Abstract
In order to study which Bartonella genotypes are circulating among small mammals in Spain, we analyzed the spleens of 395 animals from three different areas—247 animals from the Basque Country (northern Spain), 121 animals from Catalonia (northeastern Spain), and 27 animals from Madrid (central Spain)—by a triplex PCR combined with a reverse line blot previously described by our group. The prevalence of Bartonella was 26.8% (106/395), and in 4.8% (19/395) of the animals more than one Bartonella genotype was detected. The study of gltA and the intergenic transcribed spacer in the positive samples demonstrated a large diversity, allowing the assignation of them into 22 genotypes. The most prevalent genotypes were 2 and 3, which are closely related to Bartonella taylorii. In addition, nine genotypes were associated with specific mammal species. Genotypes close to the zoonotic Bartonella grahamii, Bartonella elizabethae, and Bartonella rochalimae were also detected. Ten genotypes showed a percentage of similarity with known Bartonella species lower than 96%, suggesting the presence of potential new species. Further studies of the impact of these pathogens on human health and especially in cases of febrile illness in Spain are strongly recommended. Furthermore, our method has been updated with 21 new probes in a final panel of 36, which represents a robust molecular tool for clinical and environmental Bartonella studies.
Bartonella spp. are Gram-negative facultative intracellular alphaproteobacteria that can infect the erythrocytes and endothelial cells of its hosts (10, 45). This zoonotic re-emerging pathogen has a complex cycle in nature, including different reservoir hosts and hematophagous arthropods that play a vector role (24, 45).
More than 30 different Bartonella species have been described thus far. In fact, after the implementation of more efficient molecular tools for detection, the number of new species is rapidly increasing. Moreover, four new species pathogenic for humans have been described in the last 3 years: Bartonella rochalimae (13), B. melophagi (38), B. tamiae (31), and “Candidatus Bartonella mayotimonensis” (36). The use of more accurate molecular tools will eventually identify additional Bartonella species causing human infection, taking into account that any Bartonella species can produce disease in humans, as has been hypothesized (36).
Among the different reservoir hosts described for Bartonella, small mammals are responsible for maintaining the highest number of species, as well as many others Bartonella detected but not yet named (18, 20, 21). Several of these rodent-Bartonella species have been linked with human disease. B. grahamii has been involved in ocular syndromes (27, 41), B. elizabethae has been detected in a case of endocarditis (9), B. vinsonii subsp. arupensis was identified in a farmer with fever and bacteremia (46) and also in a patient with endocarditis (14), and B. washoensis was isolated from a patient with fever and myocarditis (32).
Data on the role of small mammals as Bartonella reservoir hosts are scarce in Spain. In the Basque Country (northern Spain), ca. 20% of small-mammal blood smears presented structures compatible with Bartonella (16). In Andalusia (southern Spain), B. tribocorum was detected in 20% of the analyzed Norwegian rats (Rattus norvegicus) and two different Bartonella genotypes (GTs) close to B. elizabethae were found in 29% of the Algerian mice (Mus spretus) studied (39). However, no data are available about other small mammal species or regions in our country.
Environmental studies are essential for identifying which Bartonella species are circulating in a specific area and for evaluating their risk for humans. Bartonella is a fastidious culture bacterium that hospitals do not include in their routine studies; therefore, molecular detection is recommended. Recently, we have developed a versatile molecular tool that allows not only the detection of a positive sample but the identification the specific Bartonella species or GT, as well as potential new variants or new species (15). In the present study, small mammals captured from three different regions in Spain have been analyzed by this method, showing an enormous variability of Bartonella circulating among the small mammal populations.
MATERIALS AND METHODS
Small mammals sampling.
Small mammals were captured in three different regions of Spain: the Basque Country (northern Spain), Catalonia (northeastern Spain), and Madrid (central Spain). The animals were anesthetized with ketamine hydrochloride (Imalgene, Merial, France), 10 mg/kg administered intramuscularly, and euthanized in a CO2 chamber. Spleen samples were obtained from them and kept at −80°C until they were tested. The animals were classified by external morphological data and skull features (4).
Bartonella detection in small mammals.
DNA was extracted from spleen samples with the QIAmp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. For the molecular detection of Bartonella, between 100 and 300 ng of DNA was amplified by a multiplex PCR, and the obtained amplicons were identified by reverse line blotting (PCR/RLB), as previously described (15). This method targets simultaneously the 16S rRNA for the generic detection of any Bartonella, the intergenic transcribed spacer (ITS) 16S-23S rRNA for species identification, and an internal control for the detection of potential PCR inhibitors.
Since new ITS sequences were identified in the present study, 16 probes were designed for their proper detection in the PCR/RLB. Moreover, three new probes were added to the method for the recently described species B. rattimassiliensis, B. phoceensis, and B. rochalimae, and two probes (S-TAY2 and S-CLAR2) were modified for efficiency purposes. A list of primers and probes for the PCR/RLB is shown in Table 1.
TABLE 1.
