Abstract
Phylogenetic, microbiological, and comparative genomic analyses were used to examine the diversity among members of the genus Caldicellulosiruptor, with an eye toward the capacity of these extremely thermophilic bacteria to degrade the complex carbohydrate content of plant biomass. Seven species from this genus (C. saccharolyticus, C. bescii, C. hydrothermalis, C. owensensis, C. kronotskyensis, C. lactoaceticus, and C. kristjanssonii) were compared on the basis of 16S rRNA gene phylogeny and cross-species DNA-DNA hybridization to a whole-genome C. saccharolyticus oligonucleotide microarray, revealing that C. saccharolyticus was the most divergent within this group. Growth physiology of the seven Caldicellulosiruptor species on a range of carbohydrates showed that, while all could be cultivated on acid-pretreated switchgrass, only C. saccharolyticus, C. bescii, C. kronotskyensis, and C. lactoaceticus were capable of hydrolyzing Whatman no. 1 filter paper. Two-dimensional gel electrophoresis of the secretomes from cells grown on microcrystalline cellulose revealed that the cellulolytic species also had diverse secretome fingerprints. The C. saccharolyticus secretome contained a prominent S-layer protein that appears in the cellulolytic Caldicellulosiruptor species, suggesting a possible role in cell-substrate interactions. Growth physiology also correlated with glycoside hydrolase (GH) and carbohydrate-binding module (CBM) inventories for the seven bacteria, as deduced from draft genome sequence information. These inventories indicated that the absence of a single GH and CBM family was responsible for diminished cellulolytic capacity. Overall, the genus Caldicellulosiruptor appears to contain more genomic and physiological diversity than previously reported, and this argues for continued efforts to isolate new members from high-temperature terrestrial biotopes.
Efforts directed at microbial deconstruction of lignocellulosic biomass for second-generation biofuels production (24) have renewed interest in previously studied high-temperature (optimal temperature [Topt], ≥70°C), carbohydrate-utilizing microorganisms from terrestrial niches. Over the past 30 years, the focus has been on establishing the upper thermal limits of life, such that hyperthermophiles (Topt, ≥80°C) have been specifically examined for clues to the intrinsic basis of thermophily. However, hyperthermophiles often originate from marine ecosystems, which typically lack crystalline cellulose. Although enzyme systems that hydrolyze β-linked polysaccharides, such as xylan (32), galactomannan (15), barley glucan (2), and laminarin (13, 14), are produced by certain heterotrophic hyperthermophiles, primary cellulases that are highly active on crystalline cellulose have not yet been identified in these microorganisms (6). But, at slightly lower optimal growth temperatures, within the range of 70 to 80°C, extremely thermophilic bacteria capable of plant biomass degradation can be isolated from terrestrial freshwater ecosystems (30, 45). In particular, the genus Caldicellulosiruptor, which belongs to the Gram-positive bacteria, contains members described as extremely thermophilic, anaerobic, cellulolytic/hemicellulolytic, low G+C content, and asporogenic (44). With optimal growth temperatures ranging from 70 to 78°C, certain species within this genus hydrolyze cellulose at the highest known temperatures for biological degradation of this complex carbohydrate (6, 22, 56). Interestingly, Caldicellulosiruptor species lack a cellulosome, which is common to cellulolytic Clostridia (3), and instead secrete discrete biomass-degrading enzymes directly into the extracellular milieu (49, 51). Members of the genus Caldicellulosiruptor are also able to coferment C5 and C6 sugars, an important aspect for consolidated bioprocessing (CBP), since both pentoses and hexoses are ultimately released during biomass deconstruction (28, 52, 57).
Although Caldicellulosiruptor species were first isolated some two decades ago, there have been only a limited number of reported efforts focusing on the microbial physiology and biochemistry of these bacteria (5, 54). However, with the genome sequences of Caldicellulosiruptor saccharolyticus (51) and Caldicellulosiruptor bescii (29) now available, the physiology of these bacteria can be examined more completely within the context of their potential role in bioenergy applications. C. saccharolyticus, the type strain of the Caldicellulosiruptor genus, first isolated from a freshwater hot spring in New Zealand, is capable of growth on cellulose, hemicellulose, and pectin (44). Recently, another finished genome of a Caldicellulosiruptor species, C. bescii (formerly Anaerocellum thermophilum [56]), became available (29), and indicated that approximately 15% of the two genomes showed significant differences (31). As other Caldicellulosiruptor species are isolated, 16S rRNA gene phylogeny has been used to place isolates within the genus (36, 45), but without the benefit of complete genome sequences for those isolates, the extent of genetic diversity is difficult to assess. In order to determine the relationship among members of the genus Caldicellulosiruptor, we have used complementary in silico and microbiological methods for determining genomic relatedness in this study. In addition to characterizing the physiological response to biomass or model biomass compounds, draft genome sequence data were examined to decipher the enzymatic basis for biomass deconstruction.
MATERIALS AND METHODS
Bacterial strains and growth on sugar substrates.
Caldicellulosiruptor species used in this study (Table 1) were obtained as axenic, freeze-dried cultures from the German Collection of Microorganisms and Cell Cultures (DSMZ [http://www.dsmz.de]), except for C. bescii, which was provided by Michael W. W. Adams (University of Georgia, Athens, GA), and C. owensensis, which was provided by James Elkins (Oak Ridge National Laboratory, Oak Ridge, TN). Freeze-dried cells were resuspended in the recommended medium and subcultured upon growth into modified DSMZ medium 640 (Trypticase, cysteine-HCl, and resazurin were not added; the carbon source was increased to 5 g/liter, and 10% [wt/vol] Na2S·9H2O was added to a final concentration of 0.5%) and grown at 70°C with orbital shaking. For 24-hour cell density measurements, each culture was subcultured in the respective carbon source three to four times in serum bottles before inoculating a 50-ml batch culture in a 125-ml serum bottle under N2 headspace for 24 h. Final cell counts were recorded using epifluorescence microscopy (25). Reported cell densities are the averages of two biological replicates for each carbon source reported. Sugars and biomass used for carbon sources included the following: d-glucose (Sigma-Aldrich, St. Louis, MO), d-xylose (Sigma-Aldrich), Avicel PH-101 (FMC), and dilute-acid-pretreated switchgrass (Panicum virgatum −20/+80 mesh fraction; pretreatment was in a Sunds reactor at the National Renewable Energy Laboratory [46]). Dilute acid-treated switchgrass was used at 5 g (wet weight)/liter, which corresponds to 1.28 ± 0.04 g (dry weight)/liter (mean ± standard deviation). In the case of cultures grown on yeast extract, only DSMZ 640 medium was used, which already includes 1 g/liter yeast extract (BD Biosciences, Difco).
