Abstract
The nitrogen-fixing symbiont of alfalfa, Sinorhizobium meliloti, is able to use myo-inositol as the sole carbon source. Putative inositol catabolism genes (iolA and iolRCDEB) have been identified in the S. meliloti genome based on their similarities with the Bacillus subtilis iol genes. In this study, functional mutational analysis revealed that the iolA and iolCDEB genes are required for growth not only with the myo-isomer but also for growth with scyllo- and d-chiro-inositol as the sole carbon source. An additional, hypothetical dehydrogenase of the IdhA/MocA/GFO family encoded by the smc01163 gene was found to be essential for growth with scyllo-inositol, whereas the idhA-encoded myo-inositol dehydrogenase was responsible for the oxidation of d-chiro-inositol. The putative regulatory iolR gene, located upstream of iolCDEB, encodes a repressor of the iol genes, negatively regulating the activity of the myo- and the scyllo-inositol dehydrogenases. Mutants with insertions in the iolA, smc01163, and individual iolRCDE genes could not compete against the wild type in a nodule occupancy assay on alfalfa plants. Thus, a functional inositol catabolic pathway and its proper regulation are important nutritional or signaling factors in the S. meliloti-alfalfa symbiosis.
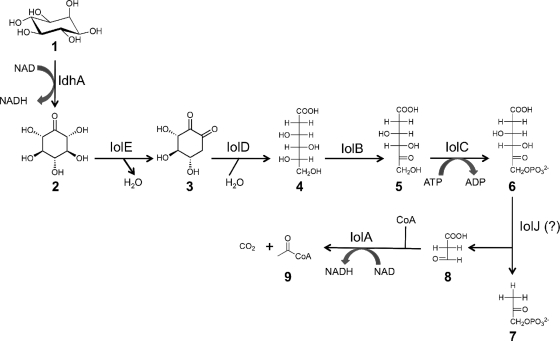
The sugar alcohol inositol, or cyclohexanehexol, occurs in several different stereoisomers, of which the myo-form (Fig. 1, compound 1) is the most abundant (1). myo-Inositol plays important structural and signaling roles in animal and plant cells (22). In the environment, myo-inositol mainly occurs in the phosphorylated form and is involved in the phosphate cycle of terrestrial and freshwater ecosystems (41). The stereoisomers d-chiro- and scyllo-inositol have recently attracted attention, because they have shown therapeutic potentials for diabetes and Alzheimer's disease, respectively (11, 21). Although there is only limited knowledge about the metabolism of d-chiro- and scyllo-inositol (25, 50), the catabolism of myo-inositol has been studied in a variety of microorganisms, including some members of the Firmicutes (17, 46, 51), Enterobacteriaceae (4, 19, 40), and Rhizobiaceae (16, 29). The myo-inositol catabolic pathway and its regulation are best understood in the Gram-positive bacterium Bacillus subtilis. The B. subtilis iol genes are organized in a divergon comprising iolABCDEFGHIJ and iolRS (47-49). In the proposed inositol catabolic pathway, the myo-inositol dehydrogenase oxidizes myo-inositol to its corresponding ketone 2-keto-myo-inositol (2KMI), which is then further catabolized by the actions of IolE, -D, -B, -C, -J, and -A (Fig. 1). The inducer of the inositol catabolic pathway in B. subtilis is the product of the IolC reaction, 2-deoxy-5-keto-d-gluconic acid 6-phosphate (DKGP; compound 6 in Fig. 1), which antagonizes the binding of the IolR repressor to the iol promoter region (51).
FIG. 1.
The proposed myo-inositol catabolic pathway (http://www.genome.jp/kegg/). Compound 1, myo-inositol (MI); compound 2, 2KMI; compound 3, 3D-(3,4/5) trihydroxycyclohexane-1,2-dione (THcHDO); compound 4, 5-deoxy glucuronic acid (5DG); compound 5, 2-deoxy-5-keto-d-gluconic acid (DKG); compound 6, DKGP; compound 7, dihydroxyacetone phosphate (DHAP); compound 8, malonic semialdehyde (MSA); compound 9, acetyl coenzyme A (acetyl-CoA). Enzymes: IdhA, myo-inositol dehydrogenase; IolE, 2KMI dehydratase; IolD, THcHDO hydrolase; IolB, 5DG isomerase; IolC, DKG kinase; IolJ aldolase (not yet identified in S. meliloti); IolA, MSA dehydrogenase.
Sinorhizobium meliloti, the nitrogen-fixing symbiont of alfalfa, can use myo-inositol as the sole carbon source (15). The idhA-encoded myo-inositol dehydrogenase had been shown to be required for myo-inositol catabolism (15), and more recently, an S. meliloti iolA mutant was reported that could not grow with myo-inositol as the sole carbon source (5). Based on comparisons with B. subtilis, a cluster of genes (iolRCDEB) was identified in the genome of S. meliloti (http://sequence.toulouse.inra.fr/S.meliloti), but their functional role has not been described. Rhizobial inositol metabolism is of special interest because of its link to the catabolism of a group of nutritional mediators in plant-bacteria interactions known as the rhizopines. Rhizopines, produced by several symbiotic S. meliloti and Rhizobium leguminosarum bv. viciae strains, are inositol derivatives, namely, scyllo-inosamine and l-3-O-methyl-scyllo-inosamine (26, 36, 38, 43). The ability to catabolize these inositol derivatives (rhizopines) has been shown to play a role in competition for nodule occupancy (35), but it seems that the ability to catabolize myo-inositol itself may also play a role in plant-bacteria interactions. For example, R. leguminosarum bv. viciae iolA and iolD mutants were reported to be strongly impaired in their ability to compete with the wild type during the nodulation process (14), and a Sinorhizobium fredii idhA mutant induced nodules with aberrant ultrastructure and showed reduced nitrogen-fixing ability (16). In contrast, an S. meliloti idhA mutant was not affected in the ability to nodulate its host plant or to fix nitrogen, but results from competition experiments have not been reported for S. meliloti (15). Here, we present a detailed analysis of the S. meliloti smc01163, iolA, and iolRCDEB genes and elucidate their roles in the catabolism of different inositol isomers and in plant-bacteria interactions.