Primers and probes used in the studya
| Target | Organism | Primer | Probe | Sequenceb | Concn (μM)c |
|---|---|---|---|---|---|
| ITS | Bartonella spp. | Bart/16-23F | 5′-bio-TTG ATA AGC GTG AGG TCG GAG G | 0.4 | |
| Bart/16-23R | 5′-bio-CAA AGC AGG TGC TCT CCC AG | 0.4 | |||
| B. taylorii | S-TAY2 | 5′-a-TAT CCA TTT CSC TTA GGC A | 3.2 | ||
| Bartonella spp.d | S-TALP | 5′-a-CAG TCC CTT TAG GTC CAT TTA ATC | 3.2 | ||
| Bartonella GT 8 | S-MUS1 | 5′-a-GTC TAT TGG ATT TAA GTG TTG | 0.4 | ||
| Bartonella GT 9 | S-MUS2 | 5′-a-GGT CCG TTT GTT AAG TGT TGG | 3.2 | ||
| Bartonella GT 21 | S-MUS3b | 5′-a-CTT TGC GAG ACT TTT TCA CTC C | 0.4 | ||
| Bartonella GT 22 | S-TOPO15b | 5′-a-CTT AAC TTG TTG AAG GCT CCC | 3.2 | ||
| Bartonella GT 6 | S-CE1MM3b | 5′-a-GAA CTC CAT ATA AAA GGC TTT AAA TAT TG | 3.2 | ||
| Bartonella GT 14-15 | S-OR3MM1 | 5′-a-AAT CAA ATT TAA GCA ATA CAA ATC | 3.2 | ||
| Bartonella GT 16 | S-R2Eb | 5′-a-AGT GCC TTT GTT AGA GAA TAC C | 3.2 | ||
| Bartonella GT 17 | S-OR3MM3 | 5′-a-AAG AAT AAA AGT CAA AAT AAT ATT G | 1.6 | ||
| Bartonella GT 18 | S-CAT9b | 5′-a-GTG TAT TAA ACG TAT CAA AGC CTC | 3.2 | ||
| B. elizabethae | S-ELIZ | 5′-a-TAA GTT CCC TTC AAG AGG ATA | 3.2 | ||
| B. doshiae | S-DOSH | 5′-a-TTT GAA CCT TCT CTC TTT AT | 3.2 | ||
| Bartonella GT 19 | S-APO38 | 5′-a- CCT TTT CTC CTT TTT AGG GGC | 3.2 | ||
| B. grahamii | S-GRAH2 | 5′-a-ATT CAA GTT GAT GAA TTT GGT TAT | 0.4 | ||
| Bartonella GT 12 | S-GU1MM1 | 5′-a- TCA AAT TGG TGA ATC TGG TTA T | 3.2 | ||
| Bartonella GT 13 | S-CAT6 | 5′-a- TAA AGA GAA GTT TGT CCA AGA G | 3.2 | ||
| Bartonella GT 20 | S-APS48 | 5′-a-ATC ACT GAA AGT TGC TCT GAG T | 1.6 | ||
| Bartonella GT 10 | S-R24Sh | 5′-a-GCT TTT CTG TTT GCC TGA GGT C | 3.2 | ||
| B. tribocorum | S-TRIB | 5′-a-TTC TAT TAA GTT TGT CAA AGG G | 0.4 | ||
| B. phoceensis | S-PHO | 5′-a-GAG AGA CGC TTT TCC CTT TGG | 1.6 | ||
| B. rattimassiliensis | S-RAT | 5′-a-CGG TGT TTT GAG GCA AAG TGC | 1.6 | ||
| B. vinsonii subsp. arupensis | S-VIN-A1 | 5′-a-ACTTGTTGGAATTGCTTAACC | 3.2 | ||
| B. vinsonii subsp. vinsonii | S-VIN-A2 | 5′-a-ATGAAAATATTGAGAGATTTG | 3.2 | ||
| B. alsatica | S-ALS | 5′-a-GCT GGT GAA ACT TGC TTA TA | 6.4 | ||
| B. quintana | S-QUIN | 5′-a-CGC TTA TCC ATT TGG TTT AA | 3.2 | ||
| B. bacilliformis | S-BACI | 5′-a-CCT ATG ATT GAT TTC TAG GC | 0.4 | ||
| B. henselae | S-HENS | 5′-a-ATC GGT TCA ATC ATA TCG CTT T | 3.2 | ||
| B. clarridgeiae | S-CLARR2 | 5′-a-ACG ATG CTA AAA GTT GCT ATA TTG | 3.2 | ||
| B. koehlerae | S-KOE | 5′-a-TTA AAT TAT ATC ACT TTG GGT CAT ACG | 0.4 | ||
| B. rochalimae | S-ZOR | 5′-a-AAC AGG GAA AAG AGC AGG CCA | 3.2 | ||
| Bartonella spp. detected in badgerd | S-TEJ | 5′-a-GAT GTT TTG TAA AAG TGC GTC G | 3.2 | ||
| B. vinsonii subsp. berkhoffii | S-VIN-B | 5′-a-TTT CGG ACA CTA TTG ATA AA | 3.2 | ||
| B. bovis | S-BOV2 | 5′-a-CGT TTT GAT AGT CTT TTG TGT TGC | 0.4 | ||
| B. chomelii, B. schoenbuchensis, B. capreoli, and B. birtlesii | S-CHOSCA | 5′-a-TTA TGA TTG CTG ATA AGT TTG CTG | 3.2 | ||
| O. grignonense | S-GRIGNO | 5′-a- GCT TTG ATA AAT GTG ATA AGG | 1.6 | ||
| 16S rRNA | Bartonella spp. | 16S-R | 5′-bio-GCC YCC TTG CGG TTA GCA CAG CA | 1 | |
| P24Emod | 5′-bio-CCT TCA GTT MGG CTG GAT C | 1 | |||
| S-BART16S | 5′-a-CTC GCC CTT AGT TGC CAG CAT T | 3.2 | |||
| TCH synthase | Cannabis sativa | CI-F | 5′-bio-ATG ATG CTG AGG GTA TGT CCT AC | 1 | |
| CI-R | 5′-bio-GTT TTC TCC TCC ACC ACC ACG | 1 | |||
| S-CI2 | 5′-a-GTG GAC ACT TTA GTG GAG GAG G | 3.2 | |||
| gltA | Bartonella spp. | GLTAF2 | 5′-GCT TTK CTG TTC CDT GTG AAG | 1 | |
| GLTAR2 | 5′-GCA AAA AGA ACA GTA AAC ATT TC | 1 | |||
| GLTAF1 | 5′-AAA ATG CTA CAA GAA ATH GG | 1 | |||
| GLTAR1 | 5′-AGC TTT TAA TGT AAT DCC DG | 1 |
The probes and primers in boldface were designed for this study. The rest of the oligonucleotides have been described previously (15).
Oligonucleotide modifications at the 5′ end. bio, biotin; a, amino link.
That is, the concentration of the oligonucleotides used in the assays.
Garcia-Esteban et al. (15).
The specificity of the new set of probes was checked with 102 genome equivalents (GE) of different Bartonella species or 102 plasmid copies of the new ITS (PCITS) found in the study (Table 1). These latter controls were built by cloning the ITS amplicons of interest with a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Further characterization of the positive samples.
Two PCRs targeting the ITS and gltA were performed with the positive samples. In the case of the ITS, the PCR was performed by reamplifying 2 μl of the initial multiplex PCR product in a 50-μl reaction volume with 10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate (Promega, Madison, WI), 1.5 U of Taq Gold DNA polymerase (Applied Biosystems, Branchburg, NJ), and 1 μM Bart/16-23F and Bart/16-23R primers (Table 1). PCR cycling consisted of an initial denaturing step of 9 min at 94°C, followed by 40 cycles of 30 s at 94°C, 1 min at 64.3°C, and 1 min 30 s at 72°C, with a final elongation step of 7 min at 72°C.
In the case of gltA, 100 to 300 ng of DNA was amplified with a nested PCR designed in the present study. First, the primers GLTAF2 and GLTAR2, which amplify a 531-bp fragment, were used. Then, 2 μl of the product was reamplified with the primers GLTAF1 and GLTAR1, yielding a final fragment of 300 bp. Both reactions were performed under the same conditions described above for the ITS, except for the annealing temperature, which was 50°C in both PCR cycles.
