Abstract
The wood protection industry has refined their products from chrome-, copper-, and arsenate-based wood preservatives toward solely copper-based preservatives in combination with organic biocides. One of these is Cu-HDO, containing the chelation product of copper and N-cyclohexyldiazenium dioxide (HDO). In this study, the fate of isotope-labeled (13C) and nonlabeled (12C) Cu-HDO incorporated in wood sawdust mixed with soil was investigated. HDO concentration was monitored by high-pressure liquid chromatography. The total carbon and the δ13C content of respired CO2, as well as of the soil-wood-sawdust mixture, were determined with an elemental analyzer-isotopic ratio mass spectrometer. The concentration of HDO decreased significantly after 105 days of incubation, and after 24 days the 13CO2 concentration respired from soil increased steadily to a maximum after 64 days of incubation. Phospholipid fatty acid-stable isotope probing (PFA-SIP) analysis revealed that the dominant PFAs C19:0d8,9, C18:0, C18:1ω7, C18:2ω6,9, C17:1d7,8, C16:0, and C16:1ω7 were highly enriched in their δ13C content. Moreover, RNA-SIP identified members of the phylum Acidobacteria and the genera Phenylobacterium and Comamonas that were assimilating carbon from HDO exclusively. Cu-HDO as part of a wood preservative effectively decreased fungal wood decay and overall microbial respiration from soil. In turn, a defined bacterial community was stimulated that was able to metabolize HDO completely.
Wood degradation in terrestrial ecosystems is a very important process with wide ecological and economical consequences. To protect wooden constructions from biological deterioration, wood preservatives are routinely applied. In Europe, roughly 6.5 million m3 of mainly coniferous wood are treated annually with water-based wood preservatives (WEI, 2010; www.wei-ieo.org/woodpreservation.html). The main targets of wood preservatives are insects and fungi such as basidiomycetes, ascomycetes, and fungi imperfecti, causing white rot, brown rot, soft rot, and staining of wood. These organisms are also known to be the major degraders of wood in most terrestrial ecosystems.
The wood protection industry has refined its products from materials containing chrome, copper, and arsenate toward preservatives based on copper combined with organic biocides. One of these is Cu-HDO, which is the chelation product of copper and N-cyclohexyldiazenium dioxide (HDO). Cu-HDO has been in use since 1988 (13) and, as part of wood preservative formulations applied in regulated concentrations, enables high durability of wood against wood-rotting fungi (13) and termites (16) both in and out of soil contact (18). With similar Cu-HDO concentrations, incubation times, and conditions, the decay of preserved wood was found to be greater in experiments using soil than in pure cultures of wood-rotting basidiomycetes, a finding that was mainly attributed to the presence of a diverse soil microbial community (10).
Soil microorganisms involved in the degradation of preserved wood must have many characteristics, such as access to wood cells and tolerance to high copper concentrations and to the cobiocide HDO. Studies of xenobiotic-degrading bacteria have revealed that members of the genera Alicycliphilus, Polaromonas, and Rhodococcus are able to degrade cyclohexanol, which is related in its chemical structure to HDO (5, 25, 35). Whether such bacterial genera are able to cope with such defined habitat conditions (e.g., presence of copper compounds) and whether they are active key players in degrading preserved wood is questionable and has not been reported so far.
To identify substrate-degrading microorganisms and to estimate their degradation rates at environmental conditions, the stable isotope probing (SIP) technique has often been applied (1, 7, 9, 19, 22, 23, 24, 26). However, pesticides have been rarely used as substrates in situ for SIP analyses since their degradation rate is low, and therefore the assimilation into microbial biomass is crucial. Cupples and Sims (7) and recently Lerch et al. (19) applied SIP of DNA (DNA-SIP) and phospholipid fatty acids (PLFA-SIP), respectively, and found that the respective herbicide was bioavailable and mineralized by the soil microbial community (7, 19). However, SIP studies of biocides routinely applied for wood protection have not been performed to date. Identification of active HDO-degrading microorganisms in environmental conditions enables a defined optimization of the biocide formulation and may lead to innovative metal-free and environmentally friendly wood preservatives.
The main objective of the present study was to investigate whether the cobiocide HDO as part of a wood preservative is bioavailable in in situ conditions and can be mineralized and assimilated by the soil microbial community. Therefore, labeling experiments were conducted where carbon atoms of the cobiocide were isotopically labeled (13C), and the results were compared to those from nonlabeled-carbon (12C) experiments. To reduce the incubation time and to extend the availability of HDO, preserved wood, including and excluding Cu-HDO, respectively, was comminuted to sawdust and mixed with soil. PLFA-SIP was applied to estimate carbon incorporation into microbial biomass. After reasonable labeling of the biomass, RNA-SIP was conducted to identify microbial key players in a cobiocide degradation process in preserved wood.
MATERIALS AND METHODS
Soil and wood samples.
The soil used in the present study was collected at the BAM technical test site Horstwalde (52°05′44, 72″N; 13°24′34, 98″E) in Brandenburg, Germany, which is located in a glade surrounded by coniferous woodland. The test site has a diverse wood-degrading fungal community, as monitored by standardized tests such as DIN EN 252. Soil samples were taken from the upper 30 cm as the A horizon at the field. The soil is a sandy loam (sand, 80.46%; clay and silt, 19.47%) with the following characteristics: 1.23% (wt/wt) organic C of dry matter (according to DIN ISO 10693), 13% (wt/wt) gravimetric water content, 11.3% (wt/wt) TC/N ratio (organic carbon), 0.3% (wt/wt) carbonate content (DIN ISO 10693), 0.022% (wt/wt) sulfur content (DIN ISO 10694), and a pH of 7.3. The grain density according to DIN 18128 was 2.62 g m−3.