TABLE 1.
Caldicellulosiruptor species available from DSMZ
| Organism | Location | G+C content (%) | Topt (°C) | DSMZ accession no. |
|---|---|---|---|---|
| C. besciia | Kamchatka, Russia | 35.2 | 75 | 6725 |
| C. saccharolyticusb | Taupo, New Zealand | 35.3 | 70 | 8903 |
| C. hydrothermalisc | Kamchatka, Russia | 36.4 | 65 | 18901 |
| C. kristjanssoniic | Hveragerdi, Iceland | 35.0 | 78 | 12137 |
| C. kronotskyensisc | Kamchatka, Russia | 35.1 | 70 | 18902 |
| C. lactoaceticusc | Hveragerdi, Iceland | 35.2 | 68 | 9545 |
| C. owensensisc | Owens Lake, CA | 36.6 | 75 | 13100 |
| C. acetigenus | Iceland | 35.7 | 70 | 7040 |
16S rRNA gene phylogenetic analysis.
16S rRNA gene sequences used for phylogenetic analyses between Caldicellulosiruptor spp. and related species were downloaded from the Ribosomal Database Project (http://rdp.cme.msu.edu) (12). Sequences used for 16S sequence identity were accessed from NCBI GenBank. Multiple sequence alignment of 16S rRNA gene sequences was conducted using Clustal W (50) as a part of the Mega 4 program (48). A 16S rRNA gene phylogenetic tree was built using the Jukes-Cantor evolutionary distance model followed by the neighbor-joining method. Bootstrap values were determined using 1,000 replicates in Mega 4 (48). Sequence identity percentages were determined using the BLASTN program (1).
Secretome isolation.
For a comparison of secretomes, each Caldicellulosiruptor species was transferred four times on modified DSM 640 medium, with either Avicel PH-101, d-xylose, or d-glucose as a carbon source (see above). Supernatant was harvested from two 500-ml batch cultures and grown for 24 h in 45-mm-diameter screw-top bottles. Briefly, the cultures were centrifuged at 5,000 rpm for 10 min to separate cells and insoluble Avicel from the medium, with the resulting supernatant filtered through a bottle-top 0.22-μm-pore-size filter (Millipore) and stored at 4°C with the addition of sodium azide (0.5%, final concentration). Sterile-filtered supernatant was further concentrated with ultrafiltration using a 10-kDa molecular mass cutoff polyethersulfone membrane (Millipore) to 100×, after which the concentrated supernatant was buffer exchanged three times into 50 mM sodium phosphate buffer (pH 7.2). Total protein concentration was estimated using the Bio-Rad protein assay reagent using the microassay for microtiter plate method per the manufacturer's protocol.
Two-dimensional gel electrophoresis of secretomes.
Buffer-exchanged supernatant corresponding to 100 or 150 μg of protein was resuspended in isoelectric focusing (IEF) buffer, per the manufacturer's recommendations {7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM dithiothreitol, and 0.2% (vol/vol) Bio-Lyte 3/10 ampholyte (Bio-Rad)}. Protein was then loaded onto a 7-cm pI 4 to 7 Bio-Rad ReadyStrip with immobilized pH gradient (IPG) for active rehydration at 50 V overnight in a Protean isoelectric focusing cell (Bio-Rad). Isoelectric focusing and equilibration of the IPG strips was done as described previously (35). The second dimension was run through a NuPAGE Novex 4-to-12% Bis-Tris gel (Invitrogen, Carlsbad, CA) using 1× 4-morpholineethanesulfonic acid (MES) buffer (Invitrogen) and 0.5% (wt/vol) low-melting-point agarose overlay. Gels were stained using GelCode Blue stain reagent (Thermo Scientific) or Silver Stain Plus (Bio-Rad) and scanned on a GS-710 densitometer (Bio-Rad). Identification of proteins from two-dimensional (2D) gels was performed by the Genomic Sciences Laboratory at NCSU. For protein identification, a 1-mm-diameter core was taken from a representative 2D gel and subjected to trypsin digestion, and subsequent identification was made after analysis by liquid chromatography (LC)-electrospray-tandem mass spectrometry (MS/MS) using a Thermo LTQ ion trap mass spectrometer (Thermo Scientific).
Genomic microarray hybridization.
Genomic DNA (gDNA) was harvested from Caldicellulosiruptor species grown to early stationary phase on modified DSMZ 640 plus cellobiose (Sigma). Cells were harvested by centrifugation at 5,000 rpm, and DNA was isolated, as previously described (18), with the addition of 100 mg/ml lysozyme (Sigma) during cell lysis. Genomic DNA was partially digested with HaeIII (New England BioLabs [NEB]), and amino allyl dUTP (Ambion) was incorporated along with deoxynucleoside triphosphates (Roche) using Klenow fragment (NEB) and random nonamers (Sigma). Labeling with Cy3 or Cy5 N-hydroxysuccinimide ester dye (GE Healthcare) was performed in 0.1 M sodium carbonate buffer, pH 9. Unincorporated dye was removed using Qiaquick spin columns according to the manufacturer's recommendations (Qiagen). Cy dye-labeled gDNA was hybridized to a C. saccharolyticus whole-genome oligonucleotide microarray chip (52) using a two-slide, dye-flip experimental design. Genomic DNA from C. saccharolyticus served as the reference control in each dye flip.
DNA microarray data analysis.
Microarray slides were scanned with a Packard BioChip Scanarray 4000 scanner (Perkin-Elmer, Waltham, MA). Signal intensity for each spot was calculated using ScanArray Express (v2.1.8; Perkin-Elmer) before importing into JMP Genomics 4.0 (SAS, Cary, NC) for normalization and application of mixed effects model analysis (43, 55). A Pearson correlation coefficient was calculated by comparing all normalized signal intensities from the tester strain versus C. saccharolyticus and was computed using Excel 2007 (Microsoft). During normalization and analysis, ratios for the tester strain versus C. saccharolyticus signal intensities were converted into a log2 format. The log2 ratios for each tester strain were plotted versus the location of the gene probe in the C. saccharolyticus genome to create the pair-wise scatter plots.
Accession numbers.
