Abstract
The ability of various subsets of poultry intestinal microbiota to protect turkeys from colonization by Campylobacter jejuni was investigated. Community subsets were generated in vivo by inoculation of day-old poults with the cecal contents of a Campylobacter-free adult turkey, followed by treatment with one antimicrobial, either virginiamycin, enrofloxacin, neomycin, or vancomycin. The C. jejuni loads of the enrofloxacin-, neomycin-, and vancomycin-derived communities were decreased by 1 log, 2 logs, and 4 logs, respectively. Examination of the constituents of the derived communities via the array-based method oligonucleotide fingerprinting of rRNA genes detected a subtype of Megamonas hypermegale specific to the C. jejuni-suppressive treatments.
Campylobacter jejuni, a spiral, flagellated epsilonproteobacterial commensal of poultry, is the predominant cause of bacterial food-borne illness in the United States, resulting in approximately 2 million cases per year. A role for endogenous poultry intestinal microbiota in competitive exclusion (CE) of Campylobacter was first investigated in 1982 (38). Since then, numerous studies have attempted to identify microbes associated with Campylobacter CE. Suspensions of intestinal bacteria, isolated from Campylobacter-free adult poultry and passaged under strict anaerobic conditions, were found to protect chicks from colonization by the pathogen (31). Bacteria derived from the scrapings of broiler intestinal mucosa were proven more effective than the earlier fecal culture, a result not surprising, as Campylobacter is known to preferentially colonize cecal crypts (4, 39). The CE function of the bacterial suspensions decreased with time in storage, however (39, 40). Evidence also indicates that CE may depend on the presence of strictly anaerobic bacteria (31). As an oxygen gradient likely occurs from the host epithelium into the luminal contents, a CE role for both mucosal and luminal microbes in concert is likely.
Attempts have been made to identify specific microbes antagonistic to Campylobacter, and initial attempts isolated mucin-dwelling organisms with in vitro antagonistic effects against the pathogen (35, 36). Recent experiments have identified numerous bacterial groups producing anti-Campylobacter bacteriocins (29, 41, 42, 44, 45). Direct treatment of market-weight birds with the therapeutic bacteriocin Enterococcus faecium E 50-52 is effective for removal of Campylobacter spp. immediately prior to slaughter (44).
Despite progress toward a solution to contamination of poultry products by Campylobacter species, incomplete or intermittent CE protection, combined with a lack of studies addressing long-term CE efficacy, indicates that the Campylobacter colonization problem is far from solved (35). In addition, risk factors for campylobacteriosis other than direct consumption of contaminated poultry include consumption of fresh vegetables and bottled water (14). Campylobacter has been found in poultry manure used to fertilize crops as well as in runoff from these farms (22, 24, 50). We believe that novel approaches for studying microbial ecology in the gut are necessary for development of intervention strategies, including competitive exclusion.
The work described here takes a functional approach to identify microbes associated with protection of the intestine from Campylobacter jejuni colonization, an approach we are calling antibiotic dissection. The cecal contents from a Campylobacter-free adult turkey were inoculated into day-old poults and the microbial communities in these poults modified by treatment with therapeutic levels of antibiotics. The resulting modified microbiota were then tested for the ability to outcompete a C. jejuni challenge, and a microbe potentially associated with C. jejuni exclusion was identified.
MATERIALS AND METHODS
Antibiotic dissection.
Beltsville White turkey poults were hatched from the Campylobacter-free USDA flock housed on the National Animal Disease Center campus, with four poults per treatment. On day 1 after hatch, an adult was culled from the Beltsville White flock. The adult turkey was anesthetized to surgical depth by intramuscular injection with telazol (6 mg/kg of body weight), ketamine (8 mg/kg), and xylazine (4 mg/kg), followed by decapitation. Ceca were removed, and cecal contents were mixed 1:1 (vol/vol) with 1× phosphate-buffered saline (PBS). Poults were randomly assigned into six treatment groups, the uninoculated control (UC), inoculated control (IC), virginiamycin (VIR)-treated, enrofloxacin (ENR)-treated, neomycin (NEO)-treated, and vancomycin (VNC)-treated groups (Table 1). Antibiotics were chosen for both relevance to the poultry industry and variety of target molecule, such that the 50S and 30S ribosomal subunits, as well as RNA polymerase (RNAP) and bacterial cell walls, were targeted (Table 1). In addition, the chosen antibiotics are known to target various combinations of aerobes, anaerobes, and Gram-positive and -negative bacteria, further suggesting that antibiotic treatment would generate varied microbial communities (Table 1). Roughly 0.3 to 0.5 ml diluted adult cecal contents per poult was administered to five of the groups by oral gavage directly into the crop. A remaining control group (UC) was sham inoculated with 1× PBS. Poults were housed in BSL-2 isolators and given UV-irradiated feed and filter sterilized water ad libitum. The UC and IC treatment groups received sterile water and food only. In an effort to achieve therapeutic treatment levels, the VIR treatment group received sterile food containing 0.3 mg/g virginiamycin, the ENR group received sterile water containing 0.05 mg/ml enrofloxacin, the NEO group received sterile water containing 0.3 mg/ml neomycin sulfate, and the VNC group received sterile water containing 0.8 mg/ml vancomycin. The therapeutic dose of enrofloxacin was determined from Baytril product inserts. Therapeutic doses of virginiamycin and neomycin were determined from primary literature describing treatment of poultry for necrotic enteritis (18) and colibacillosis (30), respectively. Vancomycin was used at the same dose as avoparcin used to treat necrotic enteritis (33). Avoparcin was not used, as it was never approved for use in the United States, having been linked to vancomycin cross-resistance (51). Individuals within each isolator were assumed to have consumed equivalent antibiotic amounts. To maintain isolation, the birds in these experiments were not weighed; however, weights taken from a separate group of poults suggest these poults were ≤80 g. Antibiotic treatment was maintained for 7 days posthatch. Days 8 through 14 posthatch constituted a resting period allowing antibiotics to be cleared from the intestinal tract. On day 15, poults were challenged via oral gavage with 5 × 103 CFU of chloramphenicol-resistant Campylobacter jejuni 11168 (28). C. jejuni 11168 was kindly donated by Qijing Zhang from the Department of Veterinary Microbiology & Preventive Medicine, Iowa State University. On day 22 posthatch, poults were killed as described above. One cecum from each poult was frozen on dry ice for storage until DNA could be isolated, and serial dilutions were made in 1× PBS from the contents of the other cecum. All animal experiments were conducted with approval by, and under the guidance of, the National Animal Disease Center institutional animal care and use committee.
