Abstract
Recombinant plasmids containing fusion proteins composed of two different modules were constructed and expressed in Escherichia coli. The modules encoded the lactase LacA (LacZ) from the thermophilic bacterium Thermoanaerobacter ethanolicus and the cellulase CelD, a cellulose-binding module (CBM) from Anaerocellum thermophilum. The CelD CBM provides a spontaneous and strong sorption of the fusion proteins onto a cellulose carrier. The enzymatic activities of both the free LacA protein and LacA-CelD CBM fusion proteins immobilized onto the cellulose carrier were assessed. The LacA activity of the fusion protein was dependent upon its position with respect to the CBM. The highest level of lactase activity and stability was observed when the lactase domain was localized at its N terminus. A continuous-flow column reactor of lactase immobilized on a cellulose carrier was constructed, and its activity was assessed. The lactose hydrolysis rate for a 150 mM (5%) solution at a flow rate of 1 reactor volume per min was 75%, which is a value optimal for further whey transformation into glucose/galactose syrup.
The glycosyl hydrolases cleave high-molecular-weight carbohydrate polymers. Many of them consist of functionally independent globular domains (modules) (3, 16, 24). After separation, such modules usually maintain their three-dimensional structure and functional activity (1, 3). Catalytic and substrate-binding modules are common. Substrate-binding modules are specific for particular carbohydrates such as cellulose, chitin, xylan, and β-glucans, etc. (4-6). The substrate-binding strength is determined by a number of factors, which include the polysaccharide moieties and their degree of polymerization and the degree of “crystallinity,” which corresponds to a density of the polysaccharides and may range from amorphous to a crystalline form (6, 29). The main function of most substrate-binding modules is to increase the concentration of catalytic subunits in the vicinity of the substrate (8, 29). Cellulose-binding modules (CBMs) have been particularly well characterized, with some CBMs being used for protein immobilization on cellulose (12, 25, 29).
In recent years, the practical use of CBMs has been established in different fields of biotechnology, and the number of published articles and patents is constantly on the rise. Three basic properties have contributed to CBMs being perfect candidates for many applications: (i) CBMs are typically independently folding units and can therefore function autonomously in chimeric proteins; (ii) the attachment matrices are abundant and inexpensive, with excellent chemical and physical properties; and (iii) the binding specificities can be controlled, which allows adaptation to existing problems. The utilization of CBMs has been extensively reported and reviewed in the literature (2, 11, 15, 17, 19, 27, 29).
The continued development of immobilized-enzyme technology is vital for the food industry. Immobilized enzymes are utilized in flow reactors for continuous hydrolysis as well as for periodic cyclical hydrolytic processes. For example, the glucosidases are used for the improvement of juice and wines (20), β-galactosidases are used for the production of lactose-free dairy products, and lipases are used for the hydrolysis of fats and lipids (22).
We propose the use of chimeric proteins immobilized on cellulose as fluid-flow microreactors. The advantages of this method include inexpensive cellulose sorbent, the high specificity of the interaction, and a high yield (close to 100%) of active enzyme during immobilization. In addition, cellulose is a nontoxic and inert material, facilitating its application in food industries (26).
The presence of lactose in whey is the major factor complicating its transformation and nutritional utilization. The production of lactose-free milk is especially important considering that the majority of the human population exhibits lactose intolerance (10).
The dietary quality of whey and milk can be improved by the enzymatic hydrolysis of lactose into its component monosaccharides, glucose and galactose (7, 9). These hydrolytic products have higher sweetness and solubility as well as being more readily fermented by microorganisms and assimilated by humans. Lactose hydrolysis by immobilized lactases (LACs) is a promising technology (17, 18), and this technique allows any lactose-containing product to be processed in an efficient manner. Its methodology utilizes mild reaction conditions that obviate the formation of undesirable hydrolysates, which decrease the product quality and are subject to extra purification. Production efficiency is achieved through the following factors: the repeated utilization of the same sorbent, improved quality of hydrolysis with a steadily high lactose conversion ratio, decreased synthesis of by-products, and absence of the enzyme in the end product. Overall, this immobilization of lactase methodology would be expected to decrease the production costs of lactose hydrolysis in whey.
