Abstract
Individual cloacal swabs of mallards (Anas platyrhynchos) and of herring gulls (Larus argentatus), as well as samples of waterbird feces obtained in 2008 and 2009, were cultivated for Escherichia coli. Isolates of E. coli were tested for susceptibilities to 12 antimicrobial agents by the disk diffusion method. Moreover, the samples were subcultivated on MacConkey agar (MCA) containing cefotaxime (2 mg liter−1) to detect E. coli with extended-spectrum beta-lactamase (ESBL) and subsequently on MCA supplemented with ciprofloxacin (0.05 mg liter−1) and MCA with nalidixic acid (20 mg liter−1) to isolate fluoroquinolone-resistant E. coli. PCR was used to detect specific antibiotic resistance genes. We found 9 E. coli isolates producing ESBL with bla genes: blaCTX-M-1 (6 isolates), blaCTX-M-9 plus blaTEM-1b (1 isolate), blaCTX-M-15 plus blaOXA-1 (1 isolate), and blaSHV-12 (1 isolate). In the isolate with blaCTX-M-15, the gene aac(6)-Ib-cr was also detected. The bla genes were harbored by transferable plasmids of the IncN and IncI1 groups. Nine quinolone-resistant E. coli isolates with qnrS genes were found and characterized. The gene qnrS was associated with a Tn3-like transposon on the IncX1 plasmid together with blaTEM-1 in two isolates. The gene qnrS was also harbored by conjugative plasmids of the IncN and IncX2 groups. Even if populations of wild birds are not directly influenced by antibiotic practice, we have demonstrated that antibiotic-resistant E. coli strains, including strains with various ESBL and qnrS genes, are found in the feces of wild birds on the coast of the Baltic Sea in Poland.
There is considerable concern about antibiotic resistance in bacteria from humans and farm animals, but the spread of resistance into wider ecosystems has received much less attention (48). Usually, isolates of the common intestinal bacterium Escherichia coli are examined to detect antibiotic resistance in populations of wild animals. Wild birds are colonized with various strains of E. coli, including strains such as E. coli O157 that are pathogenic for humans (83). Fecal strains of E. coli resistant to antibiotics have been found at various prevalences in wild bird populations. In particular, bird populations sympatric to areas inhabited by people and areas with a high density of livestock were colonized with antibiotic-resistant E. coli strains possibly selected by the antibiotic practice in humans and domestic animals. Antibiotic-resistant E. coli isolates have been found in corvids (Corvus corone, C. frugilegus, C. macrorhynchos, Pica pica, and Pyrrhocorax pyrrhocorax) (3, 46, 48, 53, 74), house sparrows (Passer domesticus) (22, 61), house martins (Delichon urbica) (73), feral pigeons (Columba livia forma domestica) (68), ducks, geese, and swans (Anas platyrhynchos, Anas acuta, Branta canadensis, and Cygnus columbianus) (17, 26, 51, 82), cormorants (Phalacrocorax auritus and P. cristatus) (20, 71), Egyptian vultures (Neophron percnopterus) (2), and, most frequently, in various gull species throughout the world (Larus argentatus, L. atricilla, L. audouinii, L. cachinans, L. crassirostris, L. glaucoides, L. hyperboreus, L. marinus, L. ridibundus, and L. vagae) (4, 7, 21, 23, 29, 66, 71, 76, 82). Corvids and gulls feeding on garbage dumps and in urbanized areas are frequently colonized with resistant strains of E. coli, and they are considered to be important reservoirs and vectors of these isolates in the environment (23, 46).
The occurrence of antibiotic resistance in E. coli from surface water and fecal pollution sources (wastewater treatment plants and bird feces) was studied in the inland Hamilton Harbor on Lake Ontario, Canada (24). Authors of the study suggested that at times, feces of wild geese, ducks, and gulls may be a more prominent contributor of resistant E. coli to a beach near Hamilton than are municipal wastewater sources.
Thousands of great cormorants (Phalacrocorax carbo), gulls (Larus argentatus, L. ridibundus, L. canus, and L. marinus) and mallards (Anas platyrhynchos) winter in Puck Bay on the Polish coast of the Baltic Sea (50). Many of them stay near the two biggest cities of this region, Gdynia and Gdansk, in harbors, municipal beaches, build-up areas, and city parks situated near the seacoast. We were concerned about antimicrobial-resistant E. coli, including strains producing extended-spectrum beta-lactamase (ESBL) and carrying class 1 and 2 integrons with resistant gene cassettes in these birds. Resistant E. coli isolates producing ESBL have been found in black-headed gulls (L. ridibundus) in the Czech Republic, yellow-legged gulls (Larus michahellis) in France, and seagulls of diverse species (not determined) in Portugal (4, 23, 66). Resistant isolates carrying class 1 and 2 integrons have been found in Canada geese (Branta canadensis) in the United States, black-headed gulls and rooks (Corvus frugilegus) in the Czech Republic, Mediterranean herring gulls (Larus cachinans) in Italy, and seagulls in Portugal (4, 17, 21, 23, 29, 46, 66). We also were concerned about strains resistant to fluoroquinolones, since such strains have emerged recently in gulls (L. audouinii, L. cachinans, L. glaucoides/L. hyperboreus, and L. ridibundus) in Italy, Portugal, Greenland, and the Czech Republic (7, 23, 29, 76).
MATERIALS AND METHODS
Locations, birds, and feces samples.
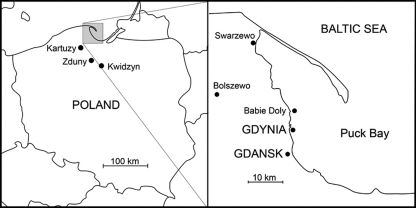
The samples were collected on the Polish coast of the Baltic Sea within or near Gdynia and Gdansk (Fig. 1). Mallards (Anas platyrhynchos) were caught in the city parks of Gdansk (n = 46) and also in Bolszewo (n = 32), Kwidzyn (n = 5), Zduny (n = 2), and Kartuzy (n = 1) during winter 2008/2009 and in November and December 2009. The latter four places are located in a region of lakes around Gdansk within a distance of up to 50 km from that city. Cloacal smears were obtained from 86 mallards in total. Herring gulls (Larus argentatus) were caught in Gdynia during winter 2008/2009 (adults) and during the nesting period (pulli) in 2009. Cloacal smears were obtained from 27 herring gulls. There is a large wave breaker within Gdynia's harbor that has no connection with the coast. On the shore at a distance of about 6 km from the harbor, there is also an old and abandoned army building standing on water near (300 m) a coast, near one of the Gdynia districts called Babie Doly. Especially inasmuch as public entry is prohibited to both of these, they are frequently used by waterbirds (gulls, cormorants, and sometimes also ducks) for resting, and many of their feces occur there. We collected samples of bird feces from both of these structures in January 2009. We obtained in total 244 samples (119 and 125 from the wave breaker and army building, respectively). A large roosting place for wintering great cormorants lies along the coast in Swarzewo, about 30 km from Gdynia. Here we collected 125 samples of cormorant feces in January 2009. Cormorants were daily moving from this roosting place to various places near the seacoast, including the sea close to Gdynia and Gdansk.