TABLE 1.
Treatment groups of poults inoculated with Campylobacter- free adult turkey intestinal contents and the antibiotics used to derive the six treatment groupsa
| Treatment group | Adult inoculum | Antibiotic target molecule | Sensitivity |
|||
|---|---|---|---|---|---|---|
| Anaerobes | Aerobes | Gram-positive bacteria | Gram-negative bacteria | |||
| UC | − | |||||
| IC | + | |||||
| VIR | + | 50S | S | R | S | R |
| ENR | + | RNAP | R | S | S | S |
| NEO | + | 30S | R | S | R | S |
| VNC | + | Cell wall | S | R | S | R |
Campylobacter culture.
Serial dilutions of cecal contents were plated on TSB agar supplemented with 10% defibrinated horse blood, 30 μg/ml cephalothin, and 5 μg/ml chloramphenicol. Plates were microaerobically incubated (5% O2, 10% CO2, and 85% N2) at 42°C for 48 h and C. jejuni colonies counted.
DNA extraction.
DNA was isolated from 200 mg cross sections of the frozen ceca such that luminal, mucosal, and intracellular microbes would be represented in the libraries. A modified QBiogene Fast Prep method was used to prepare DNA (QBiogene, Carlsbad, CA). Microbial cells were lysed in Boom L6 buffer by shaking with lysing matrix A at 5 m/s in a Fast Prep FP120 device for 30 s as described previously (6). DNA from the lysates was purified using phenol-chloroform extraction and ethanol precipitation.
ARISA.
Automated ribosomal intergenic spacer analysis (ARISA) was performed as described previously, using primer set ITSF (5′-GTCGTAACAAGGTAGCCGTA-3′) and ITSReub (5′-GCCAAGGCATCCACC-3′) (7, 9, 16). Primer ITSReub was 5′ end labeled with the 6-carboxyfluorescein (FAM) fluorochrome (Operon, Valencia, CA). Each 20-μl PCR mixture contained 4 ng template DNA. Similarity matrices were generated using the Dice coefficient, and cluster analysis was performed with the Mega 4.0 software program via the unweighted-pair group method using average linkages (UPGMA) (46).
OFRG analysis.
Oligonucleotide fingerprinting of rRNA genes (OFRG) analysis was performed as previously described by Valinsky et al. (48). Briefly, bacterial 16S rRNA gene clone libraries were constructed in Escherichia coli DH5α using a USER Friendly cloning kit (New England Biolabs, Ipswich, MA) and PCR primers 27F (5′-GGGAAAGUAGRRTTTGATYHTGGYTCAG-3′) and 1492R (5′-GGAGACAUGBTACCTTGTTACGACTT-3′) (5, 27). PCR was performed in 20-μl reaction mixtures containing 50 mM Tris (pH 8.3), 2.5 mM MgCl2, 250 μM deoxynucleoside triphosphates (dNTPs), 0.5 mg/ml bovine serum albumin (BSA), 400 nM forward and reverse primers, 1 μl fecal DNA, and 1.75 U Taq DNA polymerase. Amplification was performed with an initial denaturation at 94°C for 5 min, cycling at 94°C for 30 s (denaturation), 48°C for 40 s (annealing), and 72°C for 60 s (extension), and a final elongation for 2 min at 72°C. The number of amplification cycles varied between 15 and 30 cycles for each DNA sample and was determined from the fewest cycles found to generate a PCR product barely visible when 5 μl was examined on an ethidium bromide-stained agarose gel. A 1,536-clone library was generated, with roughly 250 clones representing each of the treatments.
The 16S rRNA gene clone library was PCR amplified using primers UserOFRGFor2 (5′-TCGAGCTCAGGCGCGCCTTAATTAAGCTGA-3′) and UserOFRGRev2 (5′-GCCAAGCTTCCTGCAGGGTTTAAACGCTGA-3′) in reaction mixtures containing 50 mM Tris (pH 8.3), 0.5 mg/ml BSA, 2.5 mM MgCl2, 250 μM dNTPs, 400 nM forward and reverse primers, 1 μl cells, and 1.75 U Taq DNA polymerase (5). Amplification was performed with an initial denaturation at 94°C for 10 min, 35 cycles of 94°C for 1 min and 72°C for 2 min, and a final elongation for 5 min at 72°C. Amplicons were arrayed onto nylon membranes with a multiblot replicator (V&P Scientific, Inc., San Diego, CA) as described previously (48). Membranes were hybridized overnight at 11°C with a set of 10-nucleotide (nt) bacterium-specific 33P-labeled DNA probes (48). Two arrays were hybridized for each probe, stripped as described previously, and rehybridized with universal probe 27F (5′-AGRRTTTGATYBTGGYTCAG-3′). Hybridizations were visualized using a Typhoon variable mode imager (Amersham Biosciences, Pittsburgh, PA), and hybridization signals were analyzed with Image Quant TL image analysis software, version 2003 (Amersham Biosciences, Pittsburgh, PA). Fingerprints containing the designations N (neither positive nor negative hybridization event), 1 (positive hybridization event), and 0 (negative hybridization event) were generated based on control clone hybridization intensities and Bayesian classification (25). Clone fingerprints containing more than 10 uncertain (N) classifications were discarded from further analysis. OFRG fingerprints were clustered using greedy clique partitioning (GCPAT) (http://alglab1.cs.ucr.edu/OFRG/gcpw.php).