Lactases are typically immobilized by chemical methods that create covalent bonds between the carrier and the enzyme (13, 21). Physical methods of immobilization involve lactase absorption or incorporation onto insoluble or semipermeable microcapsules (30). Both of these immobilization techniques have their drawbacks in that they require high-purity enzymes and multistep purification. In addition, chemical immobilization partially inactivates the enzyme. Furthermore, toxic reagents and carriers are often used in the chemical immobilization of lactase, which complicates their utilization in the food industry without an additional purification step.
Lactases from thermophilic microorganisms are promising for the milk industry due to their high thermostability, affinity for the substrate, and efficiency at the normal pH of whey (13, 18).
In this work, fusion proteins of thermostable LAC and CBM are characterized. One protein was selected for use in the construction of an efficient column continuous-flow system for the hydrolysis of lactose in solution. The fusion protein was immobilized on cellulose through the CBM of the Anaerocellum thermophilum endoglucanase CelD (NCBI accession no. Z77855), which provides a high affinity for cellulose carriers. The cellulose-immobilized recombinant thermostable LAC possessed a high level of activity toward lactose.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmid An10 containing the CelD gene was cloned previously from the thermophilic anaerobic bacterium A. thermophilum (NCBI accession no. Z77855). Escherichia coli strain M15[pREP4] and expression vector pQE13 (Qiagen) were used to clone and express the recombinant genes. The cultivation of recombinant cells and overexpression were done as recommended by the manufacturer (Qiagen).
DNA manipulations.
Plasmid DNA was isolated from E. coli cells by using a QIAprep Spin Miniprep kit (Qiagen) according to the manufacturer's instructions. Endonuclease hydrolysis, ligation, agarose gel electrophoresis, and other manipulations were performed according to standard techniques (23). Restriction endonucleases were purchased from Fermentas (Lithuania) and Promega.
Construction of a multimodal sequence with the required number of modules.
The pQE expression vectors were used to produce bimodal recombinant proteins containing the cellulose-binding and lactase modules. The designed protocol of cloning is applicable to produce any other multimodular proteins. It includes the construction of the resulting sequence from the modules of the following structure: BamHI site-module sequence-BglII site. Since the cohesive ends of the BamHI (5′-G↓GATCC-3′) and BglII (5′-A↓GATCT-3′) sites after hydrolysis are the same, the modules can be assembled in any order.
Lactase gene cloning.
To clone the lactase gene into the pQE13 expression vector, the LacA-coding sequence was obtained by PCR using Thermoanaerobacter ethanolicus chromosomal DNA as a template and oligonucleotide primers 5′-GGAGGCGGGATCCATGGGTAGAGATGTATTG-3′ (forward) and 5′-AACCCATCTAGAGGGTGGGTTTTTAAGATCTTATTGACT-3′ (reverse) (BamHI, XbaI, and BglII sites are underlined). The product as well as vector pQE13 were digested with BamHI and XbaI and ligated. The primary structure of the resulting 4,691-bp plasmid pLAC was confirmed by sequencing.
Construction of a plasmid with a spacer sequence.
A mutual influence of modules is possible in multidomain proteins, which can affect its three-dimensional structure and consequently decrease or suppress its catalytic activity. Accordingly, a synthetic DNA fragment coding for Gly-Ser repeats was cloned into the BamHI and HindIII sites of pQE13 to yield the 3,471-bp plasmid pRsp with the Gly-Ser spacer (sp). The selected length and amino acid sequence of the spacer provide its flexibility and hydrophilic properties. Such a spacer holds away the artificially connected modules at a distance sufficient for their independent functioning. Two complementary oligonucleotides were synthesized for a spacer encoding alternating Gly and Ser residues (BglII, BamHI, and HindIII sites are underlined, and the encoded amino acid sequence is given): PGSGSGSGSGSGA 5′-GATCCCCGGGTTCTGGCTCCGGCTCTGGTTCCGGTTCTGGCGCCAGATCTA-3′ 3′-GGGCCCAAGACCGAGGCCGAGACCAAGGCCAAGACCGCGGTCTAGATTCGA-5′ The annealed fragment was cloned into the BamHI and HindIII sites of pQE13. The primary structure of the resulting plasmid, pRsp, was confirmed by sequencing.
