Abstract
Staphylococcal food poisoning (SFP) is one of the most prevalent causes of food-borne illness throughout the world. SFP is caused by 21 different types of staphylococcal enterotoxins produced by Staphylococcus aureus. Among these, staphylococcal enterotoxin B (SEB) is the most potent toxin and is a listed biological warfare (BW) agent. Therefore, development of immunological reagents for detection of SEB is of the utmost importance. High-affinity and specific monoclonal antibodies are being used for detection of SEB, but hybridoma clones tend to lose their antibody-secreting ability over time. This problem can be overcome by the use of recombinant antibodies produced in a bacterial system. In the present investigation, genes from a hybridoma clone encoding monoclonal antibody against SEB were immortalized using antibody phage display technology. A murine phage display library containing single-chain variable-fragment (ScFv) antibody genes was constructed in a pCANTAB 5E phagemid vector. Phage particles displaying ScFv were rescued by reinfection of helper phage followed by four rounds of biopanning for selection of SEB binding ScFv antibody fragments by using phage enzyme-linked immunosorbent assay (ELISA). Soluble SEB-ScFv antibodies were characterized from one of the clones showing high affinity for SEB. The anti-SEB ScFv antibody was highly specific, and its affinity constant was 3.16 nM as determined by surface plasmon resonance (SPR). These results demonstrate that the recombinant antibody constructed by immortalizing the antibody genes from a hybridoma clone is useful for immunodetection of SEB.
Staphylococcus aureus is one of the most prevalent causative agents of food-borne illness throughout the world. The illness occurs following ingestion of staphylococcal enterotoxins (SEs) produced by S. aureus in the contaminated food. The symptoms of food poisoning include nausea, vomiting, abdominal cramps, and diarrhea. Twenty-one different types of SEs, i.e., staphylococcal enterotoxin A (SEA) to staphylococcal enterotoxin E (SEE) and staphylococcal enterotoxin G (SEG) to staphylococcal enterotoxin V (SEV), have already been discovered (33, 40). The immunomodulatory effects of SEs, such as immunosuppression, enhancement of endotoxic shock, and induction of cytokine release, can be attributed to the fact that SEs are potent T-cell mitogens. The ability of SEs to induce proliferation of T cells is somewhat similar to conventional antigen presentation by major histocompatibility complex (MHC) class II molecules to T-cell receptors (TCRs). However, unlike conventional antigens, the T-cell mitogenicity of SEs does not require antigen processing and lacks the normal specificity to the TCR for specific epitopes in response to conventional antigens. This bypass of normal TCR specificity for conventional T-cell epitopes results in the stimulation of a substantial proportion of the total T-cell population, and therefore, staphylococcal enterotoxins are referred to as superantigens. The stimulation of T cells leads to overproduction of cytokines, causing clinical symptoms that include fever, hypotension, and even death in severe cases (3, 30, 39).
Among SEs, SEB is the most potent toxin secreted by S. aureus (3, 35). This is a single polypeptide containing 239 amino acids with a molecular mass of 28 kDa. SEB is highly resistant to proteases, boiling temperature, and extremes of pH because of its compact tertiary structure (5, 27). In humans, 3.5 μg of SEB ingested by the oral route causes emesis (5). SEB is extremely toxic by inhalation, and as little as 30 ng is sufficient to cause fever, respiratory complaints (cough, dyspnea, and retrosternal discomfort or chest pain), and gastrointestinal symptoms. Severe intoxication results in pulmonary edema, adult respiratory distress syndrome (ARDS), shock, and death (28, 37, 42). Although exposure to SEB by the inhalation route is not a common feature of S. aureus infection, the possibility of it makes SEB a candidate weapon for biological terrorism and, hence, it is a listed biological warfare agent (21, 26). Therefore, its quick and unambiguous detection is of paramount importance.
The availability of specific and high-affinity antibodies is the major bottleneck in the development of an immunodetection system for SEB. Any immunological detection system for SEB requires specific and high-affinity antibodies, but SEB being a superantigen leads to the generation of low-titered polyclonal antibodies. The problem is further aggravated if SEB is contaminated even slightly with other, undesired proteins, leading to nonspecific antiserum. Polyclonal antibodies are generated using SEB purified by conventional protein purification methods, which do not result in SEB that is sufficiently pure for generation of specific and sensitive antibodies (6, 22). Alternatively, hybridoma technology has also been used to generate monoclonal antibodies against SEB, but hybridoma clones tend to lose their antibody-secreting ability over time (11, 23). In recent times, the advent of recombinant DNA and gene amplification technology has made it possible to clone desired antibody genes in bacteria by using antibody phage display technology. Such immortalization of antibody genes has made it technically feasible to produce a monoclonal antibody called single-chain variable-fragment (ScFv) antibody quickly in bacterial culture. These ScFv antibody molecules can further be genetically manipulated for improved specificity and affinity (8). The cost of their production is very low, and they can be fused with a marker molecule for immunological detection of several bacterial and viral agents (43). However, construction of such ScFv molecules for detection of SEB has not been reported so far. Therefore, the objective of the present study was to construct a recombinant antibody (ScFv) against SEB for use in immunological detection.
MATERIALS AND METHODS
Materials.
All chemicals and organic solvents were reagent grade or better. Plasmid pCANTAB 5E, Escherichia coli strains TG1 and HB2151, M13K07 helper phage, mouse anti-M13 horseradish peroxidase (HRP)-conjugated antibody, mouse anti-E tag HRP-conjugated antibody, and restriction enzymes SfiI and NotI were procured from GE Healthcare UK Limited (Buckinghamshire, United Kingdom). Cell culture media, reagents, and fetal bovine serum (FBS) were purchased from Sigma-Aldrich Inc. (St. Louis, MO), unless otherwise specified. Goat anti-mouse HRP-conjugated antibody was purchased from Dako Denmark A/S (Rødovre, Denmark). PCR amplification primers listed in Table 1 were synthesized by Microsynth AG (Balgach, Switzerland). Test antigen, i.e., recombinant SEB (r-SEB; molecular mass, ∼30.5 kDa) was prepared earlier in the laboratory. All DNA manipulations, if not described, were carried out by standard procedures (38).