Sequence analysis.
Clone sequencing was performed using primers UserOFRGFor2 (5′-TCGAGCTCAGGCGCGCCTTAATTAAGCTGA-3′), UserOFRGRev2 (5′-GCCAAGCTTCCTGCAGGGTTTAAACGCTGA-3′), 530F (5′-GTGCCAGCMGCCGCGG-3′), and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) (5, 27). Sequences were assembled and edited using Lasergene software (DNASTAR, Madison, WI). The sequences were compared to those in public databases by use of NCBI BLAST and Ribosomal Database Project II (RDP-II) (11). Megamonas hypermegale sequences were aligned using ClustalX, and phylogenetic analysis was performed using the neighbor-joining algorithm in Mega 4.0 with 1,000 bootstrap replications (46, 47). Sequence similarities were determined using PHYLIP DNAdist and distance-based operational taxonomic unit and richness determination (DOTUR) (15, 34).
Real-time PCR.
Real-time SYBR PCR amplification was performed with an iCycler IQ5 optical system according to the manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA). Reaction mixtures were composed of 1× iQ SYBR green supermix and 400 nM each primer. Megamonas-specific primers were designed for this study using the PRISE software program (17). The Megamonas type I (5′-ACTAAAGGAGGCCTAGTC-3′) and type II (5′-TCTAAAGGAGGCCTCTGAA-3′) forward primers were paired with the reverse primer MhypR2 (5′-CCCTAACAACAGAACTT-3′). Thermocycling required an initial denaturation at 94°C for 3 min, 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min, and a final elongation for 5 min at 72°C.
Species-specific amplification signals were normalized against the universal bacterial 16S copy number quantified using SYBR universal bacterial primers 27F (5′-AGRRTTTGATYBTGGYTCAG-3′) and 342R (5′-CTGCTGCSYCCCGTAC-3′) (27). Real-time PCR with the universal primers was performed with the M. hypermegale cycling parameters mentioned above, except a 56°C annealing temperature was used.
Intestinal response to antibiotic treatment.
An experimental repeat (no C. jejuni colonization was obtained) generated intestinal samples checked for overt intestinal morphological changes due to antibiotic treatment. At necropsy, 1- to 2-cm lengths of ceca were removed from the animals and placed directly into 10% formamide. Tissue samples were allowed to fix for 6 h and then transferred to 70% ethanol. Cross sections of approximately 4 μm were processed in low-melting-point paraffin and stained with hematoxylin and eosin. Images were captured using a Nikon Eclipse e400 microscope with an Optronics Magnafire camera (Galeta, CA). Morphometric measurements of villus height and villus section area were determined with the aid of Image-Pro Plus software (Media Cybernetics, Bethesda, MD). Measurements were taken for the four longest villi per animal, and means of villus height and area were determined for each animal and treatment.
Statistics.
Student's t test, analysis of variance (ANOVA), Tukey's pairwise comparison, and Spearman's D nonparametric rank order test were performed using the Paleontological Statistics (PAST) software package for education and data analysis (20).
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank under accession numbers FJ440020 to FJ440103 and FJ489243 to FJ489251.
RESULTS
Antibiotic intake.
Therapeutic antibiotic doses were achieved or exceeded for the virginiamycin (therapeutic dose of 0.04 g antibiotic/kg live bird weight), neomycin (therapeutic dose of 0.07 g/kg of live bird weight), and vancomycin (therapeutic dose of 0.02 g/kg live bird weight) groups (Table 2). Birds in the enrofloxacin (therapeutic dose of 0.05 g antibiotic/kg live bird weight) group consumed roughly a quarter of the therapeutic dose.
TABLE 2.
Estimated amounts of antibiotic ingested per day per bird
| Treatment group | Amt of antibiotic (mg) ingested per bird on: |
Wk 1 daily avg (mg) | ||||
|---|---|---|---|---|---|---|
| Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | ||
| VIR | 0.32 | 4.6 | 1.8 | 0.6 | 2.3 | 1.9 |
| ENR | 0.13 | 0.3 | 1.3 | 1.6 | 1.8 | 1 |
| NEO | NAa | 7.5 | 7.5 | 11.3 | 9.8 | 9 |
| VNC | 6.7 | 13.2 | 15.0 | 34.7 | 25.3 | 19 |
Water consumption was not measured.
Campylobacter quantification.
Campylobacter jejuni levels in the ceca of 21-day-old poults were quantified by plate counts (Fig. 1). Virginiamycin-derived microbiota resulted in an 18-fold C. jejuni colonization increase over the level for the control treatments, whereas enrofloxacin-, neomycin-, and vancomycin-derived microbiota reduced C. jejuni colonization 1 log, 2 logs, and 4 logs, respectively (Fig. 1). ANOVA and Tukey's least significant difference (LSD) analyses indicated that the virginiamycin increase was statistically significant (P < 0.001).