FIG. 1.
Map of the Polish coast of the Baltic Sea, where the samples were collected. Mallards (Anas platyrhynchos) originated at Gdansk, Bolszewo, Kwidzin, Zduny, and Kartuzy, herring gulls (Larus argentatus) from Gdynia, and samples of waterbird feces from Gdynia, Babie Doly, and Swarzewo.
Nonselective antibiotic-resistant E. coli isolation and characterization.
Individual cloacal swabs and samples of feces were transported to the laboratory and placed overnight in buffered peptone water (BPW) at 37°C and then cultivated for E. coli on selective chromogenic medium for E. coli and coliform bacteria (CM0956; Oxoid, United Kingdom). One colony of each plate was tested for susceptibilities to antimicrobial agents by the disk diffusion method in accordance with the Clinical and Laboratory Standards Institute (16). Susceptibilities to the following antibacterial substances were tested: amoxicillin-clavulanic acid (30 μg), ampicillin (10 μg), cephalothin (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), nalidixic acid (30 μg), streptomycin (10 μg), sulfamethoxazole-trimethoprim (25 μg), sulfonamide compounds (300 μg), and tetracycline (30 μg) (Oxoid). In E. coli isolates found to be resistant to one or more of the antibiotics listed above, PCR was used to detect specific antibiotic resistance genes, integrase genes int1 and int2, and gene cassettes within class 1 and 2 integrons (47). The presence of the genes tetA, tetB, tetC, tetD, tetE, tetG, blaTEM, blaSHV, blaOXA-1 like, cat, cmlA, floR, sul1, sul2, sul3, strA, int1, int2, variable regions of class 1 and class 2 integrons, dhfr1, dhfr12, dhfr17, aadA1, aadA2, aadA5, estX, and sat1 and sat2 were tested with primers and under conditions as described elsewhere (47). All resistant E. coli strains were identified by the API 10S test (bioMerieux, France).
Selective isolation of ESBL and quinolone-resistant isolates.
All samples from BPW were subsequently enriched in MacConkey broth and subcultivated on MacConkey agar (MCA) containing cefotaxime (2 mg liter−1) to detect E. coli strains with ESBL (84) and subsequently on MCA supplemented with ciprofloxacin (0.05 mg liter−1) to isolate fluoroquinolone-resistant E. coli. Samples of waterbird feces from BPW were enriched in MacConkey broth and also subcultivated on MCA containing nalidixic acid (20 mg liter−1) to detect E. coli strains resistant to nalidixic acid and possibly other quinolones.
PCR for ESBL/quinolone resistance genes.
The colonies grown on MCA with cefotaxime were examined using the double-disk synergy test (DDST) for the production of ESBL (16, 80). The genes responsible for the ESBL phenotype (blaTEM, blaSHV, and blaCTX-M) were identified by PCR as described elsewhere (5, 45, 64). All the PCR products were further analyzed using sequencing (ABI 310 genetic analyzer; Applied Biosystems). The colonies grown on MCA with ciprofloxacin or nalidixic acid were tested for the plasmid-encoded quinolone resistance genes qnrA, qnrB, qnrC, qnrD, qnrS, qepA, and aac(6′)-Ib by PCR as described elsewhere (9, 11, 25, 41, 57, 63, 70, 78, 85), and MICs for nalidixic acid and ciprofloxacin were determined by the agar dilution method in accordance with Clinical and Laboratory Standards Institute standards (16). In all the isolates positive for plasmid-harbored quinolone resistance genes with high MIC levels for nalidixic acid and ciprofloxacin, the gyrA and parC genes were detected by PCR and analyzed by sequencing. All isolates resistant to ampicillin and/or amoxicillin-clavulanic acid, as determined during nonselective isolations, were tested also by DDST for the production of ESBL. All isolates resistant to nalidixic acid and/or ciprofloxacin, as determined during nonselective isolation, were also tested for the presence of the qnrA, qnrB, qnrC, qnrD, qnrS, qepA, and aac(6′)-Ib genes.
Molecular characterization of ESBL/quinolone-resistant isolates.
Antimicrobial susceptibilities of the ESBL-positive and quinolone-resistant E. coli isolates were examined by the disk diffusion method, and the isolates were tested for additional antibiotic resistance genes (see above). Identification of E. coli phylogenetic groups and determination of the presence of the cib gene for colicin production were performed using PCR (15). The isolates were typed by XbaI pulsed-field gel electrophoresis (PFGE) (12). Those samples with no band differences were designated indistinguishable and possibly epidemiologically linked.
Transferability of antibiotic resistance genes.
Transferability of ESBL and plasmid-encoded quinolone resistance genes was tested by conjugation. Plate-mating experiments were done using plasmid-free, rifampin and nalidixic acid-resistant E. coli MT102RN and Salmonella enterica serovar Typhimurium SL5325 as recipients (in the case of a donor strain having resistance to quinolone, recipients resistant to rifampin and sodium azide were used) (1).
Plasmid analysis.
For plasmid analysis, transformants with a single plasmid carrying ESBL or quinolone resistance genes were prepared. Primarily plasmid DNA was extracted using the Qiagen Plasmid Midi kit (Qiagen, Germany). Plasmid DNA was introduced into competent E. coli Top10 (Invitrogen) by chemical transformation, followed by selection of transformants on LB agar supplemented with cefotaxime (2 mg liter−1) or ciprofloxacin (0.05 mg liter−1). The presence of relevant bla and quinolone resistance genes in transformants was confirmed by PCR. The size of plasmids with ESBL or quinolone resistance genes from transformants was designated by S1-PFGE. Plasmids were characterized by replicon typing and restriction fragment length polymorphism (RFLP) analysis. Plasmid DNA from transformants was digested with EcoRV and HincII and then subjected to gel electrophoresis in a 1% agarose gel for 5 h at 100 V/cm. Plasmids were replicon typed as previously described (8). The presence of Tn3-like transposons in qnrS transformants was tested by PCR (1).
RESULTS
Antimicrobial-resistant E. coli in mallards, herring gulls, and waterbird feces.
In total, 65 suspected E. coli isolates were obtained from 86 samples from mallards. Antibiotic resistance was detected in 23 E. coli isolates. The prevalence of mallards with antimicrobial-resistant E. coli was 27% (23 resistant E. coli isolates/86 mallards tested). Three of the resistant isolates showed resistance to ciprofloxacin (but plasmid-encoded quinolone resistance genes were not detected). Isolates resistant to ampicillin and amoxicillin-clavulanic acid were tested for ESBL production by DDST, and three of them were found to produce ESBL. Phenotypes and resistance genes, including integrons and gene cassettes, are summarized in Table 1. With selective cultivation on MCA with cefotaxime, one ESBL-producing E. coli isolate was detected (upon further analysis, however, this isolate was found to be identical to an isolate obtained by nonselective cultivation from the identical cloacal swab sample) (Table 2). Resistance to ciprofloxacin was found in 25 isolates by selective cultivation, and the qnrS gene was detected in four of these (Table 3).