TABLE 1.
Primers used for amplification of variable heavy (VH), variable light (VL), and ScFv antibody genes
| Primer name | Description or sequence (5′→3′) |
|---|---|
| VH reverse | Equimolar mixture of primers VHR 1, VHR 2, VHR 3, and VHR 4 |
| VHR 1 | GAG GAA ACG GTG ACC GTG GT |
| VHR 2 | GAG GAG ACT GTG AGA GTG GT |
| VHR 3 | GCA GAG ACA GTG ACC AGA GT |
| VHR 4 | GAG GAG ACG GTG ACT GAG GT |
| VL reverse | Equimolar mixture of primers VLR 1, VLR 2, VLR 3, and VLR 4 |
| VLR 1 | ACG TTT KAT TTC CAG CTT GG |
| VLR 2 | ACG TTT TAT TTC CAA CTT TG |
| VLR 3 | ACG TTT CAG CTC CAG CTT GG |
| VLR 4 | ACC TTG GAC AGT CAG TTT GG |
| RS VH For mix | Equimolar mixture of primers RS VHF 1 and RS VHF 2 |
| RS VHF 1 | GCG GCC CAG CCG GCC ATG GCC GAG GTB CAG CTB CAG CAG TCa |
| RS VHF 2 | GCG GCC CAG CCG GCC ATG GCC CAG GTG CAG CTG AAG SAR TCa |
| Lin VH Rev mix | Equimolar mixture of primers Lin VHR 1, Lin VHR 2, Lin VHR 3, and Lin VHR 4 |
| Lin VHR 1 | GGA ACC GCC GCC ACC AGA GCC ACC ACC GCC GGA CGA GGA AAC GGT GAC CGT GGT |
| Lin VHR 2 | GGA ACC GCC GCC ACC AGA GCC ACC ACC GCC GGA CGA GGA GAC TGT GAG AGT GGT |
| Lin VHR 3 | GGA ACC GCC GCC ACC AGA GCC ACC ACC GCC GGA CGC AGA GAC AGT GAC CAG AGT |
| Lin VHR 4 | GGA ACC GCC GCC ACC AGA GCC ACC ACC GCC GGA CGA GGA GAC GGT GAC TGA GGT |
| Lin VL For mix | Equimolar mixture of primers Lin VLF 1 and Lin VLF 2 |
| Lin VLF 1 | TCT GGT GGC GGC GGT TCC GGT GGC GGT GGC GAY ATC CAG CTG ACT CAG CC |
| Lin VLF 2 | TCT GGT GGC GGC GGT TCC GGT GGC GGT GGC GAY ATT GTT CTC WCC CAG TC |
| RS VL Rev mix | Equimolar mixture of primers RS VLR 1, RS VLR 2, RS VLR 3, and RS VLR 4 |
| RS VLR 1 | GAG TCA TTC TGC GGC CGC ACG TTT KAT TTC CAG CTT GGb |
| RS VLR 2 | GAG TCA TTC TGC GGC CGC ACG TTT TAT TTC CAA CTT TGb |
| RS VLR 3 | GAG TCA TTC TGC GGC CGC ACG TTT CAG CTC CAG CTT GGb |
| RS VLR 4 | GAG TCA TTC TGC GGC CGC ACC TTG GAC AGT CAG TTT GGb |
Bold bases indicate SfiI site.
Bold bases indicate NotI site.
Generation of hybridoma.
Eight-week-old female BALB/c mice were immunized subcutaneously with SEB at 3-week intervals. The priming dose consisted of 150 μg r-SEB per mouse in Freund's complete adjuvant. The booster consisted of 75 μg antigen in Freund's incomplete adjuvant. Mice were test bled, and the antiserum was checked by enzyme-linked immunosorbent assay (ELISA) to determine the response. Mice were given a booster of 150 μg antigen 3 days prior to fusion. Spleen cells were collected and fused with mouse myeloma cells as described by Harlow and Lane (13). Culture supernatant from the clones was tested in ELISA against 1 μg antigen. Positive clones were expanded and repeatedly tested for antibody secretion by ELISA. Monoclonal antibody culture supernatants were also used for determining the isotype by ELISA. The hybridoma clone 2F6D9G2 was selected and maintained in Dulbecco's modified Eagle's medium supplemented with gentamicin (50 μg/ml) and 10% (vol/vol) FBS for RNA isolation.
Amplification of antibody variable-region genes.
Total RNA was extracted from hybridoma cells by using TRI reagent (Sigma-Aldrich, Inc., St. Louis, MO) per the manufacturer's instructions. cDNA was synthesized by reverse transcription (Ominiscript RT kit; Qiagen GmbH, Hilden, Germany) using variable heavy (VH)- and variable light (VL)-chain reverse primers (Table 1) against the conserved 3′ end of antibody variable-region genes. RS VH For mix and Lin VH Rev mix and Lin VL For mix and RS VL Rev mix degenerate primers were used for the amplification of VH and VL chains, respectively (Table 1). The PCR protocol consisted of initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation (94°C, 1 min), annealing (50°C, 1 min), and extension (72°C, 2 min) and a final extension at 72°C for 10 min. PCR products were resolved by agarose gel electrophoresis and eluted using the QIAquick gel extraction kit.
Construction of SEB-ScFv expression vector.