All soil samples were sieved through a 2-mm mesh and kept in a plastic bucket at about 15°C in darkness prior to experimental use. The soil was mixed 1:5 (wt/wt) with sterilized vermiculite to enhance the water-holding capacity of the soil and to enable a stable soil pH. Pine (Pinus sylvestris L.) sapwoods originated from the county Brandenburg (Germany). The trees were felled in September 2004. The wood was shredded and sieved, and particles (i.e., sawdust) with a size between 1 and 2 mm (±0.5 mm) were sterilized by gamma radiation of 70 kGy (Gammaservice, Radeberg, Germany).
Experimental setup.
Sapwood samples were amended with wood preservative formulation, including (i) 2 mg of nonenriched Cu-HDO (12C-Cu-HDO treatment [Dr. Wolman GmbH, Sinzheim, Germany]) g−1, (ii) 2 mg of 13C-enriched Cu-HDO (99% 13C enrichment of each carbon in the cyclohexyl ring of Cu-HDO [Dr. Wolman GmbH]; 13C-Cu-HDO treatment) g−1, or (iii) 2 ml of double-distilled H2O g−1 as a control. These sapwood samples were stored for 1 week at 22°C and 65% relative air humidity to remove the solvent (water) from each treatment. Subsequently, 10-g sapwood samples with a moisture content of ca. 9% were mixed accurately with 10 g of soil-vermiculite to receive a homogenized mixture, which will be referred to as wood-soil-vermiculite mixture (WSV mixture; 5:1:4 wood-soil-vermiculite [wt/wt/wt]). In all, 72 independent WSV mixtures were incubated at the same time and from the same batch at 20°C and at a relative air humidity of 65% in darkness. Three WSV mixtures were sampled destructively per treatment after a respective incubation time (three treatments multiplied by eight incubation times [directly and after 1, 2, 4, 8, 16, 32, 64, and 105 days of incubation] × three replicate measurements). Samples were incubated in gas-tight sealed bottles to avoid loss of respired CO2. The O2 concentration of the incubation was measured throughout the incubation. O2 was added if the O2 concentration was <17%. The entire incubation period was characterized by a constant pH of 7.3.
Microbial respiration.
The WSV mixture of five replicate samples of each treatment was placed independently in a glass tube (200-mm length, 40-mm inner diameter) of an infrared gas analyzer (ADC, Hoddesdon, United Kingdom) and incubated at 20°C in darkness. Glass tubes were sealed by rubber stoppers at the bottom and top and 7 g of sterilized glass wool (Roth, Karlsruhe, Germany) at the bottom to allow the continuous flow of sterile-filtered ambient air. As a control, one glass tube was filled with glass wool only. All glass tubes were flushed periodically by moistened air to maintain water content of ca. 45% (wt/wt). The CO2 concentration of each glass tube was automatically analyzed 17 times per day by the gas analyzer. Sterilized air (Midisart; Sartorius, Göttingen, Germany) was conveyed through the samples to aerate them and to transport respiration products to the detector of the gas analyzer with a flow rate of 100 ml min−1. All water and CO2 was removed from the air previously by flowing through silica gel and soda lime (both from Roth, Karlsruhe, Germany).
Extraction and quantification of Cu-HDO.
Cu-HDO was extracted from 10 g of each of the 72 WSV mixtures (Cu-HDO treatments and controls). Each sample was mixed with 28 ml of methanol (Merck, Darmstadt, Germany) and 15 ml of 0.05 M KH2PO4 (Merck). To reduce the pH from 7 to 3.5, up to 100 μl of 85% phosphoric acid (Merck) was added. Samples were sonicated by using a Sonorex Super 10P (Bandelin, Berlin, Germany) at 40°C for 60 min to release the Cu-HDO. After sedimentation of the WSV mixture at 4°C overnight, 2 ml of supernatant was filtered through a 0.45-μm-pore-size PTFE filter (Minisart SRP 15; Sartorius). HDO was quantified by high-pressure liquid chromatography (Agilent, Waldborn, Germany) equipped with a Luna C18(2)-column (length, 75 mm; inner diameter, 4.6 mm; particle size, 3 μm) at 40°C. Cu-HDO was eluted with a methanol buffer (45% methanol and 55% KH2PO4 [pH 2.5]) at a flow rate of 1 ml min−1 and detected by a UV detector at 229 nm. The Cu-HDO concentration was calculated based on a calibration curve of Cu-HDO as a standard by using Chrom Gate software (Knauer, Berlin, Germany).
Differences between treatments or time of incubation were tested by using Student's t test at a significance level of P < 0.05 (Excel software; Microsoft, Redmond, WA).
δ13 determination of respired CO2.
Microbial respired CO2 of 20 g of WSV mixture in sealed 100-ml glass bottles was precipitated by 0.01 mol of barium hydroxide solution as described earlier (31) in four replicates for each treatment. Precipitated barium carbonate was separated from liquid barium hydroxide by centrifugation (5 min, 4,300 × g, room temperature) and thereafter dried at 43°C. The δ13C value in CO2 was measured with an elemental analyzer (EA)-isotopic ratio mass spectrometer (IRMS) system (EA: Vario EL III; Elementar Analysensysteme GmbH, Hanau, Germany; IRMS: IsoPrime; GV Instruments, Manchester, United Kingdom). Approximately 1 to 3 mg of each barium carbonate sample was weighed into tin cups and burned under a supply of oxygen (35 ml min−1, 90 s) at 1,150°C. Combustion gases were carried by helium with a flow of 200 ml min−1 to the IRMS, and ionized CO2 was measured as ionic currents by a multicollector system composed of three Faraday cups. The intensities of the ionic currents of masses 44, 45, and 46 were used for the determination of isotopic ratios R(13C/12C), calibrated to the VPDB scale through CO2 reference gas and indicated in per mill (‰): δ13C = {[R(13C/12C)sample/R(13C/12C)standard] − 1} × 1,000.