TABLE 1.
Antibiotic-resistant Escherichia coli obtained by nonselective isolation and numbers of strains with some resistance patterns from mallards, herring gulls, and waterbird feces
| Antibiotic resistance phenotypea | Antibiotic resistance gene(s)b | No. of isolates from: |
||
|---|---|---|---|---|
| Mallards | Herring gulls | Waterbird feces | ||
| A | blaTEM | 3 | ||
| Gn | NT | 1 | ||
| Na | ND | 1 | 1 | |
| S | aadA1 | 1 | ||
| S | ND | 6 | 1 | |
| ANa | blaTEM | 1 | ||
| GnS | aadA1 | 1 | ||
| GnS | ND | 1 | ||
| SSu | strA, sul2 | 1 | 1 | |
| ANaS | blaTEM, strA | 1 | ||
| ANaT | blaTEM, tetA | 1 | ||
| AST | blaTEM, strA, tetB | 1 | ||
| ANaST | blaTEM, strA, tetB | 2 | ||
| AAcCfCipNa | blaCTX-M-1 | 1 | ||
| ACipNaSxtT | blaTEM, tetA, tetB, int1; I1, 1.5 kb: dhfr1-aadA1 | 1 | ||
| NaSSuSxtT | sul1, tetA, int1; I1, 2.2 kb: aadA1 | 1 | ||
| AAcCfCipNaS | blaTEM, aadA1 | 2 | ||
| ACfSSuSxtT | blaCTX-M-1, strA, sul2, aadA1, tetB | 1 | ||
| ACCfSSuSxtT | blaCTX-M-1, cat, strA, sul1, sul2, tetB, int1; I1, 2.5 kb: dhfr1-aadA1 | 1 | ||
| Total | 23 | 3 | 5 | |
A, ampicillin; Ac, amoxicillin-clavulanic acid; C, chloramphenicol; Cf, cephalothin; Cip, ciprofloxacin; Gn, gentamicin; Na, nalidixic acid; S, streptomycin; Su, sulfonamides; Sxt, sulfamethoxazole-trimethoprim; T, tetracycline.
ND, not determined; NT, not tested; I1, class 1 integron.
TABLE 2.
Characterization of ESBL-producing Escherichia coli isolates from wild birds on the Baltic Sea coast of Poland
| Isolate no. | Origin | Antibiotic resistance phenotypec | ESBL bla genes | Antibiotic resistance genes | XbaI PFGEe | Presence of colicin (the gene cib) | Phylogenetic group | Conjugation to E. coli | Conjugation to Salmonella | Plasmid incompatibility groupe | RFLP pattern (EcoRV) | Plasmid size (kb) | Additional genes on ESBL-carrying plasmid |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 PL | Mallard | AAcCfCipNa | blaCTX-M-1 | a | − | A | + | + | |||||
| 5 PL | Mallard | ACfSSuSxtT | blaCTX-M-1 | strA, aadA1, sul2, tetB | b | − | B1 | + | + | IncN | I | 35 | |
| 78 PLa | Mallard | ACCfSSuSxtT | blaCTX-M-1 | cat, strA, sul1, sul2, tetB, int1, I1,d 2.5 kb, dhfr1-aadA1 | c | − | B2 | − | − | ||||
| 17 LA CEF | Herring gull | ACfSu | blaCTX-M-1 | sul2 | e | + | A | + | + | IncI1 | II | 90 | cib, sul2 |
| 11 LA CEF | Herring gull | ACfGnSuSxt | blaCTX-M-1 | sul1, int1, I1, 2.0 kb, dhfr12-aadA2 | d | − | B2 | + | + | ND | III | 80 | int1, I1, 2.0 kb, dhfr12-aadA2 |
| 6 LA CEF | Herring gull | ACfSSuSxtT | blaCTX-M-9 | blaTEM-1b, strA, aadA1, sul2, tetD | ND | − | D | − | − | ||||
| 88 | Waterbird feces | ACf | blaSHV-12 | int1, I1, 2.1 kb, aadA2 | g | − | A | + | + | IncI1 | IV | 95 | |
| 96 | Waterbird feces | ACfNaSuSxtT | blaCTX-M-1 | blaTEM-1b, dhfr17, aadA5, sul2, tetA, int1 | h | − | B1 | + | + | IncI1 | V | 100 | sul2, int1, dhfr17, aadA5 |
| 8Nb | Waterbird feces | ACfCipNaSSuSxtT | blaCTX-M-15 | blaOXA-1, aac(6′)-Ib-cr, sul1, tetA, int1, I1, 1.7 kb, dhfr17-aadA5 | M | − | B2 | + | − |
Two identical isolates (designed by PFGE) obtained from one bird using nonselective isolation and selective isolation on MacConkey agar with cefotaxime (2 mg/liter),
The strain was isolated by selective cultivation on MCA with nalidixic acid (20 mg/liter).
A, ampicillin; Ac, amoxicillin-clavulanic acid; C, chloramphenicol; Cf, cephalothin; Cip, ciprofloxacin; Gn, gentamicin; Na, nalidixic acid; S, streptomycin; Su, sulfonamides; Sxt, sulfamethoxazole-trimethoprim; T, tetracycline.
Class 1 integron.
ND, not detected.
TABLE 3.
Characterization of Escherichia coli with qnr genes from wild birds on the Baltic Sea coast of Poland
A, ampicillin; Ac, amoxicillin-clavulanic acid; Cf, cephalothin; Cip, ciprofloxacin; Gn, gentamicin; Na, nalidixic acid; S, streptomycin; Su, sulfonamides; Sxt, sulfamethoxazole-trimethoprim; T, tetracycline. Determined by the disk diffusion method.
I1, class 1 integron.
In total, 18 suspected E. coli isolates were detected in 27 samples from herring gulls. Three E. coli isolates were antibiotic resistant (Table 1). The prevalence of herring gulls with antimicrobial-resistant E. coli was 11% (3/27). While one isolate was resistant to ciprofloxacin, plasmid-encoded quinolone resistance genes were not detected. Three ESBL-producing E. coli isolates were found by selective isolation with cefotaxime (Table 2). Using selective isolation with ciprofloxacin, 20 E. coli isolates was detected; the qnrS gene was detected in three of these (Table 3).
From 365 waterbird feces, 58 suspected E. coli isolates were obtained by nonselective cultivation. Antibiotic resistance was detected in five E. coli isolates (Table 1). The prevalence of waterbird feces with antimicrobial-resistant E. coli was 1.4% (5/365). By selective cultivation with cefotaxime, three ESBL-producing E. coli isolates were detected (Table 2). Using selective cultivation with ciprofloxacin, 23 E. coli isolates were found, and 2 of these were positive for qnrS. Another 5 isolates were obtained by selective cultivation on MCA with nalidixic acid; 1 of these isolates was positive for a mutant variant aac(6′)-Ib-cr, and moreover, ESBL production was detected in this isolate (Table 2).