FIG. 1.
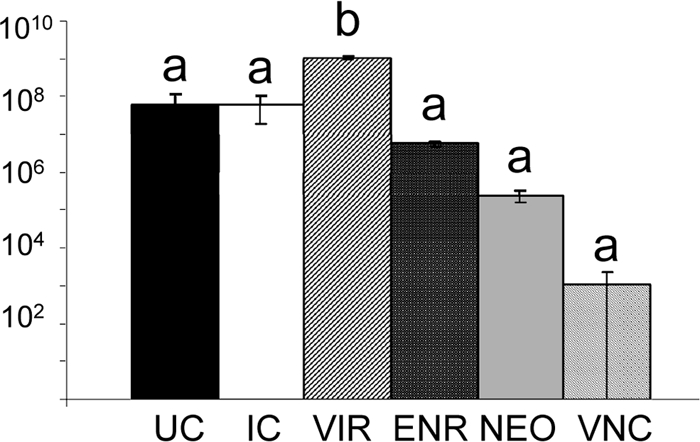
Campylobacter jejuni plate counts from trial 2. Six treatments are represented: poults that received no adult cecal content (UC), poults that received adult cecal contents only (IC), and poults that received both cecal contents and therapeutic levels of one antibiotic, either virginiamycin (VIR), enrofloxacin (ENR), neomycin (NEO), or vancomycin (VNC). C. jejuni load was measured for four poults per treatment, with the exception of the VNC treatment (three poults). ANOVA generated an F statistic of 63.4 and a P value of <0.001. Tukey's least significant difference test identified two statistically significant groups (a and b), as noted above the data bars. Error bars indicate standard errors of the means.
ARISA results.
Principal coordinates analysis of the microbiota detected four clusters (Fig. 2). One cluster was composed of samples from the IC, VIR, ENR, and NEO treatments. The other clusters were each composed of samples from individual treatments. The primary axis separated the IC, VIR, ENR, and NEO samples from the VNC, UC, and day-old-poult samples, indicating the greatest difference between these two groups. The secondary and tertiary axes separated the VNC, UC, and day-old-poult samples into different quadrants, indicating significant separation between the microbiota in these three groups (see Fig. S1 in the supplemental material).
FIG. 2.
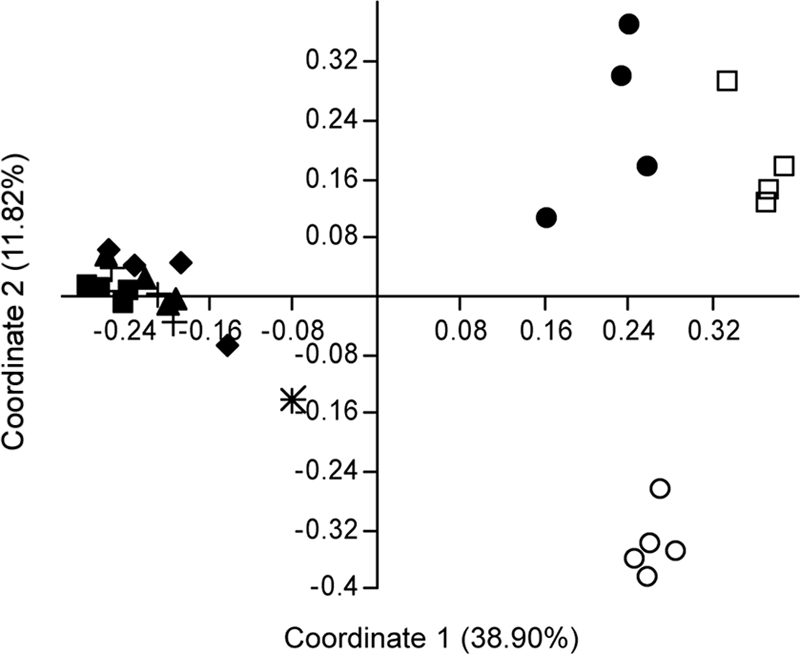
ARISA fingerprint analyses of trials 1 and 2 combined. The Dice similarity index was used to estimate differences between fingerprints from 3-week-old poults. Treatments include the following: d1, poults killed at day 1 posthatch; UC, uninoculated controls; IC, controls inoculated with Campylobacter-free adult intestinal contents; ENR, inoculated and enrofloxacin-treated poults; VIR, inoculated and virginiamycin-treated poults; NEO, inoculated and neomycin-treated poults; and VNC, inoculated and vancomycin-treated poults. “Adult” indicates the Campylobacter-free adult intestinal inoculum. “Negative” indicates a no-template ARISA reaction. Principal component analysis was prepared from Dice similarity indices via the Paleontological Statistics software package. Microbiota from all 28 poults and the adult turkey inoculum are represented. ✳, adult; ○, day 1 posthatch; □, uninoculated control; +, inoculated control; ▴, virginiamycin derived; ▪, enrofloxacin derived; ♦, neomycin derived; and •, vancomycin derived. Alternate views of this graph are available in the Fig. S1 in the supplemental material.
OFRG results.
