Abstract
Bovine isolates of Streptococcus agalactiae (n = 76), Streptococcus dysgalactiae subsp. dysgalactiae (n = 32), and Streptococcus uberis (n = 101) were analyzed for the presence of different integrative and conjugative elements (ICEs) and their association with macrolide, lincosamide, and tetracycline resistance. The diversity of the isolates included in this study was demonstrated by multilocus sequence typing for S. agalactiae and pulsed-field gel electrophoresis for S. dysgalactiae and S. uberis. Most of the erythromycin-resistant strains carry an ermB gene. Five strains of S. uberis that are resistant to lincomycin but susceptible to erythromycin carry the lin(B) gene, and one has both linB and lnuD genes. In contrast to S. uberis, most of the S. agalactiae and S. dysgalactiae tetracycline-resistant isolates carry a tet(M) gene. A tet(S) gene was also detected in the three species. A Tn916-related element was detected in 30 to 50% of the tetracycline-resistant strains in the three species. Tetracycline resistance was successfully transferred by conjugation to an S. agalactiae strain. Most of the isolates carry an ICE integrated in the rplL gene. In addition, half of the S. agalactiae isolates have an ICE integrated in a tRNA lysine (tRNALys) gene. Such an element is also present in 20% of the isolates of S. dysgalactiae and S. uberis. A circular form of these ICEs was detected in all of the isolates tested, indicating that these genetic elements are mobile. These ICEs could thus also be a vehicle for horizontal gene transfer between streptococci of animal and/or human origin.
Mobile genetic elements (MGE) play a major role in horizontal gene transfer (HGT) in bacteria (27). Among them, integrative and conjugative elements (ICEs) (also called conjugative transposons) are self-transmissible elements that are widespread in bacteria and are characterized by both integrative and conjugative properties (14, 15). In addition to the genes involved in their mobility, regulation, or maintenance, they carry other determinants, in particular antibiotic resistance genes. Many ICEs have been described in streptococci, especially in Streptococcus pneumoniae and Streptococcus pyogenes (6, 20). Most of them belong to the Tn916-Tn1545 family and carry the widely disseminated tetracycline-resistance determinant tet(M), as well as additional resistance determinants such as the erythromycin resistance genes erm(B) or mef(A) (52). The erm(B) gene encodes a methylase that generates a posttranscriptional methylation of the 23S rRNA (33), consequently resulting in cross-resistance to macrolides, lincosamides, and streptogramin B antibiotics (MLSB phenotype). In contrast, the mef(A) gene encodes a drug efflux pump affecting only 14- and 15-member macrolides (M phenotype) (52).
Recently, the analysis of the eight sequenced genomes in the Streptococcus agalactiae species revealed two novel groups of ICEs: one group integrated in the rplL gene, which encodes the 50S ribosomal subunit protein L7/L12 (ICE_Sa2603_rplL of S. agalactiae 2603 V/R and related elements) and the other in a tRNA lysine (tRNALys) gene (ICE_Sa2603_tRNALys of S. agalactiae 2603 V/R and related elements) (11). Both carry a transposon with a mer operon encoding a mercuric reductase and a transport system for mercury, which constitutes the principal mechanism of bacterial resistance to this heavy metal (3). Additionally, members of the first group exhibit a conjugation module related to one of the large conjugative transposons, Tn5252, described in S. pneumoniae, which carries a chloramphenicol resistance gene and forms a composite element called Tn5253 when aggregated to a Tn916-related element (20). The elements of the first group also carry genes related to orf11 to -13 of ICE Tn5252, which are also found on the mega element, a genetic element which carries genes encoding Mef(E) and Msr(D) macrolide efflux pumps (52). Consequently, these two groups of ICEs are possible vehicles for antibiotic and heavy metal resistance genes among streptococci.
S. agalactiae colonizes and infects both humans and animals. It is in particular one of the pathogens regularly involved in bovine mastitis along with S. dysgalactiae subsp. dysgalactiae and Streptococcus uberis (8). These two species are found in the environment of dairy cattle and as such are more difficult to eradicate than the obligate parasite S. agalactiae (10). Exchanges of DNA likely occur in their common niche, and ICEs could be a vehicle for these transfers.
In this work, we looked for the presence of three ICE groups likely associated with antibiotic and heavy metal resistance—namely, Tn916 and elements related to ICE_Sa2603_rplL and ICE_Sa2603_tRNALys—in a collection of S. agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis strains isolated from bovine mastitis. These isolates were also examined for the presence of tetracycline, macrolide, lincosamide, and heavy metal resistance genes. Finally, we tested the mobility of these ICEs to see if they can mediate gene transfer between these different bacterial species.
MATERIALS AND METHODS
Bacterial isolates.
A total of 209 strains (101 S. uberis, 76 S. agalactiae, and 32 S. dysgalactiae subsp. dysgalactiae strains) were included in this study. S. dysgalactiae and S. uberis strains were selected from a larger collection for their resistance to erythromycin (4 S. dysgalactiae isolates and 75 S. uberis isolates) or tetracycline (32 S. dysgalactiae isolates and 62 S. uberis isolates) (see Table S1 in the supplemental material). They were isolated from milk samples collected from cows with clinical or subclinical mastitis between 1984 and 2008 in 22 different geographic areas in France. Isolates were identified by conventional methods: Gram-staining cell morphology, colony morphology and hemolysis on Columbia agar supplemented with 5% defibrinated horse blood (bioMérieux), catalase activity, and API20 Strep tests (bioMérieux, Marcy l'Etoile, France). Identification of S. uberis was confirmed by PCR using species-specific primers (45) (see Table S2 in the supplemental material).