MATERIALS AND METHODS
Microbiological methods.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in LB medium (37). Antibiotic concentrations for E. coli were 50 μg/ml ampicillin (Ap), 30 μg/ml chloramphenicol (Cm), 15 μg/ml gentamicin (Gm), 25 μg/ml kanamycin (Km), 25 μg/ml spectinomycin (Sp), and 10 μg/ml tetracycline (Tc). S. meliloti cultures were grown at 28°C. Rich media for S. meliloti were tryptone yeast (TY) (3) or LB medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LBMC); minimal media were minimal M medium (34) with 0.1% KNO3 or 0.1% NH4Cl as sole nitrogen source for the catabolism studies and enzyme assays and GTS minimal medium (18) for the selection of exconjungants. Carbon sources were added to the minimal media at a final concentration of 0.2% unless otherwise indicated. Antibiotic concentrations for S. meliloti were 15 μg/ml Gm, 200 μg/ml Km, 250 μg/ml streptomycin (Sm), 200 μg/ml Sp, and 10 μg/ml Tc. For the catabolism studies, S. meliloti strains were inoculated 1:100 from TY precultures into liquid minimal M medium. Cultures were grown on a shaking incubator, and the growth was determined spectrophotometrically at 600 nm after 3, 5, and 7 days. Catabolism studies were carried out in duplicate, and values represent the averages of two independent experiments ± the standard errors of the means (SEM).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 37 |
| HB101 | supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 37 |
| Sinorhizobium meliloti strains | ||
| 1021 | Wild type, Smr derivative of SU47 | 23 |
| 2011 | Wild type, Smr derivative of SU47 | 24 |
| TIDHA | 1021 idhA::Tn5-56, Smr Kmr | 15 |
| T63 | 1021 smc01163::Ω, Smr Spr | This study |
| TIOLC | 1021 iolC::pVO155, Smr Kmr | This study |
| TIOLD | 1021 iolD::pVO155, Smr Kmr | This study |
| TIOLE | 1021 iolE::pVO155, Smr Kmr | This study |
| TIOLB | 1021 iolB::pVO155, Smr Kmr | This study |
| WIDHA | 2011mTn5STM.5.11.A04, idhA::gus, Smr Kmr | 28 |
| W63-1 | 2011mTn5STM.2.07.F05, smc01163::gus, Smr Kmr | 28 |
| W63-2 | 2011mTn5STM.5.07.B01, smc01163::gus, Smr Kmr | 28 |
| WIOLR | 2011mTn5STM.4.13.C12, iolR::gus, Smr Kmr | 28 |
| WIOLC | 2011mTn5STM.1.11.A02, iolC::gus, Smr Kmr | 28 |
| WIOLD | 2011mTn5STM.1.13.D10, iolD::gus, Smr Kmr | 28 |
| WIOLE | 2011mTn5STM.1.01.B03, iolE::gus, Smr Kmr | 28 |
| WIOLA | 2011mTn5STM.4.03.B06, iolA::gus, Smr Kmr | 28 |
| WGLYA | 2011mTn5STM.4.01.D11, glyA2::gus, Smr Kmr | 28 |
| Plasmids | ||
| pVO155 | Vector with promoterless gusA gene, Apr Kmr | 27 |
| pJQ200SK | Suicide vector with sacB gene, Gmr | 31 |
| pGEM-T | Cloning vector, Apr | Promega |
| pCR2.1-TOPO | Cloning vector, Apr Kmr | Invitrogen |
| pHP45Ω | Source for Ω fragment, Apr Smr Spr | 30 |
| pRK2013 | mobtra, Kmr | 12 |
| pRK600 | mobtra, Cmr | 13 |
| pJZ1 | pVO155 containing 301-bp fragment of iolC, Kmr | This study |
| pJZ2 | pVO155 containing 270-bp fragment of iolD, Kmr | This study |
| pJZ3 | pVO155 containing 226-bp fragment of iolE, Kmr | This study |
| pJZ4 | pVO155 containing 399-bp fragment of iolB, Kmr | This study |
| pPK63-1 | pJQ200SK containing 1,121-bp fragment of smc01163, Gmr | This study |
| pPK63Ω | pPK63-1 containing Ω, Gmr Smr Spr | This study |
| pTE3 | Broad-host-range expression vector, Tcr | 10 |
| pIOLC | pTE3 containing 2,412-bp fragment of iolC | This study |
| pIOLD | pTE3 containing 2,776-bp fragment of iolD | This study |
| pIOLE | pTE3 containing 945-bp fragment of iolE | This study |
| pIOLB | pTE3 containing 881-bp fragment of iolB | This study |
Preparation of 2-keto-myo-inositol.
The 2KMI used in this study was synthesized using Gluconobacter oxydans (Acetobacter suboxydans ATCC 621) according to the methods described by Carter et al. (7) with the following adaptations: G. oxydans was grown on sorbitol agar containing 2.5% sorbitol, 0.5% yeast extract, 0.3% peptone, and 1.5% agar or in sorbitol broth containing 10% sorbitol and 0.5% yeast extract. G. oxydans was inoculated 1:100 from an overnight preculture into oxidation medium containing 3% myo-inositol, 0.5% yeast extract, and 0.1% sorbitol. The oxidation was carried out at 28°C for 4 days. Bacteria were removed from the medium via centrifugation at 6,000 × g. The crude product was concentrated and recrystallized from a water-methanol mixture to afford the clean 2KMI.
2-Keto-myo-inositol.
Melting point 199°C (lit. m.p. 201°C); 1H NMR (400 MHz, D2O): δ 4.38 (d, J = 10.2 Hz, 2 H), 3.78 (t, J = 9.5 Hz, 1 H), 3.45 to 3.34 (m, 2 H). The melting point was determined in open capillaries using a Thomas-Hoover Unimelt instrument. The nuclear magnetic resonance (NMR) spectrum was recorded using a 400-MHz Jeol Eclipse nuclear magnetic resonance instrument.
DNA manipulations and microbiological methods.
Preparation of plasmid DNA, DNA digests, agarose gel electrophoresis, cloning, and transformation of E. coli cells were performed following established protocols (37). Di- and triparental conjugations were performed according to the methods reported by Rossbach and de Bruijn (33).
Construction of S. meliloti mutants.
Internal DNA fragments of the individual iolCDEB genes and a 1,121-bp DNA fragment that contained smc01163 were PCR amplified from cultures of S. meliloti 1021 with the primers listed in Table S1 of the supplemental material. The iolCDEB and the smc01163 PCR products were initially cloned into the pGEM-T or pCR2.1-TOPO vectors (Table 1). The individual iolCDEB inserts were recloned into the insertion vector pVO155 (Table 1) and smc01163 was cloned into the sacB-containing suicide vector pJQ200SK (Table 1). An Ω Sm/Sp fragment replaced the 341-bp NruI fragment of smc01163, resulting in plasmid pPK63::Ω. The pVO155 vectors carrying the internal fragments of the iolBCDE genes and pPK63::Ω were conjugated into S. meliloti 1021 with the helper plasmids pRK600 and pRK2013 (Table 1), respectively. The insertion events of the pVO155 derivatives were selected by plating the conjugation mixture onto LBMC Sm Km. The double homologous recombination event with pPK63::Ω was selected for by plating on GTS Sm Sp, followed by counterselection on TY Sm Sp containing 5% sucrose and screening for the absence of the suicide vector on TY Sm Gm. The correct insertions of pVO155 in the individual iolCDEB mutants, as well as of the Ω fragment in the smc01163 deletion mutant, were confirmed by PCR. The transposon (mTn5-STM) mutants of S. meliloti strain 2011 were provided by Anke Becker (28).
Construction of plasmids for complementation analysis.