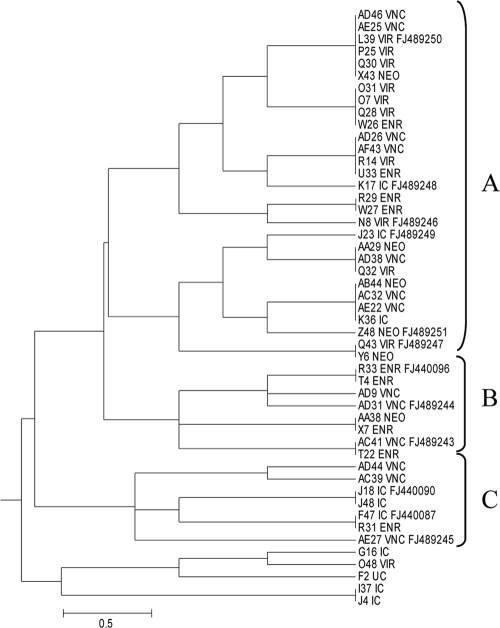
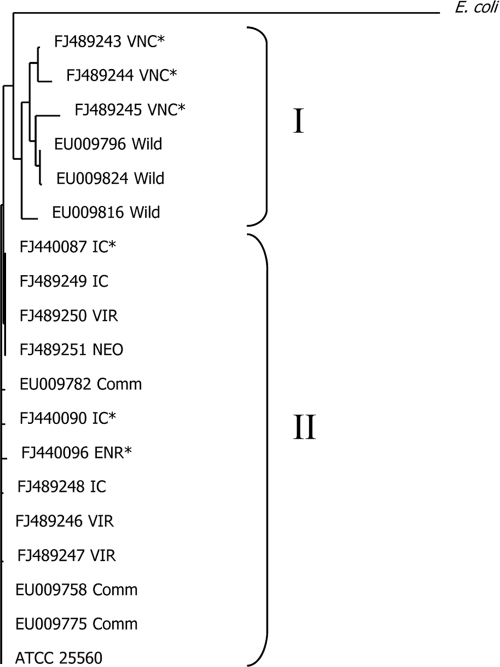
Oligonucleotide fingerprinting of rRNA genes was used to analyze 1,536 16S clones from antibiotic dissection trial 2. Clones fell primarily into the Clostridiales, Bacteroidetes, and Proteobacteria taxa (see Fig. S2 in the supplemental material). Two small clusters were composed of clones from the C. jejuni-suppressive treatment but not the C. jejuni-conducive treatment. Sequence analysis indicated that the cluster was composed of Megamonas hypermegale 16S genes (Fig. 3, clusters B and C). An adjacent cluster (cluster A) was composed of 16S clones from both conducive and suppressive treatments. Sequence analysis of the Megamonas 16S clones identified two subtypes, type I, containing Megamonas clones from the VNC treatments, and type II, containing clones from the IC, VIR, ENR, and NEO treatments (Fig. 2 and 3). These two subtypes were distinct operational taxonomic units (OTUs) at the 97% sequence similarity level as determined by DOTUR analysis and had both previously been detected in turkey cecal microbiota (Fig. 4) (37).
FIG. 3.
Dendrogram generated from fingerprints of Megamonas hypermegale 16S genes cloned from six microbiota treatments. Fingerprints were clustered using the greedy clique partitioning algorithm. Labels consist of the clone identifier followed by the treatment designation. The sequenced clones display the appropriate GenBank accession number. Clusters B and C are composed of clones from Campylobacter jejuni-protective treatments. Cluster A includes Megamonas clones from all treatments. A full dendrogram of all 1,200 fingerprints is provided in Fig. S2 in the supplemental material.
FIG. 4.
Megamonas hypermegale sequence analysis. Type I, Megamonas 16S clones derived from the vancomycin treatment and isolated from wild turkeys in a previous publication. Type II, Megamonas clones derived from the inoculated control, virginiamycin, enrofloxacin, and neomycin treatments and isolated from 18-week-old commercially raised turkeys in a previous publication. Types I and II are different at the 97% sequence identity level, as determined by DOTUR analysis. Sequences denoted by an asterisk are derived from Fig. 3, clusters B and C.
Megamonas hypermegale quantification.
Real-time PCR was used to quantify M. hypermegale in the cecal contents (Table 3 ). Megamonas spp. were relatively abundant in all antibiotic dissection treatments that received the adult inoculum (Table 3). Type I and type II Megamonas hypermegale subtypes showed different responses to the antibiotic treatments, and Student's t test indicated significantly higher loads of M. hypermegale type II than of type I in all but the IC and VNC treatments (Fig. 4 and Table 3). Spearman D nonparametric rank order tests between C. jejuni and Megamonas type I (P = 0.188) and between C. jejuni and Megamonas type II (P = 0.323) did not identify a correlation.
TABLE 3.
Quantification of Megamonas hypermegale types I and II in ceca of poults from six antibiotic dissection treatments
| Treatment group | No. of real-time target genes/10,000 16S gene copies (SE)a |
Pb | |
|---|---|---|---|
| M. hypermegale type I | M. hypermegale type II | ||
| UC | 0 | 2.2 (0.7) a | 0.0300 |
| IC | 325 (188) a | 5,493 (2,659) a | 0.1006 |
| VIR | 13 (3) a | 2,381 (542) a | 0.0047 |
| ENR | 0 (0.01) a | 2,282 (661) a | 0.0136 |
| NEO | 32 (10) a | 4,088 (1,212) a | 0.0155 |
| VNC | 978 (164) b | 93 (5.1) a | 0.0058 |
One-way ANOVA generated an F statistic of 13.55 and a P value of 0.0001 for M. hypermegale type I and an F statistic of 2.706 and a P value of 0.0563 for M. hypermegale type II. Tukey's least significant difference test identified two statistically significant groups (a and b).
