Abstract
We report the screening of 16,000 synthetic compounds for induction and inhibition of quorum sensing in a Pseudomonas putida N-acylated l-homoserine lactone (AHL) sensor strain engineered with the LasR transcriptional activator. LasR controls virulence gene expression in the opportunistic pathogen Pseudomonas aeruginosa and is of significant interest as a therapeutic target. Nine compounds that inhibit and 14 compounds that induce LasR activity were identified in our high-throughput screen.
Quorum sensing (QS) is a mode of microbial communication that is essential to controlling activities associated with pathogenesis, symbiosis, and interactions of key ecological importance (3, 12). Many proteobacteria exhibit QS behavior through the exchange of N-acylated l-homoserine lactone (AHL) signaling molecules in a cell-density-dependent manner to regulate gene expression (15). Archetypal AHL-mediated QS systems consist of LuxR-type transcriptional regulators and LuxI-type synthases that produce AHLs. Members of the LuxR family interact with a cognate AHL in a concentration-dependent manner to regulate the expression of genes.
The role of QS in Pseudomonas aeruginosa pathogenicity has been demonstrated by mutant analysis in multiple host-pathogen systems (6, 10). The linkage between QS and the production of virulence factors in P. aeruginosa predicts that inhibition of AHL-mediated gene expression could render these bacteria nonpathogenic (5). Studies on the rational design of QS inhibitors have focused on modifying the basic structure of the AHL (4, 9). High-throughput screening (HTS) of small-molecule libraries is another approach for the identification of compounds that have novel chemical scaffolds for both the development of chemical probes and eventual therapeutic drug design. Previous HTS of a sizeable corporate compound library identified two inhibitors structurally similar to AHLs and one nonnative inducer of LasR (7, 8). The inducer, triphenyl derivative TP-1, was structurally unrelated to AHLs and exhibited higher stability, and thus more prolonged activity, than N-(3-oxo-dodecanoyl)-l-homoserine lactone (3-oxo-C12-HSL). This result serves to validate the HTS approach for the identification of nonnative modulators of QS. Here, we report complementary HTS efforts for the identification of novel small molecules that are modulators of the LasR QS circuit and structurally dissimilar to native AHLs.
Small molecule modulators of QS are frequently identified using bacterial reporter strains (1, 11). We used Pseudomonas putida strain F117(pKRC12), an AHL-deficient strain that has been engineered to produce green fluorescent protein (GFP) upon activation of LasR by N-(3-oxo-decanoyl)-l-homoserine lactone (3-oxo-C10-HSL) and 3-oxo-C12-HSL (13, 14). Our HTS approach used 3-oxo-C10-HSL as the inducer of LasR-mediated signaling to identify compounds that either inhibit or induce GFP expression. We speculated that 3-oxo-C10-HSL binds LasR with weaker affinity than 3-oxo-C12-HSL, which would enable the simultaneous identification of compounds that either reduce or increase the activity of LasR. Due to its ready availability and relative structural diversity, we selected the 16,000-member Chembridge DIVERSet (ChemBridge Corporation, San Diego, CA) collection for our HTS. This library was screened at a final compound concentration of 3.3 μM. Bacterial cells were added to the compounds approximately 30 min prior to the addition of 3-oxo-C10-HSL at a final concentration of 100 nM, the 50% effective concentration (EC50) of the compound in the P. putida F117(pKRC12) AHL reporter strain. Additional information regarding reporter assays and bacterial strains is provided in the supplemental material. Compounds that reduced or increased the fluorescence by a factor ≥3 standard deviations from the mean values for the control, 3-oxo-C10-HSL, were chosen for further analysis and subjected to additional screening.