Construction of a plasmid with a Gly-Ser spacer and the CBM.
The celD DNA fragment containing the CBM was obtained from the previously constructed plasmid An10 by PCR using the following primers, corresponding to the ends of the desired region (BamHI, HindIII, and BglII sites are underlined): forward primer 5′-AAAGAAGGATCCAATGCACCTTTAGGCG-3′ and reverse primer 5′-CCTCAAAAAAGCTTTAGGTAGATCTAACATTATCTATATAC-3′. This PCR fragment and pRsp DNA were digested with BamHI plus HindIII and BglII plus HindIII, respectively, and the 563-bp fragment of the celD gene was ligated into the 3,465-bp fragment of pRsp. The primary structure of the resulting 4,028-bp plasmid pspCBM was confirmed by sequencing.
Construction of the fusion proteins LACspCBM and CBMspLAC.
For the construction of the plasmid containing the lacA (lacZ) gene, the Gly-Ser spacer, and the CBM, pLAC was digested with BglII and BglI, while pspCBM was digested with BamHI and BglI. The primary structure of the resulting 6,254-bp plasmid, pLACspCBM, was confirmed by sequencing.
Plasmid pCBMspLAC, coding for a fusion protein with the N-terminal CBM, was constructed in a similar way. The primary structure of the resulting plasmid, pCBMspLAC, was confirmed by sequencing.
Gene expression and recombinant protein purification.
Recombinant proteins with a histidine tag were purified from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli cultures by affinity chromatography on Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) according to the manufacturer's instructions. The protein concentration was measured by using a Bio-Rad protein assay kit. The protein molecular weight and purity were evaluated by SDS-PAGE (14).
Assay for lactase activity of recombinant proteins.
The enzyme activity was determined by the capacity to hydrolyze lactose to yield glucose and galactose (9, 30). In the LAC activity assay, the reaction mixture (0.5 ml) included the enzyme and 4% lactose in 50 mM potassium phosphate buffer (pH 6.0). The glucose produced was quantified by using glucose oxidase/peroxidase kit 510-A (Sigma). To estimate the activity of chimeric cellulose-immobilized lactase, an aliquot of cellulose with immobilized enzyme was added to the above-described reaction mixture. The protein concentration was measured by using a Bio-Rad protein assay kit. The pure cellulose carrier without protein was used as a control.
Activity units were defined as the amount of enzyme that released 1 μmol glucose per min. Each activity measurement was repeated at least three times, with the standard error of the mean being within 5%. The Michaelis constant (Km) and Vmax were determined from the dependence of activity on the substrate concentration (lactose, 1 to 80 mM) and were calculated by using Origin 6.1 software (OriginLab).
Characterization of the recombinant proteins.
The temperature dependence of the recombinant protein activity was studied by using 50 mM potassium phosphate buffer at pH 6.0. The pH dependence of the enzyme activity was studied with Tris-citrate buffers within the pH range of 4.5 to 9.5 at 60°C.
The thermostability of recombinant proteins was studied by their incubation at different temperatures in the absence of the substrate. After different periods of time, the enzymes were sampled for the lactase assay.
The influence of several selected substances on the activity of recombinant enzymes was studied. Either 5% of galactose or 5 to 10 mM Ca or Mg ions in phosphate-citrate buffer (pH 6.0) was added to the reaction mixture. The enzyme activity in parallel samples without any additives was accepted to be 100%.
Fusion protein immobilization and purification on cellulose.
Induced producer cells were harvested by centrifugation at 2,830 × g for 20 min, resuspended in 1% Triton X-100, incubated with 1 μg/ml lysozyme on ice for 30 min, and disintegrated by sonication. The lysate was centrifuged at 12,000 × g for 20 min, and the supernatant was mixed with 5 ml of water-swollen cellulose granules (Sigma) and incubated at 4°C for 6 h. Cellulose with absorbed protein was repeatedly washed in 10 volumes of 1% Triton X-100 with intensive mixing. The purity of the immobilized protein was tested by using protein electrophoresis with SDS-PAGE gels (14). The strength of immobilization was also estimated by using SDS-PAGE after washing of sorbent with 100 volumes of buffers with different pHs (4 to 11), temperatures (0°C to 75°C), and concentrations of NaCl (0 to 5 M). A total of 10 to 15 μl of immobilized cellulose was boiled in the sample buffer (14) for 2 min and placed into the gel. The control used was the same quantity of the carrier with immobilized protein before washing.
