Abstract
Detection of specific genetic markers can rapidly identify the presence of enteric viruses in groundwater. However, comparison of stability characteristics between genetic and infectivity markers is necessary to better interpret molecular data. Human adenovirus serotype 2 (HAdV2), in conjunction with MS2 phages or GA phages, was spiked into raw groundwater microcosms. Viral stability was periodically assessed by both infectivity and real-time PCR methods. The results of this yearlong study suggest that adenoviruses have the most stable persistence profile and an ability to survive for a long time in groundwater. According to a linear regression model, infectivity reductions of HAdV2 ranged from 0.0076 log10/day (4°C) to 0.0279 log10/day (20°C) and were significantly lower than those observed for phages. No adenoviral genome degradation was observed at 4°C, and the reduction was estimated at 0.0036 log10/day at 20°C. Occurrence study showed that DNA of human adenoviruses could be observed in groundwater from a confined aquifer (7 of the 60 samples were positive by real-time PCR), while no fecal indicators were detected. In agreement with the persistence of genetic markers, the presence of adenoviral DNA in groundwater may be misleading in term of health risk, especially in the absence of information on the infective status.
Groundwater is an important source of drinking water in many regions of the world. Usually, groundwater is considered to have a more stable composition and a higher microbial quality than those of surface waters. This is partly due to the slow filtration of the water through layers of soil and sediments, removing pathogenic microorganisms and many chemical compounds (19). Nevertheless, recent studies have pointed out the role of groundwater as a source of outbreaks in countries of different economic levels (10, 13, 26, 31). Among waterborne pathogens, enteric viruses seem to have a high potential to reach aquifers, and the genome of enteroviruses, noroviruses, rotaviruses, or hepatitis A viruses has been detected in many groundwater supplies, including private household wells, municipal wells, and unconfined aquifers (2, 4, 11). In these studies, the authors identify nearby surface waters or septic tanks as the most probable sources of contamination. More surprisingly, enteroviral genome as well as infectious enteroviruses was detected within a confined aquifer (5), and coliphages were identified in an isolated sandstone aquifer at a depth of 91 meters (37).
The presence of enteric viruses may be explained by their physical-chemistry characteristics. These viruses are extremely small (20 to 100 nm), readily passing through sediment pores that trap larger pathogens such as bacteria and protozoa (36). Some viruses also show surface properties that allow very high rates of diffusion through soils (18, 35). Another explanation is linked to the survival of viral particles. Infectious enteric viruses commonly persist longer in the environment than do fecal bacteria (39). Further, the viral genome has a longer persistence than does an infectious particle (14). Many of these last considerations have been reported by using the MS2 phage, an F-specific RNA phage (FRNAPH), as a surrogate of enteric viruses. F-specific RNA phages were commonly chosen as models for evaluating transport of pathogenic viruses in soil and aquifers and as viral fecal indicators (28). On the other hand, host specificity displayed by their four genogroups also renders F-specific RNA phages capable of providing information on sources of fecal pollution (20). According to the numerous studies performed thus far, F-specific RNA phages of genogroups II (GA-like) and III are generally associated with human pollution, whereas those of genogroups I (MS2-like) and IV are associated with animal pollution, although there are some exceptions.
Among waterborne viruses, human adenoviruses (HAdV) are nowadays described as emerging pathogens (25) and are considered to be highly resistant in water (8). High quantities of adenovirus DNA were recently detected in polluted surface waters (16, 32, 34, 41), and this genetic marker seems to be a very long-lasting indicator of fecal pollution in surface waters (21). Nevertheless, there is still a significant lack of data on the occurrence and persistence of these viruses in groundwater (30).
In that context, we first evaluated the survival and persistence of viruses in groundwater microcosms inoculated with human adenovirus serotype 2 (HAdV2) and two F-specific RNA bacteriophages (MS2 and GA) by using both real-time reverse transcription-PCR (RT-PCR) and cell culture. Further, our study was also designed to provide the first data on occurrence of both human adenoviruses and F-specific RNA phages in different confined and unconfined aquifers located in France. The occurrence study was assessed in situ in four different sampling sites belonging to two aquifers by using real-time PCR (n = 60). Our data should help researchers to better understand the implications of detection of viruses, especially adenoviruses, by molecular tools in groundwater samples.
MATERIALS AND METHODS
Sample sites.
In this study, groundwater from two French aquifers of different types was sampled. The first is described as a confined aquifer. In this aquifer, water circulates very slowly by gravity through a permeable layer, mainly made of sand and gravel, protected by a layer of clay and boulders several tens of meters thick. The second can be described as an unconfined aquifer. Water filters through up to hundreds of meters of sand in the upper part. Fractured rocks collect this gently filtered water as a drain in the lower part of the system. Four sampling sites (S1 to S4) were defined. Three sampling sites (S1, S2, and S3), corresponding to three distinct groundwater types, were located on the first aquifer at depths ranging from 25 to 130 meters. One sampling site (S4) was located on the second aquifer at a depth of 80 meters. For all groundwater samples, physical and chemical parameters (pH and some ion concentrations) were measured in situ. Values are presented in Table 1.