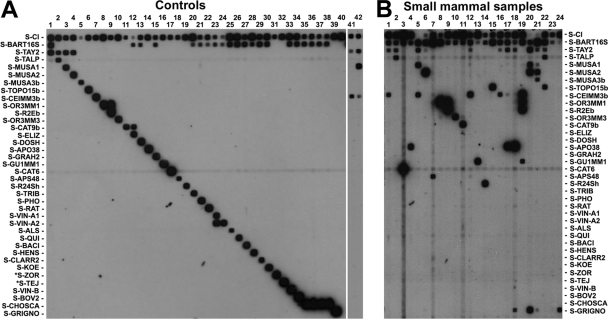
For positive samples whose hybridization pattern in the RLB was different from the ones obtained with the controls (Fig. 1A) and suggested the presence of more than one Bartonella GT, the ITS amplicons were cloned with the TOPO TA cloning kit (Invitrogen), and 10 clones were analyzed by PCR/RLB. Coinfection was confirmed in samples whose clones showed a hybridization pattern, which explains the hybridization results observed in the original sample.
FIG. 1.
Reverse line blotting results. (A) Hybridization signal obtained with different controls. Lanes: 1, 102 GE B. taylorii CIP 107028; 2, 102 plasmid copies with the insert ITS (PCITS) Bartonella spp. from a Spanish mole (15); 3, 102 PCITS from GT 8; 4, 102 PCITS from GT 9; 5, 102 PCITS from GT 21; 6, 102 PCITS from GT 22; 7, 102 PCITS from GT 6; 8, 102 PCITS from GT 14; 9, 102 PCITS from GT 16; 10, 102 PCITS from GT 17; 11, 102 PCITS from GT 18; 12, 102 GE B. elizabethae CIP 103761; 13, 102 GE B. doshiae CIP 107026; 14, 102 PCITS from GT 19; 15, 102 GE B. grahamii CIP 107024;16, 102 PCITS from GT 12; 17, 102 PCITS from GT 13; 18, 102 PCITS from GT 20; 19, 102 PCITS from GT 10; 20, 102 GE B. tribocorum CIP 105476; 21, 102 GE B. phoceensis CIP 107707; 22, 102 GE B. rattimassiliensis CIP 107705; 23, 102 GE B. vinsonii subsp. arupensis CIP 106848; 24, 102 GE B. vinsonii subsp. vinsonii CIP 103738; 25, 102 GE B. alsatica CIP 105477; 26, 102 GE B. quintana CIP 103739; 27, 102 GE B. bacilliformis CIP: 77.27; 28, 102 GE B. henselae CIP 103737; 29, 102 GE B. clarridgeiae CIP 104772; 30, 102 GE B. koehlerae CIP107025; 31, 102 PCITS B. rochalimae from a Spanish fox (15); 32, 102 PCITS Bartonella spp. from a Spanish badger (15); 33, 102 GE B. vinsonii subsp. berkhoffii CIP 104960; 34, 102 GE B. bovis CIP 106692; 35, 102 GE B. schoenbuchensis CIP 107819; 36, 102 GE B. capreoli CIP 106691; 37, 102 GE B. chomelii CIP 107869; 38, 102 GE B. birtlesii CIP 106294; 39, 102 PCITS Ochrobactrum sample PVR70-01Sh; 40, negative PCR control; 41, 102 PCITS from GT 2; 42, 102 PCITS from GT 4. Lanes where clones were tested have no hybridization signals with the S-BART16S probe because the control only contains the ITS target. (B) Examples of hybridization reactions with several small mammal samples.
Sequencing and analysis.
PCR products were run in 1% low-melting agarose (Pronadisa, Torrejón de Ardoz, Spain), and the bands of interest were purified by using the QIAquick gel extraction kit (Qiagen) and sequenced with the Big-Dye terminator cycle sequencing kit (Applied Biosystems, Branchburg, NJ) according to the manufacturer's instructions.
ITS and gltA sequences were aligned with reference sequences from GenBank (http://www.ncbi.nlm.nih.gov) with the Multi-Alignment Fast Fourier Transform (MAFFT) method (26). Pairwise distance matrices for the aligned sequences were determined by using the Kimura two-parameter method (28) with MEGA4 software (42), and phylogenetic trees were constructed applying the neighbor-joining algorithm (7) with the internal-branch test for evaluation of their topology. Dendrograms were collapsed by using a cutoff bootstrap value of 50. The percent similarities between the gltA and ITS sequences identified here and the type strain sequences of each Bartonella species were calculated using the MEGA4 software. Sequences were also analyzed by BLAST (1; http://blast.ncbi.nlm.nih.gov/blast.cgi) to identify the closest relative.
Nucleotide sequence accession numbers.
The sequences obtained in the present study have been deposited in GenBank under the following accession numbers (ITS, gltA): GT 1 (HM596431, HM596455), GT 2 (HM596432, HM596466), GT 3 (only ITS HM596433), GT 4 (HM596434, HM596454), GT 5 (only ITS HM596435), GT 6 (HM596436, HM596462), GT 7 (HM596437, HM596469), GT 8 (HM596438, HM596461), GT 9 (HM596439, HM596457), GT 10 (HM596440, HM596458), GT 11 (only gltA HM596453), GT 12 (HM596441, HM596463), GT 13 (HM596442, HM596467), GT 14 (only ITS HM596443), GT 15 (HM596444, HM596465), GT 16 (HM596445, HM596452), GT 17 (HM596446, HM596464), GT 18 (HM596447, HM596468), GT 19 (HM596448, HM596460), GT 20 (HM596449, HM596459), GT 21 (HM596450, HM596456), GT 22 (only ITS HM596451), and sample PVR71-02Sh (only ITS HM596470).
RESULTS
Presence of Bartonella spp. in small mammals.
We analyzed 395 animals (Table 2) captured in three different regions of Spain: 247 from the Basque Country, 121 from Catalonia, and 27 from Madrid. Small mammals were assigned to nine different species (Table 2), with Apodemus sylvaticus (wood mouse) the species with the highest number of studied animals (220/395, 55.7%).
TABLE 2.