Characterization of ESBL-producing E. coli isolates.
Nine ESBL-producing E. coli isolates were used for further characterization (see Table 2). The most prevalent gene in these ESBL-producing isolates was blaCTX-M-1 (6 isolates). The gene blaCTX-M-9, blaCTX-M-15, or blaSHV-12 was detected in the remaining 3 isolates. In the isolate with blaCTX-M-15, the gene aac(6′)-Ib-cr, responsible for resistance to aminoglycosides and fluoroquinolones, was detected. Most of the isolates were multiresistant, and four isolates contained a class 1 integron with gene cassettes. All the isolates showed unique XbaI-PFGE profiles. Most of the plasmids containing the bla genes for ESBL production were transferable by conjugation to E. coli and/or Salmonella. Transformants or transconjugants with single plasmids carrying the blaCTX-M-1 gene were obtained from 5 isolates. Plasmid DNA purified from these transformants was used for replicon typing and RFLP performed by EcoRV (Fig. 2) and HincII digestions. The strain 5PL, from mallards, harbored the 35-kb conjugative plasmid (RFLP type I) of the IncN group. The gene blaCTX-M-1, in another 3 isolates from a herring gull (isolate 17 LA CEF) and waterbird feces (isolates 88 and 96), was located on the 90- to 100-kb conjugative plasmids of the IncI1 group. RFLP profiles of the IncI1 plasmids showed many band similarities. A plasmid 80 kb in size from the E. coli 11 LA CEF transformant was negative for the incompatibility group by the method used and showed the unique RFLP profile.
FIG. 2.

RFLP analysis of plasmid DNA harboring genes for ESBL or quinolone resistance designed by EcoRV. Lane 1, E. coli 18 PL CIP (qnrS); lane 2, E. coli 6 LA CIP (qnrS); lane 3, E. coli 11 (qnrS); lane 4, E. coli 58 PL CIP (qnrS); lanes 5 and 12, λ DNA-HindIII Digest (Biolabs, New England); lanes 6 and 13, 2-log DNA marker (Biolabs); lane 7, E. coli 5 PL (blaCTX-M-1); lane 8, E. coli 11 LA CEF (blaCTX-M-1); lane 9, E. coli 17 LA CEF (blaCTX-M-1); lane 10, E. coli 88 (blaSHV-12); lane 11, E. coli 96 (blaCTX-M-1).
Characterization of quinolone-resistant E. coli isolates.
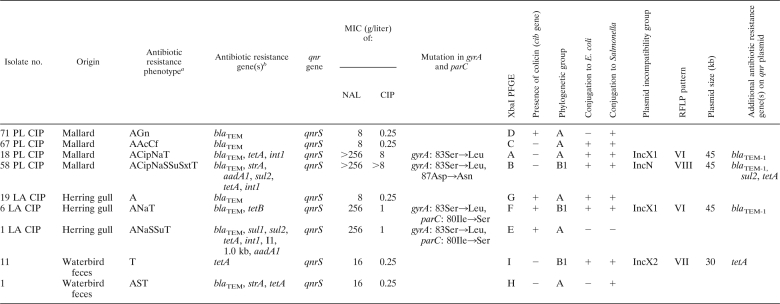
Nine quinolone-resistant isolates with the qnrS genes were characterized (Table 3). Four isolates with high MIC levels of quinolones were examined for chromosomal mutations in the genes gyrA and parC. Mutation of Ser83 to Leu in GyrA and 80Ile to Ser in ParC were the most prevalent. A class 1 integron 1 kb in size was identified in one multiresistant isolate from a herring gull. All the isolates showed unique XbaI PFGE profiles and belonged to the A (6 isolates) or B1 (3 isolates) phylogenetic group as determined by multiplex PCR. Most of the qnrS plasmids were transferable by conjugation to E. coli and/or Salmonella. Transformants or transconjugants with a single qnrS plasmid were obtained only from 4 isolates and were used for further testing of the Inc groups and EcoRV (Fig. 2) and HincII plasmid DNA profiling. The isolates 18 PL CIP from a mallard and 6 LA CIP from a herring gull harbored the same 45-kb conjugative plasmid (RFLP type VI) of the X1 incompatibility group. The gene qnrS was associated with the Tn3-like transposon on this IncX1 plasmid together with blaTEM-1 in both the isolates. A plasmid 30 kb in size from a transformant of E. coli strain 11 belonged to the IncX2 group and showed the unique RFLP profile. The qnrS gene in the isolate 58 PL CIP from a mallard was located on a 45-kb conjugative IncN plasmid. The RFLP profile of this plasmid differed from the profiles of other IncN plasmids harboring blaCTX-M-1 genes detected in this study.
DISCUSSION
Even if populations of wild birds are not directly influenced by antibiotic practice, the acquired resistance in bacteria isolated from wild waterbirds occurs in various prevalences. Studies of resistant E. coli isolates have been mostly based on phenotype characterization of these isolates (20, 24, 26, 51, 58, 71, 75, 76, 82), and the genes determining resistance to old-generation antibiotics were sporadically detected (17, 21, 22, 29). Isolates of E. coli from birds on the Polish coast of the Baltic Sea were commonly resistant to old-generation antibiotics, and they were highly variable in phenotype and genotype characteristics. Resistances to streptomycin, ampicillin, and tetracycline were recently found in waterbirds elsewhere in Europe (4, 21, 23, 29). In the United States, resistance to streptomycin and ampicillin has been frequently seen in E. coli isolates from Canada geese (17, 51).
Isolates of E. coli with class 1 integrons and gene cassettes were found in waterbirds on the Polish coasts of the Baltic Sea. The gene cassettes contained the genes aadA1, aadA2, dhfr1-aadA1, dhfr12-aadA2, and dhfr17-aadA5. In Portugal, E. coli strains with class 1 and 2 integrons have been isolated from gulls (66). In Italy, E. coli isolates with class 1 integrons have been found in Mediterranean herring gulls (29). In the Czech Republic, E. coli isolates with class 1 and 2 integrons have been found in black-headed gulls (21, 23). We may summarize that the integrons are commonly found in E. coli isolates from waterbirds living in European areas inhabited by people and that the gene cassettes detected within these integrons are highly variable even though the genes aadA1 and dhfr1 are found regularly.