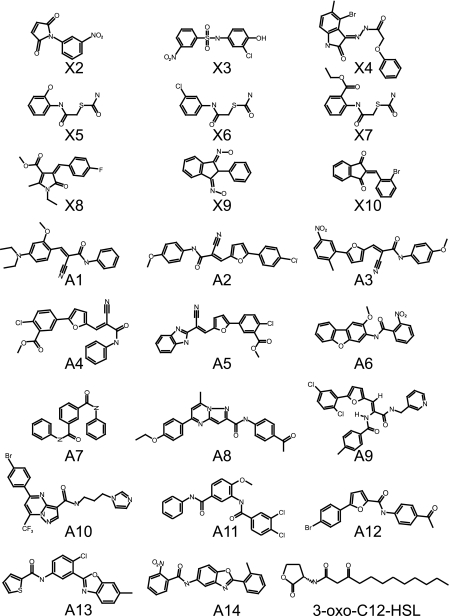
We identified nine antagonists (X2 to X10) and 14 agonists (A1 to A14) of LasR-mediated QS (Fig. 1). Antagonists designated X5, X6, and X7 share structural similarities and contain a thiocarbamate linked to an aniline ring through an amide bond with various functional group substitutions. In addition, some of the agonists share structural similarities and comprise two closely related families of compounds. Agonists A7 and A11 both contain two amide bonds that constitute a triphenyl backbone that displays varied functionality on the phenyl rings. Agonists A2, A3, and A4 share a scaffold with a furanyl ring, cyano group, and amide bond. Compounds A1 and A5 are also structurally similar, with the exception of the furanyl group from A1 and the lack of an amide bond in A5. Overall, the identification of antagonists and agonists that share structural similarities is a strong indicator that the HTS was robust and demanded some degree of structural specificity. The antagonists identified by HTS differed structurally from previously described antagonists resembling 3-oxo-C12-HSL (Fig. 1).
FIG. 1.
Structures of the antagonists (X2 to X10) and agonists (A1 to A14) identified in this study. Also shown is the structure of the native ligand of LasR, N-(3-oxo-dodecanoyl)-l-homoserine lactone (3-oxo-C12-HSL).
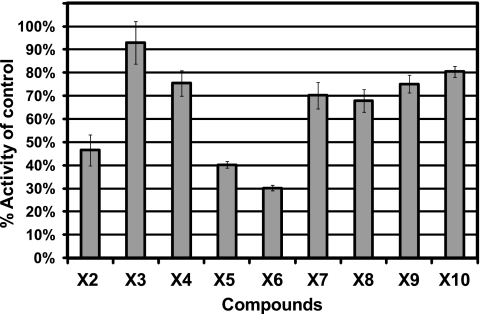
To evaluate the activity of antagonists against the natural AHL ligand of LasR, 3-oxo-C12-HSL was used as the inducer in antagonism assays (Fig. 2). Three of the antagonists inhibited biosensor activity by more than 50% at a concentration of 20 μM against 3-oxo-C12-HSL (at 50 nM). Notably, bacterial growth was not affected by the addition of the antagonists at the concentration tested (data not shown). When the 3-oxo-C12-HSL concentration was increased 20-fold, inhibition was not detected, suggesting competitive interaction between the antagonists and 3-oxo-C12-HSL for LasR binding (see Fig. S1 in the supplemental material). Loss of inhibitory activity at the higher concentration suggests that the antagonists are outcompeted when 3-oxo-C12-HSL saturates the receptor and provides further evidence that the antagonists interact specifically with LasR.
FIG. 2.
Antagonism screening data. Shown is the activity of P. putida F117(pKRC12) after 4 h when induced by 50 nM 3-oxo-C12-HSL and challenged with a 20 μM concentration of each antagonist, X2 to X10. The values reported are the mean specific fluorescence activity of each compound normalized as a percentage of the control for four replicates. Error bars indicate the standard deviation.
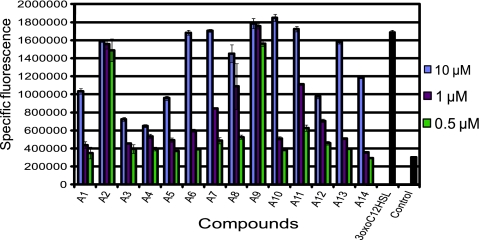
To further characterize the agonists, we measured activity at various concentrations. At a final concentration of 10 μM, all 14 of the agonists activated expression of the biosensor in the absence of exogenous AHL (Fig. 3). Moreover, the activity of agonists A2 and A9 was comparable to natural ligand 3-oxo-C12-HSL at 10× the concentration (500 nM). Interestingly, these two compounds were structurally less similar than other agonists identified in this study (such as A1 to A5 and A7 and A11).
FIG. 3.
Agonism screening data. Shown are the activities of compounds A1 to A14 compared to that of 3-oxo-C12-HSL at final concentrations of 0.5 μM (green bars), 1 μM (red bars), and 10 μM (blue bars) to determine their ability to activate LasR-regulated gene expression in P. putida F117(pKRC12). 3-Oxo-C12-HSL (50 nM) was included for comparison of agonist activities to the natural ligand. Dimethyl sulfoxide (DMSO) was included as a negative control. Specific fluorescence was determined after 4 h of growth, and the values reported are the means of three replicates. Error bars indicate the standard deviation.