Development of a continuous-flow reactor.
The sorbent with the immobilized LACspCBM protein was used to produce a continuous-flow reactor with an inner diameter of 4.6 mm and a sorbent layer height of 6.2 mm (the sorbent volume was 410 μl). The reactor temperature was controlled by an air bath (LKB). Lactose (150 mM or 5%, which corresponds to the whey concentration) in 10 mM ammonium acetate (pH 6.5) served as the substrate. The substrate flow was generated by a peristaltic pump (Gilson, France) at an empirically determined flow rate. The first 2 ml passed through the reactor was not sampled, while the subsequent 1 ml was taken for chromatographic analysis. Lactose conversion was evaluated by normal-phase high-performance liquid chromatography (HPLC). An isocratic chromatograph (Gilson) was equipped with a refractometric detector and a Zorbax NH2 analytical column (4.6 by 250 mm). An acetonitrile-water (70:30, vol/vol) mix was used as a mobile phase. Chromatography data were analyzed by Ecochrom (Russia) and ChromMan (ACDLabs, Canada) software.
RESULTS AND DISCUSSION
Previously, we cloned (9) and sequenced the thermostable lactase gene lacA from Thermoanaerobacter ethanolicus (also named lacZ). The recombinant enzyme was highly stable and active (9, 30). The cellulose-binding module of the CelD cellulase from the thermophilic anaerobic bacterium A. thermophilum was cloned and sequenced (NCBI accession no. Z77855). This module demonstrates the ability for a practically irreversible binding of cellulose. The aim of this work was to construct and study a chimeric enzyme with the activity of lactase that was also able to strongly bind cellulose carriers. Two fusion proteins with modules in different relative positions were designed with a flexible spacer of sufficient length to not disturb the properties of each module. Three recombinant plasmids were constructed on the basis of expression vector pQE13: pLAC, pLACspCBM, and pCBMspLAC.
Analysis of lactase properties of LACspCBM and CBMspLAC fusion proteins.
The biochemical properties of the chimeras were compared to that of lactase (Table 1). The LACspCBM chimera was more stable and more active than CBMspLAC, so the former was chosen for further work.
TABLE 1.
Comparison of LacA and the fusion proteins LACspCBM and CBMspLACa
| Protein | Topt (°C) | pHopt | Km (mM) | Vmax (U/mg protein) | Mean Vmax/Km (U/mg·mM) ± SEM | % thermostability at 55°C for 24 h |
|---|---|---|---|---|---|---|
| LAC | 75-80 | 5.7-6.0 | 30 ± 1.5 | 480 ± 24 | 16 ± 0.8 | 80 |
| LACspCBM | 65-70 | 5.7-6.0 | 29 ± 1.5 | 440 ± 22 | 15 ± 0.75 | 70 |
| LACspCBM-cellulose | 65-75 | 5.7-6.0 | 29 ± 1.5 | 460 ± 23 | 16 ± 0.8 | 80 |
| CBMspLAC | 60-65 | 6.5-6.8 | 59 ± 2.9 | 316 ± 16 | 5 ± 0.25 | 11 |
Topt, optimal temperature; pHopt, optimal pH.
No differences in the effects of several selected substances were observed for the engineered lactases. Galactose inhibited the source lactase and fusion protein immobilized on cellulose. Ca2+ and Mg2+ ions (5 to 10 mM) had no effect on their activity.
Single-step immobilization on cellulose and purification of fusion proteins.