The emergence and wide dissemination in recent years of E. coli resistant to broad-spectrum cephalosporins due to production of ESBL and/or E. coli resistant to fluoroquinolones due to a presence of plasmid-encoded quinolone resistance genes are of great concern and represent a serious problem for the treatment of infectious diseases (59, 79). These strains are also spread into the environment and can colonize wild mammals as well as wild birds, especially omnivorous gulls (4, 18, 23, 29, 47, 66, 67). We have proven the occurrence of E. coli isolates producing ESBL, as well as isolates with plasmid-encoded fluoroquinolone resistance, in mallards, herring gulls, and waterbird feces on the Polish coast of the Baltic Sea. Moreover, most isolates showed resistance to multiple antibiotics and contained class 1 integrons. Coresistance to non-beta-lactam antibiotics in the ESBL-producing Enterobacteriaceae is commonly described (60). Such multiresistant ESBL-producing strains are a serious problem since they severely limit therapeutic options against human and animal infections (69).
Isolates of E. coli producing ESBL found in mallards, in herring gulls, and in waterbird feces had the genes blaCTX-M-1 (most isolates), blaCTX-M-9, blaCTX-M-15, and blaSHV-12. The extended-spectrum beta-lactamase CTX-M-1 seems to be the most prevalent ESBL in animal E. coli strains throughout Europe (6, 30, 77). The various ESBLs have been found in E. coli from wild waterbirds on the Atlantic coast of Europe (66), on the coast of the Mediterranean Sea (4), and in the European inland (23). The spectrum of genes associated with the production of ESBL in Polish isolates was closer to the spectrum of genes in isolates from birds originating from the Mediterranean Sea and the European inland than to the spectrum of genes found in E. coli isolates from birds originating on the European Atlantic coast. Both gulls and mallards occurring in European areas inhabited by people are commonly colonized with E. coli strains resistant to broad-spectrum cephalosporins and having various bla genes responsible for the ESBL phenotype.
In the present study, blaCTX-M-1 was found harbored on conjugative plasmids of the incompatibility groups IncN and IncI1. The association between blaCTX-M-1 and IncN plasmids was recently described (49, 52, 55). IncN plasmids are broad-host-range plasmids that might contribute to the further spread of isolates producing CTX-M-1-like enzymes among other members of the Enterobacteriaceae. The blaSHV-12 gene in the E. coli isolate from a waterbird was harbored on an IncN conjugative plasmid. The association of SHV-12 and plasmids of this incompatibility group has been previously described (28, 49). In addition to CTX-M-1 and SHV-12, other types of ESBL have been found to be connected with IncN plasmids (28, 33). This indicates their role in disseminating ESBL genes. The IncI1 plasmids have been found to be more frequent in pathogenic than in commensal avian and human E. coli strains (35). These plasmids are also characterized by association with virulence and antibiotic resistance genes that can be involved in positive selection of these plasmids (35). Plasmids belonging to IncI1 carrying blaCTX-M-1 have recently been found in E. coli and Salmonella isolates throughout Europe (19, 28, 30, 47, 49).
The beta-lactamase CTX-M-15 is one of the most important ESBLs in human medicine. E. coli isolates producing this enzyme have emerged worldwide as an important cause of community-onset urinary tract and bloodstream infections (65). CTX-M-15 is often associated with OXA-1, TEM-1 beta-lactamases, and Aac(6′)-Ib-cr, a variant of an aminoglycoside-modifying enzyme that is responsible for reduced susceptibility to both aminoglycosides and fluoroquinolones (70). Similar findings were seen in the waterbird isolate from our study. This ESBL has been recently identified in the clone ST131, which is responsible in large measure for an international epidemic caused by CTX-M-15-producing E. coli (72). The pandemic dissemination of the clone ST131 producing CTX-M-15 was linked to the epidemic's narrow host range of IncFII plasmids (49, 62).
We found E. coli isolates with plasmid-encoded quinolone resistance associated with the qnrS gene in mallards, herring gulls, and waterbird feces on the Polish coast of the Baltic Sea. Moreover, one E. coli isolate from waterbird feces producing ESBL harbored the gene aac(6)-Ib-cr. All these findings of E. coli with genes responsible for fluoroquinolone resistance in wild birds are described here for the first time. Only recently, E. coli strains determined to be resistant to ciprofloxacin based on their resistance phenotype have been noted in waterbirds (23, 29, 76). The qnrS gene was first found in Shigella flexneri during 2003 in Japan (32), and together with the qnrB gene it has been identified as the most prevalent plasmid-mediated fluoroquinolone resistance gene (42). This gene has been detected in quinolone-resistant human, poultry, and swine E. coli and Salmonella isolates (10, 31, 34, 86).
In our study, qnrS genes were carried by plasmids of the N, X1, and X2 incompatibility groups. The association of IncN conjugative plasmids with qnrS genes was recently described (28, 37). IncX plasmids have been implicated in the acquisition and dissemination of transferable resistance in pathogenic bacteria (81).
IncX1 plasmids have recently been found to be associated with ESBL genes, such as blaTEM-52 (1, 47), and with increased production of biofilm (54, 56), which could impart to the strains carrying them a greater potential for spreading and surviving in the environment. In our study, two E. coli isolates with identical conjugative IncX1 plasmids harboring the qnrS and blaTEM-1 genes inside the Tn3-like transposon, from a mallard and a herring gull sampled in different areas of the Polish coast (the distance between the areas is about 20 km), were found. Both isolates showed high levels of resistance to quinolones and contained chromosomal mutations in the gyrA and/or parC gene. However, the isolates showed different macrorestriction profiles by PFGE and belonged to different phylogenetic groups, A and B1. This suggests a possible role for horizontal spreading of the IncX1 plasmid in the E. coli population of wild birds on the Polish coast. The association of plasmid-mediated qnrS genes with plasmids of this incompatibility group was recently described (13). The qnrS1 gene has been found to be located on a 45-kb nonconjugative IncX1 plasmid in one poultry E. coli isolate from Italy. The qnrS1 gene of this isolate was associated with a Tn3-like transposon containing the blaTEM-1 gene, as previously described for an animal isolate of Salmonella enterica serovar Infantis (39, 40). Analysis of the IncX1 plasmid carrying this element showed it to resemble that of a plasmid identified in Salmonella Dublin (14). Those authors have suggested that genetic exchange among Salmonella and E. coli has occurred, thus indicating the existence of a potential animal reservoir for the qnr genes. Further investigation of our IncX1 plasmids harboring qnrS and blaTEM-1 in E. coli from wild birds needs to be undertaken to find a possible association with the results presented by the studies mentioned above.
In one waterbird E. coli isolate, the qnrS gene was found to be located on a 30-kb conjugative plasmid of the IncX2 group. To our knowledge, this is the first report of qnrS genes on plasmids of the X2 incompatibility group. We think that our IncX2 plasmid harboring the qnrS gene could be related to the pR6K plasmid (36). Plasmids of this group are not very common, and the pR6K plasmid is the only IncX2 member described to date (36).