Data from the microarray-based comparative genomic hybridization (M-CGH) experiments are available through the Gene Expression Omnibus (GEO) database at NCBI (http://www.ncbi.nlm.nih.gov/geo/). The C. saccharolyticus microarray platform used in this study is available under accession number GPL6681. The experimental series accession number is GSE23606. Normalized (via analysis of variance) log2 ratios plus raw data used in this study for each dye flip can be found under the following experimental sample accession numbers: GSM578915, GSM578916, GSM578917, GSM578918, GSM578919, and GSM578920. For the glycoside hydrolase (GH), polysaccharide lyase, and carbohydrate-binding motif (CBM) annotations, finished and draft genome sequence data were provided by the Department of Energy Joint Genome Institute, with annotations for the finished genome sequences of C. bescii (CP001393) (29), C. hydrothermalis (CP002219), and C. owensensis (CP002216) and draft genome sequences of C. kristjanssonii (AEKC00000000), C. kronotskyensis (AEKB00000000), and C. lactoaceticus (AEKD00000000) completed at the Oak Ridge National Laboratory. Annotation for the finished genome of C. saccharolyticus (CP000679) was completed manually (51). These finished and preliminary annotations were examined for potential enzymes of interest, which were then cataloged based on homology to other CAZy annotated enzymes, using BlastP (1), InterProScan (27), and the CAZy database (http://www.cazy.org) (10). CAZy annotated enzymes for C. saccharolyticus and C. bescii were accessed directly from the CAZy database.
RESULTS AND DISCUSSION
The genus Caldicellulosiruptor.
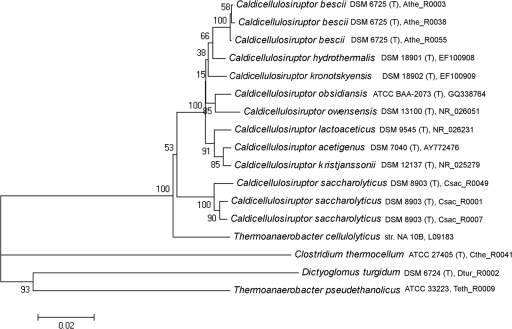
Since the first description of a Caldicellulosiruptor species (45), to date 33 16S rRNA gene sequences from various Caldicellulosiruptor species and isolates have been deposited in NCBI's GenBank and collected in the Ribosomal Database Project (12; http://rdp.cme.msu.edu). Of the deposited 16S rRNA gene sequences, only nine species have been described in the literature, and eight are available from a culture collection (Table 1). Members of this genus are typically isolated from biomass in freshwater geothermal springs, with the one exception being C. owensensis, which was isolated from sediment at Owens Lake, CA (Table 1) (26). At the time our study was initiated, only two Caldicellulosiruptor genome sequences were publically available, for C. saccharolyticus (51) and C. bescii (56). On a genomic level, the G+C nucleotide content of Caldicellulosiruptor species is low (∼35 to 36%) and has resulted in their placement in the Gram-positive phylum Firmicutes (45), which also includes other celluloytic species, such as Clostridium thermocellum (39). However, taxonomic classification within this branch of the thermophilic Firmicutes relies heavily on 16S rRNA gene phylogeny (33) and phenotype. Alternatively, common concatenated protein sequences were created to construct a phylogenetic tree, which placed C. saccharolyticus with the cellulolytic members of the genus Clostridium (21). Alignment of 16S rRNA gene sequences to construct a phylogenetic tree of the nine deposited Caldicellulosiruptor species revealed that species tend to cluster based on geographical location (Fig. 1) and that 16S rRNA gene sequence identity within the genus ranges from 94.8 to 99.4%. According to 16S rRNA analysis, C. saccharolyticus is the most phylogenetically divergent member of the genus Caldicellulosiruptor (94.4 to 96.6% rRNA identity), which may relate to it being the sole representative from New Zealand included in the analysis. However, when rRNA sequences from other New Zealand isolates were added to the phylogenetic tree, C. saccharolyticus still remained divergent from both groups (data not shown).
FIG. 1.
Neighbor-joining 16S rRNA phylogenetic tree of Caldicellulosiruptor species and members from related genera. The scale represents 0.02 substitution per nucleotide position. Bootstrap values are based on 1,000 replicates. T, type strain.
Regardless, 16S phylogenetic distances can only capture a small portion of the true genetic diversity in closely related species (47). Previous work concerning the genetic diversity within the genus Caldicellulosiruptor concentrated mainly on the cellulolytic and xylanolytic enzymes of C. saccharolyticus (4) and on other New Zealand Caldicellulosiruptor isolates, such as Caldicellulosiruptor sp. Tok7B.1 (19) and Caldicellulosiruptor sp. Rt69B.1 (38). While 16S phylogenetic analysis is useful for quickly assigning new isolates to taxonomic groups, it is clear that a more comprehensive measure of genetic relatedness is needed, especially in light of the increased interest in microbes capable of crystalline cellulose hydrolysis at high temperatures.
Microarray-based genomic hybridization.
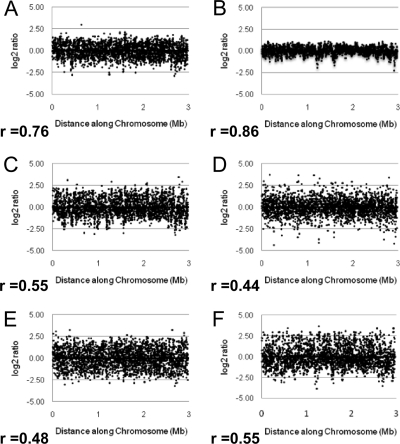
In order to further explore the genetic diversity of publicly available Caldicellulosiruptor species, seven species obtained from the German Collection of Microorganisms and Cell Cultures were surveyed for genetic similarity to C. saccharolyticus by using M-CGH. Since the first publicly available genome for a Caldicellulosiruptor species was for C. saccharolyticus, a whole-genome oligonucleotide chip had previously been designed (51) and was used for the M-CGH in our study. Genomic DNA from six additional Caldicellulosiruptor species was hybridized to the C. saccharolyticus oligonucleotide chip, using genomic DNA from C. saccharolyticus as a control in each dye flip. Plotting log2 hybridization ratios (least squares mean [LSM] intensity values of each test species versus C. saccharolyticus, with a log2-based conversion) of each species, provided a visual comparison of genetic divergence from C. saccharolyticus (Fig. 2A to F). As log2 hybridization ratios approach zero, those open reading frames, and by extension the organism, are considered to be more genetically similar to C. saccharolyticus. The more genetically divergent the species tested are from C. saccharolyticus (based on M-CGH analysis), the more the plots of log2 ratios will deviate from the horizontal axis (17). In addition, pair-wise scatter plots of hybridization intensities from unknown versus reference strains have previously been used to assess levels of diversity between strains (11). In this study, pair-wise scatter plots of LSM intensity values were used to compare the hybridization profile of each tester species versus C. saccharolyticus to experimentally calculate genetic distance. For each tester species, genetic distance was reported as a Pearson correlation coefficient (r), a measure of how close the pair-wise scatter plots of the probe intensity of the tester strain versus C. saccharolyticus converged to a straight line (Fig. 2A to F).