Antimicrobial susceptibility testing.
Bacteria were tested for their susceptibility to macrolides, lincosamide, and tetracycline by the disk diffusion method on Mueller-Hinton agar as recommended by the CA-SFM (Antibiogram Committee of the French Society of Microbiology). The following breakpoints were used: for erythromycin, susceptible, >22 mm, and resistant, <19 mm; for spiramycin, susceptible, ≥18 mm, and resistant, <14 mm; for lincomycin, susceptible, ≥21 mm, and resistant, <17 mm; and for tetracycline, susceptible, ≥19 mm, and resistant, <17 mm (17). The constitutive MLSB phenotype was characterized by the cross-resistance to macrolides and lincomycin, whereas the inducible phenotype was determined by the D-shaped induction zone of lincomycin and spiramycin resistance visible on plates (47). Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 29212 were used as quality control strains. Antibiotic discs were purchased from MAST Diagnostic (Amiens, France).
MLST of S. agalactiae strains.
Multilocus sequence typing (MLST) was performed by sequencing seven housekeeping genes (adhP, pheS, atr, glnA, sdhA, glcK, and tkt) according to Jones et al. (31) (see Table S2 in the supplemental material). Sequence types (ST) were defined using the S. agalactiae MLST website (http://pubmlst.org/sagalactiae/) developed by Man-Suen Chan and Keith Jolley (30). Sequence types not previously described in the database were deposited in the S. agalactiae MLST database, and new ST numbers were assigned. Relationships between isolates were inferred using eBURST v3 (http://eburst.mlst.net), developed by E. Feil and colleagues (26). In MLST, isolates of an expanding founding strain (founding ST) initially have the same allelic profile, but diversification results in the appearance of variants in which one of the MLST loci has changed (6/7 alleles shared defining single-locus variants [SLVs]). Further diversification generates double-locus variants (DLVs) of the founding ST, triple-locus variants, etc. The BURST algorithm identifies these clusters of closely related STs descended from the founding ST (clonal complexes [CCs]) within bacterial populations, infers the founding ST of each clonal complex, and displays the likely pattern of recent evolutionary descent of all STs within the clonal complex from this predicted founder. On the eBURST diagram, clonal complexes are typically radial, with the founding ST at the center linked by lines to all its SLVs, which may themselves be linked to DLVs of the founder, and so on (Fig. 1).
FIG. 1.
eBURSt diagram (26) showing the relationships between the S. agalactiae bovine strains (appearing as black triangles on the figure) analyzed by MLST (31).
Analysis of the S. uberis and S. dysgalactiae strains by pulsed-field gel electrophoresis (PFGE).
Cells from S. dysgalactiae and S. uberis strains were grown at 37°C aerobically with agitation (150 rpm) in brain heart infusion (BHI) broth (Difco) to an A600 of approximately 0.6. Cells from 20 ml of culture were collected by centrifugation at 4,500 × g for 10 min, washed with cold TE-sucrose (25 mM Tris HCl, 25 mM EDTA [pH 8.0], 10% [wt/vol] sucrose), and resuspended in the same solution to reach an A600 of 2.0. Equal volumes of cell suspension and molten 1.5% (wt/vol) low-melting-temperature agarose (Seakem agarose; TEBU, Le Perray-en-Yvelines, France) were mixed and dispensed into molds on ice. Once solidified, the gel blocks were incubated in lysis buffer (TE-sucrose with lysozyme from chicken egg white at 2 mg/ml) at 37°C for 6 h. The lysis buffer was then replaced by ESP buffer (0.5 M EDTA [pH 8.0], 1% [wt/vol] sodium dodecyl sulfate, 2.5 mg/ml of protease from Streptomyces griseus [Sigma]), and the gel blocks were incubated at 50°C overnight. Gels blocks were then incubated in TE buffer (10 mM Tris HCl, 1 mM EDTA [pH 8.0]) for 1 h at 50°C. This step was repeated four times. For long-term storage, blocks were kept at 4°C in this buffer.
For restriction endonuclease digestion of genomic DNA, slices were incubated in a 300-μl volume (containing the appropriate endonuclease buffer) with 60 U SmaI (New England Biolabs, Ozyme, St-Quentin-en-Yvelines, France) and incubated for 3 h at 25°C.
Separation of DNA fragments was achieved by clamped homogeneous electric field (CHEF) electrophoresis using a CHEF-Mapper XA (Bio-Rad Laboratories). Digested samples were electrophoresed through 1% (wt/vol) agarose (Seakem agarose; TEBU) gels in 0.5× Tris-borate-EDTA (TBE). Electrophoresis was carried out at 6 V cm−1 for 20 h (14°C) with a pulse time increasingly linearly from 1 to 20 s, as described by Baseggio et al. (4). SmaI-digested DNA of S. agalactiae strain A909 (50) was used as the molecular size standard in all gels.
PCR for detection of resistance genes.
Multiplex PCR was used to detect simultaneously three macrolide [erm(A), erm(B), and mef(A/E)] as well as two tetracycline [tet(M), tet(O)] resistance determinants in streptococci, as described by Malhotra-Kumar and collaborators (36) (see Table S2 in the supplemental material). PCRs to detect the lnuD and linB genes, which encode a lincosamide nucleotidyltransferase responsible for the inactivation of lincosamides, were performed on S. uberis isolates presenting resistance to lincomycin only, as already published (9, 39). The tet(S) gene was searched for by PCR in tetracycline-resistant strains which do not harbor the other genes which encode ribosomal protection proteins [tet(M) and tet(O)] (see Tables S1 and S2 in the supplemental material). The presence of a mercuric reductase gene, merA, was searched for by PCR using the SAG2023-Fwd and SAG2023-Rev primers (Table S2), designed from the S. agalactiae 2603 V/R genome sequence (GenBank accession no. NC_004116). The sequence of the merA gene is conserved at these positions (130 to 148 and 779 to 796) in other streptococci (S. mitis B6, GenBank accession no. AJ582646; and S. parasanguinis 17910 and 18110, GenBank accession no. AJ 582647 and AJ 582648, respectively); however, since the use of these primers to amplify the merA gene has not been validated, it is possible that existing merA genes have not been amplified in some strains.