Fusion proteins containing the cellulose-binding module of the Anaerocellum thermophilum cellulase CelD and the LacA module were used to produce a sorbent for a flow reactor in one stage. This represents the simultaneous purification, concentration, and immobilization of a recombinant protein on a cellulose carrier. This procedure eliminates 90% of E. coli cellular proteins. The cellulose-binding properties of LACspCBM and CBMspLAC were tested by passing 100 volumes of buffers with different pHs and NaCl concentrations at different temperatures through the columns with immobilized proteins, after which the immobilized protein was quantified. Both proteins demonstrated irreversible binding to cellulose (they were not washed off the sorbent at pH 4 to 11, at 0°C to 75°C, and with 0 to 5 M NaCl). Thus, the CBM position in the fusion protein had no effect on its cellulose-binding properties, which can be attributed to the central localization of the CBM module in the CelD molecule.
Use of the fusion protein LACspCBM immobilized on cellulose to hydrolyze lactose.
Known methods of thermostable lactase immobilization involve covalent bonding between the enzyme and organic sorbents requiring a preliminary purification of the enzyme. Immobilization on inorganic sorbents such as silica-alumina (13) relies on complex methods involving toxic substances. Previously, we immobilized lactase from T. ethanolicus on aldehyde silochrome (29), with a high yield of active immobilized enzyme, although it required preliminary multistep purification of the enzyme. The method of lactase immobilization proposed in this work includes only one stage, does not require preliminary enzyme purification, provides a high yield of active enzyme, and excludes the use of any toxic reagents.
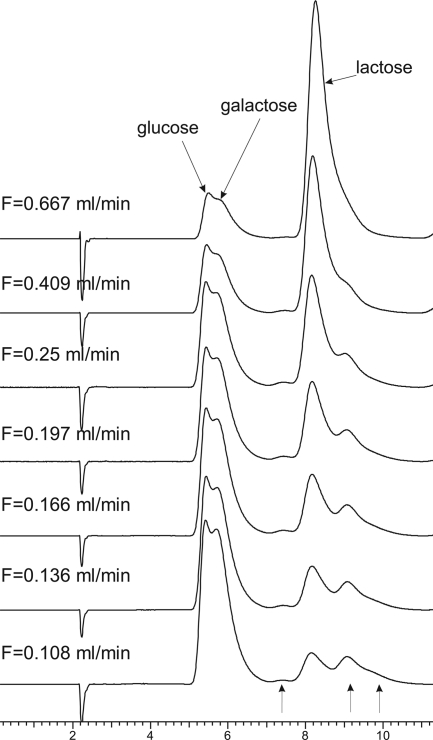
A continuous-flow column minireactor was constructed from a chromatography column filled with sorbent with immobilized fusion protein. The dependence of lactose conversion on the substrate flow rate was studied. Figure 1 presents chromatograms of eluates from the column loaded with 150 mM lactose in 10 mM acetate buffer (pH 6.5) at 45°C with different flow rates. As the flow decelerated, lactose hydrolysis became clearly more complete, and the proportion of the peaks corresponding to the major hydrolysis products, glucose and galactose, increased. The peak assignment to the major hydrolysis products was confirmed by the analysis of mixed hydrolytic products and standard solutions of glucose and galactose. In addition to the major peaks, there were three more peaks, which can be assigned to a disaccharide and two trisaccharides according to their chromatographic retentions. These extra peaks might be attributed to lactase-catalyzed transglycosylation (10).
FIG. 1.
Chromatographic analysis of lactose (150 mM in 10 mM acetate buffer [pH 6.5]) hydrolysis products at 45°C and with different flow rates (F). Three additional peaks discussed in the text are indicated by the arrows at the bottom of the figure.
Similar tests were conducted at different temperatures in the range of 40°C to 70°C. The data obtained were used to select the conditions providing efficient lactose hydrolysis that sufficed to improve the quality of whey and milk. At 70°C, the LACspCBM-based reactor hydrolyzed 70% of 150 mM (5%) lactose (which corresponds to the lactose concentration in cow's milk) at a substrate flow rate of 1.2 column volumes per min. Under the same conditions, a flow rate of 0.64 column volumes per minute provided 90% lactose hydrolysis. The passage of 10,000 column volumes through the reactor at a flow rate of 1 column volume per min did not decrease the efficiency of lactose hydrolysis. The reactor retained its lactose hydrolysis performance after storage at room temperature for 6 months.