FIG. 2.
Log2 ratios for genomic DNA based on M-CGH analysis of tested Caldicellulosiruptor species versus C. saccharolyticus. (A) C. bescii; (B) C. hydrothermalis; (C) C. kristjanssonii; (D) C. kronotskyensis; (E) C. lactoaceticus; (F) C. owensensis. Log2 ratios indicate the fold hybridization differences between the tester and reference samples, converted to a log2 format. r is the corresponding Pearson correlation coefficient, quantifying the linear relationship between LSM hybridization levels of each Caldicellulosiruptor species tested versus C. saccharolyticus.
Of all the species tested, C. hydrothermalis had a hybridization profile most similar to C. saccharolyticus (r, 0.86) (Fig. 2B), followed by C. bescii (r, 0.76) (Fig. 2A), whereas C. kristjanssonii, C. kronotskyensis, C. lactoaceticus, and C. owensensis were not as closely related (r, 0.55, 0.44, 0.48, and 0.55, respectively) (Fig. 2C to F). In the case of C. bescii compared to C. saccharolyticus, a higher Pearson correlation coefficient was expected, since extensive similarity had been noted when comparing their finished genomes (29, 31). In the case of the other Caldicellulosiruptor species, the experimental (M-CGH) data indicated a greater amount of genetic divergence from C. saccharolyticus (r, 0.86 to 0.44) than the in silico measure (16S rRNA) would indicate (94.4 to 96.6%). Unlike 16S phylogeny, geographical localization seems to have no effect on similarity of hybridization profiles or Pearson correlation coefficients, since C. bescii, C. hydrothermalis, and C. kronotskyensis were all isolated from geothermal hot springs in the Kamchatka Peninsula (Table 1; Fig. 2A, B, and D). Use of the M-CGH technique is a potentially powerful tool to assess genetic diversity of new Caldicellulosiruptor isolates from the environment. Overall, it appears that the genus Caldicellulsiruptor is more genetically diverse than would be expected if only 16S rRNA sequence identity were taken into consideration. The question is then whether the different species use similar enzymes and metabolic strategies for biomass deconstruction.
Caldicellulosiruptor growth physiology.
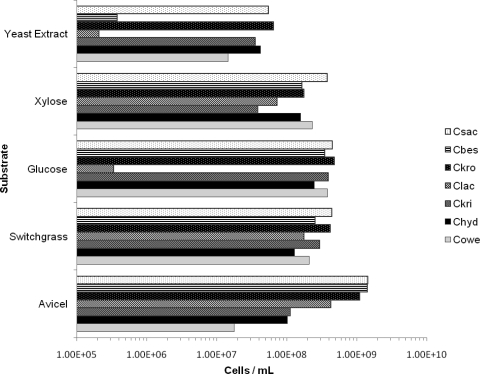
Growth on various sugar substrates has been reported in Caldicellulosiruptor species isolation studies (8, 22, 26, 36, 37, 41, 44, 56). However, a direct comparison across the entire genus with respect to carbohydrate utilization has not been done. To conduct such a comparison, the seven Caldicellulosiruptor species were grown on various simple (C5 and C6) and complex sugars, in addition to dilute-acid-pretreated biomass. Cell density measurements at 24 h were calculated for Caldicellulosiruptor species on Avicel, pretreated switchgrass, glucose, xylose, and yeast extract. Cell density measurements on yeast extract served as a control, since the growth medium contained no supplemental carbohydrate source. All Caldicellulosiruptor species were previously reported to grow on xylose, and the only significant difference in xylose growth observed here was that the cell density of C. saccharolyticus was 10-fold higher than that of C. kristjanssonii (Fig. 3). Consistent with the isolation study of C. lactoaceticus (37), no growth on glucose was observed with this species, while the rest of the species were capable of growth on glucose. Similar to C. lactoaceticus, some cellulolytic Clostridia spp. have shown similar substrate specializations, such as C. thermocellum, which will preferentially grow on cellobiose versus glucose as a carbon source and requires long acclimation times to grow on glucose (34).
FIG. 3.
Comparisons of growth of Caldicellulosiruptor spp. on selected substrates. All substrates were present at 5 g/liter in modified DSMZ 640 medium except for yeast extract, which was present at 1 g/liter in modified DSMZ 640 medium. The species order was ranked with respect to growth levels on Avicel. Species abbreviations: Cbes, C. bescii; Chyd, C. hydrothermalis; Ckri, C. kristjanssonii; Ckro, C. kronotskyensis; Clac, C. lactoaceticus; Cowe, C. owensensis; Csac, C. saccharolyticus.
When dilute-acid-pretreated biomass (switchgrass) was used as a growth substrate, all species were able to grow to cell densities higher than 108 cells/ml, indicating that the spectrum of polysaccharides present in the biomass (∼50% glucan and 8% xylan [46]) support growth of all of the Caldicellulosiruptor species tested. It is interesting that pretreated switchgrass slightly inhibited growth in comparison to growth on Avicel in some cases (C. saccharolyticus, C. bescii, C. kronotskyensis, and C. lactoaceticus), yet in other cases it stimulated growth (C. kristjanssonii and C. owensensis). Inhibition of growth on pretreated switchgrass by the more cellulolytic Caldicellulosiruptor spp. could result from the presence of sugar dehydration products and phenolics. In the case of the less cellulolytic Caldicellulosiruptor spp., stimulation of growth by pretreated switchgrass could be related to the easier accessibility of polysaccharides (xylan and amorphous cellulose, for example), which are more suitable for growth, even though growth inhibitors are presumably present (23).
The most striking difference between the Caldicellulosiruptor species was their ability to grow on microcrystalline cellulose (Avicel) (Fig. 3). It is a defining characteristic of the genus to grow on crystalline cellulose (44), although three species (C. saccharolyticus, C. bescii, and C. kronotskyensis) grew to cell densities roughly 10- and 100-fold higher on this substrate than some of the other members (C. kristjanssonii, C. hydrothermalis, and C. owensensis). Growth on Avicel may not be a definitive indication of an organism's capacity to hydrolyze the crystalline areas of cellulose, since Avicel also contains less-ordered, amorphous regions as well (42).
Secretome analysis.