PCR and nested PCR for detection of ICEs and their excised circular forms.
PCR was used to detect genes characteristic of the three ICE families: the integrase gene of ICE_Sa2603_rplL (an ICE integrated in the rplL gene of S. agalactiae 2603 V/R), the two integrase genes of ICE_Sa2603_tRNALys (a composite ICE integrated in a tRNA lysine gene of S. agalactiae 2603 V/R), and the relaxase gene (orf20) of Tn916. Analysis of the ICEs found in the sequenced genomes of S. agalactiae (11) indicated that the conjugation module of Tn916 can be associated with different integration modules. We thus chose to use the relaxase gene included in the conjugation module of Tn916 instead of the integrase gene to analyze the presence of related elements in our strain collection. PCRs to detect the ICEs were performed in 25-μl mixtures using 0.5 U of Taq DNA polymerase (New England Biolabs) and specific primers (see Table S2 in the supplemental material) according to the manufacturer's specifications. After an initial denaturation step of 5 min at 95°C, PCR was performed for 30 cycles consisting of 30 s of denaturation at 95°C, 30 s of annealing at a temperature depending on the primer set (5°C lower than the melting temperature of primers), and 1 min/kb of extension at 72°C, with a final extension step of 7 min at 72°C.
Nested PCRs were performed to amplify the fragment containing the attI attachment site resulting from the excision of the ICE integrated in the rplL and tRNA lysine genes according to the method described by Manganelli et al. (37) (Fig. 2A). Before testing excision by nested PCR, each primer was first tested individually by PCR using primers located in the upstream gene or downstream tRNALys gene to see if annealing occurs (see Table S2 in the supplemental material). For nested PCR, 25-cycle PCRs were carried out with outer primers (Table S2), and then 1 μl of each reaction mixture was used as template for a second 30-cycle PCR with an inner set of primers (Fig. 2A). Several sets of primers were tested (Table S2).
FIG. 2.
Nested-PCR analysis of the excision of two ICEs detected in bovine S. agalactiae strains. The principle of the detection of the circular form of ICEs by nested PCR using two sets of primers (one set of outer primers in black and one set of inner primers in gray) is indicated (A), as well as the results of nested PCR to detect the circular form of ICE-tRNALys in 9 strains in parallel with a positive control (S. agalactiae strain 515) (B) and of ICE-rplL in 18 strains in parallel with a positive control (S. agalactiae strain 2603 V/R) (C).
Filter mating experiments to detect Tn916 interspecies transfer.
We took advantage of the presence of a tetracycline resistance determinant on the Tn916 ICE to evaluate its mobility. A rifampin- and streptomycin-resistant mutant of S. agalactiae strain Nem16 was obtained by selection on increasing amounts of antibiotics. The analysis of the sequenced genome of this strain confirmed the absence of Tn916 elements in this strain, enabling its use as the recipient strain in the conjugation experiments. At least five strains of each species were selected among the strains carrying Tn916 to be used as donor strains in conjugation experiments (S. agalactiae strains 1667, 2584, 3288, 3464, and 4084; S. dysgalactiae strains 16072, 16213, 19103, 20385, 20386, 20387, 20402, and 20597; and S. uberis strains 14809, 19612, 20549, 20568, and 21086).
The filter matings were performed according the method described by Bellanger et al. (5), except that BHI broth and Trypticase soy agar plates were used and cells were incubated at 37°C. Conjugation experiments using S. dysgalactiae strains were made under aerobic conditions to improve the growth of the donor cells. Donor and recipient control plates were included to discard the possibility of spontaneous mutation.
To confirm that the cells that grew on the triple-antibiotic selective medium were not spontaneously resistant donor or recipient cells, transconjugants were characterized by PCR using a set of primers specific for S. agalactiae (40), a set of primers specific for the Nem316 strain (via amplification of the integrase gene of a genetic element, CIME_NEM316_tRNALeu, found in this strain) (11), and a set of primers specific for tet(M), (tet(M)Fwd and tet(M)Rev (see Table S2 in the supplemental material). To exclude a mobilization of CIME_NEM316_tRNALeu in the S. agalactiae donor cells, transconjugants were also characterized by multiple-locus variant repeat assay, as described recently by Radtke et al. (42).
RESULTS
Diversity of the bovine isolates of S. agalactiae, S. uberis, and S. dysgalactiae.
Molecular typing of the 76 S. agalactiae strains using the MLST scheme revealed that 47 strains were assigned to previously described sequence types (STs), whereas 27 strains showed new STs. These new sequence types were entered in the S. agalactiae MLST database (ST415 to ST434). Two strains (20165 and 20168; see Table S1 in the supplemental material) were not typeable due to a lack of amplification of the glcK gene (although different primers and conditions of amplification were tested) but appear to be related to the ST310 group. Overall, the strains display 35 different sequence types. The three predominant STs were ST23 (n = 14), ST310 (n = 8), and ST250 (n = 6). The analysis of the single- and double-locus variants of the sequence types allowed the grouping of most of the strains into seven clonal complexes (CCs), namely, CC1 (n = 14), CC2 (n = 9), CC6 (n = 4), CC17 (n = 2), CC19 (n = 1), CC23 (n = 21), and CC61/67 (n = 22) (Fig. 1).