DNA fragments containing the complete open reading frames (ORFs) of the wild-type iolC, iolD, iolE, and iolB genes, including their ribosomal binding sites, were PCR amplified from a liquid S. meliloti 2011 culture with primers that were engineered to contain either PstI or NsiI at their 5′ end and BamHI or BglII sites at their 3′ ends (see Table S1 in the supplemental material). The PCR products were cloned into the broad-host-range expression vector pTE3 (Table 1), bringing the PCR products in correct orientation under the control of the Salmonella trp promoter, which allows constitutive expression in S. meliloti (10). The resulting plasmids carrying the individual iolCDEB genes (Table 1) as well as pTE3 as an empty vector control were introduced into the individual S. meliloti 1021 and 2011 iolC, iolD, iolE, and iolB mutants via triparental mating (33). The presence of the wild-type and of the mutated iol genes in the mutant strains was confirmed with PCR.
β-Glucuronidase assays.
The β-glucuronidase assays were optimized based on the methods of Wilson et al. (44). Precultures of S. meliloti strains were inoculated 1:100 in 5 ml minimal M medium containing NH4Cl as N source and either myo-inositol, 2KMI, glycerol, glucose, or succinate as C sources. Aliquots (350 μl) of mid-exponential-phase cultures (optical density at 600 nm [OD600], 0.7 to 1) were harvested by centrifugation at 4,500 × g for 10 min. The pellet was resuspended in 350 μl GEB buffer (50 mM sodium phosphate buffer [pH 7], 0.6% β-mercaptoethanol, 10 mM EDTA, 1% Triton X-100, 0.1% sodium lauryl sarcosine). After an initial equilibration period of 15 min at 37°C, 35 μl of 20 mM 4-nitrophenyl-β- d-glucuronide (PNPG) was added to the cell lysate. A 100-μl aliquot of the reaction mix was transferred into 800 μl of a 400 mM NaCO3 stop solution after 5, 10, and 15 min. Cell debris was removed by centrifugation at 16,000 × g for 30 s, and the OD405 was determined spectrophotometrically. The reaction rate was expressed in nmol of p-nitrophenol produced per min per OD600 unit, ± the SEM. The values represent the means of two independent experiments, and each assay was carried out in duplicate.
NAD(H)-dependent dehydrogenase assays.
S. meliloti precultures were inoculated 1:100 into 500-ml Erlenmeyer flasks containing 100 ml minimal M medium with NH4Cl as the N source, glycerol as C source, and either 0.02% myo-inositol or 2KMI as inducer. Late-exponential-phase cultures (OD600, 1 to 1.25) were harvested via centrifugation at 6,000 × g, washed with 40 mM HEPES buffer (pH 7) containing 10 mM β-mercaptoethanol, and resuspended in 5 ml 40 mM HEPES buffer (pH 7). Cell extracts were prepared with a sonicator at 50 W with three 30-s sonication periods (Misonix XL-2020 [Farmingdale, NY]). The myo-, scyllo-, and d-chiro-inositol dehydrogenase activities were determined at room temperature (21 to 23°C). Each reaction mix (1 ml) contained 50 mM NH4Cl, 50 mM Na2CO3, 100 μl cell extract, and 0.4 mM NAD+. A baseline of background reduction of NAD+ in the absence of substrate was established at a wavelength of 340 nm for slope correction. The increase in absorbance (A340) in the presence 25 mM myo-, scyllo-, or d-chiro-inositol was monitored for 3 min. The protein content of the cell extracts was determined with a Bradford assay (Pierce Coomassie Plus: the Better Bradford assay kit; Thermo Fisher Scientific, Rockford, IL). The specific myo-, scyllo-, and d-chiro-inositol dehydrogenase activities are expressed as nmol NAD+ reduced min−1 mg of protein−1, ± the SEM. The values represent the means of two independent experiments, each of them performed in duplicate, unless otherwise indicated.
Competition assay for nodule occupancy.
Axenic alfalfa (Medicago sativa) plants were prepared by germination from surface-sterilized seeds on folded Whatman filter paper in 20 ml of nitrogen-free B&D growth medium (6) in 25-mm-diameter tubes as described previously (36). Plants were grown at room temperature under a cycle of 16 h of light and 8 h of dark. Before inoculation, rhizobial cultures were pelleted, washed, and resuspended in sterile deionized H2O, and their optical density at 600 nm was determined spectrophotometrically. Mixed cultures of the S. meliloti 2011 wild-type and individual mutant strains were prepared in a 1:1 ratio based on the OD600 values. In addition, the 1:1 input ratio was verified via serial dilution and plating on TY Sm and TY Sm Km. Seven-day-old alfalfa seedlings were inoculated with 1 ml of the mixed cultures or 1 ml of the wild-type or the individual mutant strains. One milliliter of sterile deionized H2O was added to control plants. The total number of nodules per plant was determined, and after 20 weeks, the nodules were harvested and surface sterilized with 70% ethanol. Plant fresh and dry weights were determined and averaged from six plants of each treatment group. Rhizobia were reisolated by homogenizing the nodules in sterile H2O, and serial dilutions were prepared. The wild-type versus the mutant strain output ratio was determined by selective plating on TY Sm and TY Sm Km. The values represent the averages of two independent studies ± the SEM, with six plants each.
RESULTS
Structure and organization of the inositol catabolism genes in S. meliloti.
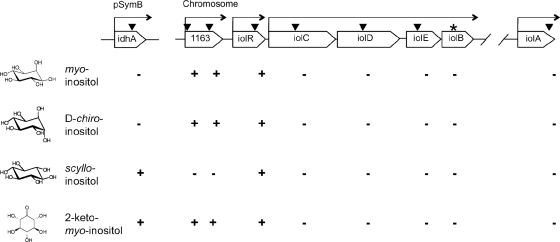
In contrast to the organization of the iol genes in B. subtilis, the inositol catabolism genes of S. meliloti are not arranged in a single gene cluster. The idhA gene is located on the pSymB plasmid, whereas the putative iolR, iolC, iolD, iolE, and iolB genes are organized in one cluster on the chromosome, all oriented in the same direction (Fig. 2). The iolA gene is located a further 400 kb away on the chromosome. An additional gene of interest, smc01163, encoding a putative dehydrogenase of the IdhA/MocA/GFO family, is located directly upstream of iolR (Fig. 2). The S. meliloti transcriptional regulator encoded by iolR belongs to the RpiR repressor family and is a homologue of the IolR regulator in Caulobacter crescentus (5). The iolCDEB genes are predicted to comprise an operon, whereas the smc01163 and iolR genes seem to be transcribed separately (www.microbesonline.org) (Fig. 2).
FIG. 2.
Ability of S. meliloti wild-type and the idhA, smc01163 (1163), iolR, iolC, iolD, iolE, iolA, and iolB mutant strains to grow with 0.2% myo-inositol, d-chiro-inositol, scyllo-inositol, or 2KMI as the sole carbon source in minimal medium. Open reading frames are depicted as open arrows. The locations of the mini-Tn5 insertions in the S. meliloti 2011 mutants are marked by vertical arrowheads. The star indicates the position of the plasmid insertion in the S. meliloti iolB mutant. Horizontal arrows above the genes indicate predicted transcriptional units. Each mutant's ability (+) or inability (−) to use inositol compounds as the sole carbon source is indicated.
Catabolism studies.