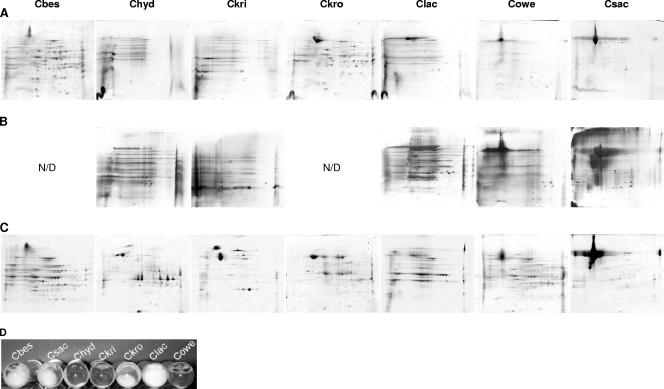
To further examine diversity within the genus Caldicellulosiruptor, especially with respect to cellulolytic ability, the Avicel-induced secretomes of various Caldicellulosiruptor species were examined and compared to their xylose-induced secretomes. Supernatant from Avicel-induced secretomes collected from all Caldicellulosiruptor species was capable of hydrolyzing carboxymethylcellulose (CMC) (data not shown), although only four species were capable of deconstructing Whatman no. 1 filter paper during growth (Fig. 4D). CMC hydrolysis implies the presence of endo-acting cellulases in all of the Caldicellulosiruptor species; however, the deconstruction of crystalline cellulose requires exo-acting cellulases (34).
FIG. 4.
2D SDS-PAGE gels of secretomes from Caldicellulosiruptor species. (A) Avicel induced, with 100 μg protein loaded; (B) Avicel induced, 150 μg protein loaded and silver stained; (C) xylose induced, 100 μg loaded; (D) filter paper deconstruction cultures. Species abbreviations: Cbes, C. bescii; Chyd, C. hydrothermalis; Ckri, C. kristjanssonii; Ckro, C. kronotskyensis; Clac, C. lactoaceticus; Cowe, C. owensensis; Csac, C. saccharolyticus. N/D, not determined.
Two-dimensional SDS-PAGE was also used to visualize the proteins secreted into the extracellular milieu during growth on Avicel and xylose, a C5 sugar capable of supporting growth for all Caldicellulosriuptor species used in this study (Fig. 3). The majority of Avicel-induced secretomes of Caldicellulosiruptor species capable of deconstructing filter paper displayed more proteins present (C. bescii, C. kronotskyensis, and C. lactoaceticus) than the less cellulolytic C. hydrothermalis, C. kristjanssonii, and C. owensensis (Fig. 4A, B, and D). The secretome of C. saccharolyticus, however, contained one predominant high-molecular-weight protein spot that was identified by LC-MS/MS as an S-layer protein (GenBank accession number YP_001181219). The microbial S-layer is a self-assembling protein coat implicated in protection from extreme environments, the creation of a Gram-positive “periplasm,” enzyme immobilization, and in some cases possibly adhesion (16). A higher total protein loading level was also used to determine if there were more protein spots present in the Avicel-induced secretomes of C. hydrothermalis, C. kristjanssonii, C. lactoaceticus, C. owensensis, and C. saccharolyticus (Fig. 4B). While some additional spots were visible using a higher total protein loading level and silver staining in all Caldicellulosiruptor species tested, only C. lactoaceticus appeared to have significantly more higher-molecular-weight protein spots visible (Fig. 4B).
Analysis of xylose-induced secretomes also demonstrated that the S-layer protein is also a predominant feature of the C. saccharolyticus secretome (Fig. 4C). Why an S-layer domain protein is the prominent feature of the C. saccharolyticus Avicel-induced (Fig. 4A), xylose-induced (Fig. 4C), or glucose-induced (data not shown) secretome is as yet unknown. Notably, in all xylose-induced secretomes a distribution of high- and low-molecular-weight protein spots was observed with the exception of C. saccharolyticus. For the more cellulolytic Caldicellulosiruptor species, their Avicel-induced secretome profiles matched closely their xylose-induced profiles (Fig. 4A and C). However, for C. hydrothermalis, C. kristjanssonii, and C. owensensis, more protein spots were observed in their xylose-induced profiles versus their Avicel-induced profiles (Fig. 4A to C).
Caldicellulosiruptor glycoside hydrolase inventory.
As mentioned above, only two (C. bescii and C. saccharolyticus) of the nine described Caldicellulosiruptor species currently have publically available genome sequences (29, 51). Due to renewed interest in biomass-degrading microbes, five additional genome sequencing projects are under way, three of which are finished (for C. hydrothermalis, C. kronotsyensis, and C. owensensis), to help pinpoint what makes some species more cellulolytic than others in the genus. Draft and finished versions of these genome sequences were examined for GH family diversity.
Previous studies looking into Caldicellulosiruptor diversity have identified glycoside hydrolases, mostly cellulases and xylanases, produced by selected species (4, 19). Here, the biomass deconstruction-related GH inventory, inferred from each draft and finished genome sequence, was examined to provide insights into the observed differential responses to microcrystalline cellulose. Based on the differential responses of each Caldicellulosiruptor species to microcrystalline cellulose, the GHs potentially involved in cellulose hydrolysis were cataloged according to CAZy GH family (10; http://www.cazy.org). Of the 11 GH families described in the literature as being endo- or exo-acting cellulases (10, 53; see the CAZy database for updates), 4 are present in some, but not all, Caldicellulosiruptor species (GH5, GH9, GH44, and GH48) (Table 2). All Caldicellulosiruptor species have at least one GH5 enzyme (potentially an endo-acting cellulase [10]) present in their genome, supporting the previously mentioned observation that all species are able to hydrolyze CMC. It is interesting that the three Caldicellulosiruptor species that did not grow well on microcrystalline cellulose (Fig. 3) and had less diverse Avicel-induced secretome profiles (Fig. 4A and B, C. hydrothermalis, C. kristjanssonii, and C. owensensis) and were missing representatives from GH9, GH44, and GH48 (Table 2, C. hydrothermalis and C. owensensis) or GH48 (Table 2, C. kristjanssonii). Both GH9 and GH48 are part of the multidomain cellulase enzyme CelA in C. bescii and C. saccharolyticus (49, 58). When catalytic activity was characterized for CelA from C. bescii, the C-terminal portion of the enzyme containing the GH48 domain was responsible for hydrolysis of microcrystalline cellulose in an exo-acting manner (58). It appears then that missing an enzyme containing a GH48 catalytic domain impedes Caldicellulosiruptor species' ability to hydrolyze and grow on crystalline cellulose. In addition to the catalytic portion of cellulase enzymes, noncatalytic CBMs involved in binding to biomass were also cataloged according to established CAZy families (7, 10). A particular crystalline cellulose-binding CBM family, CBM3, is present in multidomain enzymes from C. bescii, C. kristjanssonii, C. kronotskyensis, and C. saccharolyticus (Table 3). One of these multidomain enzymes, CelA, has been cloned and characterized in both C. saccharolyticus and C. bescii, and in the case of C. bescii it has been shown to have activity on crystalline cellulose (49, 58). In the case of a free cellulase from Clostridium thermocellum, CelI, the presence of a C-terminal CBM3 module was determined to be essential for crystalline cellulose hydrolysis (20). The absence of CBM3 from the genomes of C. hydrothermalis and C. owensensis, which also lack exo-acting cellulases, indicates that the CBM3 family may be an additional determinant for a Caldicellulosiruptor species' ability to hydrolyze crystalline cellulose.