The diversity of the S. uberis and S. dysgalactiae isolates was examined by the PFGE method. The restriction patterns observed for the isolates were very diverse especially for the S. uberis strains. Only a few strains of S. dysgalactiae exhibited a similar restriction pattern with less than seven differences over the 15 to 20 band patterns, suggesting that they are genetically related. The other strains displayed a unique band pattern with more than seven differences indicative of a genetic diversity if applying the criteria of Tenover et al. (49) (see Fig. S1 in the supplemental material as an example of the observed diversity).
Resistance to macrolide, lincosamide, and tetracycline and associated resistance genes.
Among the 76 S. agalactiae strains tested, 8 were resistant to erythromycin, spiramycin, and lincomycin and thus showed an MLSB phenotype (inducible in half of the isolates). These strains (except one) were also resistant to tetracycline. Overall, 29 strains of S. agalactiae were resistant to this antibiotic (Table 1). All of the resistant strains displayed an MLSB phenotype of resistance (constitutive or inducible), except for five strains of S. uberis that were resistant to lincomycin but susceptible to macrolides (L phenotype) (Table 1).
TABLE 1.
Distribution of macrolide, lincosamide, and tetracycline resistance genes among bovine isolates of S. agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis
| Organism (n) | Erythromycin resistance phenotypea | No. of isolates | No. of isolates with resistance gene: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| erm(B) | erm(A) | mef(A) | linB | lnuD | tet(M) | tet(O) | tet(S) | |||
| S. agalactiae (76) | cMLSB | 4 | 4 | 2 | 2 | |||||
| iMLSB | 4 | 2 | 2 | 1 | 3 | |||||
| eryS | 68 | 13 | 8 | 1 | ||||||
| S. dysgalactiae (32) | cMLSB | 2 | 2 | 1 | ||||||
| iMLSB | 2 | 2 | 2 | |||||||
| eryS | 28 | 1 | 5 | 1 | 4 | |||||
| S. uberis (101) | cMLSB | 52 | 52 | 2 | 19 | |||||
| iMLSB | 23 | 23 | 2 | 16 | ||||||
| L | 5 | 5 | 1 | 2 | 1 | |||||
| eryS | 21 | 17 | 3 | |||||||
cMLSB, constitutive phenotype of macrolide and lincosamide resistance; iMLSB, inducible phenotype of macrolide and lincosamide resistance; eryS, susceptible to erythromycin (and spiramycin); L, resistant to lincomycin but susceptible to erythromycin.
Resistance genes were searched by multiplex PCR. All of the erythromycin-resistant strains carried an erm(B) gene, except two S. agalactiae strains which carry an erm(A) resistance determinant (see Table S1 in the supplemental material). In addition, one S. dysgalactiae strain tested positive for the mef(A) gene, although the strain is susceptible to macrolides (Table 1; see Table S1 in the supplemental material). The five S. uberis strains resistant to lincomycin but susceptible to erythromycin presented the linB resistance determinant, and one had both the linB and lnuD genes.
Two genes were found to be almost equally associated with tetracycline resistance: the tet(M) gene was slightly predominant in the S. agalactiae and S. dysgalactiae strains, whereas the tet(O) gene appeared more frequent in the S. uberis strains (Table 1; see Table S1 in the supplemental material). Three strains of S. agalactiae, 5 strains of S. uberis, and 23 strains of S. dysgalactiae are resistant to tetracycline but did not carry the tet(M) or tet(O) gene. Among these strains, one strain of S. agalactiae, three strains of S. uberis, and four strains of S. dysgalactiae carried the tet(S) gene. The specificity of the PCR was confirmed by sequencing of one PCR fragment. Two strains of S. agalactiae and four strains of S. uberis carried both the erm(B) gene and the tet(M) gene.
The presence of the mer operon, encoding resistance to the heavy metal mercury (48), was also examined by detection of the merA gene by PCR. Accordingly, mercury resistance could be suspected only in four strains of S. agalactiae which gave a positive signal (see Table S1 in the supplemental material).
Prevalence of ICEs in the collection of S. agalactiae, S. uberis, and S. dysgalactiae bovine strains.
Sixteen, 26, and 9 strains of S. agalactiae, S. uberis, and S. dysgalactiae, respectively, carried the relaxase gene of Tn916 (Table 2; see Table S2 in the supplemental material). All of the strains carrying the Tn916-related element were resistant to tetracycline [except for one S. uberis strain, although it carries a tet(M) gene], but not all give a positive signal for the tet(M) resistance gene. Indeed, two strains of S. uberis (strains 16216 and 19127) and three strains of S. dysgalactiae (16212, 16213, and 19103) carried the relaxase gene of Tn916 but carry the tet(S) gene (see Table S1 in the supplemental material). Two tetracycline-resistant strains (S. uberis 19128 and S. dysgalactiae 20597) tested positive for the Tn916 relaxase gene but gave no amplification for the tet(M), tet(O), and tet(S) genes (Table S1). The strains of S. agalactiae and S. uberis that tested positive for the erm(B) and tet(M) genes also carried the Tn916 element. We thus tested if the Tn916 element carried the erm(B) gene by PCR by combining primers located in the tet(M) gene or the relaxase gene of Tn916 and primers located in the erm(B) gene. No specific PCR product was obtained.
TABLE 2.