To conduct a complete study of the roles of the predicted iol genes in the catabolism of different inositol stereoisomers, mutants of S. meliloti strain 2011 were obtained that contained mTn5-STM transposon insertions in the idhA, iolA, and the individual iolRCDE genes, as well as two mutant strains with different insertions in the smc01163 gene (28). As part of a comparative analysis, mutants were also constructed in the S. meliloti 1021 strain, specifically, insertions in smc01163 and the individual iolCDEB genes. The 1021 and 2011 strains are both streptomycin-resistant derivatives of S. meliloti SU47 wild type (23, 24), but years of culturing in different laboratories have resulted in minor differences between these two strains (20, 42).
When grown in minimal medium with myo-inositol as the sole C source, both wild-type strains grew to an OD600 of around 1.2, while their corresponding idhA, iolC, iolD, iolE, iolB, and iolA mutants did not grow (Fig. 2; see also Fig. S2A in the supplemental material). The iolR and the three different smc01163 mutants grew to similar optical densities as the wild type (shown for W63-1 and W63-2 in Fig. 2; also shown for W63-1 in Fig. S2A). In a control experiment, all strains were able to grow with glucose as the sole C source (data not shown). Hence, the idhA, iolA, and iolCDEB genes are essential for myo-inositol catabolism in S. meliloti. Since the iolCDEB genes are predicted to form an operon, the mTn5-STM transposon and the plasmid insertions in the iolC, iolD, and iolE genes could have polar effects. We cloned the individual iolCDEB genes under the control of a constitutive promoter in the broad-host-range expression vector pTE3 (Table 1). The resulting plasmids were conjugated into the respective S. meliloti 1021 iolC, iolD, iolE, and iolB mutants as well as into the strain 2011 iolC, iolD, and iolE mutants. The mutant strains containing the plasmids with the individual iolC, iolD, iolE, and iolB genes were able to grow with myo-inositol as the sole C source, whereas the same strains carrying the empty vector as control could not (see Fig. S3 in the supplemental material). The growth of the mutant strains containing the plasmid with the iolC, iolD, and iolE genes was delayed compared to the wild types or the iolB mutant strain containing the iolB-carrying plasmid (see Fig. S3). Thus, we conclude that the mTn5-STM transposon and the plasmid insertions seem to allow a low level of expression of the downstream iol genes, probably due to a weak read-through from the integrated kanamycin resistance genes.
We investigated which other inositol isomers can be catabolized by S. meliloti. Strains 1021 and 2011 were grown with commercially available scyllo-, muco-, allo-, d-chiro-, and l-chiro-inositol as sole C source (see Fig. S1 in the supplemental material). Both wild-type strains could grow with scyllo- or d-chiro-inositol (shown for strain 2011 in Fig. S2B and C in the supplemental material), but they were not able to use l-chiro-, muco-, or allo-inositol (data not shown). The roles of the iol genes in the catabolism of scyllo- and d-chiro-inositol were further investigated. The idhA and the iolR mutants could grow with scyllo-inositol, but the iolC, iolD, iolE, iolB, and iolA mutants could not (Fig. 2; see also Fig. S2B). Interestingly, all three smc01163 mutants were unable to grow with scyllo-inositol as the sole C source (see Fig. S2B, W63-1). Thus, the smc01163 gene product appears to be essential for the catabolism of scyllo-inositol. d-chiro-Inositol was used as the sole C source by the iolR and the smc01163 mutants but not by the idhA, iolC, iolD, iolE, iolB, and iolA mutants (Fig. 2; see also Fig. S2C), suggesting that the idhA-encoded myo-inositol dehydrogenase facilitates the oxidation of both myo- and d-chiro-inositol.
We also tested the first proposed intermediate in the myo-inositol catabolic pathway, 2KMI, which was synthesized in our laboratory (see Materials and Methods). The iolC, iolD, iolE, iolB, and iolA mutants did not catabolize 2KMI, but the idhA, iolR, and smc01163 mutants could (Fig. 2; see also Fig. S2D). The fact that the idhA mutant grew on 2KMI as the sole C source while the iolA and iolCDEB mutants failed to do so strongly supports the notion that, also in S. meliloti, 2KMI is the product of the myo-inositol dehydrogenase (IdhA) reaction.
Regulation of S. meliloti inositol catabolism.
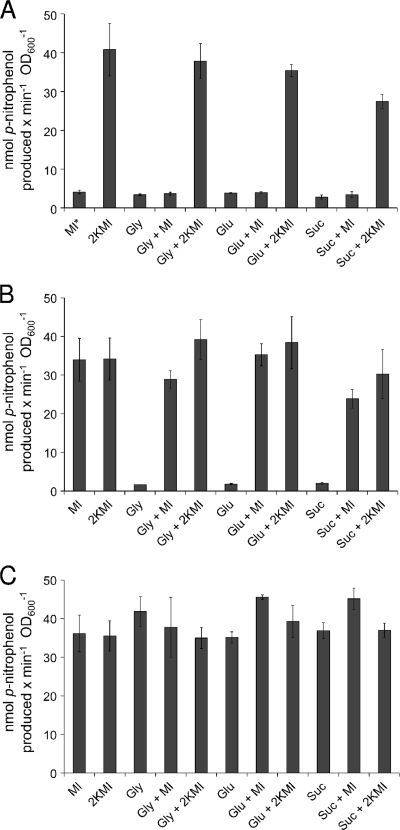
The 2011 idhA, smc01163, iolR, iolD, and iolE mutants contain the mTn5-STM::gusA transposon in the same orientation as the respective genes, creating a transcriptional fusion and therefore allowing us to investigate the regulation of the inositol genes by measuring the β-glucuronidase activity (28). Mutant strains were grown in minimal medium with either myo-inositol, 2KMI, glycerol, glucose, or succinate as carbon source, or in combinations, to analyze the effects of different carbon sources. Cells were harvested and solubilized, and their β-glucuronidase activities were determined. The wild-type strain, which does not contain a gusA gene, served as a negative control and did not exhibit any detectable β-glucuronidase activity (data not shown). The expression of the idhA gene in its corresponding mutant was not inducible by myo-inositol but was induced in the presence of 2KMI with all carbon sources tested (Fig. 3A). This finding indicates that not myo-inositol itself but either 2KMI or a later pathway intermediate functions as an inducer in S. meliloti, as has been shown for B. subtilis (51). The expression of smc01163 in its corresponding mutant was induced by myo-inositol or 2KMI with all carbon sources tested (Fig. 3B). We did not notice any major catabolite repression effect when glycerol, glucose, or succinate was present in the medium (Fig. 3A and B). The iolR gene, encoding the RpiR-like repressor, was constitutively expressed in its corresponding mutant, since high β-glucuronidase activities were displayed under all growth conditions, even for growth without inducer (Fig. 3C). It is interesting that only very low β-glucuronidase activities were observed in the iolD and iolE mutants in the presence or absence of the inducers myo-inositol or 2KMI (between 2 and 3 nmol min−1 OD600 unit−1). This further supports the notion that neither myo-inositol nor 2KMI, but a later pathway intermediate which cannot be synthesized by the iolD or iolE mutant, serves as the true inducer in S. meliloti.