TABLE 2.
Biomass deconstruction-related GH families present in selected Caldicellulosiruptor spp.
| Organism | Presence of indicated GH familya able to hydrolyze: |
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellulose |
Xylan |
Mannan |
Xyloglucan |
Pectin |
||||||||||||||||||||||||||||||||
| 1 | 3 | 5 | 9 | 44 | 48 | 94 | 5 | 10 | 11 | 39 | 43 | 51 | 67 | 2 | 5 | 26 | 36 | 1 | 3 | 5 | 16 | 29 | 31 | 35 | 44 | 74 | 95 | 28 | 78 | 88 | 105 | P1b | P3b | P9b | P11b | |
| C. bescii | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||
| C. hydrothermalis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||||||
| C. kristjanssonii | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||
| C. kronotskyensis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| C. lactoaceticus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||
| C. owensensis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||||||
| C. saccharolyticus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||
For each polysaccharide, the GH families (numbers listed beneath each polysaccharide) that hydrolyze the polysaccharide (indicated by bold symbols) or the resulting products (shown by regular, nonbold symbols) are listed. The GH and polysaccharide lyase family numbers, as designated by the CaZY database (http://www.cazy.org) are listed beneath each polysaccharide.
P, polysaccharide lyase family, as designated in the CaZY database.
TABLE 3.
Selected CBM families that bind to biomass
| Organism | Presence of indicated CBM family/substrate utilizeda |
|||||||
|---|---|---|---|---|---|---|---|---|
| CBM3/cellulose | CBM6/xylan β-glucan | CBM9/xylan | CBM22/xylan | CBM27/mannan | CBM28/cellulose β-glucan | CBM36/xylan | CBM54/xylan | |
| C. bescii | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| C. hydrothermalis | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| C. kristjanssonii | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| C. kronotskyensis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| C. lactoaceticus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| C. owensensis | ✓ | ✓ | ✓ | |||||
| C. saccharolyticus | ✓ | ✓ | ✓ | ✓ | ✓ | |||
CBM families are noted by their family number, as designated in the CaZY database, along with the substrate to which they have been reported to bind.
Other glycoside hydrolases potentially involved in biomass deconstruction were also identified, including those involved in hemicellulose (xylanases, mannanases, and xyloglucanases) and pectin hydrolysis. The genomes of the Caldicellulsiruptor species sequenced collectively contain four out of six described GH families that hydrolyze the β-1,4-xyloside linkages of xylan, three out of four described GH families that hydrolyze the β-1,4-mannoside linkages of mannan, and four out of five described xyloglucanase GH families that hydrolyze the β-1,4-glucan linkages (Table 2) (10). In addition, many different CBM families capable of binding to xylan (Table 3, CBM6, -9, -22, and -36) are present in all Caldicellulosiruptor species, and in the case of C. hydrothermalis and C. lactoaceticus a CBM family (CBM 27) involved in binding to mannan is present (Table 3). All Caldicellulosiruptor species sequenced can hydrolyze the backbone of all three major types of hemicellulose (Table 2). It is interesting that for the above-mentioned cellulase and hemicellulase GH families, only C. kronotskyensis posseses all families collectively present in the genus Caldicellulosiruptor. Having such a wealth of biomass deconstruction-related GH families present in the genome of C. kronotskyensis indicates that it is well suited for CBP, especially when a biomass feedstock contains a range of heteropolysaccharides. In addition, all Caldicellulosiruptor species produce GHs and polysaccharide lyases (PL) capable of degrading galacturonan and rhamnogalacturonan, the two major components of pectin (9). Overall, for hemicellulose- and pectin-degrading enzymes, no two Caldicellulosiruptor species have the same GH/PL profile, indicating that each has evolved a specific strategy for handling complex biomass hydrolysis. This raises the prospect for exploiting this nonredundant biomass-degrading capability in Caldicellulosiruptor species communities for CPB. Additionally, with the discovery of novel multidomain GH enzymes in the draft genome sequences, additional efforts at isolating yet-to-be-discovered Caldicellulosiruptor species are warranted.
Summary.
Although 16S rRNA phylogeny suggests that the members of the genus Caldicellulosiruptor are closely related, experimental data using M-CGH indicated that there is a great deal of genetic diversity on a whole-genome scale across the genus. In fact, the use of M-CGH can be a useful tool for identifying new isolates of Caldicellulosiruptor species. With additional whole-genome sequences becoming available, a genus-wide oligonucleotide microarray could also be designed to quickly identify the presence of desirable genes encoding GHs and CBMs in novel Caldicellulosiruptor isolates (40). When the growth physiologies of Caldicellulosiruptor species were compared, all species were capable of robust growth on pretreated switchgrass biomass, an important characteristic for CBP. Interestingly, all species showed differential abilities to grow on microcrystalline cellulose, the main polysaccharide component of biomass. In addition, differential secretome profiles indicated that each Caldicellulosiruptor species uses a different strategy for interactions with complex polysaccharides and simple sugars in their environment. The production of copious amounts of an S-layer protein from one of the cellulolytic members of the genus also hints at possible noncellulosomal methods of substrate interaction along the cell surface. When preliminary draft genome sequences were searched for enzymes involved in biomass hydrolysis, some of the differential responses to crystalline cellulose appeared to be the result of the presence or lack of GH enzymes involved in cellulose hydrolysis. Cataloging biomass degradation-related GH families from the genus Caldicellulosiruptor provided a preliminary look into the genomic diversity of the genus Caldicellulosiruptor. Further analysis of the roles of biomass-degrading related GH families in the sequenced Caldicellulosiruptor genomes will also help to pair synergistic partners for multispecies CBP. Additional comparative genomics studies are under way that aim to look more comprehensively at the most thermophilic cellulose-hydrolyzing genus discovered to date.