Presence of Bartonella GTs in small mammals
| Probe | GTa | No. of positive small mammal species (% positive)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| As (n = 220) | Af (n = 3) | Mg (n = 16) | Mdo (n = 34) | Ms (n = 30) | Sc (n = 14) | Mde (n = 1) | Cr (n = 56) | Te (n = 21) | Total (n = 395) | ||
| 16S rRNA | Generic Bartonella | 69 (31.4) | 2 (66.7) | 3 (18.8) | 3 (8.8) | 1 (3.3) | 5 (35.7) | 8 (14.3) | 15 (71.4) | 106 (26.8) | |
| S-TAY | GT 1 | 2 (0.9) | 2 (0.5) | ||||||||
| S-TAY + S-CE1MM3b | GT 2/3 | 27 (12.3) | 1 (33.3) | 1 (6.3) | 29 (7.3) | ||||||
| S-TAY + S-MUS1 + S-CEIMM3b | GT 4/5 | 6 (2.7) | 1 (6.3) | 1 (2.9) | 8 (2.0) | ||||||
| S-TAY + S-MUS1 | GT 8 | 7 (3.2) | 7 (1.8) | ||||||||
| S-TAY + S-TALP | GT 7 | 2 (5.9) | 13 (61.9) | 15 (3.8) | |||||||
| S-CE1MM3b | GT 9 | 1 (0.5) | 1 (0.3) | ||||||||
| S-CAT6 | GT 13 | 7 (12.5) | 7 (1.8) | ||||||||
| S-APO38 | GT 19 | 4 (1.8) | 4 (1.0) | ||||||||
| S-TAY + S-MUS2 | GT 9 | 1 (7.1) | 1 (0.3) | ||||||||
| S-APS48 | GT 20 | 2 (0.9) | 2 (0.5) | ||||||||
| S-OR3MM1 | GT 14/15 | 2 (0.9) | 2 (0.5) | ||||||||
| S-OR3MM1+S-R2EB | GT 16 | 2 (0.9) | 2 (0.5) | ||||||||
| S-OR3MM3 | GT 17 | 1 (0.5) | 1 (0.3) | ||||||||
| S-CAT9+S-ELIZ | GT 18 | 1 (3.3) | 1 (0.3) | ||||||||
| S-GU1MM1 | GT 12 | 1 (0.5) | 1 (0.3) | ||||||||
| S-R24SH | GT 10 | 1 (1.8) | 1 (0.3) | ||||||||
| S-TOPO15b | GT 22 | 1 (4.8) | 1 (0.3) | ||||||||
| S-TAY + S-CE1MM3b + S-APO38 | GT 2/3 and GT 19* | 7 (3.2) | 1 (33.3) | 1 (6.3) | 9 (2.3) | ||||||
| S-TAY + S-MUS1 + S-CEIMM3b + S-APO38 | GT 4/5 and GT 19* | 2 (0.9) | 2 (0.5) | ||||||||
| S-TAY + S-CE1MME3b + S-APS48 | GT 2/3 and GT 20* | 1 (0.5) | 1 (0.3) | ||||||||
| S-TAY + S-MUS1 + S-CE1MM3b + S-APS48 | GT 4/5 and GT 20* | 1 (0.5) | 1 (0.3) | ||||||||
| S-OR3MM1 + S-RE2B + GU1MM1 | GT 12 and GT 16* | 1 (0.5) | 1 (0.3) | ||||||||
| S-TAY + S-MUS1+ S-MUS2 | GT 8 and GT 9 * | 3 (21.4) | 3 (0.8) | ||||||||
| S-TAY + S-MUS1+ S-MUS2+ S-MUS3 | GT8, GT 9, and GT2* | 1 (7.1) | 1 (0.3) | ||||||||
| S-TAY + S-TALP + S-TOPO15b | GT 7 and GT 22* | 1 (4.8) | 1 (0.3) | ||||||||
| No signal with ITS probes | Bartonella spp. | 2 (0.9) | 2 (0.5) | ||||||||
| S-GRIGNO | O. grignonense | 12 (5.5) | 5 (14.7) | 3 (21.4) | 20 (5.1) | ||||||
That is, the genotype (GT) associated with each combination of hybridization signals obtained in the RLB. GTs 2/3, 4/5, and 14/15 cannot be distinguished by RLB. *, the presence of coinfections was checked by clones (see Materials and Methods).
Small mammal species: As, Apodemus sylvaticus (wood mouse); Af, A. flavicollis (yellow-neck mouse); Mg, Myodes glareolus (bank vole); Mdo, Mus domesticus (domestic mouse); Ms, M. spretus (Algerian mouse); Sc, Sorex coronatus (Millet shrew); Mde, Microtus duodecimcostatus (Mediterranean pinevole); Cr, Crocidura russula (common shrew); Te, Talpa europaea (mole). The number of animals studied (n) is indicated in parentheses with each abbreviation.
A total of 26.8% (106/395) of the small mammals were infected with Bartonella according to the PCR/RLB results (Table 2). All of the animal species were found to be infected except for the one specimen of Microtus duodecimcostatus (Mediterranean pine vole). By animal species, the percentage of infection ranged from 3.3% (1/30) in Mus spretus (Algerian mouse) to 71.4% (15/21) in Talpa europaea (mole).
Initial identification of Bartonella GTs in small mammals.
The reactivity observed in the PCR/RLB with the ITS-specific probes of our initial panel (15) identified 64 (60.4%) animals that presented hybridization with S-TAY (probe for B. taylorii), 16 (15.1%) animals with both S-TAY and S-TALP, and 26 (25.4%) small mammals that presented hybridization only with the generic probe for 16S rRNA. This latter hybridization pattern suggested the presence of potential new species or variants in these 26 samples, different from those included in our initial panel of probes.
Sequence analysis of the positive samples.