PLFA-SIP.
To determine the δ13C value in PLFAs, ∼6.5 g of the WSV mixture was extracted from each treatment after 0, 16, 32, 64, and 105 days of incubation in three independent replicate measurements. All lipids were extracted in a mixture of 80 ml of methanol and 80 ml of chloroform by using a modified Bligh and Dyer extraction method (6). The solvents were removed by means of a rotary evaporator under reduced pressure at 50°C until it had just reached dryness. The residues were solved in 500 μl of chloroform and fractionated to neutral lipids, glycol, and phospholipids by column chromatography on a column (DSC-Si, 6-ml reservoir, 500 mg of sorbent [Sigma-Aldrich]) by sequential elution with chloroform, acetone, and methanol (36, 40). Fatty acids of the phospholipid fraction were released by a mild alkaline methanolysis and methylated for 2 h at 100°C in 0.5 ml of a solution containing chloroform, concentrated HCl, and methanol (1:1:10 [vol/vol/vol]) (12, 36). The individual fatty acid methyl esters were identified by using gas chromatography-mass spectrometry. PLFA amounts were determined based on peak areas in gas chromatograms with n-hexadecanoic acid methyl ester as an external standard, and their stable carbon-isotope compositions were determined by using an isotope ratio mass spectrometer (Finnigan MAT 252) coupled to an HP 5890 gas chromatograph with a Finnigan standard combustion interface (1). To calculate isotope ratios (δ13C) for the PLFAs, δ13C values of the fatty acid methyl esters (FAMEs) were corrected for the 13C content of the carbon atom of the methyl group that was added during methanolysis (3). The conversion into FAMEs occurred with 1 ml of fresh 0.2 M NaOH for 15 min at 37°C. The neutralization of the samples was performed by the addition of 2 ml of hexane, 0.3 M acetic acid, and 2 ml of double-distilled H2O. From the δ13C values of the PLFA extracted from the 13C-labeled soils and those from the unlabeled soils (control), the amount of 13C incorporation into each PLFA was calculated.
RNA-SIP.
After 105 days of incubation, reasonable amounts of 13C were incorporated into microbial PLFAs. Therefore, three replicates of 12C- and 13C-Cu-HDO treatment, respectively, were selected for RNA extraction. Aliquots of the soil were shock-frozen in liquid nitrogen and stored at −80°C until further analysis. Nucleic acids were extracted as described previously (27). RNA was prepared from the primary extract by digestion with RNase-free DNase (Promega, Mannheim, Germany) according to the method of Noll et al. (27). Complete removal of DNA was verified by PCR, and total RNA was quantified in 96-well microtiter plates using a Ribogreen RNA quantification kit (Invitrogen, Karlsruhe, Germany) and a synergy plate reader (Bio-Tek Instruments, Bad Friedrichshall, Germany) as specified by the manufacturer. The RNA contents were approximately 1.8 and 2.1 mg per g soil from the 12C- and 13C-Cu-HDO treatments, respectively.
Gradient centrifugation of ∼500 ng of RNA was carried out in cesium trifluoroacetate (CsTFA; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) (22). Gradients were fractionated from bottom to top by displacing the gradient medium with RNase- and DNase-free water at the top of the tube (22, 23) by using a perfusor system at a flow rate of 0.75 ml min−1. The density of each fraction was determined by refractometry (AR200; Reichert, Depew, NY). RNA was precipitated by mixing the sample with 2-propanol. The RNA pellets were resuspended in 25 μl of Tris-EDTA buffer (pH 7.4; Sigma-Aldrich) for further analysis.
Terminal restriction fragment length polymorphism (T-RFLP), cloning, and sequence analysis were carried out according to the method of Noll et al. (26), with minor modifications. Briefly, bacterial small-subunit rRNA using the 27f/907r primer pair (17) were amplified by reverse transcription-PCR (RT-PCR), and PCR products were restricted using the restriction enzyme MspI (Promega) as described earlier (27). For T-RFLP analysis, the forward primer was 5′-terminally labeled with FAM (5-carboxyfluorescein) and examined with an ABI Prism 3710 DNA sequencer (Applied Biosystems, Weilheim, Germany). Based on the T-RFLP patterns, the rRNA from fractions with buoyant densities of 1.851 g ml−1 of the 12C-Cu-HDO treatment and of 1.861 g ml−1 of the 13C-Cu-HDO treatment were chosen for a cloning and sequencing approach. RT-PCR products without a FAM label from both fractions were cloned into Escherichia coli DH5α (Promega) by using a pGEM-T cloning kit (Promega) according to the manufacturer's instructions. In all, 86 clones derived from each density fraction were randomly selected, and 16S rRNA segments were sequenced by GATC Biotech (Constance, Germany). After aligning the sequences using ARB, the chimeric structures were detected by separately analyzing the phylogeny of terminal stretches at the 5′ and 3′ ends (21). Phylogenetic placement and assignment to the terminal restriction fragments (T-RFs) were achieved by inclusion of the clone sequences and relevant public domain reference sequences into a publicly available database of 16S rRNA gene sequences, which were organized as a maximum-parsimony tree.