The capacity for a virtually irreversible association between the fusion protein and cellulose sorbent and the high activity of the target protein make possible single-step fusion protein purification, concentration, and immobilization of active LacA on cellulose sorbent (28). The simplicity of the single-step immobilization and purification of the fusion lactase as well as the high specific activity and stability of the enzyme suggest its biotechnological utilization for lactose hydrolysis in dairy products.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (project no. 09-04-00204).
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Abbott, D. W., E. Ficko-Blean, A. L. van Bueren, A. Rogowski, A. Cartmell, P. M. Coutinho, B. Henrissat, H. J. Gilbert, and A. B. Boraston. 2009. Analysis of the structural and functional diversity of plant cell wall specific family 6 carbohydrate binding modules. Biochemistry 48:10395-10404. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:379-386. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. Cellulose decomposing prokaryotes and their enzyme systems, p. 234-315. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY.
- 4.Blake, A. W., L. McCartney, J. E. Flint, D. N. Bolam, A. B. Boraston, H. J. Gilbert, and J. P. Knox. 2006. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J. Biol. Chem. 281:29321-29329. [DOI] [PubMed] [Google Scholar]
- 5.Boraston, A. B., B. W. McLean, J. M. Kormos, M. Alam, N. R. Gilkes, C. A. Haynes, P. Tomme, D. G. Kilburn, and R. A. Warren. 1999. Carbohydrate-binding modules: diversity of structure and function, p. 202-211. In H. J. Gilbert, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 6.Carrard, G., A. Koivula, H. Soderlund, and P. Beguin. 2000. Cellulose binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. U. S. A. 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dabrowski, S., J. Maciunska, and J. Synowiecki. 1998. Cloning and nucleotide sequence of the thermostable β-galactosidase gene from Pyrococcus woesei in Escherichia coli and some properties of the isolated enzyme. Mol. Biotechnol. 10:217-222. [DOI] [PubMed] [Google Scholar]
- 8.Ficko-Blean, E., and A. B. Boraston. 2009. N-Acetylglucosamine recognition by a family 32 carbohydrate-binding module from Clostridium perfringens NagH. J. Mol. Biol. 390:208-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fokina, N. A., and G. A. Velikodvorskaya. 1997. Cloning and expression of the gene for thermostable beta-galactosidase from Thermoanaerobacter ethanolicus in Escherichia coli: purification and properties of the product. Mol. Gen. Mikrobiol. Virusol. 2:34-36. (In Russian.) [PubMed] [Google Scholar]
- 10.Gekas, V., and M. Lopez-Leiva. 1985. Hydrolysis of lactose: a literature review. Process Biochem. 20:2-11. [Google Scholar]
- 11.Greenwood, J. M., E. Ong, N. R. Gilkes, R. A. Warren, R. C. Miller, Jr., and D. G. Kilburn. 1992. Cellulose-binding domains: potential for purification of complex proteins. Protein Eng. 5:361-365. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, S., J. Ahn, S. Lee, T. G. Lee, S. Haam, K. Lee, I.-S. Ahn, and J.-K. Jung. 2004. Evaluation of cellulose-binding domain fused to a lipase for the lipase immobilization. Biotechnol. Lett. 26:603-605. [DOI] [PubMed] [Google Scholar]
- 13.Ladero, M., M. T. Perez, A. Santos, and F. Garcia-Ochoa. 2003. Hydrolysis of lactose by free and immobilized beta-galactosidase from Thermus sp. strain T2. Biotechnol. Bioeng. 81:241-252. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Levy, I., and O. Shoseyov. 2002. Cellulose binding domains: industrial and biotechnological application. Biotechnol. Adv. 20:191-213. [DOI] [PubMed] [Google Scholar]