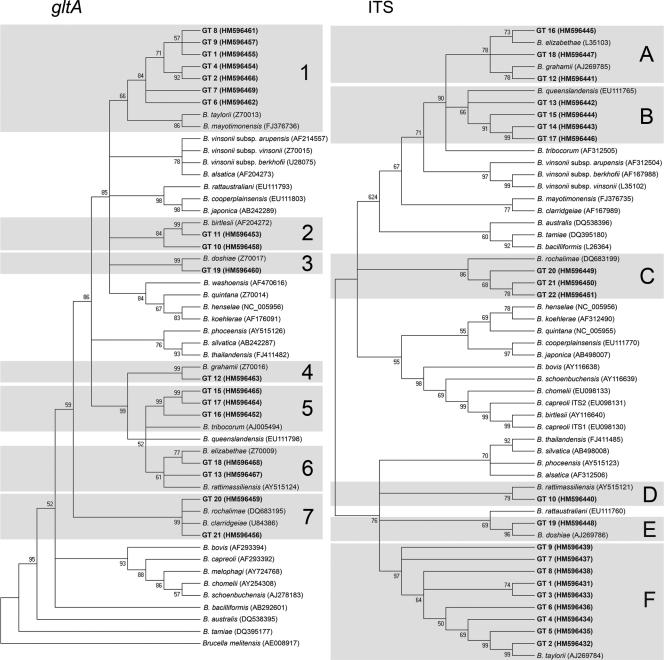
From the 106 positive samples, 63 (59.4%) were further characterized by sequencing the ITS and the gltA. These samples were selected according to the hybridization pattern and the small mammal species from which they were obtained. Analyzing the sequences, 22 Bartonella GTs were assigned in the present study with correlative numbers (Table 3). The percentage of similarity of the gltA sequence (260 bp, excluding the primers) with known Bartonella type strains ranged from 91.9 to 100%. GT 9 (91.9% with B. vinsonii subsp. arupensis) and GT 10 (91.9% with B. birtlesii) showed the lowest values (Table 3). Moreover, some remarkable hits identified by BLAST were as follows. GTs 15 to 17 showed 99 to 100% similarity to Bartonella from fleas in Portugal (AY877422 and AY877423), GTs 20 and 21 showed 97 to 98% similarity to voles from western Siberia (EF682090), GT 10 showed 97% similarity to a Bartonella strain from a small mammal captured in equatorial Africa (FJ851111), GT 8 showed 98% similarity to a Bartonella strain from a Sorex araneus in the United Kingdom (EF031548) and, finally, GT 13 showed a similarity of 98% to a Bartonella strain from a Suncus murinus in China (FJ464239). In the phylogenetic analysis, these gltA sequences were placed along the dendrogram in seven different clades that we designated 1 to 7: B. taylorii/“Candidatus Bartonella mayotimonensis”, B. birtlesii, B. doshiae, B. grahamii, B. tribocorum, B. elizabethae/B. rattimassiliensis, and B. clarridgeiae/B. rochalimae, respectively (Fig. 2).
TABLE 3.
GTs identified in small mammals
| GT |
Bartonella species (% similarity)a |
Reactivity with the ITS probes |
||
|---|---|---|---|---|
| gltA | ITS | Pattern | Laneb | |
| 1 | B. vinsonii subsp. arupensis (93.5) | B. taylorii (98.9) | S-TAY | 1 |
| 2 | B. taylorii (94.2) | B. taylorii (97.8) | S-TAY + S-CE1MM3b | 41 |
| 3 | ND | B. taylorii (93.5) | S-TAY + S-CE1MM3b | NS |
| 4 | B. taylorii (94.6) | B. taylorii (97.8) | S-TAY+ S-MUS1+ S-CE1MM3b | 42 |
| 5 | ND | B. taylorii (98.4) | S-TAY+ S-MUS1+ S-CE1MM3b | NS |
| 6 | B. vinsonii subsp. arupensis (94.5) | B. taylorii (99.0) | S-CE1MM3b | 7 |
| 7 | B. vinsonii subsp. vinsonii (95.0) | B. taylorii (73.9) | S-TAY + S-TALP | 2 |
| 8 | B. vinsonii subsp. vinsonii (93.8) | B. taylorii (91.0) | S-TAY + S-MUS1 | 3 |
| 9 | B. vinsonii subsp. arupensis (91.9) | B. taylorii (90.2) | S-TAY + S-MUS2 | 4 |
| 10 | B. birtlesii (91.9) | B. vinsonii subsp. berkhoffii (67.9), B. cooperplainsense (67.0) | S-R24Sh | 19 |
| 11 | B. birtlesii (100.0) | ND | NAc | NA |
| 12 | B. grahamii (100.0) | B. grahamii (98.8) | S-GUIMM1 | 16 |
| 13 | B. tribocorum (95.8) | B. elizabethae (77.4), B. tribocorum (77.1) | S-CAT6 | 17 |
| 14 | ND | B. elizabethae (89.3), B. grahamii (89.3) | S-OR3MM1 | 8 |
| 15 | B. elizabethae (97.3) | B. elizabethae (89.6), B. grahamii (89.6) | S-OR3MM1 | NS |
| 16 | B. elizabethae (96.9) | B. elizabethae (85.3) | S-OR3MM1+S-R2Eb | 9 |
| 17 | B. elizabethae (96.9) | B. grahamii (91.9) | S-OR3MM3 | 10 |
| 18 | B. elizabethae (97.3) | B. elizabethae (95.0) | S-ELIZ+S-CAT9 | 11 |
| 19 | B. doshiae (96.2) | B. doshiae (89.6) | S-APO38 | 14 |
| 20 | B. rochalimae (95.8) | B. rochalimae (65.4) | S-APS48 | 18 |
| 21 | B. rochalimae (96.2) | B. rochalimae (71.5) | S-MUS3b | 5 |
| 22 | ND | B. tribocorum (73.5), B. rochalimae (73.0) | S-TOPO15b | 6 |
The highest percent similarity found with the indicated Bartonella species is indicated in parentheses. ND, not determined (a sequence could not be obtained).
The corresponding lane in Fig. 1A with an example of this hybridization pattern. NS, not shown.
NA, not applicable.
FIG. 2.
Phylogenetic relationships of Bartonella spp. detected in this study. Dendrograms of gltA and ITS were built with reference sequences obtained from GenBank. The accession numbers are indicated in parentheses. The clades found in the study are shaded.
Overall, the ITS showed a lower percentage of similarity among type strains, compared to gltA sequences, ranging from 67 to 99% (Table 3). Apart from the more similar Bartonella described species, some high-similarity hits were found by BLAST. GTs 13 to 17 showed 97 to 99% similarity to Bartonella from an M. spretus captured in southern Spain (EU218552), GTs 20 to 22 showed 75 to 89% similarity to a Bartonella from a vole captured in western Siberia (EF682087), GT 6 showed 100% similarity to a Bartonella from an A. flavicollis from Slovenia (DQ155391), GT 19 had 99% similarity to a Bartonella from an A. sylvaticus from the United Kingdom (AJ269792) and, finally, GT 9 showed 98% with a Bartonella from an S. araneus captured in the United Kingdom (EF031550). In the ITS dendrogram the sequences were placed in six clades (A to F). Although the distribution of some of the GTs was similar compared to the gltA dendrogram (Fig. 2), like GTs located in the B. taylorii, B. doshiae, and B. rochalimae clades, the other GT had a different position in the dendrogram.
Identification of Bartonella GTs in small mammals with the new probes.