All clone sequences were deposited in the EMBL, GenBank, and DDBJ databases under the accession numbers HM146328 to HM146414.
RESULTS
Effect of the biocide Cu-HDO on the microbial community.
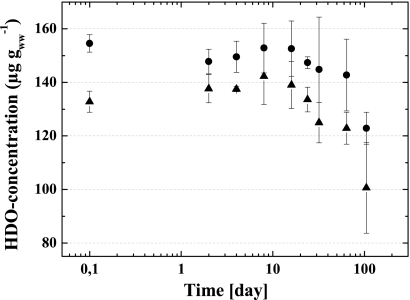
The HDO concentrations of the 13C-Cu-HDO- and 12C-Cu-HDO-amended WSV microcosms decreased significantly after 85 days of incubation (Fig. 1). The HDO concentration steadily decreased, and ca. 20% of the initial HDO concentration was degraded after 105 days of incubation.
FIG. 1.
HDO concentration of 13C-Cu-HDO (▴) and 12C-Cu-HDO (•) amended to WSV microcosms over incubation time. Means ± the standard deviations (SD) of three replicate measurements are shown. ww, wet weight.
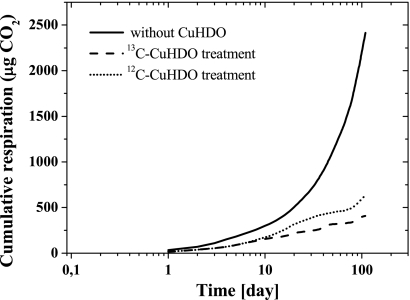
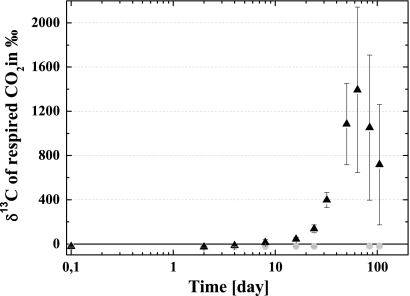
The presence of Cu-HDO in WSV significantly decreased the overall soil respiration compared to samples without Cu-HDO (Fig. 2). Although the δ13C levels of the microbial respired CO2 of the 12C-Cu-HDO and control treatments were constant during the incubation period, the δ13C level for the 13C-Cu-HDO treatment increased significantly over time, reaching a maximum after 64 days of incubation (Fig. 3). While the δ13C levels of the respired CO2 of the four independent replicate measurements differed from each other widely, each incubation had its maximum in 13C enrichment after 64 days of incubation.
FIG. 2.
Cumulative CO2 respiration in 13C-Cu-HDO- and 12C-Cu-HDO-amended and nonamended WSV microcosms during incubation, calculated by summation of the daily averages. Means ± the SD of five replicate measurements are shown.
FIG. 3.
δ13C ratio of respired CO2 of 13C-Cu-HDO (▴) and 12C-Cu-HDO (•) amended to WSV microcosms during incubation. Means ± the SD of four replicate measurements are shown.
Incorporation of 13C into PLFA and 16S rRNA.
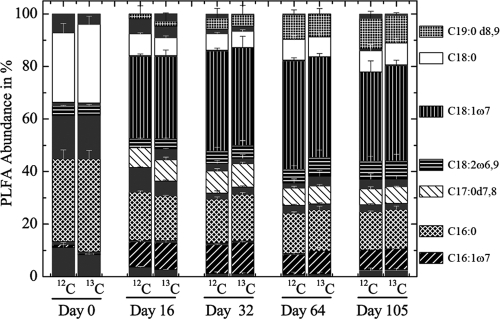
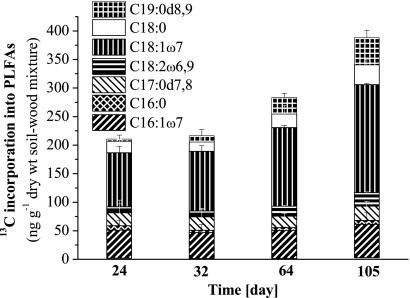
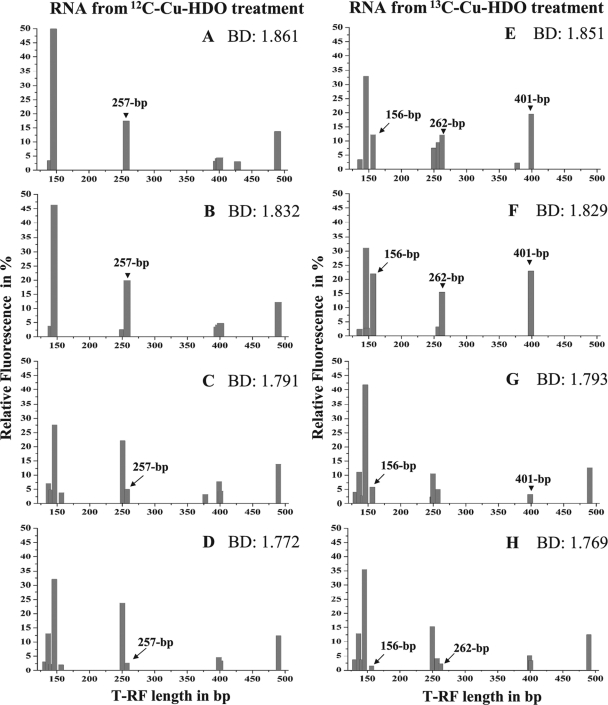
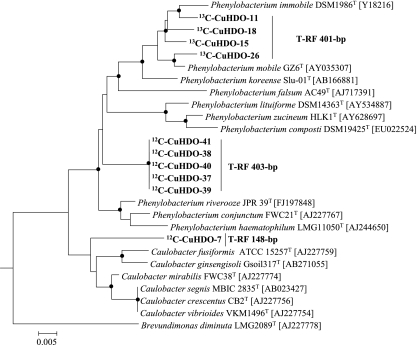
To gain more information on the soil microbial community that metabolized the cobiocide HDO in WSV, PLFAs and their δ13C levels were analyzed successively. The initial PLFA community shifted, and the PLFAs C19:0d8,9, C18:0, C18:1ω7, C18:2ω6,9, C17:1d7,8, C16:0, and C16:1ω7 became dominant (Fig. 4). The majority of these dominant PLFAs, reflecting ca. 70% of the entire PLFA diversity, were highly isotopically labeled, and their isotopic enrichment increased during the incubation period (Fig. 5). After 105 days of incubation, the isotopic labeling was highest, and comparative 16S rRNA-SIP analysis of the 12C-Cu-HDO and 13C-Cu-HDO treatments was conducted to obtain more detailed phylogenetic information. Amplification of archaeal 16S rRNA or fungal ITS RNA in buoyant densities of >1.80 g ml−1 was not