TABLE 1.
Hydrogeological and physicochemical characteristics of groundwaters
| Parameter | Sampling site |
|||
|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | Site 4 | |
| Depth (m) | Artesian | 110-130 | 25-43 | 80 |
| Flowthrough time | >2-3 yr | >5 yr | >2-3 yr | 3-6 mo |
| pH | 7.2 | 7.1 | 7.2 | 7 |
| Calcium (mg/liter) | 80 | 82 | 88 | 10 |
| Magnesium (mg/liter) | 25 | 28 | 27 | 8 |
| Sodium (mg/liter) | 5 | 8 | 6 | 11 |
| Sulfates (mg/liter) | 10 | 14 | 11 | 10 |
| Bicarbonates (mg/liter) | 355 | 365 | 370 | 70 |
Fecal indicators.
The classical fecal indicators were employed to evaluate the microbiological quality of the groundwater. Escherichia coli, total coliforms, and fecal coliforms were counted from 250 ml of groundwater by a membrane filter procedure in accordance with the International Organization for Standardization (ISO) standard (24). PFU of somatic coliphages were counted by the double-agar layer technique according to the ISO procedure (23). These phages were initially concentrated by membrane filtration from about 4 liters of groundwater.
Viral stock preparation.
Two representative F-specific RNA bacteriophage strains, MS2 and GA, belonging to genogroup I and genogroup II, respectively, were included in the present study. MS2 phages were obtained from a culture collection (ATCC 15597), and GA phages were provided by J. Jofre (University of Barcelona). The phages were replicated separately according to the standard procedure (22) without the chloroform step, using Escherichia coli Hfr K-12 (ATCC 23631) as the bacterial host. After replication, the phage suspension was centrifuged (3,000 × g for 20 min) and the supernatant was decontaminated by filtration through a sterile membrane filter (0.22 μm). Initial concentrations of viral stocks were about 1010 PFU/ml. The stocks were then stored at 4°C.
The human adenovirus serotype 2 strain (Health Protection Agency Culture Collections [HPACC]; reference no. NCPV213) was propagated in human lung carcinoma cell line A549 (HPACC; reference no. ECACC86012804) growing in minimum essential medium (MEM; Sigma) containing 5% fetal bovine serum (Invitrogen) and 1% l-glutamine (Sigma). A549 cells support the replication of most of the human adenovirus serotypes, except the fastidious serotypes 40 and 41. Viral stock was prepared by three freezing-thawing cycles followed by centrifugation (175 × g for 5 min) and filtration of the supernatant through a sterile membrane filter (5 and 0.45 μm). The stock was then stored at −80°C. Virus quantities were expressed as most probable number of cytopathic units (MPNCU). The initial concentration of adenoviral stock was about 6 × 107 MPNCU/ml.
Experimental procedures. (i) Stability of phages and adenoviruses in artificially seeded microcosms.
Microcosms consisted of raw groundwater collected at site S1. This water was sent to the laboratory within 24 h and stored in the dark at 4°C until inoculation of viruses. Two microcosms were constructed in 2-liter sterile glass bottles, each containing 1 liter of raw groundwater. Each microcosm was inoculated either with a mixture of MS2 phage and HAdV2 or with a mixture of GA phage and HAdV2. Suspensions were homogenized by a 30-min stirring. The final concentration of all virus species was 106 PFU or MPNCU/ml. This high concentration was chosen to obtain several experimental points for the estimation of the decay rates. To test the effect of temperature on survival and persistence of viruses, each viral microcosm was then dispatched in two series of 4.5-ml sterile polypropylene tubes (4 ml per tube) and stored in the dark at 4 or 20°C. Except temperature and viral species, all variables applied to the two microcosms were similar, including groundwater source, volume, darkness, and quantity of viral particles. At the time point of analysis, one tube per microcosm and per temperature was tested by cell culture and real-time reverse transcription-PCR (RT-PCR). The duration of the persistence experiment was about 400 days.
(ii) Occurrence in groundwater.
Viruses were concentrated from groundwater by a membrane filtration procedure using a ZetaPlus Virosorb 1MDS (Cuno; reference no. NM04711-1MDS) 47-mm electropositive, charge-modified, glass and cellulose membrane. The membrane was introduced into a metallic cartridge (Sartorius; reference no. SM 16249) with a supplementary metallic structure allowing reduction of water pressure and thus slowing membrane deterioration. To detect indigenous enteric viruses, a 50-liter volume of groundwater was filtered in situ at a filtration rate of approximately 100 liters/h. The filtration devices were then immediately sent to the laboratory, and total nucleic acid extraction was performed on the membrane. The occurrence of genome belonging to HAdV and the four genogroups of FRNAPH was tested by real-time RT-PCR. A total of 60 samples were collected and analyzed for bacteriological parameters, coliphages, and adenoviruses over a 6-month monitoring period. Groundwater sampling was conducted in 2007 (October, November, and December) and 2008 (February, March, and April). Moreover, on each sampling date, an additional filtration device was sent to the sampling site. It was not submitted to water filtration, but the membrane was tested as a control for all the viral analyses. Each sample (groundwater sample or control) was analyzed twice successively and was considered positive if both analyses were positive. In the case of a positive sample, the PCR product was also submitted to sequencing (Beckman Coulter Genomics, United Kingdom) to check the specificity.