Acknowledgments
We gratefully acknowledge Lynne Goodwin at the DOE Joint Genome Institute and Loren Hauser, Bob Cottingham, and David Graham at ORNL for providing information on the genome sequences of C. hydrothermalis, C. kristjanssonii, C. kronotskyensis, C. lactoaceticus, and C. owensensis. We also thank Owen Jacobs and Dhaval Mistry for technical assistance in obtaining physiological data. FMC kindly donated Avicel PH-101 used in the physiological studies.
This research was supported by the Bioenergy Science Center, a U.S. DOE Bioenergy Research Center funded by the Office of Biological and Environmental Research.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, M. W., L. E. Driskill, W. Callen, M. A. Snead, E. J. Mathur, and R. M. Kelly. 1999. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes beta-1,4 bonds in mixed-linkage (1-→3),(1-→4)-beta-d-glucans and cellulose. J. Bacteriol. 181:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 4.Bergquist, P., D. G. Moreland, D. M. Daniel, V. S. J. Te'o, J. S. David, and W. M. Hugh. 1999. Molecular diversity of thermophilic cellulolytic and hemicellulolytic bacteria. FEMS Microbiol. Ecol. 28:99-110. [Google Scholar]
- 5.Bielen, A. A. M., K. Willquist, J. Engman, J. van der Oost, E. W. J. van Niel, and W. M. K. Servé. 2010. Pyrophosphate as a central energy carrier in the hydrogen-producing extremely thermophilic Caldicellulosiruptor saccharolyticus. FEMS Microbiol. Lett. 307:48-54. [DOI] [PubMed] [Google Scholar]
- 6.Blumer-Schuette, S. E., I. Kataeva, J. Westpheling, M. W. Adams, and R. M. Kelly. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210-217. [DOI] [PubMed] [Google Scholar]
- 7.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredholt, S., J. Sonne-Hansen, P. Nielsen, I. M. Mathrani, and B. K. Ahring. 1999. Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int. J. Syst. Bacteriol. 49:991-996. [DOI] [PubMed] [Google Scholar]
- 9.Caffall, K. H., and D. Mohnen. 2009. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344:1879-1900. [DOI] [PubMed] [Google Scholar]
- 10.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter, B., G. Wu, M. J. Woodward, and M. F. Anjum. 2008. A process for analysis of microarray comparative genomics hybridisation studies for bacterial genomes. BMC Genomics 9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vos, W. M., S. W. Kengen, W. G. Voorhorst, and J. van der Oost. 1998. Sugar utilization and its control in hyperthermophiles. Extremophiles 2:201-205. [DOI] [PubMed] [Google Scholar]
- 14.Driskill, L. E., M. W. Bauer, and R. M. Kelly. 1999. Synergistic interactions among beta-laminarinase, beta-1,4-glucanase, and beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus during hydrolysis of beta-1,4-, beta-1,3-, and mixed-linked polysaccharides. Biotechnol. Bioeng. 66:51-60. [PubMed] [Google Scholar]
- 15.Duffaud, G. D., C. M. McCutchen, P. Leduc, K. N. Parker, and R. M. Kelly. 1997. Purification and characterization of extremely thermostable beta-mannanase, beta-mannosidase, and alpha-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl. Environ. Microbiol. 63:169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelhardt, H. 2007. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 160:115-124. [DOI] [PubMed] [Google Scholar]
- 17.Forquin, M. P., H. Duvergey, C. Proux, V. Loux, J. Mounier, S. Landaud, J. Y. Coppee, J. F. Gibrat, P. Bonnarme, I. Martin-Verstraete, and T. Vallaeys. 2009. Identification of Brevibacteriaceae by multilocus sequence typing and comparative genomic hybridization analyses. Appl. Environ. Microbiol. 75:6406-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geslin, C., M. Le Romancer, G. Erauso, M. Gaillard, G. Perrot, and D. Prieur. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi.” J. Bacteriol. 185:3888-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs, M. D., R. A. Reeves, G. K. Farrington, P. Anderson, D. P. Williams, and P. L. Bergquist. 2000. Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40:333-340. [DOI] [PubMed] [Google Scholar]
- 20.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, R. S., and B. Gao. 2009. Phylogenomic analyses of clostridia and identification of novel protein signatures that are specific to the genus Clostridium sensu stricto (cluster I). Int. J. Syst. Evol. Microbiol. 59:285-294. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton-Brehm, S. D., J. J. Mosher, T. Vishnivetskaya, M. Podar, S. Carroll, S. Allman, T. J. Phelps, M. Keller, and J. G. Elkins. 2010. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heer, D., and U. Sauer. 2008. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb. Biotechnol. 1:497-506. [DOI] [PMC free article] [PubMed]
- 24.Himmel, M. E., S. Y. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. [DOI] [PubMed] [Google Scholar]
- 25.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, C. Y., B. K. Patel, R. A. Mah, and L. Baresi. 1998. Caldicellulosiruptor owensensis sp. nov., an anaerobic, extremely thermophilic, xylanolytic bacterium. Int. J. Syst. Bacteriol. 48:91-97. [DOI] [PubMed] [Google Scholar]
- 27.Hunter, S., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bork, U. Das, L. Daugherty, L. Duquenne, R. D. Finn, J. Gough, D. Haft, N. Hulo, D. Kahn, E. Kelly, A. Laugraud, I. Letunic, D. Lonsdale, R. Lopez, M. Madera, J. Maslen, C. McAnulla, J. McDowall, J. Mistry, A. Mitchell, N. Mulder, D. Natale, C. Orengo, A. F. Quinn, J. D. Selengut, C. J. A. Sigrist, M. Thimma, P. D. Thomas, F. Valentin, D. Wilson, C. H. Wu, and C. Yeats. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res. 37:D211-D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadar, Z., T. de Vrije, G. E. van Noorden, M. A. Budde, Z. Szengyel, K. Reczey, and P. A. Claassen. 2004. Yields from glucose, xylose, and paper sludge hydrolysate during hydrogen production by the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl. Biochem. Biotechnol. 113-116:497-508. [DOI] [PubMed] [Google Scholar]
- 29.Kataeva, I. A., S. J. Yang, P. Dam, F. L. Poole II, Y. Yin, F. Zhou, W. C. Chou, Y. Xu, L. Goodwin, D. R. Sims, J. C. Detter, L. J. Hauser, J. Westpheling, and M. W. Adams. 2009. Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J. Bacteriol. 191:3760-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kublanov, I. V., A. A. Perevalova, G. B. Slobodkina, A. V. Lebedinsky, S. K. Bidzhieva, T. V. Kolganova, E. N. Kaliberda, L. D. Rumsh, T. Haertle, and E. A. Bonch-Osmolovskaya. 2009. Biodiversity of thermophilic prokaryotes with hydrolytic activities in hot springs of Uzon Caldera, Kamchatka (Russia). Appl. Environ. Microbiol. 75:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, D. L. 2010. Functional genomics analysis of extremely thermophilic fermentative microorganisms from the archaeal genus Pyrococcus and bacterial genus Caldicellulosiruptor. Ph.D. thesis. North Carolina State University, Raleigh, NC.