The specificities of the antagonists and agonists for LasR and additional LuxR-type proteins were evaluated in biosensor strains with different LuxR-type proteins and host backgrounds. The agonists were active and exhibited the same relative trend of activity in P. putida F117(pKRC12) and Escherichia coli DH10B(pKRC12), which both contain the LasR transcriptional regulator. These results suggest that agonist activity is independent of host background. Agonists were not active in the modified ΔlasR reporter strain, P. putida F117(pKRC12ΔlasR), indicating that the agonists interact directly with LasR. As expected, neither the agonists nor the antagonists were active in AHL reporters that detect short-chain AHLs with different LuxR-type transcriptional regulators, such as LuxR, TraR, and CviR, suggesting that these nonnative ligands are specific for LasR.
To understand how nonnative agonists and antagonists could be accommodated by the ligand binding site, we performed computational docking studies (see the supplemental materials) using the reported X-ray crystal structure of the N-terminal domain of LasR (Protein Data Bank code 2UV0) (2). Modeling of the thiocarbamate antagonists showed overall agreement between activity and the ability to mimic the natural ligand. In contrast, the most active agonist acts on LasR either by binding to an allosteric site of LasR or by significantly changing the conformation of the binding pocket of the natural ligand and may provide a strategy for design of new compounds that modulate the activity of LasR through alternative interactions.
In summary, we have identified nine nonnative antagonists and 14 nonnative agonists of LasR-mediated gene regulation via HTS. Notably, these compounds are structurally distinct from other known agonists and antagonists, hold promise as new scaffolds for the development of chemical tools to probe the mechanisms of QS in P. aeruginosa, and could, with further development, provide a pathway for the design of novel antivirulence agents.
Supplementary Material
Acknowledgments
We are grateful to Ronald Binder for generously providing synthetic AHLs and Leo Eberl, Michael Givskov, Max Teplitski, and Barbara Iglewski for providing plasmids and strains. We are also grateful for the technical assistance and services provided by the Keck-UWCCC Small Molecule Screening Facility.
Research in the Handelsman lab is supported in part by the NIH, Howard Hughes Medical Institute, and the USDA-Hatch Program. Research in the Blackwell lab is supported in part by the NIH (AI063326), the Burroughs Welcome Fund, Johnson & Johnson, DuPont, and the Greater Milwaukee Foundation. H.E.B. is an Alfred P. Sloan Foundation Fellow. G.D.G was supported by an American Chemical Society Division of Medicinal Chemistry predoctoral fellowship.
Footnotes
Published ahead of print on 8 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottomley, M. J., E. Muraglia, R. Bazzo, and A. Carfi. 2007. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 282:13592-13600. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 4.Geske, G. D., J. C. O'Neill, and H. E. Blackwell. 2008. Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria. Chem. Soc. Rev. 37:1432-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hentzer, M., and M. Givskov. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Invest. 112:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lesprit, P., F. Faurisson, O. Join-Lambert, F. Roudot-Thoraval, M. Foglino, C. Vissuzaine, and C. Carbon. 2003. Role of the quorum-sensing system in experimental pneumonia due to Pseudomonas aeruginosa in rats. Am. J. Respir. Crit. Care Med. 167:1478-1482. [DOI] [PubMed] [Google Scholar]
- 7.Muh, U., B. J. Hare, B. A. Duerkop, M. Schuster, B. L. Hanzelka, R. Heim, E. R. Olson, and E. P. Greenberg. 2006. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc. Natl. Acad. Sci. U. S. A. 103:16948-16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muh, U., M. Schuster, R. Heim, A. Singh, E. R. Olson, and E. P. Greenberg. 2006. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 50:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni, N., M. Li, J. Wang, and B. Wang. 2009. Inhibitors and antagonists of bacterial quorum sensing. Med. Res. Rev. 29:65-124. [DOI] [PubMed] [Google Scholar]
- 10.Pearson, J. P., M. Feldman, B. H. Iglewski, and A. Prince. 2000. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 68:4331-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen, T. B., and M. Givskov. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296:149-161. [DOI] [PubMed] [Google Scholar]
- 12.Rice, S. A., M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Bacterial signals and antagonists: the interaction between bacteria and higher organisms. J. Mol. Microbiol. Biotechnol. 1:23-31. [PubMed] [Google Scholar]
- 13.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Hoiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 14.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarwood, J. M., E. M. Volper, and E. P. Greenberg. 2005. Delays in Pseudomonas aeruginosa quorum-controlled gene expression are conditional. Proc. Natl. Acad. Sci. U. S. A. 102:9008-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.