FIG. 3.
β-Glucuronidase activities of the S. meliloti idhA (A), smc01163 (B), and iolR (C) gusA reporter gene fusions in the respective mutant strains. The reaction rate is expressed in nmol p-nitrophenol produced per minute per OD600 unit. Cultures were grown in minimal medium containing 0.2% of the following carbon sources: myo-inositol (MI), 2KMI, glycerol (Gly), glucose (Glu), succinate (Suc), or combinations thereof. Bars represent the averages of two independent experiments, and error bars denote SEM. MI* indicates that the idhA mutant did not grow with myo-inositol as the sole carbon source in minimal medium, but the residual β-glucuronidase activity is probably due to the carryover of cells from the TY preculture.
Determination of myo-inositol dehydrogenase activity.
For the determination of the myo-inositol dehydrogenase activity, an NAD(H)-dependent dehydrogenase assay was used. Cultures were grown in minimal medium containing glycerol as C source and with either myo-inositol or 2KMI as inducer. The specific myo-inositol dehydrogenase activities of the wild types, the idhA, the smc01163, and the individual iolCDEB mutant strains were low without prior induction (0.2 to 21 nmol min−1 mg of protein−1) (Table 2). Upon induction with myo-inositol, the 2011 and 1021 wild types displayed myo-inositol dehydrogenase activities of 103 and 131 nmol min−1 mg of protein−1, respectively (Table 2). When grown with 2KMI as inducer, the wild-type cells exhibited ∼30% lower myo-inositol dehydrogenase activities (Table 2). Regardless of whether the idhA mutant was induced with myo-inositol or 2KMI, its myo-inositol dehydrogenase activity was basically abolished (Table 2), which confirmed the results of Galbraith et al. (15), who found that the idhA gene, located on the pSymB plasmid, encodes the myo-inositol dehydrogenase. All three mutants with insertions in smc01163 exhibited myo-inositol dehydrogenase activities comparable to the wild type (Table 2, W63-1). The iolR mutant displayed a 4- to 5-fold-higher myo-inositol dehydrogenase activity than the wild type, even when not induced (Table 2). The iolC, iolD, iolE, and iolB mutants, however, exhibited very low myo-inositol dehydrogenase activities (Table 2), regardless of whether they were grown with or without myo-inositol or 2KMI as inducers, indicating that all four gene products (IolC, IolD, IolE, and IolB) are needed for inducer production.
TABLE 2.
Specific myo-inositol dehydrogenase activities of S. meliloti wild-type and mutant strains
| Strain | Relevant genotype |
myo-Inositol dehydrogenase sp act (nmol/min/mg of protein) |
||
|---|---|---|---|---|
| Uninduced | 2KMI induced | myo-Inositol induced | ||
| 2011 | Wild type | 5 ± 0.3 | 65 ± 9 | 103 ± 4 |
| 1021 | Wild type | 21 ± 4 | 79 ± 10 | 131 ± 7 |
| WIDHA | idhA | 2 ± 0.3 | 0.6 ± 0.3 | 0.5 ± 0 |
| W63-1 | smc01163 | 10 ± 4 | 55 ± 5 | 103 ± 42 |
| WIOLR | iolR | 522 ± 2 | 337 ± 6 | 410 ± 24 |
| WIOLC | iolC | 0.2 ± 0.1 | 0.4 ± 0.1 | 8 ± 0 |
| WIOLD | iolD | 2 ± 2 | 5 ± 1 | 5 ± 0 |
| WIOLE | iolE | 0.4 ± 0.2 | 0 ± 0 | 5 ± 0 |
| TIOLB | iolB | 5 ± 1 | 2 ± 0.3 | 0.4 ± 0.2 |
Determination of scyllo-inositol dehydrogenase activity.
The results of the catabolism studies suggest that the dehydrogenase encoded by smc01163 functions as a scyllo-inositol dehydrogenase. Thus, the scyllo-inositol dehydrogenase activities of the wild type and the smc01163 mutants were investigated. Based on our findings that the smc01163 gene was inducible by myo-inositol and 2KMI (see above), wild-type and mutant strains were grown in minimal medium with glycerol as C source and with myo-inositol or 2KMI as inducer. The specific scyllo-inositol dehydrogenase activities of the uninduced wild type and the smc01163 mutants were very low (10 and 5 nmol min−1 mg of protein−1, respectively). Upon induction with myo-inositol, the 2011 and 1021 wild types displayed scyllo-inositol dehydrogenase activities of 69 and 91 nmol min−1 mg of protein−1, respectively, while the three smc01163 mutants showed between 58% and 75% of the wild-type scyllo-inositol dehydrogenase activity (Table 3). The results of the scyllo-inositol dehydrogenase assay were unexpected. The smc01163 mutants were unable to use scyllo-inositol as sole C source, suggesting that the smc01163 gene encodes the scyllo-inositol dehydrogenase and the mutants would not exhibit any scyllo-inositol dehydrogenase activity. Nevertheless, we obtained the same results for all three smc01163 mutants, which were constructed independently in two different genetic backgrounds. The scyllo-inositol dehydrogenase activities of the iolR and iolE mutants were also determined. The iolR mutant displayed an almost-7-fold-higher scyllo-inositol dehydrogenase activity than the wild type (Table 3). The scyllo-inositol dehydrogenase activity of the iolE mutant was very low (Table 3). When the scyllo-inositol dehydrogenase activity was determined from cultures grown with 2KMI as inducer, similar results were obtained (Table 3).
TABLE 3.
Specific scyllo-inositol dehydrogenase activities of S. meliloti wild-type and mutant strains
| Strain | Relevant genotype |
scyllo-Inositol dehydrogenase sp act (nmol/min/mg of protein) |
|
|---|---|---|---|
| 2KMI induced | myo-Inositol induced | ||
| 2011 | Wild type | 74 ± 2 | 69 ± 1 |
| 1021 | Wild type | NDa | 91 ± 2 |
| WIOLR | iolR | ND | 477b |
| W63-1 | smc01163 | 50 ± 5 | 40 ± 1 |
| W63-2 | smc01163 | 39b | 52b |
| T63 | smc01163 | ND | 62 ± 1 |
| WIOLE | iolE | ND | 3 ± 0.5 |
ND, not done.
Value is based on one experiment.
Determination of d-chiro-inositol dehydrogenase activity.
The 2011 wild type and the corresponding idhA and iolR mutants were subjected to a d-chiro-inositol dehydrogenase assay. The wild type displayed a d-chiro-inositol dehydrogenase activity of 52 ± 0.2 nmol min−1 mg of protein−1, while the idhA mutant showed only marginal dehydrogenase activity in the presence of d-chiro-inositol (0.3 ± 0.1 nmol min−1 mg of protein−1). The d-chiro-inositol dehydrogenase activity of the WIOLR mutant was increased 4-fold (235 ± 10 nmol min−1 mg of protein−1).
Competition assay for nodule occupancy.