PCR-amplified VH and VL gene fragments containing linker overhangs were joined together by overlap extension PCR using equimolar concentrations of VH and VL gene fragments. RS VH For mix and RS VL Rev mix were used for the ScFv construction. The optimum PCR conditions have been described above, except annealing (63°C for 1 min). The assembled ScFv gene was resolved on a 1.5% agarose gel, and the DNA band was eluted using a QIAquick gel extraction kit. The ScFv gene was digested with SfiI and NotI restriction enzymes and ligated into an SfiI- and NotI-linearized phagemid vector, pCANTAB 5E. Electrocompetent E. coli TG1 cells were transformed with the resulting phagemid vector, pCANTAB 5E-ScFv, by electroporation. Transformed TG1 cells were recovered in 1 ml of 2× yeast extract-tryptone (YT) medium containing 2% (vol/vol) glucose and incubated at 37°C for 1 h. Transformed cells were then plated on super optimal broth (SOB) agar plates containing 20 g/liter glucose and 100 μg/ml ampicillin followed by overnight incubation at 30°C. Colonies obtained on these plates were scraped and recovered with 2× YT medium. A portion of this culture mixture was diluted to an optical density at 600 nm (OD600) of ∼0.5 in 2× YT-AG (2× YT medium containing 100 μg/ml ampicillin and 2% [vol/vol] glucose) for biopanning. Glycerol stocks were prepared from the rest of the scraped cells and stored at −80°C.
Rescue of phagemid library and biopanning of phage antibody library.
Phage particles displaying ScFv were rescued by infection with helper phage (M13K07). Briefly, the above diluted culture was grown and helper phage was added at a multiplicity of infection (MOI) of 20. After incubation for 30 min without shaking and incubation for another 30 min with shaking, the culture was centrifuged at 1,000 × g for 10 min. The entire cell pellet was gently resuspended in 10 ml of 2× YT-AK medium (2× YT medium containing 100 μg/ml ampicillin and 50 μg/ml kanamycin). After overnight incubation at 37°C with shaking at 250 rpm, the culture was again centrifuged at 1,000 × g for 20 min at room temperature and the supernatant containing recombinant phage particles was collected. The phage particles were precipitated with polyethylene glycol (PEG)-NaCl on ice for 1 h and collected by centrifugation at 10,000 × g for 20 min at 4°C. Precipitated phage particles were resuspended in 2× YT medium, and their titer was determined for subsequent biopanning experiments.
The ScFv phage library was biopanned for clones binding SEB in a 25-cm2 culture flask. For biopanning, the phage rescuing was done as described above. The resuspended phage particles were diluted with 10 ml of 10% MPBS (phosphate-buffered saline [PBS] containing 10% [wt/vol] skim milk and 0.01% sodium azide) and incubated at room temperature for 15 min. Diluted recombinant phage particles (20 ml) were added to the SEB-coated flask followed by blocking with 10% MPBS for 2 h at 37°C. The flask was washed 10 times with PBST (PBS containing 0.05% [vol/vol] Tween 20) and another 10 times with PBS. Log-phase TG1 cells (10 ml) were added to the flask and incubated for 1 h with shaking at 37°C for reinfection with phage particles. The enriched library was then plated on SOB plates containing ampicillin (100 μg/ml) and glucose (2%), rescued, and used for further panning cycles. Further cycles were carried out essentially as described above, with increasing stringency and an increasing number of wash cycles. After four rounds of biopanning, 20 clones were randomly selected for further analysis.
Screening for SEB-specific binding clones by phage ELISA.
SEB binding clones were selected by phage ELISA. Rescue of individual phage colonies was carried out as described above. A Maxisorp ELISA plate was coated with SEB in bicarbonate buffer (pH 9.6). After blocking with 4% MPBS, approximately 1012 phage particles were added and incubated for 1 h at room temperature. Bound phage particles were detected after incubation with a 1:2,000 dilution of anti-M13 HRP-conjugated antibody. The colorimetric reaction was performed with ABTS [2,2′-azino-bis(3-ethyl-benzothiazoline-6-sulfonic acid)] as the enzyme substrate. Bovine serum albumin (BSA)-coated wells were used as a negative control. Clones were considered positive if they demonstrated at least twice the signal developed in the negative control.
Expression of soluble anti-SEB ScFv antibodies and screening by Western blotting.
To produce soluble anti-SEB ScFv, a strong positive recombinant phage clone was made to infect log-phase E. coli HB2151 cells. Expression of soluble anti-SEB ScFv was induced by isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 1 mM). The culture was induced for 7 h at 30°C followed by centrifugation at 1,500 × g for 20 min. Cell pellet was resuspended in ice-cold 1× TES (0.2 M Tris hydrochloride, 0.5 mM ethylenediaminetetraacetic acid, 0.5 M sucrose) at the rate of 2% of initial culture volume. Subsequently, ice-cold 1/5× TES was added at the rate of 3% of initial culture volume and the mixture was incubated on ice for 30 min to induce a mild osmotic shock. The contents were centrifuged at 12,000 × g for 10 min. The supernatant containing the soluble antibodies from the periplasm was transferred to fresh tubes and stored at −20°C. Periplasmic extract containing anti-SEB ScFv antibody was resolved on a 12% SDS-PAGE gel and stained with Coomassie brilliant blue R-250. Proteins resolved by SDS-PAGE were blotted on a polyvinylidene difluoride (PVDF) membrane. After the membrane was blocked with 5% MPBS, anti-SEB ScFv was detected by incubating the blot with anti-E-tag HRP-conjugated antibodies. The blot was developed using diaminobenzidine (DAB)-H2O2 solution. Purification of ScFv antibodies from the periplasmic extract of E. coli was carried out using anti-E-tag affinity column (GE Healthcare UK Limited, Buckinghamshire, United Kingdom) according to the manufacturer's instructions.
Antigen binding assay for anti-SEB ScFv antibodies.
The SEB binding assay for the anti-SEB ScFv antibodies was done by both ELISA and Western blotting. For direct measurement of antigen binding activity, ELISA plate wells were coated with 250 ng SEB in bicarbonate buffer (pH 9.6). After blocking with 5% MPBS, different dilutions of anti-SEB ScFv antibodies were incubated for 1 h at room temperature. Untransformed HB2151 periplasmic extract was used as a negative control. Bound SEB-ScFv was detected with anti-E-tag HRP-conjugated antibodies.