Mallards and gulls are identified as important bioindicators, as reservoirs and vectors of E. coli strains resistant to old-generation antibiotics, broad-spectrum cephalosporins, and fluoroquinolones, and as a potential melting pot for the development of new resistance types. We regard the common occurrence of these isolates as alarming. It provides evidence that these E. coli isolates, selected for in humans and domestic animals, are colonizing wild waterbirds and that via these birds, E. coli isolates important from a public health viewpoint can be spread into the environment. Although waterbirds do not naturally come into contact with antimicrobials, they are omnivorous and often search for food in agricultural, rural, and urban areas and in communal garbage dumps and sewage water treatment plants. Thus, they can easily be infected with resistant bacteria from domestic animal and human sources. Urban waters, too, can possibly be a source of antibiotic-resistant isolates inasmuch as these waters harbor higher percentages of resistant E. coli strains than do rural waters (38).
Waterbirds excrete large quantities of fecal coliforms, and they are theoretically capable, through their feces, of affecting drinking and recreational water quality (27, 43, 44). It is important to keep these birds away from swimming areas by limiting food supplies. Further, it is important to educate owners of beaches and restaurants, as well as the general public. Keeping beaches clean and prohibiting the feeding of birds are simple ways of limiting potential problems. The occurrence of resistant bacteria in populations of wild waterbirds where there is no selective pressure can imply that such resistance will be difficult to displace.
Acknowledgments
We thank George Jacoby (Lahey Clinic Inc., Burlington, MA), Lina Cavaco and Henrik Hasman (National Food Institute, Copenhagen, Denmark), Ming-Gui Wang (Institute of Antibiotics, Huashan Hospital, Shanghai, China), and Surbhi Malhotra-Kumar (University of Antwerp, Antwerp, Belgium) for positive-control strains. Our thanks also go to Katerina Dibdakova, Eva Suchanova, Marketa Machalkova, and Katerina Albrechtova for their excellent laboratory work. We thank the Veterinary Research Institute (Brno, Czech Republic) for providing equipment for pulsed-field gel electrophoresis.
This study was funded by grant no. MSM6215712402 from the Ministry of Education, Youth and Sports of the Czech Republic and grant no. P502/10/P083 from the Czech Science Foundation.
Wild birds were caught according to the permission given to Wlodzimierz Meissner by the Polish General Director for Environmental Protection, no. 187/2009.
Footnotes
Published ahead of print on 15 October 2010.
REFERENCES
- 1.Bergenholtz, R. D., M. S. Jorgensen, L. H. Hansen, L. B. Jensen, and H. Hasman. 2009. Characterization of genetic determinants of extended-spectrum cephalosporinases (ESCs) in Escherichia isolates from Danish and imported poultry meat. J. Antimicrob. Chemother. 64:207-209. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, G., J. A. Lemus, J. Grande, L. Gangoso, J. M. Grande, J. A. Donazar, B. Arroyo, O. Frias, and F. Hiraldo. 2007. Geographical variation in cloacal microflora and bacterial antibiotic resistance in a threatened avian scavenger in relation to diet and livestock farming practices. Environ. Microbiol. 9:1738-1749. [DOI] [PubMed] [Google Scholar]
- 3.Blanco, G., J. A. Lemus, and J. Grande. 2009. Microbial pollution in wildlife: linking agricultural manuring and bacterial antibiotic resistance in red-billed choughs. Environ. Res. 109:405-412. [DOI] [PubMed] [Google Scholar]
- 4.Bonnedahl, J., M. Drobni, M. Gauthier-Clerc, J. Hernandez, S. Granholm, Y. Kayserr, A. Melhus, G. Kahlmeter, J. Waldenstrom, A. Johansson, and B. Olsen. 2009. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. Plos ONE 4:e5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinas, L., M. Zaraga, Y. Saenz, F. Ruiz-Larrea, and C. Torres. 2002. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinas, L., M. A. Moreno, T. Teshager, Y. Saenz, M. C. Porrero, L. Dominguez, and C. Torres. 2005. Monitoring and characterization of extended-spectrum beta-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob. Agents Chemother. 49:1262-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camarda, A., E. Circella, D. Pennelli, A. Madio, G. Bruni, V. Lagrasta, G. Marzano, E. Mallia, and E. Campagnari. 2006. Wild birds as biological indicators of environmental pollution: biotyping and antimicrobial resistance patterns of Escherichia coli isolated from Audouin's gulls (Larus audouinii) living in the Bay of Gallipoli (Italy). Ital. J. Anim. Sci. 5:287-290. [Google Scholar]
- 8.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 9.Cattoir, V., L. Poirel, V. Rotimi, C. J. Soussy, and P. Nordmann. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394-397. [DOI] [PubMed] [Google Scholar]
- 10.Cavaco, L. M., N. Frimodt-Moller, H. Hasman, L. Guardabassi, L. Nielsen, and F. M. Aarestrup. 2008. Prevalence of quinolone resistance mechanisms and associations to minimum inhibitory concentrations in quinolone-resistant Escherichia coli isolated from humans and swine in Denmark. Microb. Drug Resist. 14:163-169. [DOI] [PubMed] [Google Scholar]
- 11.Cavaco, L. M., H. Hasman, S. Xia, and F. M. Aarestrup. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovars Kentucky and Bovismorbificans of human origin. Antimicrob. Agents Chemother. 53:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC. 2004. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. National Molecular Network for Foodborne Disease Surveillance, CDC, Atlanta, GA.
- 13.Cerquetti, M., A. Garcia-Fernandez, M. Giufre, D. Fortini, M. Accogli, C. Graziani, I. Luzzi, A. Caprioli, and A. Carattoli. 2009. First report of plasmid-mediated quinolone resistance determinant qnrS1 in an Escherichia coli strain of animal origin in Italy. Antimicrob. Agents Chemother. 53:3112-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu, C., Y. Feng, A. C. Chien, S. Hu, C. H. Chu, and C. H. Chiu. 2008. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339-343. [DOI] [PubMed] [Google Scholar]
- 15.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. 2008. Performance standards for antimicrobial susceptibility testing, 8th informational supplement. CLSI, Wayne, PA.