Student's t test.
Morphometric measurements.
Neither cecal villus height nor villus area differed significantly between treatments (data not shown).
DISCUSSION
In the current work, antibiotic treatment was used to generate subsets of the microbiota from a Campylobacter-free turkey flock, and these subsets were tested in vivo for competitive exclusion capability. The four antibiotics were chosen for their historic use in commercial poultry production and/or their various modes of action (Table 1). In addition, these antibiotics are controversial because of their potential influence on human health. Virginiamycin is a growth-promoting antibiotic commonly used in poultry production in the United States as well as a therapeutic treatment for necrotic enteritis and aflatoxosis (1, 2). However, virginiamycin also causes cross-resistance to quinupristin-dalfopristin, the drug of last resort that is used to treat patients with vancomycin-resistant Enterococcus faecium or methicillin-resistant Staphylococcus aureus infections (8, 12). Use of the antibiotic avoparcin for growth promotion was banned in Europe in 1997 because a statistically significant association exists between the use of the antibiotic in poultry production and the occurrence of vancomycin-resistant Enterococcus species (26). Subsequent to the ban, significant reductions have been observed in the poultry carriage rates of vancomycin-resistant enterococci (49). Enrofloxacin (Baytril) is a fluoroquinolone used to treat poultry for Escherichia coli and Pasteurella infections. Treatment of poultry with various fluoroquinolones, including enrofloxacin, induces ciprofloxacin cross-resistance in a variety of intestinal pathogens (23). Ciprofloxacin is the drug of choice for treatment of campylobacteriosis; however, recent increases in ciprofloxacin resistance are reducing the utility of this drug for efficacious treatment (13, 52).
C. jejuni colonization was inhibited to various levels by differently derived microbial subsets. Morphometric analysis of the gross structure of the cecal villi did not differ between treatments; however, fingerprint analysis of the antibiotic-derived consortia indicated that community composition was unique to each of the treatments (Fig. 2). Communities harbored by day-old poults, vancomycin-treated poults, and the 3-week-old uninoculated controls were all different from one another and very different from the other three antibiotic-derived communities (Fig. 2). As initial cecal communities are known to be dominated by lactobacilli, clostridia, and enterobacteria, principal component analysis (PCA) and clustering results are consistent with predicted vancomycin resistance shown by aerobes (53). In contrast, ARISA patterns from virginiamycin-derived communities clustered with the adult and inoculated control (IC) profiles, suggesting predominantly anaerobic communities (Fig. 2). This result was a bit unexpected, as virginiamycin and vancomycin are predicted to have similar ranges of activity (Table 1). However, the necessarily limited scope of antibiotic sensitivity testing for different bacterial species appears to have masked the true sensitivity ranges in vivo.
A repeat of the animal trial was performed to determine the repeatability of community selection using antibiotics in vivo. Fingerprint analysis of the antibiotic-derived consortia indicated that community composition was unique to each of the trials (see Fig. S3 in the supplemental material). It is not surprising to note that, independent of antibiotic treatment, ultimate microbiota composition is dependent on the initial community. This is demonstrated by the clustering of communities by experiment (e.g., clusters I and II).
In 1979, M. hypermegale was suggested as a competitive exclusion strain against Salmonella enterica (3). On the basis of 16S sequence, M. hypermegale, previously named Bacteroides hypermegas, was recently reassigned to the phylum Firmicutes, family Acidaminococcaceae (21, 32). M. hypermegale cells are large, up to 15 μm long, are obligately anaerobic, require fermentable sugars, and produce acetic and propionic acids (21). This analysis of the antibiotic-derived communities indicated two small clusters of Megamonas hypermegale 16S genes containing only clones from C. jejuni-suppressive treatments (Fig. 3, clusters B and C). DOTUR analysis of the Megamonas sequences indicated two subtypes at the 97% sequence identity level (Fig. 4). One type was composed of M. hypermegale sequences from the vancomycin-derived community, and the second type contained M. hypermegale sequences from the remaining treatments. The Megamonas hypermegale type strain, ATCC 25560, belongs to Megamonas type II (Fig. 4) (10). Examination of Megamonas sequences from a previous study indicated the presence of Megamonas type I in wild turkeys (GenBank accession no. EU009816, EU009796, and EU009824) and Megamonas type II in commercial birds (GenBank accession no. EU009775, EU009758, and EU009782) (Fig. 4).
To confirm whether M. hypermegale colonization correlated with C. jejuni suppression, real-time quantification was performed on the antibiotic-treated populations. Megamonas type I was significantly enriched in the VNC (C. jejuni suppressive) treatments compared to the levels for the other treatments, while Megamonas type II was (not significantly) suppressed (Table 3). Spearman D analysis of C. jejuni and Megamonas loads revealed no direct correlation. However, the analysis was performed across antibiotic treatments. If Megamonas is one of a group of organisms involved in CE, this correlation may have been obscured. Further perusal of the antibiotic dissection data, perhaps by combining trial results and metagenomic analysis, is needed to identify more microbes associated with C. jejuni exclusion. In addition, it will be necessary to isolate both Megamonas types and perform in vivo competition experiments against C. jejuni, as only the presence of M. hypermegale type I appears to correlate with C. jejuni suppression.
The preliminary results described here suggest a correlation between vancomycin-derived microbiota, including M. hypermegale type I, and C. jejuni suppression. In addition, a correlation is suggested between virginiamycin-derived cecal microbiota and the enhanced ability of C. jejuni to colonize (Fig. 1). In light of growth-promoting virginiamycin use on commercial farms in the United States, it will be interesting to determine whether type I Megamonas strains are virginiamycin sensitive.