Based on these preliminary results we designed 21 ITS probes, indicated in Materials and Methods, for improving the RLB (Table 1, probes in boldface) and distinguishing the GTs identified above. To check the specificity of the updated method, the 36 ITS probes were tested against genomic DNA from different Bartonella GTs, as well as with the cloned controls, with the purpose of determining the accuracy of their differentiation by hybridization (Fig. 1A). We were able to differentiate all of the GTs detected in the study except GTs 2 and 3, GTs 4 and 5, and GTs 14 and 15 that shared the same hybridization pattern (Table 3); therefore, these will be referred from now on as GTs 2/3, 4/5, and 14/15, respectively, since they cannot be differentiated by PCR/RLB.
Afterward, all of the samples (n = 395) were tested again with the updated method. An example of these results is shown in Fig. 1B. The most frequently found Bartonella were GTs 2/3, detected in 10.1% (40/395) of the animals, followed by GT 7 in 4.1% (16/395) and GT 19 in 3.8% (15/395) of the studied animals (Table 2). On the other hand, some Bartonella GTs were quite infrequent, such as GT 10, GT 17, or GT 18, which were detected in only one specimen each. Moreover, 4.8% (19/395) of the animals showed coinfections with at least two different Bartonella GTs, which were assessed by analysis of the cloned PCR products and PCR/RLB. The most common combinations were GTs 2/3 and 19, detected in 2.3% (9/395) of the animals (Table 2).
The small mammal species that presented a larger variability of Bartonella was A. sylvaticus. Interestingly, some GTs were found to be associated with some small mammal species such as GTs 9 and 21 with Sorex coronatus, GT 18 with M. spretus, GTs 10 and 13 with Crocidura russula, GTs 7 and 22 with T. europaea, or GTs 2 to 5 and 19 with Apodemus spp. and Myodes glareolus (Table 2).
The updated method with the new probes designed in the present study allowed us to perfectly identify all of the samples, including the 26 positive samples that initially reacted only with the generic probe S-BART16S, except for two animals. A further characterization of these specimens was performed, although only in one of them (GT 11) was it possible to sequence gltA, showing a perfect match with B. birtlesii (Table 3 and Fig. 1B, lane 23).
Finally, five small mammals presented only a faint hybridization signal with the S-BART16S probe (Fig. 1B, lane 24). However, they were not considered Bartonella-infected animals because the ITS sequence, identical in all of them (GenBank accession no. HM596470) showed a 99% similarity with the ITS of Ochrobactrum grignonense (AJ242581). Therefore, a specific probe for this ITS was designed (Table 1) and used in the updated method. This microorganism was detected in 5.1% (20/395) of the animals and specifically in 14.7% (5/34) of Mus domesticus (Table 3).
DISCUSSION
The use of molecular tools in epidemiological studies is essential for a fastidious culture bacterium such as Bartonella. The method designed previously by us (15) has been updated with 21 ITS new probes in a final panel of 36, allowing the detection of new variants and potentially new species among the small mammals that could not be identified by a traditional approach. Only two samples could not be properly identified by this method, probably due to the low number of the microorganisms present in the samples. Using this method, we determined which Bartonella GTs are circulating in the small mammal populations in three areas of Spain.
Bartonella spp. had infected 26.8% of the animals studied. This percentage is similar to the values found in other studies: 26% in Japan (22), 28% in Denmark (12) and Spain (39), 30% in Greece (43) and France (17), and 31% in Poland (44).
As has been shown in the studies described above, the variability of Bartonella GTs in small mammals is large, and the detection of potentially new species is frequent. Likewise, we have identified 22 different GTs in our study, which were distributed along the gltA dendrogram in seven different clades, which represents a high variability. Moreover, it has been proposed that a similarity percentage of less than 96% of a gltA fragment of 321 bp suggests the presence of a new species (34). In our study, although the gltA fragment analyzed was slightly smaller, 10 GTs had a percent similarity of less than 96%. The isolation and further characterization of these GTs will allow us to determine their exact taxonomic positions.
In the case of the ITS, the percent similarity to the described Bartonella species was lower compared to the gltA, as was previously found (34). It is known that the ITS hypervariability prevents the accurate alignment of the sequences (3), and the ITS phylogenetic information should be interpreted with caution. However, this hypervariability allowed us to design highly specific probes to differentiate the Bartonella GTs.
B. grahamii and B. taylorii are well distributed in Eurasia. However, in some countries, such as Sweden (19), China (37), or Japan (22), B. grahamii is the predominant species, whereas B. taylorii is predominant in Poland (44) or Greece (43). In the present study, GTs 2/3, which are closely related to B. taylorii, were the most frequently detected Bartonella GTs.
The percentage of infection in moles (T. europaea) was the highest among all of the small mammals studied, with GT 7 being the most prevalent GT in this species. This is the first study on the prevalence of Bartonella in moles. B. talpae is the only species that has been associated with this animal species until now (45). However, no type strain or molecular data are available from this species, and GT 7 cannot yet be assigned to this Bartonella species until a type strain is isolated and well characterized.
GTs 15 to 18, which were closely related to B. elizabethae, were found less frequently infecting small mammals in the present study. Interestingly, they presented a high similarity with Bartonella identified in rodent fleas from Portugal (11) and M. spretus in Andalusia, southern Spain (39), indicating that these GTs are well distributed in the Iberian Peninsula. However, the percentage of infection in M. spretus was quite low (3%) compared to the percentage found for this species in Andalusia (29%). This was probably due to a more efficient cycle of transmission in southern Spain than in the rest of the country, although more studies are needed to confirm this point.
Whereas some GTs (2/3, 4/5, or 19) were detected in several animal species, other GTs (10, 12, 13, 18, 19, or 22) were only found in one species. Some of these associations have already been proposed in other studies. Indeed, some of our GTs demonstrated a high similarity to GTs detected in the United Kingdom in shrews (6, 19) or in Algerian mice in Spain (39). This host association could be related to the higher copy number of genes for putative host-adaptability factors that have been identified in the genome of B. grahamii (2, 40) and are likely to be present in other rodent Bartonella strains. Another explanation could be the existence of restricted cycles between some vectors and animals, such as a GT transmitted by a specific arthropod species. A previous study in the Basque Country (16) found an association between the Palaelopsylla soricis flea and the Echinonyssus soricis mite with S. coronatus and the Polyplax reclinata louse with C. russula (16). These associations could be responsible for the transmission of the specific GTs found in shrews. In contrast, also in the Basque Country, other vectors, such as the Ctenophtalmus baeticus avernus flea, were less host specific (16) and could be responsible of the transmission of Bartonella GTs present in different small mammals such as GTs 2/3. In fact, Ctenophtalmus nobilis is a competent vector for B. taylorii (closely related to GTs 2/3) and B. grahamii (5). Additional studies are needed to confirm the role of these arthropods as vectors in the transmission of these GTs.