Extraction of viral RNA and DNA.
The following two protocols were used to extract the viral genome, depending on the experiment. (i) In the persistence experiments, viral RNA (FRNAPH) and DNA (HAdV2) were extracted at the same time using the QIAamp viral RNA mini kit (Qiagen) according to the manufacturer's instructions. Extraction was performed on 140 μl of seeded water; the final volume was 100 μl. The extracts were immediately stored at −80°C. (ii) In occurrence experiments, the filter membrane was directly incubated for 10 min at room temperature under slight agitation (200 rpm) in a 50-ml-volume conical tube containing 10 ml of NucliSens lysis buffer (bioMérieux). The membrane was then removed, and the nucleic acids were extracted with a NucliSens magnetic extraction kit (bioMérieux) in accordance with the supplier's instructions. The elution step was carried out twice successively, and the 100 μl of extracted nucleic acids was immediately stored at −80°C.
Real-time PCRs.
For human adenovirus detection, the conserved region of the hexon gene was used as the target area. The real-time PCR was performed according to the assay previously described by Hernroth et al. (17) with degenerated primers and probe Ad(ACDEF). By using a standard curve generated by serial dilutions of known amounts of a linearized plasmid containing the target region, the real-time PCR sensitivity has been estimated as 5 to 10 genome copies/reaction.
F-specific RNA phages were amplified with genogroup-specific RT-PCR primers and probes by using the protocol described earlier (33). The limits of detection for various FRNAPH ranged from 1 to 10 PFU/ml for MS2, 0.01 to 0.1 PFU/ml for GA and SP, and 0.1 to 1 PFU/ml for phage Qβ.
All real-time PCR amplifications were performed on an ABI Prism 7700 Sequence Detector using SDS v.1.6.3 software (Applied Biosystems). RNase-DNase-free water was used as a negative control during reverse transcription and PCRs while genomes from MS2, GA, Qβ, and SP phages (genogroups I to IV, respectively) and HAdV2 were used as positive controls.
Enumeration of infectious viruses.
(i) F-specific RNA phages were quantified by the double-agar layer plaque assay method (22), using Escherichia coli K-12 Hfr as host. Phage concentration was then expressed in PFU per milliliter. (ii) Infectious HAdV2 was quantified in 96-well microplaques by using A549 cells. Fifty-microliter portions of three successive logarithmic dilutions of each sample were added to 200 μl of preparation containing 1 × 104 A549 cells/ml in MEM containing 2% fetal bovine serum, 1% l-glutamine, and 1% antibiotic-antimycotic solution (penicillin-streptomycin-amphotericin B). Each dilution was seeded in 40 wells. The plates were incubated in 5% CO2 at 37°C for 11 days and then examined for cytopathic effects. The final result of each sample analyzed was expressed as the geometric mean of the most probable number of cytopathic units (MPNCU) per milliliter calculated for two independent replicates.
Statistical analysis.
For the persistence experiment, data were analyzed as previously described (40). Briefly, regression analysis was used to model the survival of each virus as a function of time. To this end, simple linear regression models were fitted to experimental data and used to estimate T90 (predicted number of days to achieve a 1-log10 reduction) reduction times. The survival function characterizing each virus under the given experimental conditions (i.e., temperature fixed at 4°C or 20°C) was modeled as log10 (Qt/Q0) = at, where Qt (Q0) is the viral density at time t (respectively 0), expressed in days. The slopes calculated for the different viruses considered express the corresponding concentration decreases in log10 units per day and were subsequently subjected to statistical comparisons. Concretely, the t test was performed on the slopes taken two-by-two to determine significant differences in persistence between viruses under the given experimental conditions. On the other hand, the results of the occurrence study were submitted to a nonparametric statistical analysis (Fisher exact test) to evaluate the significance of the level of viral contamination associated with each sampling site. The calculations were performed using XLSTAT software (Addinsoft, France).
RESULTS AND DISCUSSION
Stability of enteric viruses in groundwater.