Distribution of integrative and conjugative elements among bovine isolates of S. agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis as indicated by the presence of the elements' relaxase gene (orf20) or integrase gene (int)
| Organism (n) | No. (%) of isolates with gene |
||
|---|---|---|---|
| Tn916 orf20 | int ICE_Sa2603_rplL | int ICE_Sa2603_tRNALys | |
| S. agalactiae (76) | 16 (21) | 56 (74) | 39 (51) |
| S. dysgalactiae (32) | 9 (28) | 25 (78) | 5 (16) |
| S. uberis (101) | 26 (26) | 99 (98) | 22 (22) |
The integrase gene of ICE_Sa2603_rplL was detected in the majority of the strains of S. agalactiae (56/76 strains tested), S. uberis (99/101), and S. dysgalactiae (25/32) included in this study (Table 2).
Twenty-eight strains of S. agalactiae carried the SAG1993 integrase gene of ICE_Sa2603_tRNALys, and 12 have the SAG1986 integrase gene. One strain harbored both integrase genes. A genetic element integrated in the tRNA lysine gene was detected not only in S. agalactiae but also in 22 strains of S. uberis and 5 strains of S. dysgalactiae (Table 2).
Mobility of the ICEs.
A nested-PCR approach was used to detect the excision of the element integrated in the tRNALys gene in the strains of S. agalactiae which carry the SAG1993 integrase gene. A circular form was observed for all nine of the strains examined (Fig. 2B), by using two pairs of specific primers. The specificity of the PCR product was confirmed by sequencing for two strains. The primers used for S. agalactiae were tested for the S. dysgalactiae and S. uberis strains but did not give any amplification. New primers could be designed after sequencing of the right extremity (which includes the integrase gene) of the ICE in two strains. However, the left extremity of the ICE appeared less conserved, and despite numerous attempts (using ligation-mediated PCR and primers designed from the genome sequence of S. uberis 0140J [GenBank accession no. NC_012004]), the sequence of the left extremity of the ICE could not be obtained.
The same approach was used to detect the excised circular form of the element integrated in the rplL gene in 18 strains of S. agalactiae (Fig. 2C; see Table S1 in the supplemental material) belonging to different clonal complexes and selected among the 56 strains that tested positive for the ICE_Sa2603_rplL integrase gene (Table S1). A circular form was detected by nested PCR in all of the S. agalactiae strains examined (Fig. 2C). The specificity of the nested-PCR product was confirmed by sequencing for two strains. The same primers were tested to detect the excised circular form of the element integrated in the rplL gene in selected strains of S. dysgalactiae (n = 25) and S. uberis (n = 25). However, none gave a PCR product, probably because of an insufficient ICE sequence conservation or truncation of the element in these strains.
Transfer of Tn916 was tested by using different donor cells of each species and a spontaneous rifampin and streptomycin mutant of S. agalactiae Nem316 strain. Transconjugants were obtained at very low frequency (1 × 10−8 to 5 × 10−8 transconjugants per CFU of donor cell, depending on the strain) for two strains of S. agalactiae (1667 and 2584), one strain of S. uberis (20549), and one strain of S. dysgalac- tiae (strain 20402). The transconjugants were confirmed by PCR using different sets of primers and by multiple-locus variant repeat assay. The results indicated that the transconjugants are cells of the Nem316 S. agalactiae strain, which has acquired the Tn916 tet(M) gene (Fig. 3).
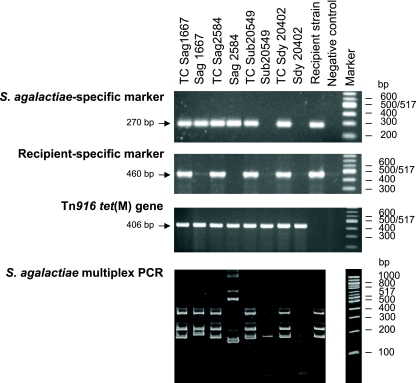
FIG. 3.
Characterization of triple-resistant (rifampin, streptomycin, and tetracycline) transconjugant clones obtained by conjugal transfer of Tn916 from the different donor cells tested to recipient strain S. agalactiae Nem316. TC Sag1667 and TC Sag2584, transconjugants obtained using S. agalactiae strains 1667 and 2584, respectively, as donor cells; TC Sub20549 and TC Sdy20402, transconjugants obtained using S. uberis 20549 and S. dysgalactiae 20402, respectively, as donor cells. PCRs were done with primers specific for the S. agalactiae species (first lane), primers specific for a genetic element found in the Nem316 recipient strain (second lane), primers specific for the tet(M) gene of Tn916 (third lane), and five sets of primers for the multiple-locus variant repeat assay (fourth lane).
DISCUSSION
We studied a collection of S. agalactiae, S. dysgalactiae, and S. uberis strains, isolated from milk of dairy cattle suffering from subclinical or clinical mastitis, for the presence of three families of integrative and conjugative elements (Tn916-, ICE_Sa2603_rplL-, and ICE_Sa2603_tRNALys-related elements) and their association with macrolide, lincosamide, tetracycline, and/or mercury resistance genes. This collection includes isolates with a high genetic diversity, as shown by the MLST and PFGE experiments.
