive immunization, mice treated with the C-terminal region-absorbed anti-rSpaA rabbit serum were protected from lethal challenge. Imada et al. showed that immunization of mice and pigs with HisSpaA342 protein of strain Fujisawa (amino acid residues 90 to 431) conferred protection against challenge with strains Fujisawa and 82-875 (serovars 1a and 2, respectively, expressing SpaA). We speculate that part if not most of the protection afforded mice actively immunized with SpaA proteins or passively immunized with the C-terminal region-absorbed anti-rSpaA.1 serum in the above-mentioned investigations likely resulted from antibodies elicited by the protection-eliciting epitopes of the α-helical domain.
Mice and pigs immunized with the purified rSpaC427 protein developed a strong humoral immune response with antibodies reactive against the rSpaC664 and rSpaC427 proteins, as well as the rSpaA and rSpaB proteins, as found by ELISA. The amount of cross-reactivity was unexpected considering the degree of sequence diversity in the α-helical N-terminal half among Spa proteins. This observation could therefore be due to the presence of common short linear peptides that act as B-cell epitopes or conformational epitopes of different primary sequence.
The immune response induced by immunization with rSpaC427 efficiently protected mice against challenge with representative strains of serovars 1 and 2, which express SpaA; of serovars 6 and 19, which express SpaB; and of serovar 18, which expresses SpaC. The data from passive-immunization studies were consistent with those obtained from active-immunization experiments. This result clearly indicated that the protection induced by immunization is not limited to strain 715 (serovar 18) that expresses a homologous Spa (SpaC). This is not surprising, since Western blot analyses revealed that anti-rSpaC427 mouse and rabbit antisera strongly reacted with native SpaA, SpaB, and SpaC proteins produced by representative strains of Spa types, and vice versa.
Moreover, it was observed that mice immunized with rSpaC253 (consisting of a proline-rich repeat region and a 20-amino-acid repeat region) did not survive lethal challenge with various serovars, thus indicating that the C-terminal 253-amino-acid domain of SpaC is not necessary for protection. Pigs immunized with rSpaC427 were completely protected against challenge with highly virulent strain Fujisawa expressing SpaA (heterologous). Strains Dolphin E-1 (serovar 6) and IV 12/8 (serovar 11) expressing SpaB (heterologous) and strain 715 (serovar 18) expressing SpaC (homologous) were not used for challenging pigs since these strains did not induce signs of acute generalized erysipelas in experimentally infected swine. A low pathogenicity of Erysipelothrix strains expressing SpaB and SpaC for pigs in experimental infection models has also been reported previously (12, 19, 20).
The mechanism by which anti-rSpaC427 antibodies passively protected mice against challenge with different E. rhusiopathiae serovars was not defined in the present study. Nevertheless, data from a report by Imada et al. (6) showed that anti-SpaA antibodies enhanced uptake and killing of the bacteria within swine macrophages after opsonization with anti-rSpaA swine antiserum. We assume that anti-rSpaC427 antibodies bind to the SpaC protein on the bacterial cell surface and act as opsonins to enhance bacterial uptake by phagocytic cells. Within the macrophages, binding of the antibodies to the bacteria triggers an oxidative burst during the ingestion and also interferes with the replication of the bacteria (17, 18).
The C-terminal region of Spas consisting of 9 to 10 highly conserved 20-amino-acid repeats has been suggested to function as an anchor for binding Spa protein tightly to the bacterial surface (8), as it was found in other surface proteins of Gram-positive bacteria (1, 5, 15, 28). The repeat region of SpaC has high sequence homology with the repeats of Streptococcus pneumoniae secretory IgA binding protein (SpsA) (48% over 200 amino acids) (5) and pneumococcal surface protein A (PspA) (46% over 200 amino acids) (27). Previous studies show that secretory immunoglobulin A and the secretory component binding activity of SpsA and the protective epitopes of PspA are located within the α-helical domain, which comprises the N-terminal half of molecule. The proline-rich domain of PspA and SpsA of S. pneumoniae has been localized within the cell wall (5, 10). Similar to the proline-rich regions of PspA and SpsA of S. pneumoniae, it could be postulated that those of Spas likely serve as a flexible tether anchoring Spas to the cell wall through the repeat region. These regions have been thought to span the cell wall and capsule layer and to contain none of protection-eliciting epitopes (26).
We have presented here results demonstrating for the first time that vaccination with the recombinant SpaC427, the N-terminal fragment of SpaC, gives an antibody response that is highly protective in mice and pigs against challenge with various serovars. From these results, we suggest that SpaC427 is a potential candidate for the development of a safe and effective vaccine for eradication of erysipelas.
Acknowledgments
We thank members of the Second Division of the Nippon Institute for Biological Science for the excellent care in the animal experiments.