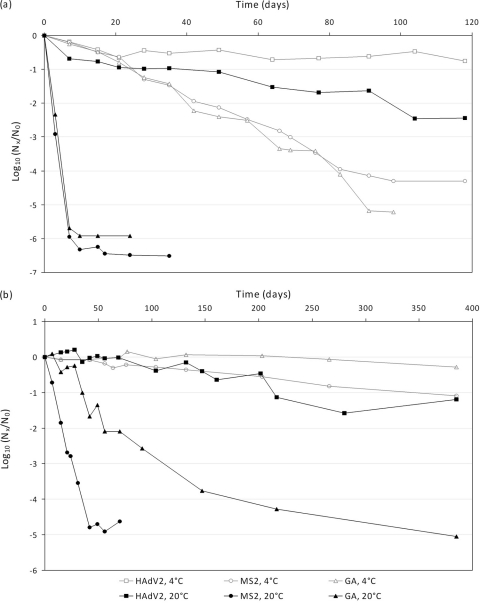
To estimate how long HAdV and FRNAPH particles survive and genetic (DNA or RNA) markers persist in an aquifer, we constructed groundwater microcosms by using defined parameters. Microcosms were fed with raw groundwater because indirect influence of the indigenous microorganism activities on viral decay has been previously suggested (15), and temperature was included as a variable because it is an important factor influencing viral stability (42). Microcosms were inoculated with either HAdV2 and MS2 phage or HAdV2 and GA phage and then stored in the dark over a 400-day period. The data obtained (Fig. 1) were analyzed by regression modeling. The slopes of the fitted log-linear regression models, the corresponding decay rates (i.e., percentage of decay per day), and the estimated T90 values (predicted number of days to achieve a 1-log10 reduction) are presented in Table 2. However, the data corresponding to HAdV2 genome degradation at 4°C could not be modeled because no decrease of this target was observed during the 385 days of the study.
FIG. 1.
Decay of virus concentration [log10 (Nx/N0)] as determined by infectivity assays (a) and real-time PCR (b) at 4°C and 20°C in groundwater microcosms. In the infectivity assay, Nx and N0 are expressed in PFU/ml for phages or MPNCU/ml for adenovirus. In the molecular assay, decay was calculated using the threshold cycle (CT) values and the slope of the standard curve: log10 (Nx/N0) = (CT0 − CTx)/slope. The value corresponding to each time point is the geometric mean of two independent replicates (n = 2).
TABLE 2.
Decay rates of MS2 phage, GA phage, and human adenovirus 2 in raw groundwater at 4 and 20°C
| Virus | Temp (°C) | Cell culture |
Real-time RT-PCR |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Slope | P valuea | Decay rate (%/day) | T90 (days) | 95% confidence limit (days) | Slope | P valuea | Decay rate (%/day) | T90 (days) | 95% confidence limits (days) | ||
| MS2 phage | 4 | −0.0428 | <10−4 | 9.4 | 23.4 | 22.5; 24.2 | −0.0029 | <10−4 | 0.7 | 344.8 | 323.9; 365.7 |
| 20 | −0.7195 | <10−4 | 80.9 | 1.4 | 1.1; 1.6 | −0.1166 | <10−4 | 23.5 | 8.6 | 8.1; 9.0 | |
| GA phage | 4 | −0.0503 | <10−4 | 10.9 | 19.9 | 19.1; 20.7 | −0.0004 | 0.019 | 0.1 | 2,500.0 | 109.9; 4,890.1 |
| 20 | −0.8081 | <10−4 | 84.4 | 1.2 | 1.1; 1.3 | −0.0278 | <10−4 | 6.2 | 35.9 | 32.8; 39.2 | |
| HAdV2 | 4 | −0.0076 | <10−4 | 1.7 | 131.6 | 100.7; 162.8 | NSb | ||||
| 20 | −0.0279 | <10−4 | 6.2 | 35.6 | 33.3; 38.4 | −0.0036 | <10−4 | 0.8 | 278.0 | 193.3; 362.2 | |
Note that use of the Bonferroni correction has only a slight effect on the outcome of the multiple testing procedure displayed in the table, since the slopes of the fitted models (excepting the GA genome at 4°C) remain highly significantly different from 0.
NS, slope value was not significantly different from 0.
As expected, a strong effect of the temperature on viral stabilities was observed. In comparing the survival rates (i.e., the slopes of the fitted models) of all the studied viruses under controlled conditions, it was shown that viruses were systematically more persistent at 4°C than at 20°C (t tests, P < 10−7). In the same way, the reduction of infectivity was faster than the disappearance of genetic markers at 4°C and 20°C, regardless of the virus considered (t tests, P < 10−12). This comparison of infectivity and genetic markers is consistent with previous studies performed with poliovirus, coxsackievirus, and feline calicivirus seeded in mineral water (14) or artificial groundwater (7).