In the present study, 4.8% of the studied animals were infected with more than one Bartonella GT. We have been able to detect these coinfections by using the PCR/RLB we have designed. The presence of coinfection has been observed in different studies performed in southern Spain (39), the United Kingdom (3, 5), the United States (30, 33), and Japan (22), and it seems to be a common event in small-mammal populations. Also, subsequent infection with different species has been observed in nature (3, 30). Although it has been shown that there is cross serological reactivity between Bartonella species, there is a lack of heterologous protection between B. henselae and B clarridgeiae or even between different B. henselae serotypes in cats (47), explaining subsequent Bartonella infections or the possibility of coinfections, such as those we observed in small mammals.
A surprising finding was the high prevalence of a bacterium closely related to O. grignonense in the studied animals. This species is an environmental microorganism (35) which is phylogenetically close to the recently described species O. pseudogrignonense and O. haematophilum that can infect humans (25). The potential zoonotic role of the agent detected in the present study is unknown, although it can infect small mammals. Our design of a specific probe for this microorganism could be useful for future clinical and environmental surveys. In addition, since the Ochrobactrum genus is closely related to Bartonella, the use of 16S rRNA as a target in Bartonella studies can produce false positives and overestimate the real prevalence of this bacterium and should be carefully surveyed in environmental studies using only this target.
Bartonella is an emerging zoonotic pathogen; the number of Bartonella species implicated in human disease is increasing rapidly. Apart from the rodent-Bartonella zoonotic species B. grahamii, B. elizabethae, B. washoensis, and B. vinsonii subsp. arupensis, there is serological evidence of rodent-associated Bartonella involved in febrile illness in the southwestern United States (23), among intravenous drug users (8) and, more recently, in patients with febrile illness in Thailand, where Bartonella spp. closely related to B. elizabethae, B. rattimassilensis, and B. tribocorum (29) were detected. In our study, GTs close to the zoonotic B. grahamii, B. elizabethae, and B. rochalimae have been detected. These data call for performing additional studies to determine the impact of these pathogens on human health and especially in the case of febrile illness for which no etiological agent has been identified.
Acknowledgments
This study was supported by grants from the Instituto de Salud Carlos III (MPY 025/09) and Fondo de Investigación Sanitaria (PI10/00051).
We thank Frank M. Hodgkins for the English correction of the manuscript.
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Berglund, E. C., A. C. Frank, A. Calteau, P. O. Vinnere, F. Granberg, A. S. Eriksson, K. Naslund, M. Holmberg, H. Lindroos, and S. G. Andersson. 2009. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 5:e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birtles, R. J., S. M. Hazel, M. Bennett, K. Bown, D. Raoult, and M. Begon. 2001. Longitudinal monitoring of the dynamics of infections due to Bartonella species in UK woodland rodents. Epidemiol. Infect. 126:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, J. C., M. Alcántara, C. Ibañez, A. Aguilar, E. Grau, S. Moreno, J. Balbotín, G. Jordán, and R. Villafuerte. 1998. Mamíferos de España. Planeta, Barcelona, Spain.
- 5.Bown, K. J., M. Bennet, and M. Begon. 2004. Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg. Infect. Dis. 10:684-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray, D. P., K. J. Bown, P. Stockley, J. L. Hurst, M. Bennett, and R. J. Birtles. 2007. Haemoparasites of common shrews (Sorex araneus) in Northwest England. Parasitology 134:819-826. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J. K. 1994. Bootstrap hypothesis tests for evolutionary trees and others dendrograms. Proc. Natl. Acad. Sci. U. S. A. 91:12293-12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comer, J. A., T. Diaz, D. Vlahov, E. Monterroso, and J. E. Childs. 2001. Evidence of rodent-associated Bartonella and Rickettsia infections among intravenous drug users from Central and East Harlem, New York City. Am. J. Trop. Med. Hyg. 65:855-860. [DOI] [PubMed] [Google Scholar]
- 9.Daly, J. S., M. G. Worthingon, D. J. Brenner, C. W. Moss, D. G. Hollis, R. S. Weyant, A. G. Steigerwalt, R. E. Weaver, M. I. Daneshvar, and S. P. O'Connor. 1993. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31:872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehio, C. 2005. Bartonella-host cell interactions and vascular tumour formation. Nat. Rev. Microbiol. 3:621-631. [DOI] [PubMed] [Google Scholar]
- 11.De Sousa, R., P. Edouard-Fournier, M. Santos-Silva, F. Amaro, F. Bacellar, and D. Raoult. 2006. Molecular detection of Rickettsia felis, Rickettsia typhi, and two genotypes closely related to Bartonella elizabethae. Am. J. Trop. Med. Hyg. 75:727-731. [PubMed] [Google Scholar]
- 12.Engbaek, K., and P. A. Lawson. 2004. Identification of Bartonella species in rodents, shrews and cats in Denmark: detection of two Bartonella henselae variants, one in cats and the other in the long-tailed field mouse. APMIS 112:336-341. [DOI] [PubMed] [Google Scholar]
- 13.Eremeeva, M. E., H. L. Gerns, S. L. Lydy, J. S. Goo, E. T. Ryan, S. S. Mathew, M. J. Ferraro, J. M. Holden, W. L. Nicholson, G. A. Dasch, and J. E. Koehler. 2007. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N. Engl. J. Med. 356:2381-2387. [DOI] [PubMed] [Google Scholar]
- 14.Fenollar, F., S. Sire, and D. Raoult. 2005. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J. Clin. Microbiol. 43:945-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Esteban, C., H. Gil, M. Rodríguez-Vargas, X. Gerrikagoitia, J. Barandika, R. Escudero, I. Jado, C. García-Amil, M. Barral, A. L. García-Pérez, M. Bhide, and P. Anda. 2008. Molecular method for Bartonella species identification in clinical and environmental samples. J. Clin. Microbiol. 46:776-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil, H. 2002. Zonas endémicas de enfermedad de Lyme en la Comunidad Autónoma del País Vasco: Estudio del papel de los micromamíferos en el mantenimiento de Borrelia burgdorferi sensu lato en el medio natural. Ph.D. thesis. Universidad de Zaragoza, Zaragoza, Spain.