successful. Bacterial T-RFLP fingerprint patterns of the 12C-Cu-HDO and 13C-Cu-HDO treatments were similar in the light-density fractions (buoyant densities <1.81 g ml−1) but differed in the heavy-density fraction (buoyant densities >1.82 g ml−1) (Fig. 6). The 156-, 262-, and 401-bp T-RFs demonstrated higher relative abundances in the high-density fraction of the 13C-Cu-HDO treatment than in the 12C-Cu-HDO treatment (Table 1). Sequences of the 13C-Cu-HDO treatment affiliated with Phenylobacterium were phylogenetically closely related to sequences of the 12C-Cu-HDO treatment but differed in their T-RF sizes (Fig. 7). The 148-bp T-RF with high relative abundances in both the 13C-Cu-HDO and the 12C-Cu-HDO treatments was assigned to the genera Sphingomonas, Caulobacter, and Bradyrhizobium. However, the abundance of sequences affiliated with Sphingomonas was higher in the clone library (Table 1).
FIG. 4.
PLFA patterns of the 12C-Cu-HDO-amended (12C) and 13C-Cu-HDO-amended (13C) WSV microcosms over time. Means + the SD of three replicate measurements are shown. PLFAs with <1% relative abundance are denoted without a pattern (ECL 20.147; ECL 19.599; ECL 18.793; C18:1ω6; C18:0i; C16:0; C17:0; C17:1ω6; C17:0a; ECL 16.114; Ct16:1ω7; C15:0a; C15:0i; C14:0; C14:1ω9; ECL 11.864; ECL 11.746).
FIG. 5.
Incorporation of 13C into abundant PLFAs over time. Means + the SD of three replicate measurements are shown.
FIG. 6.
Bacterial 16S rRNA cDNA T-RFLP patterns of the buoyant density fractions of 12C-Cu-HDO-amended (A to D) and 13C-Cu-HDO-amended (E to H) WSV microcosms after 105 days of incubation. 13C-labeled T-RFs are indicated by their lengths and marked by arrows (156, 262, and 401 bp). Only T-RFs with a relative abundances of >3% are indicated. CsTFA (cesium trifluoroacetate) buoyant densities (BD; in g ml−1) of fractions are indicated.
TABLE 1.
Assignment of T-RFs to bacterial taxa based on 13C-Cu-HDO and 12C-Cu-HDO treatments
| Taxonomic affiliation | No. of clones for treatmenta: |
T-RF size(s) (bp) | |
|---|---|---|---|
| 13C-Cu-HDO | 12C-Cu-HDO | ||
| Acidobacteria | 4 | 262 | |
| Alphaproteobacteria | |||
| Acidosphera | 2 | 74, 127 | |
| Bradyrhizobium | 2 | 148 | |
| Caulobacter | 7 | 148, 403 | |
| Haphomicrobium | 1 | 2 | 399 |
| Kaistina | 2 | 1 | 78 |
| Leisingera | 1 | 138 | |
| Sphingomonas | 30 | 20 | 148 |
| Phenylobacterium | 4 | 401 | |
| Betaproteobacteria | |||
| Achromobacter | 1 | 444 | |
| Comamonas | 1 | 156 | |
| Ralstonia | 2 | 429 | |
| Gammaproteobacteria | |||
| Beggiatoa/Nevskia | 1 | 490 | |
| Colwellia | 1 | 151 | |
| Pseudomonas | 2 | 491 | |
| Verrucomicrobia | 1 | 496 | |
Samples for the cloning and sequencing are derived from the 1.851 and 1.861 g ml−1 buoyant density fractions of the 12C-Cu-HDO and 13C-Cu-HDO treatments, respectively.
FIG. 7.