On the one hand, the two phages exhibited similar survival rates, while MS2 genome persisted less than did the GA genome. More specifically, Fig. 1a shows that the reduction of infectivity of MS2 and GA reached more than 5 log10 in 7 days at 20°C. MS2 and GA phages were detectable for 35 and 24 days, respectively. At this last temperature, the two infectious survival profiles were the same (t test, P = 0.307). At 4°C, the infectivity seemed to decrease slower for MS2 than for GA (t test, P = 10−7); nevertheless, the corresponding two T90 values were quite similar (i.e., 19.9 days for GA and 23.4 days for MS2). Only MS2 phage inactivation has previously been studied in groundwater, and no data are available on the GA phage. Our T90 values were in the range of those reported by others. Yates et al. (42) were the first to report data on MS2 inactivation in groundwater. From their results, the T90 values were calculated to be about 37 and 3 days at 4°C and 23°C, respectively. More recently, similar data were described with T90 values of over 20 and 11 days at 4°C and 25°C, respectively (3). Along with the significant interest in molecular methods for enteric virus detection, it is also necessary to better understand persistence of the genetic marker in groundwater. Here, we showed for the first time that the decrease in RNA genome reached 1.10 log10 for MS2 and 0.29 log10 for GA in 385 days, at 4°C. At 20°C, the MS2 genome was no longer detectable after 50 days and the decrease of GA genome reached 5.05 log10 in 385 days (Fig. 1b). Thus, GA RNA was more persistent than was MS2 RNA in our groundwater microcosms (t tests, P < 10−15). Persistence profiles of FRNAPH genomes have been previously compared (27) in filtered seawater microcosms incubated away from direct sunlight at ambient temperature (21 to 23°C) over a 7-day period. Under these unfavorable conditions, a more rapid genome degradation was observed, but the values were quite similar for MS2 and GA phages. Indeed, the T90 values reached 2.9 and 2.1 days for MS2 and GA, respectively.
On the other hand, HAdV2 was the most stable of the three viruses tested by both cell culture and real-time PCR, and differences were evident between HAdV2 and the two FRNAPH (t tests, P < 10−12). Unlike MS2 and GA, adenoviruses were detected throughout the monitoring period and the reduction of infectivity within 120 days reached only 2.4 and 0.7 log10 at 20°C and 4°C, respectively (Fig. 1a). The reported differences in the virus survival times may be due to several factors, including the type and the structure of virus, the viral strain employed, and the external conditions of the experiment, as well as the aggregation status of the virus and the adsorption of the virus to particulate material or the inner surface of the container. To our knowledge, only two studies have focused on the survival of human adenoviruses in groundwater (6, 8). The T90 values (i.e., about 153 and 38 days at 4 and 23°C, respectively) described by Enriquez et al. (8) for infectious adenovirus serotype 41 closely resemble our findings, which suggest that adenoviruses have significant survival rates in groundwater. In contrast, Charles et al. (6) reported a rapid decrease in the infectivity of HAdV2 at 12°C, with a reduction of 4.2 log10 over the initial 21 days of the study. Nevertheless, these authors also observed that infectious HAdV2 could still be detected over a 364-day period. The present work provides the first investigation on quantitative evaluation of genome persistence for one serotype of human adenovirus. In our microcosms, adenovirus genome was detected throughout the monitoring period (400 days). No decrease was observed at 4°C, and we estimated a T90 value of 278 days at 20°C, showing that adenoviral DNA was very stable in groundwater. Our results support the observations by Charles et al. (6), who reported that HAdV2 genome at 12°C was detectable for 672 days of a 728-day study, using qualitative PCR. The longer persistence of the adenovirus genetic marker, compared to that of phage, could be, at least in part, related to the nature of DNA/RNA molecules, since double-stranded genome (such as that of adenovirus) is known to be generally more stable than is single-stranded genome (9).
Occurrence of bacterial fecal indicators, viral fecal indicators, and adenoviruses in raw groundwater.
Water quality is normally estimated by the presence of fecal bacterial indicators. In the investigated groundwater, no bacterial indicator (Escherichia coli and fecal and total coliforms) or somatic coliphage could be isolated during the whole monitoring period, regardless of the aquifers or the sampling sites. This observation attests to the good microbiological quality and the high protection level of the selected aquifers with regard to sources of fecal pollution. Nevertheless, genomes of HAdV and FRNAPH were observed on several occasions. Of the 60 groundwater samples collected for the viral analyses, only the genome of phages belonging to genogroup I was detected, once (Table 3). This positive result was associated with a sample from site S1. Phage occurrence observed at site S1 (1/15) is low, and the three other sites of our study did not have any phage genome. Even if comparison is not obvious, it is interesting to underline that Powell et al. (37) reported positive samples (2/12 and 1/10) from two groundwater sites located on a confined aquifer. Another previous study performed on groundwater of different aquifers in various geographical areas (29) did detect infectious FRNAPH in semiconfined aquifers. In these two studies (29, 37), infectious coliphages were always concomitantly present with bacterial indicators. In pathogenic virus analyses, seven samples were positive for the presence of adenoviral genome (Table 3). These seven adenovirus-positive samples were taken from sites S1, S2, and S4, with 1, 4, and 2 positives, respectively. Site S3 was virus negative throughout the monitoring period. In the seven adenovirus-positive samples, only serotype 41 was identified. While there are numerous reports of enteroviruses identified in groundwater by using cell culture or RT-PCR methods (1, 2, 5), only one recent study observed the presence of adenoviral genome. Futch et al. (12) investigated five wells along the main reef of the Florida Keys for adenovirus and enterovirus presence. Eight samples (32%) and four wells (80%) were positive for adenoviruses. They reported that adenovirus DNA was found at a significantly greater frequency from all sample types than was enterovirus RNA. In the present study, 11.7% of groundwater samples and 75% of sampling sites were adenovirus positive, a smaller proportion than that in the previously quoted work. The hydrogeological characteristics of the groundwater studied can certainly explain this difference because a low-permeability layer overlies confined aquifers, commonly limiting the migration of microbial contaminants into the aquifer.