- 17.Cole, D., D. J. V. Drum, D. E. Stallknecht, D. G. White, M. D. Lee, S. Ayers, M. Sobsey, and J. J. Maurer. 2005. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 11:935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa, D., P. Poeta, Y. Saenz, L. Vinue, B. Rojo-Bezares, A. Jouini, M. Zarazaga, J. Rodriguez, and C. Torres. 2006. Detection of Escherichia coli harbouring extended-spectrum β-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 58:1311-1312. [DOI] [PubMed] [Google Scholar]
- 19.Dierikx, C., A. van Essen-Zandbergen, K. Veldman, H. Smith, and D. Mevius. 2010. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 145:273. [DOI] [PubMed] [Google Scholar]
- 20.Dobbin, G., H. Hariharan, P.-Y. Daoust, S. Hariharan, S. Heaney, M. Coles, L. Price, and C. A. Muckle. 2005. Bacterial flora of free-living double-crested cormorant (Phalacrocorax auritus) chicks on Prince Edward Island, Canada, with reference to enteritic bacteria and antibiotic resistance. Comp. Immunol. Microbiol. Infect. Dis. 28:71-82. [DOI] [PubMed] [Google Scholar]
- 21.Dolejska, M., A. Cizek, and I. Literak. 2007. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 103:11-19. [DOI] [PubMed] [Google Scholar]
- 22.Dolejska, M., D. Senk, A. Cizek, J. Rybarikova, O. Sychra, and I. Literak. 2008. Antimicrobial resistant Escherichia coli isolates in cattle and house sparrows on two Czech dairy farms. Res. Vet. Sci. 85:491-494. [DOI] [PubMed] [Google Scholar]
- 23.Dolejska, M., B. Bierosova, L. Kohoutova, I. Literak, and A. Cizek. 2009. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106:1941-1950. [DOI] [PubMed] [Google Scholar]
- 24.Edge, T. A., and S. Hill. 2005. Occurrence of antibiotic resistance in Escherichia coli from surface waters and fecal pollution sources near Hamilton, Ontario. Can. J. Microbiol. 51:501-505. [DOI] [PubMed] [Google Scholar]
- 25.Everett, M. J., Y. F. Jin, V. Ricci, and L. J. Piddock. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallacara, D. M., C. M. Monahan, T. Y. Morishita, and R. W. Wack. 2001. Fecal shedding and antimicrobial susceptibility of selected bacterial pathogens and a survey of intestinal parasites in free-living waterfowl. Avian Dis. 45:128-135. [PubMed] [Google Scholar]
- 27.Fogarty, L. R., S. K. Haack, M. J. Wolcott, and R. I. Whitman. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 94:865-878. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Fernandez, A., G. Chiaretto, A. Bertini, L. Villa, D. Fortini, A. Ricci, and A. J. Carattoli. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61:1229-1233. [DOI] [PubMed] [Google Scholar]
- 29.Gionechetti, F., P. Zucca, F. Gombac, C. Monti-Bragadin, C. Lagatolla, E. Tonin, E. Edalucci, L. A. Vitali, and L. Dolzani. 2008. Characterization of antimicrobial resistance and class 1 integrons in Enterobacteriaceae isolated from Mediterranean herring gulls (Larus cachinans). Microb. Drug Resist. 14:93-99. [DOI] [PubMed] [Google Scholar]
- 30.Girlich, D., L. Poirel, A. Carattoli, I. Kempf, M. F. Lartigue, A. Bertini, and P. Nordmann. 2007. Extended-spectrum beta-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl. Environ. Microbiol. 73:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunell, M., M. A. Webber, P. Kotilainen, A. J. Lilly, J. M. Caddick, J. Jalava, P. Huovinen, A. Siitonen, A. J. Hakanen, and L. J. Piddock. 2009. Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob. Agents Chemother. 53:3832-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopkins, K. L., E. Liebana, L. Villa, M. Batchelor, E. J. Threlfall, and A. Carattoli. 2006. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins, K. L., L. Wootton, M. R. Day, and E. J. Threlfall. 2007. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J. Antimicrob. Chemother. 59:1071-1075. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, T. J., Y. M. Wannemuehler, S. J. Johnson, C. M. Logue, D. G. White, C. Doetkott, and L. K. Nolan. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73:1976-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, C. S., D. J. Osborne, and J. Stanley. 1993. Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J. Gen. Microbiol. 139:735-741. [DOI] [PubMed] [Google Scholar]
- 37.Karah, N., L. Poirel, S. Bengtsson, M. Sundqvist, G. Kahlmeter, P. Nordmann, A. Sundsfjord, O. Samuelsen, and Norwegian Study Group on PMQR. 2010. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in Escherichia coli and Klebsiella spp. from Norway and Sweden. Diagn. Microbiol. Infect. Dis. 66:425-431. [DOI] [PubMed] [Google Scholar]
- 38.Kaspar, C. W., J. L. Burgess, I. T. Knight, and R. R. Colwell. 1990. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can. J. Microbiol. 36:891-894. [DOI] [PubMed] [Google Scholar]
- 39.Kehrenberg, C., S. Friederichs, A. de Jong, G. B. Michael, and S. Schwarz. 2006. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J. Antimicrob. Chemother. 58:18-22. [DOI] [PubMed] [Google Scholar]
- 40.Kehrenberg, C., A. de Jong, S. Friederichs, A. Cloeckaert, and S. Schwarz. 2007. Molecular mechanisms of decreased susceptibility to fluoroquinolones in avian Salmonella serovars and their mutants selected during the determination of mutant prevention concentrations. J. Antimicrob. Chemother. 59:886-892. [DOI] [PubMed] [Google Scholar]
- 41.Kim, H. B., C. H. Park, C. J. Kim, E. C. Kim, G. A. Jacoby, and D. C. Hooper. 2009. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob. Agents Chemother. 53:639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavilla, S., J. J. Gonzalez-Lopez, M. Sabate, A. Garcia-Fernandez, M. N. Larrosa, R. M. Bartolome, A. Carattoli, and G. Prats. 2008. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J. Antimicrob. Chemother. 61:291-295. [DOI] [PubMed] [Google Scholar]
- 43.Levesque, B., P. Brousseau, P. Simard, E. Dewailly, M. Meisels, D. Ramsay, and J. Joly. 1993. Impact of the ring-billed gull (Larus delawarensis) on the microbiological quality of recreational water. Appl. Environ. Microbiol. 59:1228-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levesque, B., P. Brousseau, F. Bernier, E. Dewailly, and J. Joly. 2000. Study of the bacterial content of ring-billed gull droppings in relation to recreational water quality. Water Res. 34:1089-1096. [Google Scholar]
- 45.Lewis, J. S., II, M. Herrera, B. Wickes, and J. E. Petterson. 2007. First report of the emergence of CTX-M-type extended spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Literak, I., R. Vanko, M. Dolejska, A. Cizek, and R. Karpiskova. 2007. Antibiotic resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett. Appl. Microbiol. 45:616-621. [DOI] [PubMed] [Google Scholar]
- 47.Literak, I., M. Dolejska, T. Radimersky, J. Klimes, M. Friedman, F. M. Aarestrup, H. Hasman, and A. Cizek. 2010. Antimicrobial resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum beta-lactamases in wild boars. J. Appl. Microbiol. 108:1702-1711. [DOI] [PubMed] [Google Scholar]
- 48.Livermore, D. M., M. Warner, L. M. C. Hall, V. I. Enne, S. J. Projan, P. M. Dunman, S. L. Wooster, and G. Harrison. 2001. Antibiotic resistance in bacteria from magpies (Pica pica) and rabbits (Oryctolagus cuniculus) from west Wales. Environ. Microbiol. 3:658-661. [DOI] [PubMed] [Google Scholar]
- 49.Marcade, G., C. Deschamps, A. Boyd, V. Gautier, B. Picard, C. Branger, E. Denamur, and G. Arlet. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 63:67-71. [DOI] [PubMed] [Google Scholar]
- 50.Meissner, W., J. Typiak, A. Kosmicki, and S. Bzoma. 2009. Numbers of waterbirds on the Bay of Gdansk in the span May 2007-April 2008. Notatki Ornitol. 50:65-72. (In Polish.) [Google Scholar]
- 51.Middleton, J. H., and A. Ambrose. 2005. Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl. Dis. 41:334-341. [DOI] [PubMed] [Google Scholar]
- 52.Moodley, A., and L. Guardabassi. 2009. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 53:1709-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamura, M., H. Yoshimura, and T. Koeda. 1982. Drug resistance and R plasmids of Escherichia coli strains isolated from six species of wild birds. Jpn. J. Vet. Sci. 44:465-471. [DOI] [PubMed] [Google Scholar]
- 54.Norman, A., L. H. Hansen, Q. She, and S. J. Sorensen. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59-74. [DOI] [PubMed] [Google Scholar]
- 55.Novais, A., R. Canton, R. Moreira, L. Peixe, F. Baquero, and T. M. Coque. 2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51:796-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ong, C. L., S. A. Beatson, A. G. McEwan, and M. Schembri. 2009. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl. Environ. Microbiol. 75:6783-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum beta lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paterson, D. L. 2006. Resistance in Gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control. 119:S20-S28. [DOI] [PubMed] [Google Scholar]
- 61.Pawiak, R., M. Mazurkiewicz, J. Molenda, J. Pinowski, and A. Wieliczko. 1989. The occurrence of Escherichia coli strains pathogenic to humans and animals in the eggs and nestlings of Passer spp., p. 139-151. In J. Pinowski, B. P. Kavanagh, and W. Górski (ed.), Nestling mortality of granivorous birds due to microorganisms and toxic substances. Proceedings of the International Symposium of the Working Group on Granivorous Birds, INTECOL. Polish Scientific Publishers, Warsaw, Poland.