Phylogenetic tree of 16S rRNA cDNA sequences (in boldface) and reference sequences of the type strains of the genera Phenylobacterium and Caulobacter and Brevundimonas diminuta. Accession numbers are given in brackets. The tree was calculated with MEGA 4.0 (neighbor-joining method, deletion of gaps/missing data, maximum-composite-likelihood model, 788 valid positions between positions 51 and 898 of the 16S rRNA gene of Escherichia coli). Dots at the nodes indicate confirmation of topology by neighbor joining and parsimony using the same data set. The outgroup was Hirschia baltica (X52909). The T-RF sizes of 16S rRNA cDNA sequences are included. Scale bar, 0.005% similarity. 12C-CuHDO, 12C-Cu-HDO amended; 13C-CuHDO, 13C-Cu-HDO-amended WSV mixture.
DISCUSSION
The addition of Cu-HDO as part of preserved wood in soil contact reduced the overall respiration activity drastically and shifted the structure of the soil microbial community significantly toward microorganisms coping with high copper and HDO concentrations. Comparative SIP analyses with labeled and nonlabeled cobiocide allowed the detection of PLFAs and RNA labeled with 13C from the assimilated HDO part. Incorporation of 13C derived from HDO was clearly demonstrated by PLFA-SIP (Fig. 4 and 5) and RNA-SIP analyses (Fig. 6). Members of the genera Phenylobacterium (401 bp), Acidobacteria (262 bp), and Comamonas (156 bp) were exclusively isotopically enriched in the 13C-Cu-HDO treatment, whereas Sphingomonas, Caulobacter, and Bradyrhizobium (148 bp) were present in both 13C-Cu-HDO and 12C-Cu-HDO treatments (Fig. 6, Table 1). Since the density gradient after ultracentrifugation of environmental RNA can be confounded by RNA with a high G+C content (9), the results obtained from both treatments, such as affiliated microorganisms to the 148-bp T-RF, are doubtful. Indeed, members of the genus Sphingomonas were predominant in T-RFLP patterns, as well as in the clone library of the heavy fractions of both the 13C-Cu-HDO and the 12C-Cu-HDO treatments. The genomic information of many members of the genus Sphingomonas indicates that they have a high G+C content (37), and their RNA transcripts may have increased the buoyant densities to 1.85 of CsTFA ml−1 rather than the expected 1.81 to 1.83 g of CsTFA ml−1 (22, 23, 26). However, the 16S rRNA sequences affiliated with Sphingomonas do not differ in their G+C contents compared to the other sequences obtained in the present study. An SIP gradient was applied to total RNA, and its G+C content also affects the placement of 16S rRNA in density fractionation as discussed previously in detail (22). Indeed, Harding et al. demonstrated that the flanking regions of functional genes of Sphingomonas elodea have a significantly higher G+C content than do the respective functional genes (14), which may also be true for the flanking sequences of the 16S rRNA.
Apart from members of the phylum Acidobacteria, all of the genera that are indicative for HDO degradation have been reported to persist or even degrade pesticides in their respective environments (28, 30, 34, 38). However, the analyzed pesticides and environmental conditions in these studies varied, and none of these pesticides had a chemical structure similar to that of HDO or potential metabolites. In addition, members of all of these genera were characterized as copper tolerant except those of the genus Phenylobacterium (8, 11, 29, 32, 39).
SIP-PLFA results showed that after 16 days of incubation a microbial community was established that was capable of incorporating 13C derived from HDO into its biomass (Fig. 4). This HDO-degrading community outcompeted the initial community structure relatively quickly, although the overall respiration rate was significantly reduced (Fig. 1). Moreover, the steady incorporation of 13C into the biomass of one defined set of bacterial PLFAs over an incubation period of 105 days (Fig. 5) suggests that a temporal stable HDO-degrading community was present and that successional shifts in the microbial community were responsible for HDO or HDO-metabolite degradation. Based on the PLFA-SIP results, a cross-feeding of HDO consumers with other microorganisms had not occurred. The rate of HDO degradation was slow (Fig. 1) and, finally, after 105 days of incubation, ca. 10% of them were highly 13C labeled (Fig. 4 and 5), and therefore RNA-SIP became feasible (24). Since RNA-SIP was carried out only once, a temporal development of the HDO-degrading community could not be compared to the PLFA-SIP results. The concentration of amended HDO had significantly decreased (Fig. 1). Therefore, we conclude that access to the biocide inside wood and/or the inhibitory concentration of the biocide decelerated the degradation rate of HDO. However, once HDO was degraded, respired 13CO2 co-occurred, suggesting that HDO and potential HDO metabolites were mineralized completely and did not accumulate.
13C-labeled PLFA community pattern were composed by a defined set of phylogenetically different microorganisms. The phylogenetic information of SIP-RNA results were mostly in line with SIP-PLFA results since the 16S rRNA phylogeny corresponded well to the respective fatty acid profiles (Fig. 4, 5, and 6) (2, 4, 20, 37). However, the PLFA data set for the phylum Acidobacteria is still too limited for a comprehensive comparison. In addition, the comparison of RNA-SIP and PLFA-SIP results was ambiguous at the species level in the genus Phenylobacterium. The isotopic highly enriched fatty acid C18:0 (Fig. 5) was described as highly characteristic for Phenylobacterium koreense Slu-01T (2), but phylogenetic analyses of the 16S rRNA data set indicated that Phenylobacterium immobile DSM 1986T was the closest relative sequence (Fig. 7). P. koreense is not able to degrade the anti-inflammatory antipyrin [1,5-dimethyl-2-phenyl-2,3-dihydro-1-H-pyrazol-3-on] or the herbicide chloridazon [5-amino-4-chloro-2-phenyl-3(2H)-pyridazinone] (4), but P. immobile can metabolize all of the carbon atoms of both compounds as the sole carbon source (20). These compounds are based on a benzene ring linked to an amino group, whereas HDO is based on a cyclohexyl structure, which is also linked to an amino group. Therefore, it is more likely that the detected environmental microorganisms were more closely affiliated with P. immobile. Degradation of pesticides or biocides by members of the phylum Acidobacteria has thus far not been described, but the present findings may contribute to the ongoing discussion of acidobacteria as key players in bioremediation processes (15, 33).