An MLSB phenotype of macrolide resistance was observed for 10% of the strains of S. agalactiae, associated with an erm(B) gene in all strains, except for two which carry an erm(A) gene. The predominance of erm(B) resistance genes in S. agalactiae was already reported by several authors (35, 38, 53). All of the macrolide-resistant strains of S. uberis and S. dysgalactiae also carry an erm(B) gene. Five strains of S. uberis displayed an L phenotype, with lincomycin resistance but susceptibility to macrolides. These strains carry a linB resistance determinant, and one has both linB and lnuD resistance determinants. The linB gene was found initially on a plasmid of Enterococcus faecium (9) but has since been described, although sporadically, in S. agalactiae (23), S. dysgalactiae subsp. dysgalactiae (43), and S. uberis (28). This suggests a horizontal gene transfer of this resistance determinant after exchange of the gene between a plasmid and an ICE. One-third of the strains of S. agalactiae were resistant to tetracycline, associated in the same proportion with a tet(M) or tet(O) gene as that for strains of S. dysgalactiae. In S. agalactiae, the tet(M) and tet(O) genes were found equally distributed between strains of CC23, primarily associated with human strains (12), and strains of CC61/67, mainly described in cattle (7). This is in contrast to what has been described in other studies in which bovine strains of S. agalactiae predominantly carry a tet(M) resistance gene (24, 25). In contrast, the tet(O) gene was predominant in the S. uberis strains. A tet(S) gene was detected in one strain of S. agalactiae, three strains of S. uberis, and four strains of S. dysgalactiae. To our knowledge, this is the first report of the presence of this resistance determinant in these species. This gene has been initially described on a conjugative plasmid of a multiresistant Listeria monocytogenes strain (18) and subsequently on a plasmid of Lactococcus lactis (51) and in the chromosome of Enterococcus faecalis (19). More recently, this gene has been detected in isolates of Streptococcus pyogenes (29), Streptococcus intermedius (32), and S. dysgalactiae subsp. equisimilis (34), suggesting a lateral gene transfer of this tetracycline resistance determinant.
The conjugative elements were searched for by PCR, using primers designed from the sequence of ICEs described previously in human strains of S. agalactiae (11). Their mobility was then evaluated by nested PCR to test excision of the elements integrated in the rplL and tRNA lysine genes and by conjugation experiments to test Tn916 transfer.
A Tn916-related element was detected in half of the S. agalactiae and S. uberis tetracycline-resistant strains and in one-third of the S. dysgalactiae tetracycline-resistant strains. This ICE was already proved to be widespread in Gram-positive bacteria (44). It is thus not surprising to find this element also in the S. dysgalactiae and in the S. uberis species. A tet(M) gene was detected by PCR in all of the S. agalactiae strains carrying the Tn916 relaxase gene. This was not the case for S. dysgalactiae and S. uberis, for which several strains gave a negative result for tet(M) gene amplification. As all of these strains are resistant to tetracycline, this could be due either to differences in the sequence of the tet(M) impeding detection of the gene by PCR or to the presence of another tetracycline resistance determinant. Indeed, Tn916-related elements carrying a tet(S) gene have been described in S. intermedius (46), and two strains of S. uberis and three strains of S. dysgalactiae were found to carry both a Tn916-related element and a tet(S) gene. There are multiple descriptions of association of tet(M) and erm(B) resistance genes on the same mobile genetic element in S. pyogenes and S. pneumoniae (52). Such association has also been reported once in S. agalactiae (52). Among the strains carrying the Tn916 relaxase gene in our collection, two S. agalactiae isolates and four S. uberis isolates also carry the erm(B) gene. However, we did not obtain a PCR product when combining primers located in the Tn916 orf20 gene and in the erm(B) gene when using genomic DNA of these strains. We evaluated the frequency of transfer of Tn916 in vitro by using an S. agalactiae strain as the recipient and different S. agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis strains as donors. A few strains of S. agalactiae and S. uberis tested enabled us to recover transconjugants with the expected triple-resistance pattern, but the frequency of transfer was very low. For S. dysgalactiae, conjugative transfer was obtained only with one strain out of the eight strains tested. All of these transconjugants obtained were confirmed by PCR to be the Nem316 strain, which has acquired the Tn916 tet(M) gene by conjugation. We do not have direct evidence of the conjugative transfer of Tn916, so we cannot discard the hypothesis of a transfer of the genes carried by Tn916 by mobilization by another conjugative element or by an Hfr-like mechanism, as described recently in S. agalactiae (13). It is possible that conjugative transfer between streptococci occurs at very low frequency, but that in vivo transfer is mediated through other species, in particular Enterococcus faecalis, which proved to easily transfer ICEs to other species (5, 41). Tn916 is a preferential target site of integration for numerous MGEs (46) and is also able to mobilize nonconjugative elements (1, 44). This family of elements thus likely plays a major role in horizontal transfer of antibiotic resistance genes between streptococcal species and other species that share the same ecological niche.