TABLE 3.
Proportions of positive samples observed for each site in the groundwater experimentsa
| Virus | No. of positive samples/total no. of samples (P value) |
|||
|---|---|---|---|---|
| Site 1 (A1) | Site 2 (A1) | Site 3 (A1) | Site 4 (A2) | |
| FRNAPH (all genogroups) | 1/60 (0.231) | 0/60 (1) | 0/60 (1) | 0/60 (1) |
| HAdV | 1/15 (0.070) | 4/15 (1.58 × 10−5) | 0/15 (1) | 2/15 (0.005) |
| FRNAPH (all genogroups) + HAdV | 2/75 (0.074) | 4/75 (0.005) | 0/75 (1) | 2/75 (0.074) |
P values corresponding to statistical comparisons with the control (0/200) using the Fisher exact test are given in parentheses. Viral occurrences significantly different from the control are in bold. Note that the values in bold remain significant (P ≤ 0.05) if the Bonferroni correction is applied to the multiple comparisons of the table. A, aquifer, FRNAPH, F-specific RNA phages; HAdV, human adenoviruses.
The relevance of the positive detections of viral genome (independently of the viral type) was examined by comparing the frequencies of positive results from each sampling site with the control. The outcomes of the statistical tests comparing the observed frequencies with the control are displayed in Table 3. The main advantage of this approach is the possibility of interpreting the presence of viral genome in accordance with the full number of analyzed samples. Note that no viral genome was detected (0/200) among all the blank tests performed (considered the control in statistical analyses), with respect to the initial defined criteria. It appears that the proportions of positive samples (FRNAPH plus HAdV2) associated with sites S1, S3, and S4 were not significantly different from the control (Fisher exact test, P > 0.07), while a significant difference was observed for site S2 (Fisher exact test, P < 0.05). This emphasized that only the water coming from site S2 may be effectively characterized by intrinsic pollution, in terms of viral genome. The determination of the transport pathways for viruses to reach the studied aquifer was not the original aim of this work. Because our information regarding possible fecal sources (i.e., the few results of FRNAPH genotyping) and viral transport in confined aquifers is too sparse, it was not feasible to determine the exact source of human adenoviruses here, and many questions remain.
All these findings imply that, in groundwater without fecal bacteria, signs of adenoviral presence could be identified, a finding which certainly merits additional, more detailed investigations in various other aquifers. Groundwater represents a dark and cool environment (about 10°C in the aquifers studied), and the stability of viruses is known to be greater under these favorable conditions. Moreover, there is some evidence that occurrence of viral particles or viral genome in groundwater is principally dependent on the capacity of the viruses to survive over a sufficient period of time. According to our results, the observation of adenovirus DNA in our confined aquifer could be partially explained by its prolonged persistence time. Additionally, because of the significant survival times of adenoviruses in groundwater, detection of their genomes in the field was not necessarily due to recent contamination, raising the question of the applicability of real-time PCR methods in detecting adenoviruses in groundwater. Although this molecular counting method is demonstrated to be particularly useful in some circumstances, it determines only one part of the virus and does not connect the genome presence and the infective status. Some authors have recently underlined the importance of detecting infectious viruses instead of viral RNA for the purpose of public health risk assessment (38). So, the above deliberations highlight the fact that in groundwater, real-time PCR results for DNA viruses should be examined with caution to avoid misinterpretation of the relative public health risks.
Acknowledgments
We acknowledge Laurent Diez (LCPME, France) for his excellent technical assistance as well as Annick Moreau and Jean-Jacques Beley from the Danone Research Center (France) for having provided the hydrological and physicochemical characteristics of groundwater and also for their kind cooperation in improving the manuscript.
Footnotes
Published ahead of print on 15 October 2010.