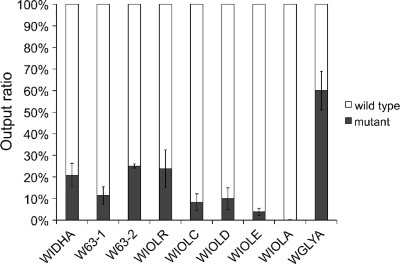
To investigate the role of S. meliloti inositol catabolism during symbiotic interactions, a competition assay for nodule occupancy was performed with the 2011 wild type and each of the idhA, smc01163, iolR, iolC, iolD, iolE, and iolA mutants. The wild type and the individual mutant strains were inoculated onto axenic alfalfa plants in a 1:1 ratio and as single inoculants as controls. All strains nodulated the host plants successfully when inoculated individually. Plants developed six nodules on average, and there was no difference between the average fresh and dry weight of the plants nodulated by the wild type (average fresh weight, 131 ± 12 g; average dry weight, 14 ± 2 g) or by the individual mutant strains (average fresh weight, 138 ± 16 g; average dry weight, 14 ± 2 g). Twenty weeks postinoculation, the nodules were harvested and rhizobia were reisolated from surface-sterilized nodules. On average, 106 bacteria were reisolated from the nodules of one plant, and the output ratio of kanamycin-resistant (mutant) to kanamycin-sensitive rhizobia (wild type) was determined by selective plating. If a gene does not play a role in the competition for nodule occupancy, the same output as input ratio (50:50) is expected. This was true for a control mutant (WGLYA) that carried an mTn5-STM insertion in an unrelated gene, since WGLYA was reisolated from the nodules with a frequency of 60%, which was similar to the input ratio of 50% (Fig. 4). In contrast, the mutants with insertions in the idhA, smc01163, iolR, iolC, iolD, iolE, or iolA genes represented only a small fraction of the rhizobia reisolated from the nodules. The values varied between 0.15% for the iolA mutant and 25% for the smc01163 mutant (Fig. 4). Thus, the idhA, smc01163, and iol mutants were outcompeted by the wild type in all cases, showing that a functional inositol catabolic pathway, the transcriptional regulator IolR, and the dehydrogenase encoded by smc01163 are all required for successful competition during alfalfa nodulation. We also determined the wild type/mutant output ratios after 10 and 15 weeks, with essentially the same results (data not shown).
FIG. 4.
Competition assay for nodule occupancy. The S. meliloti 2011 idhA (WIDHA), smc01163 (W63-1 and W63-2), iolR (WIOLR), iolC (WIOLC), iolD (WIOLD), iolE (WIOLE), iolA (WIOLA), and glyA2 (WGLYA) mutant strains were inoculated on alfalfa plants in a 1:1 ratio with the wild type. After 20 weeks nodules were harvested and surface sterilized, and rhizobia were reisolated from the nodules. The wild-type versus the mutant output ratio of the reisolated rhizobia was determined via selective plating. Bars represent the averages of two independent experiments representing nodules from six plants each. Error bars denote SEM.
DISCUSSION
We have shown that the iolA and iolCDEB genes are essential not only for myo-inositol but also for scyllo- and d-chiro-inositol catabolism in S. meliloti. It is interesting that myo- and especially scyllo-inositol serve as excellent carbon sources for S. meliloti, since the wild types grew to an OD600 of >1 in minimal medium, which is comparable to growth with other C sources, such as glucose, glycerol, or succinate. This is in contrast to B. subtilis, which does not grow as efficiently with myo- or scyllo-inositol as with glucose (25). Similarly, a lag time of 60 h has been reported for Salmonella enterica when grown with myo-inositol (19).
The idhA-encoded myo-inositol dehydrogenase acts on myo- and d-chiro-inositol.
The idhA mutant could not grow with myo- or with d-chiro-inositol as the sole C source. As confirmation, cell extracts of the idhA mutant did not display any detectable dehydrogenase activity when myo- or d-chiro-inositol was offered as substrate in the enzyme assay. Thus, we conclude that the initial dehydrogenation of myo- as well as of d-chiro-inositol is carried out by the idhA-encoded dehydrogenase. This is not without precedent; the purified myo-inositol dehydrogenase of B. subtilis has been shown to oxidize both myo- and d-chiro-inositol (8). The S. meliloti idhA mutant was able to use scyllo-inositol as the sole C source, indicating that there is at least one other dehydrogenase involved in the oxidation of scyllo-inositol and that scyllo-inositol is probably not a substrate for the myo-inositol dehydrogenase. In fact, scyllo-inositol could not react with the purified myo-inositol dehydrogenase of B. subtilis (32), and our preliminary data revealed that scyllo-inositol is not a substrate for the idhA gene product overexpresssed in E. coli (P. R. A. Kohler, unpublished observation).
Inositol catabolism in S. meliloti requires induction through a pathway intermediate.
The results from the β-glucuronidase and NAD(H)-dependent dehydrogenase assays clearly demonstrated that functional idhA, iolC, iolD, iolE, and iolB genes are required for induction of the iol genes. Thus, the inositol catabolism genes are not necessarily induced by inositol but by a later pathway intermediate. This is comparable to B. subtilis, in which the binding of DKGP to the negative regulator IolR antagonizes the transcriptional repression of the iol genes (51). DKGP is the fifth intermediate in the pathway and the product of the IolC reaction (Fig. 1). Usually, myo-, scyllo-, and d-chiro-inositol occur together in soil (41), and that may explain the advantage a common pathway intermediate would offer as an inducer over a specific inositol isomer.
IolR negatively regulates the activities of the myo- and scyllo-inositol dehydrogenases.
The iolR mutant was able to grow with myo-, scyllo-, and d-chiro-inositol and 2KMI as sole C sources, demonstrating that the iolR gene is not a structural gene in the inositol catabolic pathway. Nevertheless, the iolR gene is required for the regulation of the myo- and scyllo-dehydrogenase activities in S. meliloti, since the activity of both enzymes was 4- to 7-fold higher in the iolR mutant than in the wild type (Tables 2 and 3). The S. meliloti IolR, like IolR from C. crescentus, belongs to the RpiR repressor family (5). In C. crescentus, a conserved DNA sequence, GGAANATNCGTTCCA, was identified in the promoter region of the iol genes as a probable IolR-binding site (5). Computational predictions revealed related motifs in S. meliloti upstream of the idhA, iolR, and iolC genes (5). Interestingly, we detected a similar sequence ∼80 bp upstream of the start codon of smc01163 (CGAATAAATATTCCA). Our biological data confirm that IolR represses the myo- and scyllo-inositol dehydrogenase activities. The presence of a putative IolR-binding site upstream of iolR and the constitutive expression of the iolR-gusA fusion in the iolR mutant indicate that IolR negatively regulates its own expression.
The smc01163 gene is essential for scyllo-inositol metabolism.