Binding of anti-SEB ScFv with SEB was also confirmed by Western blotting. For this, 1 μg SEB was run on a 12% SDS-PAGE gel and blotted on a PVDF membrane. After being blocked with 5% MPBS, the blot was incubated with anti-SEB ScFv antibodies. The blot was then incubated with anti-E-tag HRP-conjugated antibody and developed using DAB-H2O2 solution.
Nucleic acid analysis of anti-SEB ScFv antibody gene.
The anti-SEB ScFv antibody gene cloned in pCANTAB 5E phagemid vector was sequenced using vector-specific primers, pCANTAB5-S1 (5′-CAACGTGAAAAAATTATTATTCGC-3′) and pCANTAB5-S6 (5′-GTAAATGAATTTTCTGTATGAGG-3′). Sequences were delineated and analyzed using LaserGene version 5.07/5.52 software (DNAStar Inc., Madison, WI) and were submitted to the NCBI GenBank. Both the heavy- and light-chain variable-region sequences were numbered per the Kabat rule (20). Canonical classes of each of the complementarity-determining regions (CDRs) were identified per the work of Chothia and Lesk (7) by using online software (http://www.bioinf.org.uk/abs/). The anti-SEB ScFv sequence was also analyzed online using the V-Quest software provided by the International ImMunoGeneTics information system (IMGT) (http://imgt.cines.fr) for identification of germ line origin of VH/VL regions (12). Homology searches for nucleotide and protein sequences were done using BLAST (1).
Affinity determination of SEB-ScFv by SPR.
The biomolecular interaction and affinity of SEB-ScFv was measured using a two-channel cuvette-based electrochemical surface plasmon resonance (SPR) system (Autolab ESPRIT; EcoChemie, Netherlands). The SPR measurement was automatically monitored by data acquisition software version 4.3.1 supplied along with the instrument. All the SPR measurements were carried out at 25°C. SEB antigen was immobilized on a carboxymethyldextran modified gold disc per the manufacturer's instructions. A flow rate of 16.5 μl/s was maintained during the SPR interaction measurements, and a set of five anti-SEB ScFv dilutions (10 nM to 1 μM) prepared in PBS were performed for 700 s. After each anti-SEB ScFv dilution, the chip was regenerated with 10 mM HCl for 120 s. The affinity constant was calculated using Kinetic evaluation software version 5.0 supplied with the SPR.
Specificity profile of anti-SEB ScFv.
The specificity of the anti-SEB ScFv antibody was determined by ELISA against staphylococcal enterotoxins (SEA, SEC1, SEC2, SEC3, and SED) that are frequently associated with staphylococcal food poisoning (SFP) outbreaks. Anti-SEB ScFv bound to antigen was detected by incubation with anti-E-tag HRP-conjugated antibody. The colorimetric reaction was performed with ABTS as the enzyme substrate.
Nucleotide sequence accession number.
The nucleotide sequence of 4PCL2 ScFv was submitted to GenBank (NCBI accession no. GQ465983).
RESULTS
Generation of mouse monoclonal antibody.
The hybridoma was generated by using a stringent screening method as described above. After immunization the titer of mouse antiserum was 1:51,200 as observed by ELISA. Ten ELISA-positive clones were obtained after fusion. After repeated subcloning, one clone, 2F6D9G2, showing an IgG1 isotype, was expanded further for ScFv construction.
PCR amplification of variable-region genes and cloning of ScFv gene.
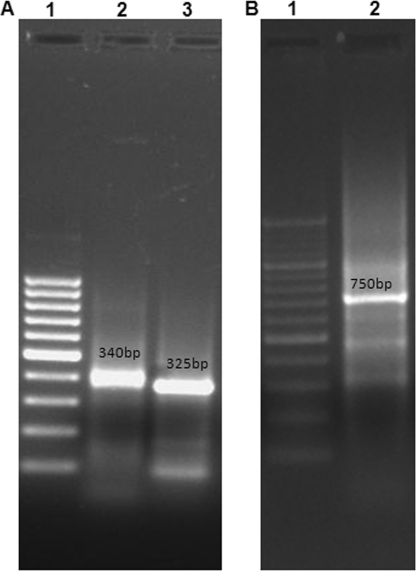
VH and VL chain genes were amplified by PCR from the 2F6D9G2 hybridoma clone. The VH gene product formed a band at ∼340 bp and the VL gene product formed a band at ∼325 bp, as revealed on an agarose gel (Fig. 1A). The VH and VL fragments containing linker overhangs were joined to generate a single-chain variable fragment at ∼750 bp (Fig. 1B).
FIG. 1.
Agarose gels showing VH, VL, and ScFv gene amplicons. (A) Lane 1, 100-bp DNA ladder (Fermentas); lane 2, VH gene amplicon (∼340 bp); lane 3, VL gene amplicon (∼325 bp). (B) Lane 1, 100-bp DNA ladder (Fermentas); lane 2, ScFv gene amplicon (∼750 bp).
ScFv library construction and biopanning.
A phage antibody library on the order of 6.5 × 1011 PFU/ml in size was obtained by cloning the ScFv genes in pCANTAB 5E phagemid vector. After four rounds of biopanning, 20 clones were randomly selected for binding with SEB antigen. One of the clones, 4PCL2, showing maximum reactivity against SEB, was selected for further studies.
Expression of soluble anti-SEB ScFv antibody.