REFERENCES
- 1.Abbaszadegan, M., M. S. Huber, C. P. Gerba, and I. L. Pepper. 1993. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbaszadegan, M., M. LeChevallier, and C. Gerba. 2003. Occurrence of viruses in US groundwaters. J. Am. Water Works Assoc. 95:107-120. [Google Scholar]
- 3.Bae, J., and K. J. Schwab. 2008. Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 74:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchardt, M. A., P. D. Bertz, S. K. Spencer, and D. A. Battigelli. 2003. Incidence of enteric viruses in groundwater from household wells in Wisconsin. Appl. Environ. Microbiol. 69:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchardt, M. A., K. R. Bradbury, M. B. Gotkowitz, J. A. Cherry, and B. L. Parker. 2007. Human enteric viruses in groundwater from a confined bedrock aquifer. Environ. Sci. Technol. 41:6606-6612. [DOI] [PubMed] [Google Scholar]
- 6.Charles, K. J., J. Shore, J. Sellwood, M. Laverick, A. Hart, and S. Pedley. 2009. Assessment of the stability of human viruses and coliphage in groundwater by PCR and infectivity methods. J. Appl. Microbiol. 106:1827-1837. [DOI] [PubMed] [Google Scholar]
- 7.De Roda Husman, A. M., W. J. Lodder, S. A. Rutjes, J. F. Schijven, and P. F. M. Teunis. 2009. Long-term inactivation study of three enteroviruses in artificial surface and groundwaters, using PCR and cell culture. Appl. Environ. Microbiol. 75:1050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enriquez, C. E., C. J. Hurst, and C. P. Gerba. 1995. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and waste water. Water Res. 29:2548-2553. [Google Scholar]
- 9.Espinosa, A. C., M. Mazari-Hiriart, R. Espinosa, L. Maruri-Avidal, E. Méndez, and C. F. Arias. 2008. Infectivity and genome persistence of rotavirus and astrovirus in groundwater and surface water. Water Res. 42:2618-2628. [DOI] [PubMed] [Google Scholar]
- 10.Fong, T. T., L. S. Mansfield, D. L. Wilson, D. J. Schwab, S. L. Molloy, and J. B. Rose. 2007. Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, Ohio. Environ. Health Perspect. 115:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fout, G. S., B. C. Martinson, M. W. N. Moyer, and D. R. Dahling. 2003. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Futch, C., W. G. Dale, and K. L. Erin. 2010. Human enteric viruses in groundwater indicate offshore transport of human sewage to coral reefs of the Upper Florida Keys. Environ. Microbiol. 12:964-974. [DOI] [PubMed] [Google Scholar]
- 13.Gallay, A., H. De Valk, M. Cournot, B. Ladeuil, C. Hemery, C. Castor, F. Bon, F. Mégraud, P. Le Cann, and J. C. Desenclos. 2006. A large multi-pathogen waterborne community outbreak linked to faecal contamination of a groundwater system, France, 2000. Clin. Microbiol. Infect. 12:561-570. [DOI] [PubMed] [Google Scholar]
- 14.Gassilloud, B., L. Schwartzbrod, and C. Gantzer. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl. Environ. Microbiol. 69:3965-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, C., and S. Toze. 2003. Influence of groundwater characteristics on the survival of enteric viruses. J. Appl. Microbiol. 95:536-544. [DOI] [PubMed] [Google Scholar]
- 16.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2007. Quantitative analysis of human enteric adenoviruses in aquatic environments. J. Appl. Microbiol. 103:2153-2159. [DOI] [PubMed] [Google Scholar]
- 17.Hernroth, B. E., A. C. Conden-Hansson, A. S. Rehnstam-Holm, R. Girones, and A. K. Allard. 2002. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Appl. Environ. Microbiol. 68:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hijnen, W. A. M., A. J. Brouwer-Hanzens, K. J. Charles, and G. J. Medema. 2005. Transport of MS2 phage, Escherichia coli, Clostridium perfringens, Cryptosporidium parvum, and Giardia intestinalis in a gravel and a sandy soil. Environ. Sci. Technol. 39:7860-7868. [DOI] [PubMed] [Google Scholar]
- 19.Howard, G., J. Bartam, S. Pedley, O. Schmoll, I. Chorus, and P. Berger. 2006. Groundwater and public health, p. 3-20. In O. Schmoll, G. Howard, J. Chilton, and I. Chorus (ed.), Groundwater for health: managing the quality of drinking-water sources. IWA Publishing, London, United Kingdom.
- 20.Hsu, F. C., Y. S. Shieh, J. van Duin, M. J. Beekwilder, and M. D. Sobsey. 1995. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 61:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hundesa, A., C. Maluquer de Motes, S. Bofill-Mas, N. Albinana-Gimenez, and R. Girones. 2006. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl. Environ. Microbiol. 72:7886-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Organization for Standardization. 2001. ISO 10705-1. Water quality—detection and enumeration of bacteriophages. Part 1: enumeration of F-specific RNA bacteriophages. International Organization for Standardization, Geneva, Switzerland.
- 23.International Organization for Standardization. 2001. ISO 10705-2. Water quality—detection and enumeration of bacteriophages. Part 2: enumeration of somatic coliphages. International Organization for Standardization, Geneva, Switzerland.
- 24.International Organization for Standardization. 2000. ISO 9308-1. Water quality—detection and enumeration of Escherichia coli and coliform bacteria. Part 1: membrane filtration method. International Organization for Standardization, Geneva, Switzerland.