An element was found integrated in the rplL gene in the majority of S. agalactiae, S. dysgalactiae, and S. uberis strains included in this study. In these three streptococcal species examined, this ICE family is even more widespread than the Tn916 family. We detected a circular form of this element in all of the isolates of S. agalactiae tested, suggesting that this MGE is functional. We were not able to test the excision of the elements found in the isolates of S. dysgalactiae and S. uberis. Such ICEs, which combine a conjugation module related to the one of Tn5252 (2) and an integrase gene allowing integration in the rplL gene, were detected in other streptococcal species, in particular S. dysgalactiae subsp. equisimilis (ICESde3396) (22) and S. pneumoniae (ICESp23FST81) (21). Genome analyses also indicate a similar element in several strains of S. suis (11, 21). These ICEs are thus widespread in streptococci. ICESp23FST81 of S. pneumoniae is a composite element comprising a Tn916 element [carrying the tet(M) tetracycline resistance gene] inserted into a Tn5252-related element. It harbors a chloramphenicol resistance gene carried by the Tn5252-related element, similar to the gene found on plasmid pC194 of S. aureus, in addition to a tetracycline resistance gene (21). ICESde3396 of S. dysgalactiae subsp. equisimilis does not carry an antibiotic resistance gene (22). However, as for related ICEs found in S. agalactiae (11), it contains several genes whose homologues are found in other MGEs that do harbor antibiotic resistance determinants, in particular the mega element, which carries genes encoding Mef(E) and Msr(D) macrolide efflux pumps (52). This mega element was also described in the human mouth-colonizing species Streptococcus salivarius (52). It is able to integrate in other MGEs, in particular those of the Tn916 family (46). This can lead to complex structures harboring multiple resistance genes. For example, composite elements described in S. pneumoniae (Tn2010 and Tn2017) harbor three macrolide resistance genes in addition to the tet(M) tetracycline resistance gene by combining three different MGEs (Tn916-MAS element-mega element and Tn916-Tn917-mega element respectively) (46, 52). The ability of these ICEs to exchange with other MGEs can make them a reservoir of antibiotic resistance genes and a source of horizontal gene transfer between streptococci.
Half of the S. agalactiae strains harbor an element integrated in a gene encoding a tRNALys. This ICE family thus appears to be widespread not only in human strains (11) but also in bovine strains of S. agalactiae. In addition, we detected a circular form of this element in all isolates of S. agalactiae tested, suggesting that this MGE is functional. Integrase genes of these elements were detected by PCR in strains of S. uberis and S. dysgalactiae, suggesting their presence in these species. However, testing for their excision was not allowed—probably due to a lack of sequence conservation at the extremities of the ICEs in these species. In addition to a related ICE, ICESt1/St3 of S. thermophilus (16), previous genome analyses detected related elements in other species, including Streptococcus mutans and S. pyogenes (11).
In addition to previous reports or evidence of ICEs integrated in the rplL and tRNALys genes in S. agalactiae, S. pyogenes, and S. suis (11), as well as S. pneumoniae (21) and S. dysgalactiae subsp. equisimilis (22), this work indicates that such elements are also largely present in S. dysgalactiae subsp. dysgalactiae and S. uberis strains. rplL and tRNALys genes thus appear as hot spots for integration of ICEs in streptococci. Excision of the elements was demonstrated in all strains tested, indicating that these elements are mobile. Although most of the strains examined carry antibiotic resistance genes, we did not observe any association between the presence of these ICEs and antibiotic resistance. However, in S. agalactiae, in silico analyses suggest that ICEs of this family carry several genes that could confer adaptation and virulence properties to the bacterium. Elucidation of their function will require deeper investigation. These ICEs could be a vehicle for horizontal gene transfer of adaptation and virulence genes within the genus Streptococcus and probably also with phylogenetically related bacteria. The present study also highlights the relevance of further dedicated epidemiological studies to determine more accurately the prevalence of ICEs within the population of streptococci in animals.
Supplementary Material
Acknowledgments
We thank Philippe Glaser, Mathieu Brochet, and Elisabeth Couvé (Institut Pasteur, Paris, France) for help with the MLST experiments and helpful discussions and Nicola Jones for assigning numbers to new STs. We thank Gérard Guédon for interesting comments on the manuscript and helpful discussions. We are grateful to Roland Leclercq for providing the lnuD control strain.
This work was supported by the Agence Nationale de Sécurité Sanitaire (Anses) and the Institut National de la Recherche Agronomique (INRA) through their joint program 2007-2008 (AIP 297 INRA/ AFSSA).
Footnotes
Published ahead of print on 15 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achard, A., and R. Leclercq. 2007. Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae. J. Bacteriol. 189:4328-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcon-Chaidez, F., J. Sampath, P. Srinivas, and M. N. Vijayakumar. 1997. Tn5252: a model for complex streptococcal conjugative transposons. Adv. Exp. Med. Biol. 418:1029-1032. [DOI] [PubMed] [Google Scholar]
- 3.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 4.Baseggio, N., P. D. Mansell, J. W. Browning, and G. F. Browning. 1997. Strain differentiation of isolates of streptococci from bovine mastitis by pulsed-field gel electrophoresis. Mol. Cell Probes 11:349-354. [DOI] [PubMed] [Google Scholar]
- 5.Bellanger, X., A. P. Roberts, C. Morel, F. Choulet, G. Pavlovic, P. Mullany, B. Decaris, and G. Guedon. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J. Bacteriol. 191:2764-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beres, S. B., and J. M. Musser. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2:e800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botrel, M. A., M. Haenni, E. Morignat, P. Sulpice, J. Y. Madec, and D. Calavas. 2010. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhone-Alpes, France. Foodborne Pathog. Dis. 7:479-487. [DOI] [PubMed] [Google Scholar]
- 9.Bozdogan, B., L. Berrezouga, M. S. Kuo, D. A. Yurek, K. A. Farley, B. J. Stockman, and R. Leclercq. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley, A. 2002. Bovine mastitis: an evolving disease. Vet. J. 164:116-128. [DOI] [PubMed] [Google Scholar]
- 11.Brochet, M., E. Couve, P. Glaser, G. Guedon, and S. Payot. 2008. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 190:6913-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brochet, M., E. Couve, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8:1227-1243. [DOI] [PubMed] [Google Scholar]
- 13.Brochet, M., C. Rusniok, E. Couve, S. Dramsi, C. Poyart, P. Trieu-Cuot, F. Kunst, and P. Glaser. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 105:15961-15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77-97. [DOI] [PubMed] [Google Scholar]
- 15.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 16.Burrus, V., Y. Roussel, B. Decaris, and G. Guedon. 2000. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 66:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CA-SFM. 2008. Recommandations du Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM), groupe de travail antibiogramme vétérinaire. Comité de l'Antibiogramme de la Société Française de Microbiologie, Paris, France. http://www.sfm.asso.fr/publi/general.php?pa=1.