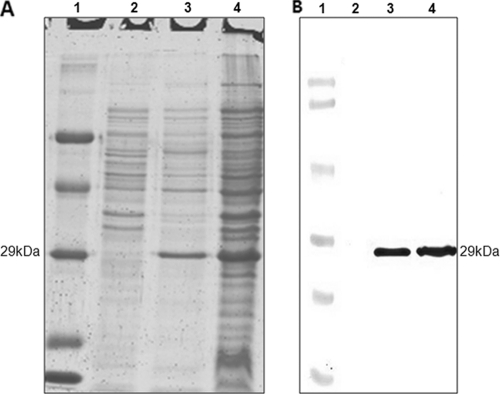
The anti-SEB ScFv antibody was expressed in soluble form in 4PCL2-infected HB2151 cells after induction with IPTG. A protein band at ∼29 kDa, corresponding to the expected size of anti-SEB ScFv, was detected in the periplasmic extract of 4PCL2-infected HB2151 cells (Fig. 2A). The presence of an anti-SEB ScFv band at ∼29 kDa shown by Western blotting using anti-E-tag HRP-conjugated antibody further confirmed its expression (Fig. 2B).
FIG. 2.
SDS-PAGE (A) and Western blot (B) profiles showing expression of anti-SEB ScFv in soluble form. (A) Lane 1, molecular mass markers (66, 45, 29, 20, and 14.2 kDa); lane 2, uninduced culture of 4PCL2-infected HB2151 cells; lanes 3 and 4, induced culture and periplasmic extract of 4PCL2-infected HB2151 cells, respectively, showing the anti-SEB ScFv band at ∼29 kDa. (B) Lane 1, molecular mass markers (97.4, 66.2, 45, 31, 26.6, and 14.4 kDa); lane 2, uninduced culture of 4PCL2-infected HB2151 cells; lanes 3 and 4, induced culture and periplasmic extract of 4PCL2-infected HB2151 cells, respectively, showing the anti-SEB ScFv band at 29 kDa.
SEB binding activity and affinity determination of anti-SEB ScFv antibody.
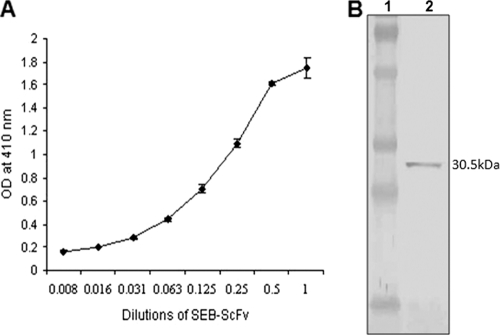
The SEB binding activities of different dilutions of anti-SEB ScFv were studied by ELISA. It was observed that with an increase or a decrease in the concentration of anti-SEB ScFv there was an increase or a decrease in binding with SEB as revealed by OD values (Fig. 3A). The effective binding of anti-SEB ScFv with SEB antigen was further confirmed by Western blotting, and the result is depicted in Fig. 3B. Further, the affinity of anti-SEB ScFv for the SEB antigen was studied by SPR using the SEB immobilized sensor chip. It was observed that an increase in the concentration of the antibody increases the SPR response linearly. The SPR data were used for the quantitative determination of the affinity constant of the anti-SEB ScFv antibody. The calculated affinity constant of anti-SEB ScFv was in the low-nanomolar range (3.16 nM) as determined by Kinetic evaluation software, version 5.0.
FIG. 3.
ELISA (A) and Western blot (B) showing binding of anti-SEB ScFv with SEB antigen. ELISA was performed with different dilutions of soluble anti-SEB ScFv antibody. Periplasmic extract of untransformed HB2151 cells was used as a negative control (OD, 0.0945). (B) Lane 1, prestained molecular mass markers (70, 55, 35, 27, and 15 kDa); lane 2, r-SEB band at 30.5 kDa showing binding with anti-SEB ScFv. The bound anti-SEB ScFv antibody was detected using anti-E-tag HRP-conjugated antibody.
Sequence analysis of variable-region genes.
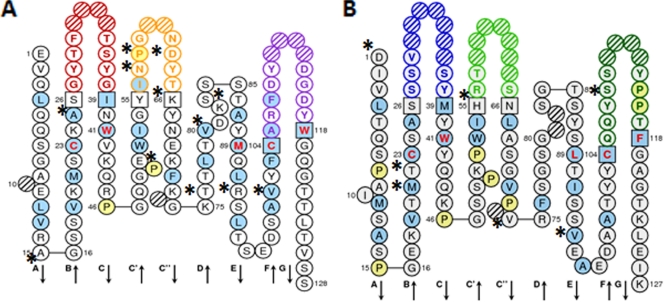
The nucleotide sequence of 4PCL2 ScFv was submitted to GenBank (NCBI accession no. GQ465983). The nucleotide and amino acid sequences of the variable heavy-chain (Fig. 4) and variable light-chain (Fig. 5) regions of 4PCL2 ScFv aligned with mouse germ line genes. Analysis of the amino acid sequences revealed that CDR L1, CDR L2, CDR L3, and CDR H1 belong to the type 1 canonical class. CDR H2 belongs to class 2, and the 7-residue-long CDR H3 has a kinked profile. A homology search using V-Quest software revealed that the heavy-chain variable region of 4PCL2 is 94.10% identical to the mouse germ line VH gene IGHV1-58*01 (IMGT accession no. AC0871666). The light-chain variable region belongs to Vκ class 4 and is 95.65% identical to mouse germ line genes IGKV4-61*01 (IMGT accession no. AJ231209) and IGKV4-68*01 (IMGT accession no. AJ231222). Figure 6 shows an IMGT “collier de perle” (pearl necklace) graphical two-dimensional (2-D) representation of 4PCL2 ScFv. Differences between 4PCL2 ScFv and the amino acids encoded by the mouse germinal genes most similar to 4PCL2 ScFv are indicated in the figure.
FIG. 4.
Homology of 4PCL2 ScFv heavy chain with mouse germ line genes. Nucleotide and amino acid sequences of 4PCL2 ScFv heavy-chain variable region aligned with the most homologous germ line V region (IGHV1-58*01, IMGT accession no. AC087166). In the case of germ line genes, only the differences in nucleotides and amino acids are shown. Amino acids are numbered according to the Kabat numbering scheme. The NCBI accession number for the sequence of 4PCL2 ScFv is GQ 465983.