- 32.Liebl, W., C. Winterhalter, W. Baumeister, M. Armbrecht, and M. Valdez. 2008. Xylanase attachment to the cell wall of the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 190:1350-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig, W., K.-H. Schleifer, and W. Whitman. 2009. Revised road map to the phylum Firmicutes, p. 1-14. In P. de Vos, G. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, K.-H. Schleifer, and W. B. Whitman (ed.), Bergey's manual of systematic bacteriology, vol. 3. Springer, New York, NY. [Google Scholar]
- 34.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madding, L. S., J. K. Michel, K. R. Shockley, S. B. Conners, K. L. Epting, M. R. Johnson, and R. M. Kelly. 2007. Role of the beta1 subunit in the function and stability of the 20S proteasome in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 189:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miroshnichenko, M. L., I. V. Kublanov, N. A. Kostrikina, T. P. Tourova, T. V. Kolganova, N. K. Birkeland, and E. A. Bonch-Osmolovskaya. 2008. Caldicellulosiruptor kronotskyensis sp. nov. and Caldicellulosiruptor hydrothermalis sp. nov., two extremely thermophilic, cellulolytic, anaerobic bacteria from Kamchatka thermal springs. Int. J. Syst. Evol. Microbiol. 58:1492-1496. [DOI] [PubMed] [Google Scholar]
- 37.Mladenovska, Z., I. M. Mathrani, B. K. Ahring, and B. K. Ahring. 1995. Isolation and characterization of Caldicellulosiruptor lactoaceticus sp. nov., an extremely thermophilic, cellulolytic, anaerobic bacterium. Arch. Microbiol. 163:223-230. [Google Scholar]
- 38.Morris, D. D., M. D. Gibbs, M. Ford, J. Thomas, and P. L. Bergquist. 1999. Family 10 and 11 xylanase genes from Caldicellulosiruptor sp. strain Rt69B.1. Extremophiles 3:103-111. [DOI] [PubMed] [Google Scholar]
- 39.Ng, T. K., T. K. Weimer, and J. G. Zeikus. 1977. Cellulolytic and physiological properties of Clostridium thermocellum. Arch. Microbiol. 114:1-7. [DOI] [PubMed] [Google Scholar]
- 40.Oh, S., D. R. Yoder-Himes, J. Tiedje, J. Park, and K. T. Konstantinidis. 2010. Evaluating the performance of oligonucleotide microarrays for bacterial strains with increasing genetic divergence from the reference strain. Appl. Environ. Microbiol. 76:2980-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onyenwoke, R. U., Y. J. Lee, S. Dabrowski, B. K. Ahring, and J. Wiegel. 2006. Reclassification of Thermoanaerobium acetigenum as Caldicellulosiruptor acetigenus comb. nov. and emendation of the genus description. Int. J. Syst. Evol. Microbiol. 56:1391-1395. [DOI] [PubMed] [Google Scholar]
- 42.Park, S., D. K. Johnson, C. I. Ishizawa, P. A. Parilla, and M. F. Davis. 2009. Measuring the crystallinity index of cellulose by solid state 13C nuclear magnetic resonance. Cellulose 16:641-647. [Google Scholar]
- 43.Pysz, M. A., S. B. Conners, C. I. Montero, K. R. Shockley, M. R. Johnson, D. E. Ward, and R. M. Kelly. 2004. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rainey, F. A., A. M. Donnison, P. H. Janssen, D. Saul, A. Rodrigo, P. L. Bergquist, R. M. Daniel, E. Stackebrandt, and H. W. Morgan. 1994. Description of Caldicellulosiruptor saccharolyticus gen. nov., sp. nov: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol. Lett. 120:263-266. [DOI] [PubMed] [Google Scholar]
- 45.Rainey, F. A., and E. Stackebrandt. 1993. 16S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol. Lett. 113:125-128. [DOI] [PubMed] [Google Scholar]
- 46.Raman, B., C. Pan, G. B. Hurst, M. Rodriguez, Jr., C. K. McKeown, P. K. Lankford, N. F. Samatova, and J. R. Mielenz. 2009. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 48.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 49.Te'o, V. S., D. J. Saul, and P. L. Bergquist. 1995. celA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl. Microbiol. Biotechnol. 43:291-296. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van de Werken, H. J., M. R. Verhaart, A. L. VanFossen, K. Willquist, D. L. Lewis, J. D. Nichols, H. P. Goorissen, E. F. Mongodin, K. E. Nelson, E. W. van Niel, A. J. Stams, D. E. Ward, W. M. de Vos, J. van der Oost, R. M. Kelly, and S. W. Kengen. 2008. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 74:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.VanFossen, A. L., M. R. Verhaart, S. M. Kengen, and R. M. Kelly. 2009. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl. Environ. Microbiol. 75:7718-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vlasenko, E., M. Schulein, J. Cherry, and F. Xu. 2010. Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour. Technol. 101:2405-2411. [DOI] [PubMed] [Google Scholar]
- 54.Willquist, K., and E. W. van Niel. 2010. Lactate formation in Caldicellulosiruptor saccharolyticus is regulated by the energy carriers pyrophosphate and ATP. Metab. Eng. 12:282-290. [DOI] [PubMed] [Google Scholar]
- 55.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 56.Yang, S. J., I. Kataeva, J. Wiegel, Y. Yin, P. Dam, Y. Xu, J. Westpheling, and M. W. Adams. 2010. Classification of ‘Anaerocellum thermophilum’ strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int. J. Syst. Evol. Microbiol. 60:2011-2015. [DOI] [PubMed] [Google Scholar]
- 57.Zeidan, A. A., and E. W. J. Van Niel. 2009. Developing a thermophilic hydrogen-producing co-culture for efficient utilization of mixed sugars. Int. J. Hydrogen Energy 34:4524-4528. [Google Scholar]
- 58.Zverlov, V., S. Mahr, K. Riedel, and K. Bronnenmeier. 1998. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 144:457-465. [DOI] [PubMed] [Google Scholar]