Supplementary Material
Acknowledgments
We acknowledge Dean C. Adams for statistical guidance and support and David P. Alt and Karen Halloum for sequencing services. We are also grateful to Virginia Montgomery for histology and Shari R. Steadham for villus measurements.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 15 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abo-Norag, M., T. S. Edrington, L. F. Kubena, R. B. Harvey, and T. D. Phillips. 1995. Influence of a hydrated sodium calcium aluminosilicate and virginiamycin on aflatoxicosis in broiler chicks. Poult. Sci. 74:626-632. [DOI] [PubMed] [Google Scholar]
- 2.Allen, P. C. 1992. Effect of virginiamycin on serum carotenoid levels and long, segmented, filamentous organisms in broiler chicks. Avian Dis. 36:852-857. [PubMed] [Google Scholar]
- 3.Barnes, E. M., C. S. Impey, and B. J. Stevens. 1979. Factors affecting the incidence and anti-salmonella activity of the anaerobic caecal flora of the young chick. J. Hyg. (Lond.) 82:263-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beery, J. T., M. B. Hugdahl, and M. P. Doyle. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bent, E., B. Yin, A. Figueroa, X. Ye, Q. Fu, Z. Liu, M. Chrobak, D. Jeske, T. Jiang, and J. Borneman. 2006. Development of a 9,600 clone array for oligonucleotide fingerprinting of rRNA genes: utilization to compare four different soil DNA extraction methods. J. Microbiol. Methods 67:171-180. [DOI] [PubMed] [Google Scholar]
- 6.Boom, R., C. Sol, M. Beld, J. Weel, J. Goudsmit, and P. Wertheim-van Dillen. 1999. Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J. Clin. Microbiol. 37:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 16:175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cato, E. P., and E. M. Barnes. 1976. Designation of the neotype strain of Bacteroides hypermegas Harrison and Hansen. Int. J. Syst. Bacteriol. 26:494-497. [Google Scholar]
- 11.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox, L. A., Jr., and D. A. Popken. 2004. Quantifying human health risks from virginiamycin used in chickens. Risk Anal. 24:271-288. [DOI] [PubMed] [Google Scholar]
- 13.DuPont, H. L., C. D. Ericsson, A. Robinson, and P. C. Johnson. 1987. Current problems in antimicrobial therapy for bacterial enteric infection. Am. J. Med. 82:324-328. [PubMed] [Google Scholar]
- 14.Evans, M. R., C. D. Ribeiro, and R. L. Salmon. 2003. Hazards of healthy living: bottled water and salad vegetables as risk factors for Campylobacter infection. Emerg. Infect. Dis. 9:1219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package), version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA.
- 16.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, Q., P. Ruegger, E. Bent, M. Chrobak, and J. Borneman. 2007. PRISE (PRImer SElector): software for designing sequence-selective PCR primers. J. Microbiol. Methods 72:263-267. [DOI] [PubMed] [Google Scholar]
- 18.George, B. A., C. L. Quarles, and D. J. Fagerberg. 1982. Virginiamycin effects on controlling necrotic enteritis infection in chickens. Poult. Sci. 61:447-450. [DOI] [PubMed] [Google Scholar]
- 19.Goodman, L. S., and A. Gilman. 1985. The pharmaceuticological basis of therapeutics, 7th ed. The Macmillan Company, New York, NY.
- 20.Hammer, O., D. A. T. Harper, and P. D. Ryan. 2001. Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4(1):4A. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- 21.Harrison, A. P., Jr., and P. A. Hansen. 1963. Bacteroides hypermegas nov. spec. Antonie Van Leeuwenhoek 29:22-28. [DOI] [PubMed] [Google Scholar]
- 22.Hill, D. D., W. E. Owens, and P. B. Tchounwou. 2005. Prevalence of selected bacterial infections associated with the use of animal waste in Louisiana. Int. J. Environ. Res. Public Health 2:84-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphrey, T. J., F. Jorgensen, J. A. Frost, H. Wadda, G. Domingue, N. C. Elviss, D. J. Griggs, and L. J. V. Piddock. 2005. Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in commercial poultry flocks before, during, and after treatment with fluoroquinolones. Antimicrob. Agents Chemother. 49:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchison, M. L., L. D. Walters, S. M. Avery, B. A. Synge, and A. Moore. 2004. Levels of zoonotic agents in British livestock manures. Lett. Appl. Microbiol. 39:207-214. [DOI] [PubMed] [Google Scholar]
- 25.Jampachaisri, K., L. Valinsky, J. Borneman, and S. J. Press. 2005. Classification of oligonucleotide fingerprints: application for microbial community and gene expression analyses. Bioinformatics 21:3122-3130. [DOI] [PubMed]
- 26.Kruse, H., B. K. Johansen, L. M. Rorvik, and G. Schaller. 1999. The use of avoparcin as a growth promoter and the occurrence of vancomycin-resistant Enterococcus species in Norwegian poultry and swine production. Microb. Drug Resist. 5:135-139. [DOI] [PubMed] [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY.