FIG. 5.
Homology of the 4PCL2 ScFv light chain with mouse germ line genes. Nucleotide and amino acid sequences of 4PCL2 ScFv light-chain variable region aligned with the most homologous germ line V region (IGKV4-61*01, IMGT accession no. AJ231209). In the case of germ line genes, only the differences in nucleotides and amino acids are shown. Amino acids are numbered according to the Kabat numbering scheme. The NCBI accession number for the sequence of 4PCL2 ScFv is GQ465983.
FIG. 6.
IMGT collier de perle graphical 2-dimensional representation of 4PCL2 ScFv. (A) Variable heavy chain (VH). (B) Variable light chain (VL). Collier de perle representations are displayed according to IMGT unique numbering. Asterisks indicate differences between 4PCL2 ScFv and the mouse germ line genes most similar to 4PCL2 ScFv. Hydrophobic amino acids (those with positive hydropathy index values, i.e., I, V, L, F, C, M, and A) and tryptophan (W) are shown in blue circles. All proline (P) residues are shown in yellow circles. The complementarity-determining region (CDR-IMGT) sequences are delimited by amino acids shown in squares (anchor positions), which belong to the neighboring framework region (FR-IMGT). Hachured circles correspond to missing positions according to IMGT unique numbering. Colors of circle outlines indicate regions: in the VH domain, red for CDR1-IMGT, orange for CDR2-IMGT, and purple for CDR3-IMGT; in the VL domain, blue for CDR1-IMGT, green for CDR2-IMGT, and turquoise for CDR3-IMGT. Residues at positions 23 (first cysteine), 41 (tryptophan), 89 (hydrophobic amino acid), 104 (second cysteine), and 118 (hydrophobic amino acid) are conserved and shown in red letters.
DISCUSSION
SFP caused by SEs is one of the most prevalent causes of gastroenteritis worldwide (4, 32). Among different types of SEs, SEB is the most potent and is also a listed biological warfare agent (21, 26). Therefore, development of immunodiagnostic reagents against SEB is of the utmost importance. Any immunological detection system for SEB requires specific and sensitive antibodies, but SEB being a superantigen leads to the production of low-titered antiserum in animal models. Contamination of SEB with other proteins results in nonspecific antiserum generation as conventional purification methods do not yield homogenously pure SEB (6, 22). The problem of nonspecific and low-titered antibodies was addressed with the generation of monoclonal antibody by using hybridoma technology. However, hybridoma clones secreting monoclonal antibody are genetically unstable and tend to lose the antibody-secreting ability in the course of time; however, the reasons for this are not very well understood (11, 23). These problems were overcome with the advent of a new molecular biology tool called antibody phage display technology. Antibody phage display technology is one of the most remarkable achievements in antibody engineering. With this technique, the repertoires of VH and VL genes are amplified and joined together by PCR and finally inserted into a phagemid (17). After transformation into E. coli, phage displaying the desired antibody fragment, called ScFv, on its surface is enriched and selected by a technique called biopanning. In this way, the antibody phage display technique links the phenotype of ScFv to its genotype, which can be cloned into an E. coli host secreting soluble ScFv antibodies (19, 25). Therefore, antibody phage display technology permits the immortalization of monoclonal antibody-encoding genes from a hybridoma clone. This technique also facilitates gene manipulation to improve the affinity of the antibody.
In the present study, we report the construction of a highly reactive and specific ScFv antibody fragment from a phage display library suitable for SEB detection. The gene encoding the monoclonal antibody reactive against SEB was immortalized by cloning in E. coli by using antibody phage display technology, and the resultant recombinant antibody was characterized. The VH and VL chain genes were amplified separately along with a 15-amino-acid-long (Gly4-Ser)3 linker molecule. Both these genes were joined by a single-step method called overlap extension PCR, which minimizes the possibility of mutations associated with multiple-step PCR (15). Usually a 15-residue hydrophilic sequence, (Gly4-Ser)3, is used for the linker, but short linker molecules from 5 to 10 amino acids result in the multimerization of ScFv molecule, leading to enhanced avidity (24, 41). The ScFv gene was cloned into pCANTAB 5E vector, and a large repertoire of antibody library (6.5 × 1011 PFU/ml) was obtained. The size of the library is dependent on and governed by the transformation efficiency, which is the major limitation of antibody phage display technology (2). Screening of the phage antibody library revealed the expression of anti-SEB ScFv on the phage surface. The pIII protein fused upstream of anti-SEB ScFv facilitates its transportation onto the phage surface. This helps in selection of ScFv molecules reactive against the desired antigen, i.e., SEB. After four rounds of biopanning, the clone 4PCL2, showing maximum binding with SEB, was selected for further studies. The recombinant phage (4PCL2) displaying anti-SEB ScFv antibody was made to infect a nonsuppressor E. coli strain, HB2151. In this E. coli strain, translation is aborted after the synthesis of ScFv due to recognition of a stop codon. This results in the production and secretion of soluble ScFv antibodies into the periplasmic location of the bacteria (16). In this study, soluble anti-SEB ScFv (∼29 kDa) was successfully expressed in the periplasmic extract of E. coli. The nucleic acid sequence analysis of the antibody heavy and light chains indicates that it belongs to the canonical class of 1-2-1-1-1 (i.e., H1-H2-L1-L2-L3). Homology search using IMGT/V-Quest revealed variation in VH and VL genes of anti-SEB ScFv from the mouse germ line genes. This variation is attributed to 14 replacements and three silent mutations in the VH gene and nine replacements and three silent mutations in the VL gene of anti-SEB ScFv. The ratio of replacement to silent mutations in the CDRs was 4/0 and 2/2 in VH and VL genes, respectively. If replacement mutation takes place randomly, the expected number of replacement mutations at CDRs would be 5.6 in VH and 3 in VL genes (18). The affinity of anti-SEB ScFv antibody is very high (3.16 nM), which shows that SEB antigen and its corresponding anti-SEB ScFv antibody interact with high affinity. Mechaly and coworkers (31) reported ScFv antibodies against Bacillus anthracis spores with an affinity in the low-nanomolar range (30 nM). On the other hand, even a picomolar level of KD (affinity constant) (41 pM) has also been reported for ricin toxin (36). Further, anti-SEB ScFv antibody did not cross-react with SEs (SEA, SEC1, SEC2, SEC3, and SED) commonly implicated in SFP outbreaks, making it useful for SEB detection.