- 17.Gundi, V. A., B. Davoust, A. Khamis, M. Boni, D. Raoult, and B. La Scola. 2004. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J. Clin. Microbiol. 42:3816-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gundi, V. A., C. Taylor, D. Raoult, and B. La Scola. 2009. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov., and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int. J. Syst. Evol. Microbiol. 59:2956-2961. [DOI] [PubMed] [Google Scholar]
- 19.Holmberg, M., J. N. Mills, S. McGill, G. Benjamin, and B. A. Ellis. 2003. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol. Infect. 130:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue, K., H. Kabeya, H. Shiratori, K. Ueda, M. Y. Kosoy, B. B. Chomel, H. J. Boulouis, and S. Maruyama. 2010. Bartonella japonica sp. nov. and Bartonella silvatica sp. nov., isolated from Apodemus mice. Int. J. Syst. Evol. Microbiol. 60:759-763. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, K., S. Maruyama, H. Kabeya, K. Hagiya, Y. Izumi, Y. Une, and Y. Yoshikawa. 2009. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg. Infect. Dis. 15:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue, K., S. Maruyama, H. Kabeya, N. Yamada, N. Ohashi, Y. Sato, M. Yukawa, T. Masuzawa, F. Kawamori, T. Kadosaka, N. Takada, H. Fujita, and H. Kawabata. 2008. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl. Environ. Microbiol. 74:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iralu, J., Y. Bai, L. Crook, B. Tempest, G. Simpson, T. Mckenzie, and F. Koster. 2006. Rodent-associated Bartonella febrile illness, southwestern United States. Emerg. Infect. Dis. 12:1081-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampfer, P., H. C. Scholz, B. Huber, E. Falsen, and H. J. Busse. 2007. Ochrobactrum haematophilum sp. nov. and Ochrobactrum pseudogrignonense sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 57:2513-2518. [DOI] [PubMed] [Google Scholar]
- 26.Katoh, K., K. Misawa, K. I. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkhoff, F. T., A. M. Bergmans, Z. A. van Der, and A. Rothova. 1999. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J. Clin. Microbiol. 37:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 29.Kosoy, M., Y. Bai, K. Sheff, C. Morway, H. Baggett, S. A. Maloney, S. Boonmar, S. Bhengsri, S. F. Dowell, A. Sitdhirasdr, K. Lerdthusnee, J. H. Richardson, and L. F. Peruski. 2010. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 82:1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosoy, M., E. Mandel, D. Green, E. Marston, D. Jones, and J. Childs. 2004. Prospective studies of Bartonella of rodents. II. Diverse infections in a single rodent community. Vector Borne Zoonotic Dis. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 31.Kosoy, M., C. Morway, K. W. Sheff, Y. Bai, J. Colborn, L. Chalcraft, S. F. Dowell, L. F. Peruski, S. A. Maloney, H. Baggett, S. Sutthirattana, A. Sidhirat, S. Maruyama, H. Kabeya, B. B. Chomel, R. Kasten, V. Popov, J. Robinson, A. Kruglov, and L. R. Petersen. 2008. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J. Clin. Microbiol. 46:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosoy, M., M. Murray, R. D. Gilmore, Jr., Y. Bai, and K. L. Gage. 2003. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41:645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. E. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 34.La Scola, B., Z. Zeaiter, A. Khamis, and D. Raoult. 2003. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 11:318-321. [DOI] [PubMed] [Google Scholar]
- 35.Lebuhn, M., W. Achouak, M. Schloter, O. Berge, H. Meier, M. Barakat, A. Hartmann, and T. Heulin. 2000. Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int. J. Syst. Evol. Microbiol. 50:2207-2223. [DOI] [PubMed] [Google Scholar]
- 36.Lin, E. Y., C. Tsigrelis, L. M. Baddour, H. Lepidi, J. M. Rolain, R. Patel, and D. Raoult. 2010. Candidatus Bartonella mayotimonensis and endocarditis. Emerg. Infect. Dis. 16:500-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, Q., J. Sun, L. Lu, G. Fu, G. Ding, X. Song, F. Meng, H. Wu, T. Yang, Z. Ren, E. Chen, J. Lin, H. Lv, and C. Chai. 2010. Detection of Bartonella species in small mammals from Zhejiang Province, China. J. Wildl. Dis. 46:179-185. [DOI] [PubMed] [Google Scholar]
- 38.Maggi, R. G., M. Kosoy, M. Mintzer, and E. B. Breitschwerdt. 2009. Isolation of Candidatus Bartonella melophagi from human blood. Emerg. Infect. Dis. 15:66-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Márquez, F. J., J. J. Rodríguez-Liebana, M. E. Pachón-Ibáñez, F. Docobo-Pérez, A. Hidalgo-Fontiveros, M. Bernabeu-Wittel, M. A. Muniain, and J. Pachón. 2008. Molecular screening of Bartonella species in rodents from south western Spain. Vector Borne Zoonotic Dis. 8:695-700. [DOI] [PubMed] [Google Scholar]
- 40.Saenz, H. L., P. Engel, M. C. Stoeckli, C. Lanz, G. Raddatz, M. Vayssier-Taussat, R. Birtles, S. C. Schuster, and C. Dehio. 2007. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat. Genet. 39:1469-1476. [DOI] [PubMed] [Google Scholar]
- 41.Serratrice, J., J. M. Rolain, B. Granel, N. Ene, J. Conrath, J. F. Avierinos, P. Disdier, D. Raoult, and P. J. Weiller. 2003. Bilateral retinal artery branch occlusions revealing Bartonella grahamii infection. Rev. Med. Intern. 24:629-630. [DOI] [PubMed] [Google Scholar]
- 42.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 43.Tea, A., S. Alexiou-Daniel, A. Papoutsi, A. Papa, and A. Antoniadis. 2004. Bartonella species isolated from rodents, Greece. Emerg. Infect. Dis. 10:963-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welc-Faleciak, R., A. Paziewska, A. Bajer, J. M. Behnke, and E. Sinski. 2008. Bartonella spp. infection in rodents from different habitats in the Mazury Lake District, Northeast Poland. Vector Borne Zoonotic Dis. 8:467-474. [DOI] [PubMed] [Google Scholar]
- 45.Welch, D. F. 2005. Family II: Bartonellaceae, p. 362-370. In K. Brenner and G. Staley (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 46.Welch, D. F., K. C. Carroll, E. K. Hofmeister, D. H. Persing, D. A. Robison, A. G. Steigerwalt, and D. J. Brenner. 1999. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J. Clin. Microbiol. 37:2598-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, K., B. B. Chomel, R. W. Kasten, C. C. Chang, T. Tseggai, P. R. Decker, M. Mackowiak, K. A. Floyd-Hawkins, and N. C. Pedersen. 1998. Homologous protection but lack of heterologous-protection by various species and types of Bartonella in specific pathogen-free cats. Vet. Immunol. Immunopathol. 65:191-204. [DOI] [PubMed] [Google Scholar]