This study was supported by a Grant-in-Aid from the Ministry of Agriculture, Forestry, and Fisheries of Japan via the Japanese Association of Veterinary Biologics.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman, M. J. 1964. Effects of vaccination on the development of arthritis in swine with erysipelas: clinical, hematologic, and gross pathologic observations. Am. J. Vet. Res. 25:589-597. [PubMed] [Google Scholar]
- 3.Galan, J. E., and J. F. Timoney. 1990. Cloning and expression in Escherichia coli of a protective antigen of Erysipelothrix rhusiopathiae. Infect. Immun. 58:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groschup, M. H., K. Cussler, R. Weiss, and J. F. Timoney. 1991. Characterization of a protective protein antigen of Erysipelothrix rhusiopathiae. Epidemiol. Infect. 107:637-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammerschmidt, S., S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol. 25:1113-1124. [DOI] [PubMed] [Google Scholar]
- 6.Imada, Y., N. Goji, H. Ishikawa, M. Kishima, and T. Sekizaki. 1999. Truncated surface protective antigen (SpaA) of Erysipelothrix rhusiopathiae serotype 1a elicits protection against challenge with serotype 1a and 2b in pigs. Infect. Immun. 67:4376-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Makino, S., K. Yamamoto, H. Asakura, and T. Shirahata. 2000. Surface antigen, SpaA, of Erysipelothrix rhusiopathiae binds to gram-positive bacterial cell surfaces. FEMS Microbiol. Lett. 186:313-317. [DOI] [PubMed] [Google Scholar]
- 9.Makino, S., K. Yamamoto, S. Murakami, T. Shirahata, K. Uemura, T. Sawada, H. Wakamoto, and H. Morita. 1998. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb. Pathog. 25:101-109. [DOI] [PubMed] [Google Scholar]
- 10.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. E. Briles. 1994. Location of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 11.Opriessnig, T., L. J. Hoffman, D. L. Harris, S. B. Gaul, and P. G. Halbur. 2004. Erysipelothrix rhusiopathiae: genetic characterization of midwest US isolates and live commercial vaccines using pulsed-field gel electrophoresis. J. Vet. Diagn. Invest. 16:101-107. [DOI] [PubMed] [Google Scholar]
- 12.Ozawa, M., K. Yamamoto, A. Kojima, M. Takagi, and T. Takahashi. 2009. Etiological and biological characteristics of Erysipelothrix rhusiopathiae isolated between 1994 and 2001 from pigs with swine erysipelas in Japan. J. Vet. Med. Sci. 71:697-702. [DOI] [PubMed] [Google Scholar]
- 13.SAS Institute, Inc. 1988. SAS/SAT user's guide, 6.03 ed. SAS Institute, Inc., Cary, NC.
- 14.Sawada, T., and T. Takahashi. 1987. Cross protection of mice and swine inoculated with culture filtrate of attenuated Erysipelothrix rhusiopathiae and challenge exposed to strains of various serovars. Am. J. Vet. Res. 48:239-242. [PubMed] [Google Scholar]
- 15.Shi, H., S. Wang, K. L. Roland, B. Gunn, and R. Curtiss III. 2010. Immunogenicity of a live recombinant Salmonella vaccine expressing pspA in neonates and infant mice born from naive and immunized mothers. Clin. Vaccine Immunol. 17:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimoji, Y., Y. Mori, and V. A. Fischetti. 1999. Immunological characterization of a protective antigen of Erysipelothrix rhusiopathiae: identification of the region responsible for protective immunity. Infect. Immun. 67:1646-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimoji, Y., Y. Yokomizo, and Y. Mori. 1996. Intracellular survival and replication of Erysipelothrix rhusiopathiae within murine macrophages: failure of induction of the oxidative burst of macrophages. Infect. Immun. 64:1789-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasa Rao, P. S., T. M. Lim, and K. Y. Leung. 2001. Opsonized virulent Edwardsiella tarda strains are able to adhere to and survive and replicate within fish phagocytes but fail to stimulate reactive oxygen intermediates. Infect. Immun. 69:5689-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi, T., T. Fujisawa, Y. Tamura, S. Suzuki, M. Muramatsu, T. Sawada, Y. Benno, and T. Mitsuoka. 1992. DNA relatedness among Erysipelothrix rhusiopathiae strains representing all twenty-three serovars and Erysipelothrix tonsillarum. Int. J. Syst. Bacteriol. 42:469-473. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi, T., N. Nagamine, M. Kijima, S. Suzuki, M. Takagi, Y. Tamura, M. Nakamura, M. Muramatsu, and T. Sawada. 1996. Serovars of Erysipelothrix strains isolated from pigs affected with erysipelas in Japan. J. Vet. Med. Sci. 58:587-589. [DOI] [PubMed] [Google Scholar]
- 21.To, H., and S. Nagai. 2007. Genetic and antigenic diversity of the surface protective antigen proteins of Erysipelothrix rhusiopathiae. Clin. Vaccine Immunol. 14:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.To, H., S. Someno, and S. Nagai. 2005. Development of a genetically modified nontoxigenic Pasteurella multocida toxin as a candidate for use in vaccines against progressive atrophic rhinitis in pigs. Am. J. Vet. Res. 66:113-118. [DOI] [PubMed] [Google Scholar]
- 23.To, H., G. Zhang, H. Otsuka, M. Ogawa, O. Ochiai, S. Nguyen, T. Yamaguchi, N. Nagaoka, M. Akiyama, K. Amano, and K. Hirai. 1996. Q. fever pneumonia in children in Japan. J. Clin. Microbiol. 34:647-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood, R. L., G. D. Booth, and R. C. Cutlip. 1981. Susceptibility of vaccinated swine and mice to generalized infection with specific serotypes of Erysipelothrix rhusiopathiae. Am. J. Vet. Res. 42:608-614. [PubMed] [Google Scholar]
- 25.Wood, R. L., and L. M. Henderson. 2006. Erysipelas, p. 629-638. In B. E. Straw et al. (ed.), Diseases of swine. Blackwell Publishing Professional, Ames, IA.
- 26.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequences analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, G., H. To, K. E. Russell, L. R. Hendrix, T. Yamaguchi, H. Fukushi, K. Hirai, and J. E. Samuel. 2005. Identification and characterization of an immunodominant 28-kilodalton Coxiella burnetii outer membrane protein specific to isolates associated with acute disease. Infect. Immun. 73:1561-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]