Reports on the construction of recombinant antibodies for the detection of food-borne pathogens (29, 34), toxins (9, 10), and biological warfare agents (9, 10, 14, 31, 36) are scanty. Recombinant Fab antibodies, isolated from a phage display library, have been used in detection of botulinum toxin (9, 10). Phage display was also utilized for the construction of ScFv antibodies against biological warfare agents like anthrax (31), Brucella melitensis (14), and ricin (36). This study on construction of the first ScFv antibody against SEB will facilitate specific detection of SEB in cases of SFP and in the event of biological terrorism.
Acknowledgments
We are grateful to the Director, DRDE Gwalior, for providing the research facilities. We are also grateful to Stefan Dübel and Michael Hust, Department of Biotechnology, Technical University of Braunschweig, Germany, for helping us to troubleshoot the problems faced during the course of the study.
Footnotes
Published ahead of print on 15 October 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Mayers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Azzazy, H. M. E., and W. E. Highsmith, Jr. 2002. Phage display technology: clinical applications and recent innovations. Clin. Biochem. 35:425-445. [DOI] [PubMed] [Google Scholar]
- 3.Baker, M. D., A. C. Papageorgiou, R. W. Titball, J. Miller, S. White, B. Lingard, J. J. Lee, D. Cavanagh, M. A. Kehoe, J. H. Robinson, and K. R. Acharya. 2002. Structural and functional role of threonine 112 in a superantigen Staphylococcus aureus enterotoxin B. J. Biol. Chem. 277:2756-2762. [DOI] [PubMed] [Google Scholar]
- 4.Balaban, N., and A. Rasooly. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Bergdoll, M. S. 1983. Enterotoxins, p. 559-598. In C. S. F. Easmon and C. Adlam (ed.), Staphylococci and staphylococcal infections. Academic Press, New York, NY.
- 6.Casman, E. P., and R. W. Bennett. 1964. Production of antiserum for staphylococcal enterotoxin. Appl. Microbiol. 12:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chothia, C., and A. M. Lesk. 1987. Canonical structures for the hypervariable loops of immunoglobulins. J. Mol. Biol. 196:901-917. [DOI] [PubMed] [Google Scholar]
- 8.Coia, G., P. J. Hudson, and R. A. Irving. 2001. Protein affinity maturation in vivo using E. coli mutator cells. J. Immunol. Methods 251:187-193. [DOI] [PubMed] [Google Scholar]
- 9.Emanuel, P. A., J. Dang, J. S. Gebhardt, J. Aldrich, E. A. Garber, H. Kulaga, P. Stopa, J. J. Valdes, and A. Dion-Schultz. 2000. Recombinant antibodies: a new reagent for biological agent detection. Biosens. Bioelectron. 14:751-759. [DOI] [PubMed] [Google Scholar]
- 10.Emanuel, P., T. O'Brien, J. Burans, B. R. DasGupta, J. J. Valdes, and M. Eldefrawi. 1996. Directing antigen specificity towards botulinum neurotoxin with combinatorial phage display libraries. J. Immunol. Methods 193:189-197. [DOI] [PubMed] [Google Scholar]
- 11.Frame, K. K., and W. S. Hu. 1990. The loss of antibody productivity in continuous culture of hybridoma cells. Biotechnol. Bioeng. 35:469-476. [DOI] [PubMed] [Google Scholar]
- 12.Giudicelli, V., D. Chaume, and M. P. Lefranc. 2004. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 32:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Hayhurst, A., S. Happe, R. Mabry, Z. Koch, B. L. Iverson, and G. Georgiou. 2003. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J. Immunol. Methods 276:185-196. [DOI] [PubMed] [Google Scholar]
- 15.Heng, C. K., T. C. Seng, N. Khalid, J. A. Harikrishna, and R. Y. Othman. 2003. Synthesis of a soluble flag-tagged single chain variable fragment (scFv) antibody targeting cucumber mosaic virus (CMV) coat protein. Asia Pac. J. Mol. Biol. Biotechnol. 11:93-100. [Google Scholar]
- 16.Hoogenboom, H. R., A. D. Griffiths, K. S. Johnson, D. J. Chriswell, P. Hudson, and G. Winter. 1991. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoogenboom, H. R., A. P. de Bruin, S. E. Hufton, R. M. Hoet, J. W. Arends, and R. C. Roovers. 1998. Antibody phage display technology and its applications. Immunotechnology 4:1-20. [DOI] [PubMed] [Google Scholar]
- 18.Ikematsu, H., Y. Ichiyoshi, E. W. Schettino, M. Nakamura, and P. Casali. 1994. VH and V kappa segment structure of anti-insulin IgG autoantibodies in patients with insulin-dependent diabetes mellitus: evidence for somatic selection. J. Immunol. 152:1430-1441. [PMC free article] [PubMed] [Google Scholar]
- 19.Jurado, P., D. Ritz, J. Beckwith, V. de Lorenzo, and L. A. Fernandez. 2002. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J. Mol. Biol. 320:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Kabat, E. A., T. T. Wu, H. M. Perry, K. S. Gottesman, and C. Foeller. 1991. Sequences of proteins of immunological interest, 5th ed. National Institutes of Health, U.S. Department of Health and Human Services, Bethesda, MD.