Cu-HDO has been shown to be an effective biocide to extend the durability of preserved wood, in particular against the main wood degraders (10, 13, 16, 18). To our knowledge, this is the first application of SIP to characterize HDO-degrading microorganisms in situ. Wood-degrading fungi were not involved in HDO degradation since PLFAs indicative for fungi (e.g., C18:2ω6,9) were not labeled or increased in their relative abundance over the incubation time, although members of the fungal community could be amplified in “light” density fractions (<1.80 g ml−1). Identified bacteria such as members of the genera Phenylobacterium and Comamonas, as well as the phylum Acidobacteria, have thus far not been linked to HDO degradation; details regarding which metabolic pathways are relevant will be of interest in upcoming studies.
Acknowledgments
We are grateful to Lydia Bräunlich for her support in the density fractionation of the RNA samples and Esther Surges for determination of the carbon isotope ratios. 12C-Cu-HDO and 13C-Cu-HDO were kindly provided by Dr. Wolman GmbH (Sinzheim, Germany).
This study was not conducted to serve any interest of Dr. Wolman GmbH.
Footnotes
Published ahead of print on 15 October 2010.
REFERENCES
- 1.Abraham, W. R., C. Hesse, and O. Pelz. 1998. Ratios of carbon isotopes in microbial lipids as an indicator of substrate usage. Appl. Environ. Microbiol. 64:4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham, W. R., A. J. Macedo, H. Lunsdorf, R. Fischer, S. Pawelczyk, J. Smit, and M. Vancanneyt. 2008. Phylogeny by a polyphasic approach of the order Caulobacterales, proposal of Caulobacter mirabilis sp. nov., Phenylobacterium haematophilum sp. nov. and Phenylobacterium conjunctum sp. nov., and emendation of the genus Phenylobacterium. Int. J. Syst. Evol. Microbiol. 58:1939-1949. [DOI] [PubMed] [Google Scholar]
- 3.Abrajano, Jr., T. A., D. E. Murphy, J. Fang, P. Comet, and J. M. Brooks. 1994. 13C/12C ratios in individual fatty acids of marine mytilids with and without bacterial symbionts. Organic Geochem. 21:611-617. [Google Scholar]
- 4.Aslam, Z., W. T. Im, L. N. Ten, and S. T. Lee. 2005. Phenylobacterium koreense sp. nov., isolated from South Korea. Int. J. Syst. Evol. Microbiol. 55:2001-2005. [DOI] [PubMed] [Google Scholar]
- 5.Auffret, M., D. Labbe, G. Thouand, C. W. Greer, and F. Fayolle-Guichard. 2009. Degradation of a mixture of hydrocarbons, gasoline, and diesel oil additives by Rhodococcus aetherivorans and Rhodococcus wratislaviensis. Appl. Environ. Microbiol. 75:7774-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Physiol. Pharmacol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 7.Cupples, A. M., and G. K. Sims. 2007. Identification of in situ 2,4-dichlorophenoxyacetic acid-degrading soil microorganisms using DNA-stable isotope probing. Soil Biol. Biochem. 39:232-238. [Google Scholar]
- 8.Cytryn, E. J., S. Jitacksorn, E. Giraud, and M. J. Sadowsky. 2008. Insights learned from pBTAi1, a 229-kb accessory plasmid from Bradyrhizobium sp. strain BTAi1 and prevalence of accessory plasmids in other Bradyrhizobium sp. strains. ISME J. 2:158-170. [DOI] [PubMed] [Google Scholar]
- 9.DeRito, C. M., G. M. Pumphrey, and E. L. Madsen. 2005. Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl. Environ. Microbiol. 71:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edlund, M.-L., and T. Nilsson. 1999. Performance of copper and non-copper based wood preservatives in terrestrial microcosms. Holzforschung 53:369-375. [Google Scholar]
- 11.Espirito-Santo, C., P. V. Morais, and G. Grass. 2010. Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl. Environ. Microbiol. 76:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredrickson, H. L., T. E. Cappenberg, and J. W. Leeuw. 1986. Polar lipid ester-linked fatty acid composition of Lake Vechten seston: an ecological application of lipid analysis. FEMS Microbiol. Lett. 38:381-396. [Google Scholar]
- 13.Göttsche, R., and H.-N. Marx. 1989. Copper-HDO: an effective substance variously applicable in wood preservation. Holz-Roh. Werkstoff. 47:509-513. [Google Scholar]
- 14.Harding, N., Y. Patel, and R. Coleman. 2004. Organization of genes required for gellan polysaccharide biosynthesis in Sphingomonas elodea ATCC 31461. J. Indust. Microbiol. Biotechnol. 31:70-82. [DOI] [PubMed] [Google Scholar]
- 15.Jones, R. T., M. S. Robeson, C. L. Lauber, M. Hamady, R. Knight, and N. Fierer. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, G.-H., W.-J. Hwang, T. Yoshimura, and Y. Imamura. 2010. Laboratory evaluation of the termiticidal efficacy of copper HDO. J. Wood Sci. 56:166-168. [Google Scholar]
- 17.Lane, D. J. 1991. 16S/23S rRNA sequencing. In E. S. M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, England.