- 62.Peirano, G., M. Costello, J. D. Pitout. 2010. Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int. J. Antimicrob. Agents 36:19-23. [DOI] [PubMed] [Google Scholar]
- 63.Perichon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by qepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pitout, J. D., A. Hossain, and N. D. Hanson. 2004. Phenotypic and molecular detection of CTX-M-beta-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 42:5715-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pitout, J. D. 2010. Infections with extended-spectrum beta-lactamase-producing enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs 70:313-333. [DOI] [PubMed] [Google Scholar]
- 66.Poeta, P., H. Radhouani, G. Igrejas, A. Goncalves, C. Carvalho, J. Rodrigues, L. Vinue, S. Somalo, and C. Torres. 2008. Seagulls of the Berlengas natural reserve of Portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl. Environ. Microbiol. 74:7439-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poeta, P., H. Radhouani, L. Pinto, A. Martinho, V. Rego, R. Rodrigues, A. Goncalves, J. Rodrigues, V. Estepa, C. Torres, and G. Igrejas. 2009. Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 49:584-588. [DOI] [PubMed] [Google Scholar]
- 68.Radimersky, T., P. Frolkova, D. Janoszowska, M. Dolejska, P. Svec, E. Roubalova, P. Cikova, A. Cizek, and I. Literak. 2010. Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. J. Appl. Microbiol. 109:1687-1695. [DOI] [PubMed] [Google Scholar]
- 69.Rice, L. B. 2009. The clinical consequences of antimicrobial resistance. Curr. Opin. Microbiol. 12:476-481. [DOI] [PubMed] [Google Scholar]
- 70.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 71.Rose, J. M., R. J. Gast, A. Bogomolni, J. C. Ellis, B. J. Lentell, K. Touhey, and M. Moore. 2009. Occurrence and patterns of antibiotic resistance in vertebrates off the Northeastern United States coast. FEMS Microbiol. Ecol. 67:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rossolini, G. M., M. M. D'Andrea, C. Mugnaioli. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14:33-41. [DOI] [PubMed] [Google Scholar]
- 73.Rybarikova, J., M. Dolejska, D. Materna, I. Literak, and A. Cizek. 2010. Phenotypic and genotypic characteristics of antimicrobial resistant Escherichia coli isolated from symbovine flies, cattle and sympatric insectivorous house martins from a farm in the Czech Republic (2006-2007). Res. Vet. Sci. 89:179-183. [DOI] [PubMed] [Google Scholar]
- 74.Sato, G., C. Oka, M. Asagi, and N. Ishiguro. 1978. Detection of conjugative R plasmids conferring chloramphenicol resistance in Escherichia coli isolated from domestic and feral pigeons and crows. Zentralbl. Bakteriol. Orig. A 241:407-417. [PubMed] [Google Scholar]
- 75.Sayah, R. S., J. B. Kaneene, Y. Johnson, and R. A. Miller. 2005. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sjolund, M., J. Bonnedahl, J. Hernandez, S. Bengtsson, G. Cederbrant, J. Pinhassi, G. Kahlmeter, and B. Olsen. 2008. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg. Infect. Dis. 14:70-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smet, A., A. Martel, D. Persoons, J. Dewulf, M. Heyndrickx, B. Catry, L. Herman, F. Haesebrouck, and P. Butaye. 2008. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 52:1238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sreedharan, S., M. Oram, B. Jensen, L. R. Peterson, and L. M. Fisher. 1990. DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J. Bacteriol. 172:7260-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strahilevitz, J., G. A. Jacoby, D. C. Hooper, and A. Robicsek. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomson, K. S., and C. C. Sanders. 1992. Detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob. Agents Chemother. 36:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Threlfall, E. J., L. R. Ward, and B. Rowe. 1986. R plasmids in Salmonella typhimurium in the United Kingdom. J. Antimicrob. Chemother. 18(Suppl. C):175-177. [DOI] [PubMed] [Google Scholar]
- 82.Tsubokura, M., A. Matsumoto, K. Otsuki, S. B. Animas, and T. Sanekata. 1995. Drug resistance and conjugative R plasmids in Escherichia coli strains isolated from migratory waterfowl. J. Wildl. Dis. 31:352-357. [DOI] [PubMed] [Google Scholar]
- 83.Wallace, J. S., T. Cheasty, and K. Jones. 1997. Isolation of Vero cytotoxin-producing Escherichia coli O157 from wild birds. J. Appl. Microbiol. 82:399-404. [DOI] [PubMed] [Google Scholar]
- 84.Wu, S., E. Chouliara, H. Hasman, A. Dalsgaard, A. Vieira, L. B. Jensen. 2008. Detection of a single isolate of CTX-M-1-producing Escherichia coli from healthy pigs in Denmark. J. Antimicrob. Chemother. 61:747-749. [DOI] [PubMed] [Google Scholar]
- 85.Yamane, K., J. Wachino, S. Suzuki, and Y. Arakawa. 2008. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 52:1564-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yue, L., H. X. Jiang, X. P. Liao, J. H. Liu, S. J. Li, X. Y. Chen, C. X. Chen, D. H. Lu, and Y. H. Liu. 2008. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet. Microbiol. 132:414-420. [DOI] [PubMed] [Google Scholar]