- 18.Charpentier, E., G. Gerbaud, and P. Courvalin. 1993. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 131:27-34. [DOI] [PubMed] [Google Scholar]
- 19.Charpentier, E., G. Gerbaud, and P. Courvalin. 1994. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Churchward, G. 2002. Conjugative transposons and related mobile elements, p. 177-191. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 21.Croucher, N. J., D. Walker, P. Romero, N. Lennard, G. K. Paterson, N. C. Bason, A. M. Mitchell, M. A. Quail, P. W. Andrew, J. Parkhill, S. D. Bentley, and T. J. Mitchell. 2009. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191:1480-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies, M. R., J. Shera, G. H. Van Domselaar, K. S. Sriprakash, and D. J. McMillan. 2009. A novel integrative conjugative element mediates genetic transfer from group G streptococcus to other β-hemolytic streptococci. J. Bacteriol. 191:2257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dogan, B., Y. H. Schukken, C. Santisteban, and K. J. Boor. 2005. Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J. Clin. Microbiol. 43:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duarte, R. S., B. C. Bellei, O. P. Miranda, M. A. Brito, and L. M. Teixeira. 2005. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 49:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 28.Haenni, M., E. Saras, S. Chaussiere, M. Treilles, and J. Y. Madec. ermB-mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet. J., in press. [DOI] [PubMed]
- 29.Hammerum, A. M., H. U. Nielsen, Y. Agerso, K. Ekelund, and N. Frimodt-Moller. 2004. Detection of tet(M), tet(O) and tet(S) in tetracycline/minocycline-resistant Streptococcus pyogenes bacteraemia isolates. J. Antimicrob. Chemother. 53:118-119. [DOI] [PubMed] [Google Scholar]
- 30.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lancaster, H., A. P. Roberts, R. Bedi, M. Wilson, and P. Mullany. 2004. Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet(S). J. Bacteriol. 186:4395-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 34.Liu, L. C., J. C. Tsai, P. R. Hsueh, S. P. Tseng, W. C. Hung, H. J. Chen, and L. J. Teng. 2008. Identification of tet(S) gene area in tetracycline-resistant Streptococcus dysgalactiae subsp. equisimilis clinical isolates. J. Antimicrob. Chemother. 61:453-455. [DOI] [PubMed] [Google Scholar]
- 35.Loch, I. M., K. Glenn, and R. N. Zadoks. 2005. Macrolide and lincosamide resistance genes of environmental streptococci from bovine milk. Vet. Microbiol. 111:133-138. [DOI] [PubMed] [Google Scholar]
- 36.Malhotra-Kumar, S., C. Lammens, J. Piessens, and H. Goossens. 2005. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob. Agents Chemother. 49:4798-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manganelli, R., L. Romano, S. Ricci, M. Zazzi, and G. Pozzi. 1995. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid 34:48-57. [DOI] [PubMed] [Google Scholar]
- 38.Marimon, J. M., A. Valiente, M. Ercibengoa, J. M. Garcia-Arenzana, and E. Perez-Trallero. 2005. Erythromycin resistance and genetic elements carrying macrolide efflux genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 49:5069-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petinaki, E., V. Guerin-Faublee, V. Pichereau, C. Villers, A. Achard, B. Malbruny, and R. Leclercq. 2008. Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob. Agents Chemother. 52:626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phuektes, P., P. D. Mansell, and G. F. Browning. 2001. Multiplex polymerase chain reaction assay for simultaneous detection of Staphylococcus aureus and streptococcal causes of bovine mastitis. J. Dairy Sci. 84:1140-1148. [DOI] [PubMed] [Google Scholar]
- 41.Puopolo, K. M., D. C. Klinzing, M. P. Lin, D. L. Yesucevitz, and M. J. Cieslewicz. 2007. A composite transposon associated with erythromycin and clindamycin resistance in group B streptococcus. J. Med. Microbiol. 56:947-955. [DOI] [PubMed] [Google Scholar]
- 42.Radtke, A., B. A. Lindstedt, J. E. Afset, and K. Bergh. 2010. Rapid multiple-locus variant-repeat assay (MLVA) for genotyping of Streptococcus agalactiae. J. Clin. Microbiol. 48:2502-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rato, M. G., R. Bexiga, S. F. Nunes, C. L. Vilela, and I. Santos-Sanches. 2010. Human group A streptococci virulence genes in bovine group C streptococci. Emerg. Infect. Dis. 16:116-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riffon, R., K. Sayasith, H. Khalil, P. Dubreuil, M. Drolet, and J. Lagace. 2001. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J. Clin. Microbiol. 39:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, A. P., and P. Mullany. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251-258. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt-Van de Leemput, E., and R. N. Zadoks. 2007. Genotypic and phenotypic detection of macrolide and lincosamide resistance in Streptococcus uberis. J. Dairy Sci. 90:5089-5096. [DOI] [PubMed] [Google Scholar]
- 48.Soge, O. O., N. K. Beck, T. M. White, D. B. No, and M. C. Roberts. 2008. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J. Antimicrob. Chemother. 62:674-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 52.Varaldo, P. E., M. P. Montanari, and E. Giovanetti. 2009. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng, X., F. Kong, H. Wang, A. Darbar, and G. L. Gilbert. 2006. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Antimicrob. Agents Chemother. 50:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.