None of the three different smc01163 mutants constructed in two different strains was able to grow with scyllo-inositol as the sole C source. Thus, the putative dehydrogenase encoded by the smc01163 gene is essential for scyllo-inositol catabolism. Nevertheless, the smc01163 mutants displayed 58 to 75% of the wild-type dehydrogenase activity when scyllo-inositol was offered as the substrate in the enzyme assay. We can exclude nonspecific NAD+ reduction through background activity, because the background NAD+-reducing activity was determined for each cell extract in the absence of the substrate for slope correction. All strains showed little NAD+-reducing background activities (1 to 5% of the myo- and scyllo-inositol dehydrogenase activities). In addition, we determined the enzyme activities in the presence of various substrate concentrations to ensure substrate specificity (data not shown).
Although smc01163 encodes a dehydrogenase essential for scyllo-inositol catabolism, this enzyme may not be the only dehydrogenase that interacts with scyllo-inositol. Recently, two scyllo-inositol dehydrogenases were identified in B. subtilis, IolX and IolW (25). Both purified enzymes reacted with scyllo-inositol, but only the iolX mutant showed impaired growth with scyllo-inositol as the sole C source (25). Interestingly, our computational analysis of the smc01163-deduced protein predicts the presence of an N-terminal signal peptide. Thus, a periplasmic location of SMc01163 is probable. We conclude that the catabolism of scyllo-inositol in S. meliloti may require at least one additional, probably cytoplasmic enzyme. This might explain the results of the growth studies compared to the results from the scyllo-dehydrogenase assays using crude cell extracts. SMc01163 seems to be essential for the initial interaction with scyllo-inositol in the periplasm, but this reaction is bypassed when the cell is lysed by sonication for the dehydrogenase assay. Another enzyme, not the one encoded by smc01163, oxidizes scyllo-inositol in the cytoplasm of S. meliloti and was detectable as scyllo-inositol dehydrogenase activity in the enzyme assay with the crude cell extract. Further work will be necessary to experimentally verify the periplasmic location of the smc01163-encoded dehydrogenase and to identify the substrate(s) and product(s) of the purified enzyme.
The inositol catabolism genes and their regulation are required for successful competition during alfalfa nodulation.
The idhA mutants of S. meliloti and R. leguminosarum bv. viciae nodulated their host plants and fixed nitrogen at the same levels as the wild-type strains (14, 15, 29). Nevertheless, the iolA and iolD mutants of R. leguminosarum bv. viciae could not compete with the wild type in a competition assay (14). Our results showed that the idhA, smc01163, iolA, and the iolRCDEB mutants of S. meliloti nodulated alfalfa successfully when inoculated onto plants individually, but in cochallenge experiments the mutants were outcompeted by the wild type. This demonstrates that a functional inositol catabolism is required for S. meliloti to successfully compete during the process of host nodulation and colonization of nodules. It is worth emphasizing that the iolR mutant also could not compete against the wild type for nodule occupancy. The inositol catabolic genes are derepressed in the iolR mutant, which should allow an even higher rate of inositol catabolism. Thus, the correct regulation of inositol catabolism genes also appears to be required for S. meliloti to successfully compete for nodule occupancy, suggesting that the role of inositol isomers extends beyond being a nutrient source.
Recently, some inositol isomers have gained great interest in the medical field because of their therapeutic potentials (11, 21). Our study contributes a better understanding of inositol metabolism and emphasizes its role in symbiotic nitrogen fixation in agriculture. Since legume crops such as alfalfa plants contain inositol (9), the presence of inositol compounds may act as one of the signals for rhizobia in communicating the presence of potential host plants and may also sustain the rhizobia while they are moving toward the plant, during root hair attachment or multiplication in the infection thread or nodule. In fact, in soybean nodules induced by Bradyrhizobium japonicum, the second and third most abundant carbohydrates after sucrose are myo- and d-chiro-inositol, with 1 to 2 mg per g of nodule fresh weight (39). Also, inositol has been found to be exuded into soil by legume plants (45). Other inositol derivatives, the rhizopines scyllo-inosamine and l-3-O-methyl-scyllo-inosamine, have been termed nutritional mediators, because they represent exclusive carbon and nitrogen sources for the rhizobial strains that carry the rhizopine catabolism genes (26, 36, 38, 43). Rhizopine and inositol catabolism are interrelated, because a functional inositol catabolic pathway was shown to be required for the catabolism of rhizopines (2, 15, 38). Clearly, inositol compounds and the ability to catabolize them play important nutritional or signaling roles in the symbiotic relationship between rhizobia and legume plants.
Supplementary Material
Acknowledgments
We thank an anonymous reviewer for improvements of the manuscript, Mariah Ramsdell and Lars Kohler for help with the preparation of 2-keto-myo-inositol, and Anke Becker and Michael Hynes for providing the S. meliloti 2011 mTn5-STM mutants and plasmid pJQ200SK, respectively.
This work was supported in part by a Dissertation Completion Fellowship and the Gwen Frostic Doctoral Fellowship from the Graduate College, by a Faculty Research and Creative Activities Award from Western Michigan University, and by the Herman Frasch Foundation.
Footnotes
Published ahead of print on 22 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, L. 1972. The cyclitols, p. 519-579. In W. Pigman and D. Horton (ed.), The carbohydrates, 2nd ed., vol. 1A. Academic Press, New York, NY. [Google Scholar]
- 2.Bahar, M., J. de Majnik, M. Wexler, J. Fry, P. S. Poole, and P. J. Murphy. 1998. A model for the catabolism of rhizopine in Rhizobium leguminosarum involves a ferredoxin oxygenase complex and the inositol degradative pathway. Mol. Plant Microbe Interact. 11:1057-1068. [DOI] [PubMed] [Google Scholar]
- 3.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 4.Berman, T., and B. Magasanik. 1966. The pathway of myo-inositol degradation in Aerobacter aerogenes: dehydrogenation and dehydration. J. Biol. Chem. 241:800-806. [PubMed] [Google Scholar]
- 5.Boutte, C. C., B. S. Srinivasan, J. A. Flannick, A. F. Novak, A. T. Martens, S. Batzoglou, P. H. Viollier, and S. Crosson. 2008. Genetic and computational identification of a conserved bacterial metabolic module. PLoS Genet. 4:e1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broughton, W. J., and M. J. Dilworth. 1971. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, H. E., R. K. Clark, Jr., B. Lytle, and G. E. McCasland. 1948. Oxidation of inositol by Acetobacter suboxydans. J. Biol. Chem. 174:415-426. [PubMed] [Google Scholar]
- 8.Daniellou, R., H. Zheng, D. M. Langill, D. A. Sanders, and D. R. Palmer. 2007. Probing the promiscuous active site of myo-inositol dehydrogenase using synthetic substrates, homology modeling, and active site modification. Biochemistry 46:7469-7477. [DOI] [PubMed] [Google Scholar]
- 9.Duke, J. 1992. Handbook of phytochemical constituents of GRAS herbs and other economic plants. CRC Press, Inc., Boca Raton, FL.