- 28.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Line, J. E., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. P. Levchuk, O. E. Svetoch, B. S. Seal, G. R. Siragusa, and N. J. Stern. 2008. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 52:1094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marrett, L. E., E. J. Robb, and R. K. Frank. 2000. Efficacy of neomycin sulfate water medication on the control of mortality associated with colibacillosis in growing turkeys. Poult. Sci. 79:12-17. [DOI] [PubMed] [Google Scholar]
- 31.Mead, G. C., M. J. Scott, T. J. Humphrey, and K. McAlpine. 1996. Observations on the control of Campylobacter jejuni infection of poultry by ‘competitive exclusion.’ Avian Pathol. 25:69-79. [DOI] [PubMed] [Google Scholar]
- 32.Morotomi, M., F. Nagai, and H. Sakon. 2007. Taxonomic note: genus Megamonas should be placed in the lineage of Firmicutes; Clostridia; Clostridiales; Acidaminococcaceae; Megamonas. Int. J. Syst. Evol. Microbiol. 57:1673-1674. [DOI] [PubMed] [Google Scholar]
- 33.Prescott, J. F. 1979. The prevention of experimentally induced necrotic enteritis in chickens by avoparcin. Avian Dis. 23:1072-1074. [PubMed] [Google Scholar]
- 34.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoeni, J. L., and M. P. Doyle. 1992. Reduction of Campylobacter jejuni colonization of chicks by cecum-colonizing bacteria producing anti-C. jejuni metabolites. Appl. Environ. Microbiol. 58:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoeni, J. L., and A. C. Wong. 1994. Inhibition of Campylobacter jejuni colonization in chicks by defined competitive exclusion bacteria. Appl. Environ. Microbiol. 60:1191-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scupham, A. J., T. G. Patton, E. Bent, and D. O. Bayles. 2008. Comparison of the cecal microbiota of domestic and wild turkeys. Microb. Ecol. 56:322-331. [DOI] [PubMed] [Google Scholar]
- 38.Soerjadi, A. S., G. H. Snoeyenbos, and O. M. Weinack. 1982. Intestinal colonization and competitive exclusion of Campylobacter fetus subsp. jejuni in young chicks. Avian Dis. 26:520-524. [PubMed] [Google Scholar]
- 39.Stern, N. J. 1994. Mucosal competitive exclusion to diminish colonization of chickens by Campylobacter jejuni. Poult. Sci. 73:402-407. [DOI] [PubMed] [Google Scholar]
- 40.Stern, N. J., N. A. Cox, J. S. Bailey, M. E. Berrang, and M. T. Musgrove. 2001. Comparison of mucosal competitive exclusion and competitive exclusion treatment to reduce Salmonella and Campylobacter spp. colonization in broiler chickens. Poult. Sci. 80:156-160. [DOI] [PubMed] [Google Scholar]
- 41.Stern, N. J., E. A. Svetoch, B. V. Eruslanov, Y. N. Kovalev, L. I. Volodina, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, and V. P. Levchuk. 2005. Paenibacillus polymyxa purified bacteriocin to control Campylobacter jejuni in chickens. J. Food Prot. 68:1450-1453. [DOI] [PubMed] [Google Scholar]
- 42.Stern, N. J., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. D. Pokhilenko, V. P. Levchuk, O. E. Svetoch, and B. S. Seal. 2006. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 50:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan, A., C. Edlund, and C. E. Nord. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101-114. [DOI] [PubMed] [Google Scholar]
- 44.Svetoch, E. A., B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. N. Borzenkov, V. P. Levchuk, O. E. Svetoch, Y. N. Kovalev, Y. G. Stepanshin, G. R. Siragusa, B. S. Seal, and N. J. Stern. 2008. Diverse antimicrobial killing by Enterococcus faecium E 50-52 bacteriocin. J. Agric. Food Chem. 56:1942-1948. [DOI] [PubMed] [Google Scholar]
- 45.Svetoch, E. A., N. J. Stern, B. V. Eruslanov, Y. N. Kovalev, L. I. Volodina, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. D. Pokhilenko, V. N. Borzenkov, V. P. Levchuk, O. E. Svetoch, and T. Y. Kudriavtseva. 2005. Isolation of Bacillus circulans and Paenibacillus polymyxa strains inhibitory to Campylobacter jejuni and characterization of associated bacteriocins. J. Food Prot. 68:11-17. [DOI] [PubMed] [Google Scholar]
- 46.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valinsky, L., A. J. Scupham, J. Della Vedova, Z. Liu, A. Figuerosa, K. Jampachaisri, B. Yin, J. Press, T. Jiang, and J. Borneman. 2004. Oligonucleotide fingerprinting of ribosomal RNA genes (OFRG), p. 569-585. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology methods, 2nd ed., vol. 1. Kluwer Academic Press, Dordrecht, Netherlands. [Google Scholar]
- 49.van den Bogaard, A. E., N. Bruinsma, and E. E. Stobberingh. 2000. The effect of banning avoparcin on VRE carriage in The Netherlands. J. Antimicrob. Chemother. 46:146-148. [DOI] [PubMed] [Google Scholar]
- 50.Vereen, E., Jr., R. R. Lowrance, D. J. Cole, and E. K. Lipp. 2007. Distribution and ecology of campylobacters in coastal plain streams (Georgia, United States of America). Appl. Environ. Microbiol. 73:1395-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wegener, H. C., F. M. Aarestrup, L. B. Jensen, A. M. Hammerum, and F. Bager. 1999. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg. Infect. Dis. 5:329-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wistrom, J., and S. R. Norrby. 1995. Fluoroquinolones and bacterial enteritis, when and for whom? J. Antimicrob. Chemother. 36:23-39. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, X. Y., and R. D. Joerger. 2003. Composition of microbiota in content and mucus from cecae of broiler chickens as measured by fluorescent in situ hybridization with group-specific, 16S rRNA-targeted oligonucleotide probes. Poult. Sci. 82:1242-1249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.