- 25.Jiang, S. C. 2006. Human adenoviruses in water: occurrence and health implications: a critical review. Environ. Sci. Technol. 40:7132-7140. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. H., D. S. Cheon, J. H. Kim, D. H. Lee, W. H. Jheong, Y. J. Heo, H. M. Chung, Y. Jee, and J. S. Lee. 2005. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. J. Clin. Microbiol. 43:4836-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirs, M., and D. C. Smith. 2007. Multiplex quantitative real-time reverse transcriptase PCR for F+-specific RNA coliphages: a method for use in microbial source tracking. Appl. Environ. Microbiol. 73:808-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leclerc, H., S. Edberg, V. Pierzo, and J. M. Delattre. 2000. Bacteriophages as indicators of enteric viruses and public health risk in groundwaters. J. Appl. Microbiol. 88:5-21. [DOI] [PubMed] [Google Scholar]
- 29.Lucena, F., F. Ribas, A. E. Duran, S. Skraber, C. Gantzer, C. Campos, A. Moron, E. Calderon, and J. Jofre. 2006. Occurrence of bacterial indicators and bacteriophages infecting enteric bacteria in groundwater in different geographical areas. J. Appl. Microbiol. 101:96-102. [DOI] [PubMed] [Google Scholar]
- 30.Mena, K. D., and C. P. Gerba. 2009. Waterborne adenovirus. Rev. Environ. Contam. Toxicol. 198:133-167. [DOI] [PubMed] [Google Scholar]
- 31.Migliorati, G., V. Prencipe, A. Ripani, C. Di Francesco, C. Casaccia, S. Crudeli, N. Ferri, A. Giovannini, M. M. Marconi, C. Marfoglia, V. Melai, G. Savini, G. Scortichini, P. Semprini, and F. M. Ruggeri. 2008. Gastroenteritis outbreak at holiday resort, central Italy. Emerg. Infect. Dis. 14:474-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muscillo, M., M. Pourshaban, M. Iaconelli, S. Fontana, A. Di Grazia, S. Manzara, G. Fadda, R. Santangelo, and G. La Rosa. 2008. Detection and quantification of human adenoviruses in surface waters by nested PCR, TaqMan real-time PCR and cell culture assays. Water Air Soil Pollut. 191:83-93. [Google Scholar]
- 33.Ogorzaly, L., and C. Gantzer. 2006. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J. Virol. Methods 138:131-139. [DOI] [PubMed] [Google Scholar]
- 34.Ogorzaly, L., A. Tissier, I. Bertrand, A. Maul, and C. Gantzer. 2009. Relationship between F-specific RNA phage genogroups, faecal pollution indicators and human adenoviruses in river water. Water Res. 43:1257-1264. [DOI] [PubMed] [Google Scholar]
- 35.Pang, L., M. Close, M. Goltz, M. Noonan, and L. Sinton. 2005. Filtration and transport of Bacillus subtilis spores and the F-RNA phage MS2 in a coarse alluvial gravel aquifer: implications in the estimation of setback distances. J. Contam. Hydrol. 77:165-194. [DOI] [PubMed] [Google Scholar]
- 36.Pedley, S., M. Yates, J. F. Schijven, J. West, G. Howard, and M. Barrett. 2006. Pathogens: health relevance, transport and attenuation, p. 49-80. In O. Schmoll, G. Howard, J. Chilton, and I. Chorus (ed.), Groundwater for health: managing the quality of drinking-water sources. IWA Publishing, London, United Kingdom.
- 37.Powell, K. L., R. G. Taylor, A. A. Cronin, M. H. Barrett, S. Pedley, J. Sellwood, S. A. Trowsdale, and D. N. Lerner. 2003. Microbial contamination of two urban sandstone aquifers in the UK. Water Res. 37:339-352. [DOI] [PubMed] [Google Scholar]
- 38.Rutjes, S. A., W. J. Lodder, A. D. van Leeuwen, and A. M. De Roda Husman. 2009. Detection of infectious rotavirus in naturally contaminated source waters for drinking water production. J. Appl. Microbiol. 107:97-105. [DOI] [PubMed] [Google Scholar]
- 39.Skraber, S., B. Gassilloud, L. Schwartzbrod, and C. Gantzer. 2004. Survival of infectious Poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and Poliovirus-1 genome. Water Res. 38:2927-2933. [DOI] [PubMed] [Google Scholar]
- 40.Skraber, S., L. Ogorzaly, K. Helmi, A. Maul, L. Hoffmann, H. M. Cauchie, and C. Gantzer. 2009. Occurrence and persistence of enteroviruses, noroviruses and F-specific RNA phages in natural wastewater biofilms. Water Res. 43:4780-4789. [DOI] [PubMed] [Google Scholar]
- 41.Xagoraraki, I., D. H. W. Kuo, K. Wong, M. Wong, and J. B. Rose. 2007. Occurrence of human adenoviruses at two recreational beaches of the Great Lakes. Appl. Environ. Microbiol. 73:7874-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]