- 21.Kamboj, D. V., A. K. Goel, and L. Singh. 2006. Biological warfare agents. Def. Sci. J. 56:495-506. [Google Scholar]
- 22.Kamboj, D. V., V. Nema, A. K. Pandey, A. K. Goel, and L. Singh. 2006. Heterologous expression of staphylococcal enterotoxin B (seb) gene for antibody production. Electron. J. Biotechnol. www.ejbiotechnology.info/content/vol9/issue5/full/3/index.html.
- 23.Kessler, N., M. Aymard, and S. Bertrand. 1993. Stability of a murine hybridoma is dependent on the clone line and culture media. In Vitro Cell. Dev. Biol. 29A:203-207. [DOI] [PubMed] [Google Scholar]
- 24.Kortt, A. A., M. Lah, G. W. Oddie, C. L. Gruen, J. E. Burns, L. A. Pearce, J. L. Atwell, A. J. McCoy, G. J. Howlett, D. W. Metzger, R. G. Webster, and P. J. Hudson. 1997. Single-chain Fv fragments of anti-neuraminidase antibody NC10 containing five- and ten-residue linkers form dimers and with zero-residue linker a trimer. Protein Eng. 10:423-433. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. H., and J. W. Kwak. 2003. Expression and functional reconstitution of a recombinant antibody (Fab') specific for human apolipoprotein B-100. J. Biotechnol. 101:189-198. [DOI] [PubMed] [Google Scholar]
- 26.Llewelyn, M., and J. Cohen. 2002. Superantigens: microbial agents that corrupt immunity. Lancet Infect. Dis. 2:156-162. [DOI] [PubMed] [Google Scholar]
- 27.Mantis, N. J. 2005. Vaccines against the category B toxins: staphylococcal enterotoxin B, epsilon toxin and ricin. Adv. Drug Deliv. Rev. 57:1424-1439. [DOI] [PubMed] [Google Scholar]
- 28.Mattix, M. E., R. E. Hunt, C. L. Wilhelmson, A. J. Johnson, and W. B. Baze. 1995. Aerosolized staphylococcal enterotoxin B-induced pulmonary lesions in rhesus monkeys (Macaca mulatta). Toxicol. Pathol. 23:262-268. [DOI] [PubMed] [Google Scholar]
- 29.Meyer, T., J. Stratmann-Selke, J. Meens, T. Schirmann, G. F. Gerlach, R. Frank, S. Dübel, K. Strutzberg-Minder, and M. Hust. 2010. Isolation of scFv fragment specific to OmpD of Salmonella typhimurium. Vet. Microbiol., in press. doi: 10.1016/j.vetmic.2010.06.023. [DOI] [PubMed]
- 30.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 31.Mechaly, A., E. Zahavy, and M. Fisher. 2008. Development and implementation of a single-chain Fv antibody for specific detection of Bacillus anthracis spores. Appl. Environ. Microbiol. 74:818-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nema, V., R. Agrawal, D. V. Kamboj, A. K. Goel, and L. Singh. 2007. Isolation and characterization of heat resistant enterotoxigenic Staphylococcus aureus from a food poisoning outbreak in Indian subcontinent. Int. J. Food Microbiol. 117:29-35. [DOI] [PubMed] [Google Scholar]
- 33.Ono, H. K., K. Omoe, K. Imanishi, Y. Iwakabe, D. L. Hu, H. Kato, N. Saito, A. Nakane, T. Uchiyama, and K. Shinagawa. 2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 76:4999-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paoli, G. C., L. G. Kleina, and J. D. Brewster. 2007. Development of Listeria monocytogenes-specific immunomagnetic beads using a single-chain antibody fragment. Foodborne Pathog. Dis. 4:74-83. [DOI] [PubMed] [Google Scholar]
- 35.Papageorgiou, A. C., H. S. Tranter, and K. R. Acharya. 1998. Crystal structure of microbial superantigen staphylococcal enterotoxin B at 1.5 Å resolution: implications for superantigen recognition by MHC class II molecules and T-cell receptors. J. Mol. Biol. 277:61-79. [DOI] [PubMed] [Google Scholar]
- 36.Pelat, T., M. Hust, M. Hale, M. P. Lefranc, S. Dübel, and P. Thullier. 2009. Isolation of a human-like antibody fragment (scFv) that neutralizes ricin biological activity. BMC Biotechnol. 9:60. http://www.biomedcentral.com/1472-6750/9/60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rusnak, J. M., M. Kortepeter, R. Ulrich, M. Poli, and E. Boudreau. 2004. Laboratory exposure to staphylococcal enterotoxin B. Emerg. Infect. Dis. 10:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russel. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Schlievert, P. M. 1993. Role of superantigens in human disease. J. Infect. Dis. 167:997-1002. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, D. Y., S. Jarraud, B. Lemercier, G. Cozon, K. Echasserieau, J. Etienne, M. L. Gougeon, G. Lina, and F. Vandenesch. 2006. Staphylococcal enterotoxins like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 74:4724-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner, D. J., M. A. Ritter, and A. J. George. 1997. Importance of the linker in expression of single-chain Fv antibody fragments: optimization of peptide sequence using phage display technology. J. Immunol. Methods 205:43-54. [DOI] [PubMed] [Google Scholar]
- 42.Ulrich, R. B., S. Sidell, T. J. Taylor, C. L. Wilhelmsen, and D. R. Franz. 1997. Staphylococcal enterotoxin B and related pyrogenic toxins, p. 621-630. In F. R. Sidell, E. T. Takafuji, and D. R. Franz (ed.), Textbook of military medicine, warfare, weaponry, and the casualty. Medical aspects of chemical and biological warfare. Office of the Surgeon General, Department of the Army, Falls Church, VA.
- 43.Wang, S. H., J. B. Zhang, Z. P. Zhang, Y. F. Zhou, R. F. Yang, J. Chen, Y. C. Guo, F. You, and X. E. Zhang. 2006. Construction of single chain variable fragment (scFv) and biscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal. Chem. 78:997-1004. [DOI] [PubMed] [Google Scholar]