- 10.Egelhoff, T. T., and S. R. Long. 1985. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein and expression of nodA in Rhizobium meliloti. J. Bacteriol. 164:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenili, D., M. Brown, R. Rappaport, and J. McLaurin. 2007. Properties of scyllo-inositol as a therapeutic treatment of AD-like pathology. J. Mol. Med. 85:603-611. [DOI] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry, J., M. Wood, and P. S. Poole. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant Microbe Interact. 14:1016-1025. [DOI] [PubMed] [Google Scholar]
- 15.Galbraith, M. P., S. F. Feng, J. Borneman, E. W. Triplett, F. J. de Bruijn, and S. Rossbach. 1998. A functional myo-inositol catabolism pathway is essential for rhizopine utilization by Sinorhizobium meliloti. Microbiology 144:2915-2924. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, G., A. H. Krishnan, Y. W. Kim, T. J. Wacek, and H. B. Krishnan. 2001. A functional myo-inositol dehydrogenase gene is required for efficient nitrogen fixation and competitiveness of Sinorhizobium fredii USDA191 to nodulate soybean (Glycine max [L.] Merr.). J. Bacteriol. 183:2595-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawsar, H. I., K. Ohtani, K. Okumura, H. Hayashi, and T. Shimizu. 2004. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 235:289-295. [DOI] [PubMed] [Google Scholar]
- 18.Kiss, G. B., E. Vincze, Z. Kalman, T. Forrai, and A. Kondorosi. 1979. Genetic and biochemical analysis of mutants affected in nitrate reduction in Rhizobium meliloti. J. Gen. Microbiol. 113:105-118. [Google Scholar]
- 19.Kroeger, C., and T. M. Fuchs. 2009. Characterization of the myo-inositol utilization island of Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272:1-17. [DOI] [PubMed] [Google Scholar]
- 21.Larner, J. 2002. d-chiro-Inositol: its functional role in insulin action and its deficit in insulin resistance. Int. J. Exp. Diabetes Res. 3:47-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewus, F. A., and P. P. N. Murthy. 2000. myo-Inositol metabolism in plants. Plant Sci. 150:1-19. [Google Scholar]
- 23.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meade, H. M., and E. R. Signer. 1977. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 74:2076-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morinaga, T., H. Ashida, and K. Yoshida. 2010. Identification of two scyllo-inositol dehydrogenases in Bacillus subtilis. Microbiology 156:1538-1546. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, P. J., N. Heycke, Z. Banfalvi, M. E. Tate, F. J. de Bruijn, A. Kondorosi, J. Tempe, and J. Schell. 1987. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc. Natl. Acad. Sci. U. S. A. 84:493-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 28.Pobigaylo, N., D. Wetter, S. Szymczak, U. Schiller, S. Kurtz, F. Meyer, T. W. Nattkemper, and A. Becker. 2006. Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:4329-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, P. S., A. Blyth, C. J. Reid, and K. Walters. 1994. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 140:2787-2795. [Google Scholar]
- 30.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 31.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow directed selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 32.Ramaley, R., Y. Fujita, and E. Freese. 1979. Purification and properties of Bacillus subtilis inositol dehydrogenase. J. Biol. Chem. 254:7684-7690. [PubMed] [Google Scholar]
- 33.Rossbach, S., and F. J. de Bruijn. 2007. Transposon mutagenesis, p. 684-708. In C. A. Reddy (ed.), Methods for general and molecular microbiology, 3rd ed. ASM, Washington, DC.
- 34.Rossbach, S., D. A. Kulpa, U. Rossbach, and F. J. de Bruijn. 1994. Molecular and genetic characterization of the (mocABCR) genes of Rhizobium meliloti L5-30. Mol. Gen. Genet. 245:11-24. [DOI] [PubMed] [Google Scholar]
- 35.Rossbach, S., B. McSpadden, M. Ganoff, and F. J. de Bruijn. 1995. Rhizobium meliloti rhizopine catabolism genes: distribution, role in competition and potential as marker gene to track microbes, p. 180-188. In M. Levin, C. Grim, and J. S. Angle (ed.), Biotechnology risk assessment. University of Maryland Biotechnology Institute, College Park, MD.
- 36.Rossbach, S., G. Rasul, M. Schneider, B. Eardly, and F. J. de Bruijn. 1995. Structural and functional conservation of the rhizopine catabolism (moc) locus is limited to selected Rhizobium meliloti strains and unrelated to their geographical origin. Mol. Plant Microbe Interact. 8:549-559. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schoffers, E., S. R. Gurung, P. R. Kohler, and S. Rossbach. 2008. Chemical synthesis of scyllo-inosamine and catabolism studies in Sinorhizobium meliloti. Bioorg. Med. Chem. 16:7838-7842. [DOI] [PubMed] [Google Scholar]
- 39.Streeter, J. G. 1987. Carbohydrate, organic acid, and amino acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol. 85:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundaram, T. K. 1972. myo-Inositol catabolism in Salmonella typhimurium: enzyme repression dependent on growth history of organism. J. Gen. Microbiol. 73:209-219. [DOI] [PubMed] [Google Scholar]
- 41.Turner, B. L., M. J. Paphazy, P. M. Haygarth, and I. D. McKelvie. 2002. Inositol phosphates in the environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357:449-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wais, R. J., D. H. Wells, and S. R. Long. 2002. Analysis of differences between Sinorhizobium meliloti 1021 and 2011 strains using the host calcium spiking response. Mol. Plant Microbe Interact. 15:1245-1252. [DOI] [PubMed] [Google Scholar]
- 43.Wexler, M., D. M. Gordon, and P. J. Murphy. 1995. The distribution of inositol rhizopine genes in Rhizobium populations. Soil Biol. Biochem. 27:531-537. [Google Scholar]
- 44.Wilson, K. J., S. G. Hughes, and R. A. Jefferson. 1992. The Escherichia coli gus operon: introduction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria, p. 7-23. In S. R. Gallagher (ed.), GUS protocols: using the GUS gene as a reporter for gene expression. Academic Press, Inc., San Diego, CA.
- 45.Wood, M., and A. P. Stanway. 2001. myo-Inositol catabolism by Rhizobium in soil: HPLC and enzymatic studies. Soil Biol. Biochem. 33:375-379. [Google Scholar]
- 46.Yebra, M. J., M. Zuniga, S. Beaufils, G. Perez-Martinez, J. Deutscher, and V. Monedero. 2007. Identification of a gene cluster enabling Lactobacillus casei BL23 to utilize myo-inositol. Appl. Environ. Microbiol. 73:3850-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida, K., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, K., H. Sano, Y. Miwa, N. Ogasawara, and Y. Fujita. 1994. Cloning and nucleotide sequencing of a 15 kb region of the Bacillus subtilis genome containing the iol operon. Microbiology 140:2289-2298. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida, K., M. Yamaguchi, T. Morinaga, M. Ikeuchi, M. Kinehara, and H. Ashida. 2006. Genetic modification of Bacillus subtilis for production of D-chiro-inositol, an investigational drug candidate for treatment of type 2 diabetes and polycystic ovary syndrome. Appl. Environ. Microbiol. 72:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, K., M. Yamaguchi, T. Morinaga, M. Kinehara, M. Ikeuchi, H. Ashida, and Y. Fujita. 2008. myo-Inositol catabolism in Bacillus subtilis. J. Biol. Chem. 283:10415-10424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.