- 16.Montanier, C., A. L. van Bueren, C. Dumon, J. E. Flint, M. A. Correia, J. A. Prates, S. J. Firbank, R. J. Lewis, G. G. Grondin, M. G. Ghinet, T. M. Gloster, C. Herve, J. P. Knox, B. G. Talbot, J. P. Turkenburg, J. Kerovuo, R. Brzezinski, C. M. G. A. Fontes, G. J. Davies, A. B. Boraston, and H. J. Gilbert. 2009. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U. S. A. 106:3065-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakkharat, P., and D. Haltrich. 2006. Lactose hydrolysis and formation of galactooligosaccharides by a novel immobilized beta-galactosidase from the thermophilic fungus Talaromyces thermophilus. Appl. Biochem. Biotechnol. 129-132:215-225. [DOI] [PubMed] [Google Scholar]
- 18.Neuhaus, W., S. Novalin, K. Klimacek, B. Splechtna, I. Petzelbauer, A. Szivak, and K. D. Kulbe. 2006. Optimization of an innovative hollow-fiber processes to produce lactose-reduced skim milk. J. Appl. Biochem. Biotechnol. 134:1-14. [DOI] [PubMed] [Google Scholar]
- 19.Ong, E., J. M. Greenwood, N. R. Gilkes, D. G. Kilburn, R. C. Miller, Jr., and A. J. Warren. 1989. The cellulose-binding domain of cellulases: tools for biotechnology. Trends Biotechnol. 7:239-243. [Google Scholar]
- 20.Ong, E., N. R. Gilkes, R. C. Miller, Jr., A. J. Warren, and D. G. Kilburn. 1991. Enzyme immobilization using a cellulose-binding domain: properties of a beta-glucosidase fusion protein. Enzyme Microb. Technol. 13:59-65. [DOI] [PubMed] [Google Scholar]
- 21.Piesecki, S., and E. Hochuli. 1993. Immobilization of β-galactosidase for application in organic chemistry using a chelating peptide. Biotechnol. Bioeng. 42:178-184. [DOI] [PubMed] [Google Scholar]
- 22.Rotticci-Mulder, J. C., M. Gustavsson, M. Holmquist, K. Hult, and M. Martinelle. 2001. Expression in Pichia pastoris of Candida antarctica lipase B and lipase B fused to a cellulose-binding domain. Protein Expr. Purif. 21:386-392. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Schwarz, W. H., V. V. Zverlov, and H. Bahl. 2004. Extracellular glycosyl hydrolases from clostridia. Adv. Appl. Microbiol. 56:215-261. [DOI] [PubMed] [Google Scholar]
- 25.Shoseyov, O., Z. Shani, and I. Levy. 2006. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomme, P., N. R. Gilkes, M. M. Guarna, C. A. Haynes, D. Hasenwinkle, E. Jervis, P. Johnson, L. McIntosh, R. A. Warren, and D. G. Kilburn. 1996. Cellulose-binding domains. Versatile affinity tags for inexpensive large-scale purification, concentration, and immobilization of fusion proteins. Ann. N. Y. Acad. Sci. 799:418-424. [DOI] [PubMed] [Google Scholar]
- 27.Tomme, P., A. Boraston, B. McLean, J. Kormos, A. L. Creagh, K. Sturch, N. R. Gilkes, C. A. Haynes, R. A. Warren, and D. G. Kilburn. 1998. Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B 715:283-296. [DOI] [PubMed] [Google Scholar]
- 28.Velikodvorskaya, G. A., V. V. Zverlov, A. S. Karyagina-Zhulina, N. V. Lavrova, V. G. Lunin, N. A. Lunina, E. M. Ryazanova, O. V. Sergienko, and T. V. Tikhonova. June 2006. Russian Federation patent 2278160.
- 29.Volkov, I., N. A. Lunina, and G. A. Velikodvorskaya. 2004. Outlooks of using substrate-binding modules of glycosyl hydrolases. Appl. Biochem. Microbiol. 40:499-504. [PubMed] [Google Scholar]
- 30.Volkov, I., N. A. Lunina, O. V. Berezina, G. A. Velikodvorskaya, and V. V. Zverlov. 2005. Thermoanaerobacter ethanolicus gene cluster containing the alpha- and beta-galactosidases genes melA and lacA, and properties of recombinant lacA. Mol. Biol. (Mosk.) 39:915-922. (In Russian.) [DOI] [PubMed] [Google Scholar]