- 18.Lebow, S. 2004. Alternatives to chromated copper arsenate (CCA) for residential construction. In Proceedings of the Environmental Impacts of Preservative-Treated Wood Conference, Gainesville, FL, 8 to 10 February 2004.
- 19.Lerch, T. Z., M.-F. Dignac, N. Nunan, G. Bardoux, E. Barriuso, and A. Mariotti. 2009. Dynamics of soil microbial populations involved in 2,4-D biodegradation revealed by FAME-based stable isotope probing. Soil Biol. Biochem. 41:77-85. [Google Scholar]
- 20.Lingens, F., R. Blecher, H. Blecher, F. Blobel, J. Eberspacher, C. Frohner, H. Gorisch, H. Gorisch, and G. Layh. 1985. Phenylobacterium immobile gen. nov., sp. nov., a gram-negative bacterium that degrades the herbicide chloridazon. Int. J. Syst. Bacteriol. 35:26-39. [Google Scholar]
- 21.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 23.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manefield, M., A. S. Whiteley, N. Ostle, P. Ineson, and M. J. Bailey. 2002. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial function. Rapid Commun. Mass Spectrom. 156:205-210. [DOI] [PubMed] [Google Scholar]
- 25.Mattes, T. E., A. K. Alexander, P. M. Richardson, A. C. Munk, C. S. Han, P. Stothard, and N. V. Coleman. 2008. The genome of Polaromonas sp. strain JS666: insights into the evolution of a hydrocarbon- and xenobiotic-degrading bacterium, and features of relevance to biotechnology. Appl. Environ. Microbiol. 74:6405-6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noll, M., P. Frenzel, and R. Conrad. 2008. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol. Ecol. 65:125-132. [DOI] [PubMed] [Google Scholar]
- 27.Noll, M., D. Matthies, P. Frenzel, M. Derakshani, and W. Liesack. 2005. Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 7:382-395. [DOI] [PubMed] [Google Scholar]
- 28.Penning, H., S. R. Sorensen, A. H. Meyer, J. Aamand, and M. Elsner. 2010. C, N, and H isotope fractionation of the herbicide isoproturon reflects different microbial transformation pathways. Environ. Sci. Technol. 44:2372-2378. [DOI] [PubMed] [Google Scholar]
- 29.Simoes, L.-C., M. Simoes, R. Oliveira, and M. J. Vieira. 2007. Potential of the adhesion of bacteria isolated from drinking water to materials. J. Basic Microbiol. 47:174-183. [DOI] [PubMed] [Google Scholar]
- 30.Smith, D., S. Alvey, and D. E. Crowley. 2005. Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol. Ecol. 53:265-273. [DOI] [PubMed] [Google Scholar]
- 31.Sturm, R. N. 1973. Biodegradability of nonionic surfactants: screening test for predicting rate and ultimate biodegradation. J. Am. Oil Chem. Soc. 50:159-167. [DOI] [PubMed] [Google Scholar]
- 32.Vânia, S. B., and V. M. Marilis. 2005. Genes involved in cadmium resistance in Caulobacter crescentus. FEMS Microbiol. Lett. 251:289-295. [DOI] [PubMed] [Google Scholar]
- 33.Verónica, M., L.-L. Arantxa, F. Cecilia, G. Myriam, G. Bernardo, V. Mónica, R.-M. Ramón, and S. Michael. 2010. Bioaugmentation with Pseudomonas sp. strain MHP41 promotes simazine attenuation and bacterial community changes in agricultural soils. FEMS Microbiol. Ecol. 71:114-126. [DOI] [PubMed] [Google Scholar]
- 34.Vieira, R., C. Silva, and A. Silveira. 2007. Soil microbial biomass C and symbiotic processes associated with soybean after sulfentrazone herbicide application. Plant Soil 300:95-103. [Google Scholar]
- 35.Weelink, S. A., N. C. Tan, H. ten Broeke, C. van den Kieboom, W. van Doesburg, A. A. Langenhoff, J. Gerritse, H. Junca, and A. J. Stams. 2008. Isolation and characterization of Alicycliphilus denitrificans strain BC, which grows on benzene with chlorate as the electron acceptor. Appl. Environ. Microbiol. 74:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 37.Wittich, R.-M., H.-J. Busse, P. Kampfer, A. J. Macedo, M. Tiirola, M. Wieser, and W.-R. Abraham. 2007. Sphingomonas fennica sp. nov. and Sphingomonas haloaromaticamans sp. nov., outliers of the genus Sphingomonas. Int. J. Syst. Evol. Microbiol. 57:1740-1746. [DOI] [PubMed] [Google Scholar]
- 38.Yang, C., Y. Li, K. Zhang, X. Wang, C. Ma, H. Tang, and P. Xu. 2010. Atrazine degradation by a simple consortium of Klebsiella sp. A1 and Comamonas sp. A2 in nitrogen enriched medium. Biodegradation 21:97-105. [DOI] [PubMed] [Google Scholar]
- 39.Yin, H., L. Cao, G. Qiu, D. Wang, L. Kellogg, J. Zhou, X. Liu, Z. Dai, J. Ding, and X. Liu. 2008. Molecular diversity of 16S rRNA and gyr B genes in copper mines. Arch. Microbiol. 189:101-110. [DOI] [PubMed] [Google Scholar]
- 40.Zelles, L., and Q. Y. Bai. 1993. Fractionation of fatty acids derived from soil lipids by solid phase extraction and their quantitative analysis by GC-MS. Soil Biol. Biochem. 25:495-507. [Google Scholar]